Abstract
Myostatin is a member of the transforming growth factor-β (TGF-β) family and a strong negative regulator of muscle growth. Here, we present the crystal structure of myostatin in complex with the antagonist follistatin 288 (Fst288). We find that the prehelix region of myostatin very closely resembles that of TGF-β class members and that this region alone can be swapped into activin A to confer signalling through the non-canonical type I receptor Alk5. Furthermore, the N-terminal domain of Fst288 undergoes conformational rearrangements to bind myostatin and likely acts as a site of specificity for the antagonist. In addition, a unique continuous electropositive surface is created when myostatin binds Fst288, which significantly increases the affinity for heparin. This translates into stronger interactions with the cell surface and enhanced myostatin degradation in the presence of either Fst288 or Fst315. Overall, we have identified several characteristics unique to myostatin that will be paramount to the rational design of myostatin inhibitors that could be used in the treatment of muscle-wasting disorders.
Keywords: Alk5, follistatin, heparin, myostatin, TGF-β
Introduction
Myostatin, also known as growth and differentiation factor-8 (GDF-8), is a transforming growth factor-β (TGF-β) family member that has been identified as a strong inhibitor of muscle growth. Myostatin knock-out mice exhibit muscles that are 2–3 times larger than those of wild-type (WT) mice (McPherron et al, 1997). In addition, transgenic overexpression of myostatin inhibitors, such as follistatin (Fst) or the dominant-negative form of the receptor ActRIIB, results in a similar phenotype (Lee and McPherron, 2001). Naturally occurring myostatin mutations have been identified in animals such as cattle, sheep, dogs, and human beings (Grobet et al, 1997; Kambadur et al, 1997; McPherron and Lee, 1997; Schuelke et al, 2004; Clop et al, 2006; Mosher et al, 2007). Although it is likely that other TGF-β family members also have some function in muscle development (Lee et al, 2005; Lee, 2007), myostatin seems to predominantly and specifically negatively regulate muscle growth. For these reasons, inhibitors targeting myostatin are actively being sought after as potential therapeutics in the treatment of muscle-wasting disorders such as muscular dystrophy and sarcopenia (Bradley et al, 2008; Tsuchida, 2008).
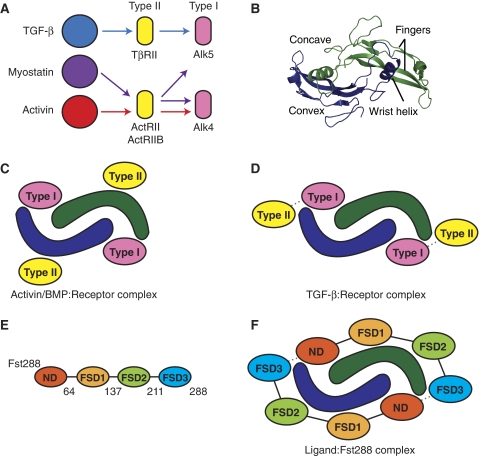
TGF-β family ligands can be subdivided into three classes based on sequence identity, receptor utilization, and shared inhibitors: TGF-β, bone morphogenetic protein (BMP)/GDF, and activin/inhibin (Innis et al, 2000). These secreted ligands are produced with a propeptide region that is later cleaved to yield disulfide-bonded homo- or heterodimers (Ling et al, 1986; Mellor et al, 2003; Shimmi et al, 2005; Lavery et al, 2008; Little and Mullins, 2009) that signal by binding two type II and two type I transmembrane serine/threonine kinase receptors. The signal is propagated inside the cell when type I receptors phosphorylate Smad proteins, which then form heteromeric complexes and accumulate in the nucleus to control gene expression (Schmierer and Hill, 2005). Different TGF-β ligands use specific subsets of receptors for signalling. For example, to activate similar Smads (Smads 2 and 3), the TGF-β class signals through the type II receptor TβRII and type I receptor Alk5 (Franzèn et al, 1993; Bassing et al, 1994), whereas the activin class uses ActRII or ActRIIB and Alk4 (Figure 1A) (Attisano et al, 1993; Carcamo et al, 1994). Crystallographic analysis has revealed that BMPs and activins bind the ectodomains of type II receptors on their convex surface and type I receptors on their concave surface (Figure 1B and C) (Kirsch et al, 2000; Greenwald et al, 2003; Thompson et al, 2003; Keller et al, 2004; Allendorph et al, 2006; Weber et al, 2007). TGF-βs also bind type I receptors on their concave surface, but bind type II receptors more distally towards the fingertip region (Figure 1D) (Hart et al, 2002; Groppe et al, 2008). In this case, the two types of receptors interact with each other, which is necessary for high-affinity ternary complex formation and signalling (Groppe et al, 2008). Although binding of all type I receptors has been shown to occur at the concave dimer surface, their orientation in this location is highly variable, especially between BMP and TGF-β type I receptors (Groppe et al, 2008). Differences are even observed between the orientations of the closely related BMP type I receptors, BMPR-IA and BMPR-IB (Kirsch et al, 2000; Keller et al, 2004; Kotzsch et al, 2009). Traditionally, myostatin has been thought to belong to the activin class of molecules, as it was originally identified to bind both the activin type II receptors, ActRIIB and ActRIIA, and the activin inhibitor Fst (Lee and McPherron, 2001). In addition to the activin type I receptor, Alk4, myostatin also signals through the TGF-β type I receptor, Alk5 (Rebbapragada et al, 2003), despite utilizing only the activin type II receptors. GDF-11, which is 89% identical at the amino-acid level to myostatin, likewise has been shown to signal through Alk5 in vivo (Andersson et al, 2006).
Figure 1.
Receptor and antagonist interactions with TGF-β family ligands. (A) Receptor specificity within the TGF-β family. Type I receptor signalling that specifically causes activation of Smads 2 and 3. Myostatin is distinctive in that it can effectively signal through both Alk4 and Alk5. (B) Architecture of TGF-β family ligands, showing myostatin. (C) During activin or BMP signalling, type II receptors typically bind on the convex surface of the ligand, whereas type I receptors bind on the concave surface. (D) In contrast, during TGF-β signalling, type II receptors bind more distally on the ligand, towards the fingertip region. There are also contacts between the type II and type I receptors. (E) Domain layout of Fst288, with the last residue of each domain indicated. (F) Schematic of ligand antagonism by Fst288. Two Fst288 molecules completely surround the ligand, blocking all four receptor-binding sites. Additional interactions occur between the ND of one Fst288 molecule and FSD3 of the other.
Ligand signalling is tightly regulated by extracellular antagonists, some of which are broad antagonists that inhibit multiple TGF-β family members, whereas others are more specific and inhibit only a few family members. Myostatin has been shown to be regulated by several antagonists including its propeptide component, decorin, GASP-1, and Fst-type molecules, including Fst-like 3 (Fstl3) and Fst isoforms Fst288 and Fst315 (Lee and McPherron, 2001; Thies et al, 2001; Hill et al, 2002, 2003; Amthor et al, 2004; Sidis et al, 2006). Fst is a multi-domain protein consisting of an N-terminal domain (ND) and three subsequent Fst domains (FSD1-3) (Figure 1E). Fst binds heparin, which localizes it to the cell surface and facilitates endocytosis of Fst-bound ligands (Ueno et al, 1987; Hashimoto et al, 1997). Fst315 has a C-terminal tail containing several acidic residues, which decrease its heparin affinity (Sugino et al, 1993). Fstl3 has a domain structure similar to that of Fst, but lacks heparin binding and the third Fst domain (Sidis et al, 2005).
Fst is a broad antagonist and also strongly inhibits the close family member of myostatin, activin A. The structure of activin A in complex with either Fst288, Fst315, or Fstl3 shows that Fst-type molecules inhibit signalling by completely surrounding the ligand with two molecules, blocking all four receptor-binding sites (Figure 1F) (Thompson et al, 2005; Lerch et al, 2007; Stamler et al, 2008). Fst also binds some members of the BMP class, albeit weakly, but does not bind members of the TGF-β class (Yamashita et al, 1995; Iemura et al, 1998). Fst binds activin A with a KD of 1.7 nM, but binds myostatin with a lower affinity of 12.3 nM KD (Nakatani et al, 2008). The molecular variations between myostatin and activin A that confer binding differences are not clear, as myostatin and activin A are 40% identical at the amino-acid level. Furthermore, studies determining which components of Fst288 are necessary for binding or antagonist specificity between ligands are ambiguous. It is widely accepted that FSD1 and FSD2 contribute significantly to binding affinity, but findings concerning the importance of the ND and whether or not it is necessary for activin A and BMP antagonism are mixed (Sidis et al, 2001; Keutmann et al, 2004; Harrison et al, 2006; Stamler et al, 2008). To our knowledge, similar binding experiments have not been performed for myostatin. However, insight into ligand specificity may emerge from studies that have altered Fst molecules by substituting FSD2 with FSD1. This creates an antagonist with high affinity for myostatin, but not activin A, presumably by disrupting the interaction with the convex ligand surface (Nakatani et al, 2007; Schneyer et al, 2008).
Here, we present the structure of myostatin in complex with Fst288. From the structure of myostatin, we have determined that the conformation of the prehelix region of myostatin is a feature that allows signalling through the TGF-β type I receptor, Alk5. We have also found evidence suggesting that Fst288 may interact more favourably with myostatin, particularly through its ND. In addition, we have discovered that binding myostatin greatly increases the affinity of Fst for heparin, and, in turn, increases binding to the cell surface and subsequent myostatin degradation. Overall, we have identified several characteristics of myostatin that make it unique from other family members. We believe that this is an important and even crucial first step in the rational design of myostatin inhibitors.
Results
The structure of myostatin
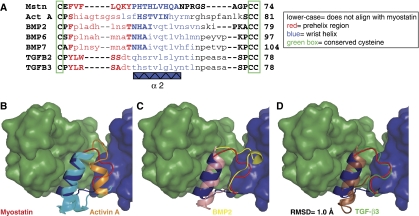
The complex of two Fst288 molecules bound to one myostatin dimer was resolved to 2.15 Å using X-ray crystallography. The myostatin in this complex represents the first known structure of this ligand. Myostatin displays the traditional TGF-β family hand-shaped architecture, with each monomer consisting of four curved beta strands or ‘fingers', a cystine knot motif in the ‘palm' region, and a major helix or ‘wrist' (Figure 1B). Two monomers come together palm-to-palm in an anti-parallel direction and form an intermolecular disulfide bond, creating the mature dimer. Comparing myostatin to activin A and TGF-β3, the root mean square deviation (RMSD) values of Cα atoms are 1.6 Å (105 residues) and 2.7 Å (102 residues), respectively, as calculated using CE and one monomer from each of 2B0U, 2PJY, and 3HH2 (Thompson et al, 2005; Groppe et al, 2008). This is illustrated in structural overlays of myostatin with other TGF-β family members, which are shown in Supplementary Figure 1. Although myostatin is structurally similar to other TGF-β family ligands, there are significant differences in the N-termini and the region preceding the wrist helix, termed the prehelix loop, which is situated in the type I receptor-binding site (Figure 2A; Supplementary Figure 1*,). It has been postulated that much of the type I receptor specificity occurs through variation in this region. Therefore, we compared the prehelix loop of myostatin with that of other TGF-β family ligands in an attempt to understand type I receptor specificity.
Figure 2.
The prehelix loop of myostatin is most similar to that of the TGF-β class. (A) Structure-based sequence alignment against myostatin. (B–D) Myostatin monomer A (myostatinA, blue) and monomer B (myostatinB, green) are shown, with monomers B aligned between ligands. Superimposed on myostatin are the wrist regions of the other ligands: activin A (2B0U) in cyan and orange, BMP2 (2H64) in pink and yellow, and TGF-β3 (2PJY) in brown and green (Thompson et al, 2005; Weber et al, 2007; Groppe et al, 2008). An RMSD value for main chain atoms of the prehelix regions of myostatin (residues 49–55) and TGF-β3 (residues 50–56) is shown.
The prehelix loop of myostatin is most similar to that of TGF-β
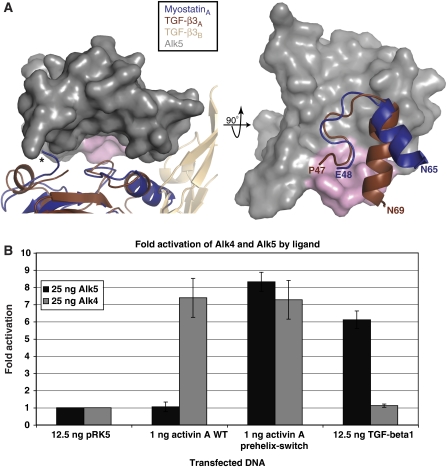
As stated earlier, myostatin, but not activin A, can effectively use the TGF-β type I receptor, Alk5, as well as the activin type I receptor, Alk4 (Rebbapragada et al, 2003). To elucidate this difference in type I receptor utilization, we compared the prehelix region of myostatin to a representative structure from each TGF-β class (Figure 2B–D). Although the amino-acid sequences and overall conformation of myostatin and activin A are similar, there is much deviation in the prehelix region (residues 49–55 and 46–55, respectively). Activin A contains a series of glycine and serine residues that cause this region to be flexible, whereas myostatin contains a number of larger hydrophobic residues (Figure 2A, red). Accordingly, these segments are structurally very different from each other (Figure 2B). There is actually significant structural variability in this region within activin A itself. For example, in the activin A:ActRIIB structures, many of the prehelix residues could not be modelled, likely because of flexibility, but in the activin A:Fst288 structure, this region forms a novel helix (Thompson et al, 2003, 2005; Greenwald et al, 2004; Harrington et al, 2006). In the myostatin:Fst288 complex structure, the prehelix region has well-defined electron density (Supplementary Figure 2). Although it is possible that myostatin similarly adopts alternate conformations in the free and bound state, it may be more rigid than activin A and, thus, less prone to multiple conformations. The prehelix loop of myostatin is also very different from that of BMPs (Figure 2C). Surprisingly, the conformation of the prehelix loop of myostatin most closely resembles that of a TGF-β class ligand (Figure 2D). In fact, the RMSD of prehelix main chain atoms of myostatin (49-55) and TGF-β3 (50-56) is 1.0 Å (calculated using COOT). Generally, the TGF-β class is thought to be dissimilar to the activin and BMP classes, as TGF-βs display a more elongated dimer conformation as well as differences in receptor assembly (Figure 1C and D; Supplementary Figure 1C) (Hart et al, 2002; Groppe et al, 2008). Conformational differences are especially apparent in the fingertip region of TGF-β, which is more extended, and in the tilt of the wrist helix (Figure 3A; Supplementary Figure 1C). Nonetheless, the prehelix region of myostatin traces very closely with that of TGF-β3 despite the fact that the rest of the ligand is divergent. This led us to hypothesize that the prehelix loop of myostatin is a feature that allows signalling through Alk5. Aligning myostatin to TGF-β3 and superimposing Alk5, it seems reasonable that this would be the case (Figure 3A). The contact of Alk5 with TGF-β3 is minimal and limited mostly to van der Waals interactions with the prehelix loop. Thus, the interaction seems to be based on the conformation of the prehelix loop rather than the types of residues it contains. We next tested this hypothesis in a cell-based assay.
Figure 3.
The conformation of the prehelix region of myostatin is a feature that allows signalling through Alk5. (A) MyostatinA and TGF-β3A are aligned, with Alk5 (grey) superimposed (2PJY (Groppe et al, 2008)). The N-terminus of myostatin is indicated (*). (B) Luciferase-reporter gene assay with the CAGA12 promoter in L17 RIB cells. Reporter activation was monitored after cells were transfected with and without Alk4 or Alk5 receptors along with various plasmids containing ligands. Substituting the prehelix region of activin A with that of myostatin creates a ligand (activin A prehelix-switch) with a greatly increased ability to signal through Alk5. Fold activation represents response to ligand with receptor transfected over response with no ligand transfected, after subtracting background of the transfected ligand without added receptor.
Switching the prehelix region of activin A with that of myostatin allows robust signalling through Alk5
To determine whether the prehelix loop allows myostatin to signal through Alk5, we replaced the prehelix region of activin A (P45-F58) with that of myostatin (E48-P56) and tested this mutant's ability to then signal through Alk5. We used a luciferase-reporter gene assay with the CAGA12 promoter in L17 RIB cells, which do not normally express Alk5 (Attisano et al, 1993). Activin A WT or mutant (activin A prehelix-switch) was transfected into cells, along with Alk5. Activin A WT shows a very slight increase in signalling in the presence of Alk5, which has been reported (Figure 3B) (Rebbapragada et al, 2003). In contrast, the activin A prehelix-switch mutant shows a greatly increased ability to signal through Alk5 (∼eight-fold). We also tested the signalling of these ligands through the other type I receptor, Alk4. Both activin A WT and activin A prehelix-switch signal robustly through Alk4 at comparable levels. We confirmed this observation by titrating increasing amounts of ligand DNA into the cells in the presence of Alk5 or Alk4 (Supplementary Figure 3). Single point mutations to alanine were also made for each residue of the prehelix loop of activin A prehelix-switch (Supplementary Figure 4). None of these mutants exhibited decreased signalling through Alk5, supporting that it is the conformation of the prehelix loop rather than the residues it contains that is important for Alk5 signalling. We have also shown that TGF-β1 signals well through Alk5, but not Alk4, as expected (ten Dijke et al, 1994). More TGF-β1 plasmid was used in comparison to the activin A plasmids, presumably because TGF-β ligands are subject to inhibition by their propeptide regions. We also attempted to assay myostatin WT, yet, despite evidence that the protein was expressed, we could not detect signalling in this assay, presumably because of strong inhibition of myostatin by its propeptide region (data not shown). Altogether, activin A can be converted into a ligand that signals robustly through Alk5 simply by swapping in the prehelix loop of myostatin.
Structure of the myostatin:Fst288 complex
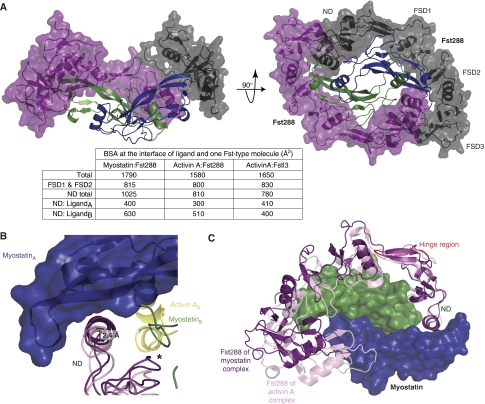
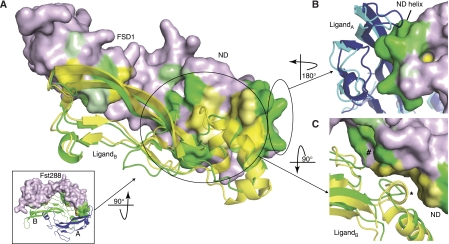
The myostatin:Fst288 complex structure contains not only the first structure of myostatin, but also represents the highest resolution complete Fst-type structure to date. Two Fst288 molecules wrap around the myostatin dimer and completely block all four receptor-binding sites, as is seen in the activin A:Fst288 complex and complexes of activin A with Fst315 and Fstl3 (Figure 4A) (Thompson et al, 2005; Lerch et al, 2007; Stamler et al, 2008). FSD1 and FSD2 contact only one myostatin monomer and bury the type II receptor-binding site (FSD1,FSD2:type II interface, Figure 1F), whereas the ND contacts both monomers and plugs the type I receptor-binding site (ND:type I interface). In addition, inter-Fst288 contacts are again seen between the ND of one molecule and FSD3 of the other, with FSD3 lacking any contact with the ligand itself. As this is the first structure of an Fst-type molecule bound to a ligand other than activin A, we carried out an in-depth comparison of the myostatin:Fst288 structure with the activin A:Fst288 structure. This has helped to pinpoint ligand and antagonist characteristics that may confer specificity to binding.
Figure 4.
Overview of the myostatin:Fst288 structure with gross comparisons to activin A:Fst-type structures. (A) Overview of the myostatin:Fst288 structure. The ND and FSD1 are removed from one chain (black) in the left panel. Interface buried surface area calculations are shown in the table. Activin A:Fstl3 complex calculations are shown for comparison. LigandA (e.g. myostatinA) refers to the ligand monomer opposite the ND, whereas ligandB refers to the monomer adjacent to the ND. (B) MyostatinA and activin AA in the myostatin:Fst288 (blue, green: purple) and activin A:Fst288 (yellow: pink) complexes are aligned. Activin AA is not shown. The Fst288 ND helix is 2.4 Å closer to myostatinA than activin AA. An asterisk indicates where the ND of Fst288 in the myostatin complex would overlap with the prehelix region of activin A. (C) NDs of one Fst288 molecule in each of the two complexes are aligned. Activin A and one Fst288 molecule from each complex are not shown. A hinge region directly following the ND in residues 64–74 of FSD1 is highlighted (red).
Fst288 forms a more intimate ND:type I receptor-binding site interaction with myostatin than activin A
To highlight areas that may contain important ligand:Fst288 interactions, buried surface area analyses of complex interfaces were carried out. Overall, more surface area is buried at the myostatin:Fst288 interface than at the activin A:Fst288 interface, with significant contributions in the former coming from the ND:type I site interaction (Figure 4A). In the myostatin complex, about 200 Å2 more surface area is buried at the ND:type I interface than at the FSD1,FSD2:type II interface. This is the first time that the ND has been identified as burying significantly more surface area than FSD1 and FSD2 (Thompson et al, 2005; Lerch et al, 2007; Stamler et al, 2008). The ND actually forms a closer contact with the type I receptor-binding site of myostatin than activin A, especially with the fingers of the monomer opposite to the ND (here on referred to as myostatinA, whereas the monomer adjacent to the ND is myostatinB, Figure 4B). In fact, the ND helix is shifted 2.4 Å closer to myostatinA than activin AA. The prehelix region of activin A would have to adopt an alternate conformation to interact with the ND in the same fashion as myostatin, as the alignment shows a clear steric conflict (Figure 4B,*). This close relationship of ND to ligand is most similar to that observed in the activin A:Fstl3 complex, in which the ND shows a similar shift inwards, burying a comparable amount of surface area with activin AA (Figure 4A) (Stamler et al, 2008). Many interactions particular to the myostatin complex occur between the ND and the type I receptor-binding site. Shape complementarity (Sc) analysis quantifies the ‘goodness of fit' between the ND and myostatin to be 0.75 compared with 0.50 between the ND and activin A, indicating that the ND interacts much more advantageously with myostatin.
Fst288 in the myostatin:Fst288 complex forms a more open conformation
When comparing the myostatin:Fst288 and activin A:Fst288 complexes, there is an apparent difference in the gross overall conformation of Fst288 itself. In binding myostatin, Fst288 adopts a more open conformation (Figure 4C). Domain motion analysis shows that there is a hinge at residues 64–74, causing an inwards rotation after the ND of ∼15° in the activin A:Fst288 complex and allowing Fst288 to take on a more clamped profile. It seems that Fst288 adopts alternate conformations to adapt to the shape of the different ligands. Alternatively, Fst288 may be able to compress activin A because of the flexibility of the ligand, which could be lacking in myostatin. Therefore, whether Fst288 itself or differences between myostatin and activin A, or possibly both, are the cause of the altered Fst288 conformation is unclear.
The fingertip region of myostatin in the type I receptor-binding site forms more extensive contacts with the ND of Fst288
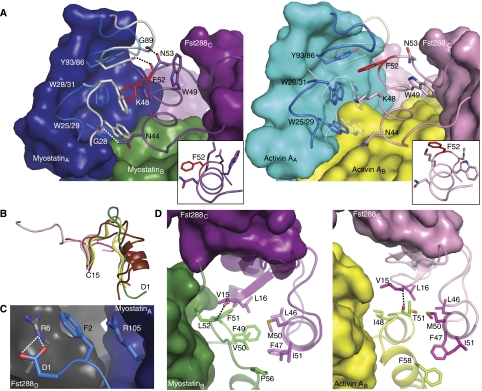
Myostatin and activin A bind many of the same regions on Fst288. However, these areas are used differently by the two ligands. There are two particular sites that merit discussion, both of which involve the ND of Fst288. There have been mixed reports concerning the importance of the ND to ligand binding (Thompson et al, 2005; Harrington et al, 2006; Harrison et al, 2006; Stamler et al, 2008). We find that the ND may be even more important to myostatin binding, as it interacts more closely with myostatin and in a way that is distinctive from activin A. This is best illustrated by examining the surfaces on Fst288 that are buried differently by myostatin versus activin A (Figure 5A). At the type I interface, the fingertip loops of myostatin clamp down on the ND helix to form contacts not present in the activin A:Fst288 complex, including three new hydrogen bonds (Figures 5B and 6A). In addition, there is actually a rotation or shift in the conformation and hydrogen bonding within the ND helix itself when comparing the two complexes (insets in Figure 6A). Interestingly, the C-terminal end of the ND helix is more extended when bound to myostatin, wherein the backbone carbonyl oxygens of F47 and K48 hydrogen bond to the i+4 residue instead of the i+3 residue observed in the activin A complex. This leads to a more tightly wound spring shape in the ND helix of the myostatin:Fst288 complex. In addition, the sidechains of N53 and W49 rearrange to help stabilize each of the two helical conformations. These differences alter the interface interactions significantly, as F52 and K48 of Fst288 splay outwards in opposite directions (insets in Figure 6A), allowing W31 of myostatin (which adopts a different rotamer compared with activin A W28) to insert between them, overall, permitting the ND to interact more closely with myostatin than it does with activin A.
Figure 5.
Buried surface area comparison between myostatin:Fst288 and activin A:Fst288. Myostatin (green and dark blue) and activin A (yellow and cyan) use different areas on Fst288. (A) NDs of one Fst288 from each complex are aligned. Inset shows myostatin in complex with one Fst288 molecule (FSD2 and FSD3 are removed) and is used for orientation. Fst288 is coloured according to which ligand buries more of its surface in specific areas. Fst288 residues that are buried more in the activin A:Fst288 complex are shaded brighter yellow, whereas those that are buried more in the myostatin:Fst288 complex are shaded brighter green. The strongest shades represent buried surface area differences of up to 45 Å2. Areas that are either not buried or buried equally by both ligands are tinted light purple. Differences in buried surface areas were calculated between the two complexes on a per residue basis. (B) The ND helix forms a closer contact with the fingertip area of myostatin (see also Figure 6A). (C) The N-terminus of myostatin forms novel contacts with Fst288 (#, see also Figure 6C), whereas activin A buries more surface area through its prehelix region (*, see also Figure 6D).
Figure 6.
Fst288 ND contact differences between myostatin and activin A. (A) Comparison of the Fst288 ND helix interactions with myostatin (left) and activin A (right). Residues shown in stick are carved out of the surface for clarity. Superimposed on myostatin and activin A are the fingertip regions of the other ligand (transparent). The ND helix is shifted closer to myostatinA as compared with activin AA, allowing additional hydrophobic interactions and three hydrogen bonds to be formed at the ND helix:fingertip interface (in both panels shown in cartoon and stick). Comparing the two complexes, a shift occurs in the conformation and hydrogen bonding within the ND helix (inset), specifically altering the position of F52 (red). Helices are shown from the same perspective, with the NDs aligned. (B) Comparison of the N-termini of different TGF-β ligands up through the first conserved cysteine. BMPs are shown in shades of pink (1M4U 28–38, 2H64 10–14, 2QCQ 4–8, and 2R53 26–31), TGF-βs in shades of brown (2PJY 1–15 and 2TGI 1–15), activin A in shades of yellow (2ARV 1–11 and 2B0U 1–11), and myostatin in green (annotated 1–15) (Daopin et al, 1992; Groppe et al, 2002, 2008; Thompson et al, 2005; Harrington et al, 2006; Allendorph et al, 2007; Weber et al, 2007; Saremba et al, 2008). (C) The N-terminus of myostatin (blue) forms a stabilizing cation-π interaction with Fst288 (grey), which is distinctive from activin A. (D) Activin A (right) fills a crevice on the ND of Fst288 with its prehelix region. Both ligands form a backbone hydrogen bond here, but myostatin (left) presents more hydrophobic residues on its prehelix loop to form additional interactions with those of Fst288.
The N-terminus and prehelix region of myostatin form an interaction with the ND of Fst288 that is distinctive from that of activin A
The N-termini of different TGF-β family ligands are quite variable, as can be seen by comparing a selection of ligands from each of the three classes (Figure 6B; Supplementary Figure 5). Myostatin is no exception, and its N-terminus forms a unique interaction with Fst288 (Figure 5C, #). Myostatin D1 forms a bidentate interaction with Fst288 R6 Nɛ (Figure 6C). In addition, a stabilizing cation-π interaction occurs in which F2 of myostatin is sandwiched by R105 of myostatin and R6 of Fst288 in a stacking fashion. No contacts are present between Fst288 and the N-terminus of activin A, and, in fact, activin A lacks the first two residues present in myostatin.
The interactions between the prehelix regions of the ligands and the ND of Fst288 are also unique. Activin A primarily fills a crevice on the ND with its prehelix region, which fits nicely into this area (Figure 5C,*). Both ligands form a backbone hydrogen bond here, with myostatin L52 bonding to Fst288 L16 and activin A T51 to Fst288 V15 (Figure 6D). However, myostatin presents more hydrophobic residues here that interact to a greater extent with those of Fst288. In addition, residue conformations of Fst288 are altered to interact differently with the two ligands. F47 of Fst288 adopts an alternate rotamer and points towards F49 and V50 of the prehelix loop of myostatin. This seems to pull in the ND and allow it to interact more closely with myostatin. This reveals the accommodating nature of Fst288 and begins to explain how it can act as a broad antagonist. In other words, interactions at the FSD1,FSD2:type II interface are virtually the same, and this is likely the site that confers high affinity, as has been suggested earlier (Harrington et al, 2006; Harrison et al, 2006). In contrast, the ND:type I interface seems to lend specificity to binding, and structural evidence points to Fst288 being more suited to interact with myostatin than activin A because of this interface. Nonetheless, the affinity of Fst288 for myostatin is lower than that for activin A, which implies that other factors influence binding.
The myostatin:Fst288 complex exhibits a unique polar electrostatic surface potential and has an increased affinity for heparin
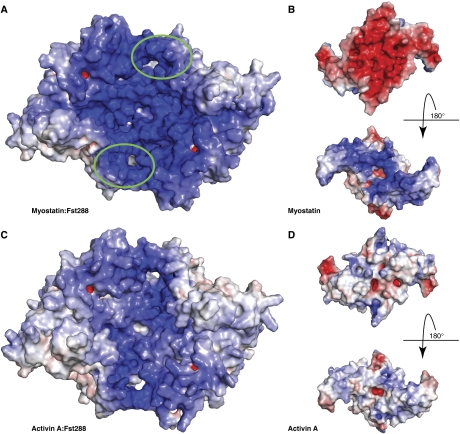
Fst288 is known to bind heparin, which has implications in localization and cellular endocytosis, as this allows it to bind cell surfaces (Hashimoto et al, 1997). To visualize the effect that binding myostatin may have on the affinity of Fst288 for heparin, we examined the electrostatic surface potential of these proteins. As seen in Figure 7A, the myostatin:Fst288 complex exhibits a surface that is remarkably electropositive, especially in comparison with the activin A:Fst288 complex (Figure 7C). This surface forms a deep continuous groove across the ligand and contains the FSD1 heparin-binding sites of both Fst288 molecules (circled in green). The myostatin ligand itself displays a surprisingly polar surface potential, with the bottom (side facing the cell surface on receptor binding) being very electronegative and the top very electropositive (Figure 7B). Again, this is especially prominent in comparison to the activin A dimer, which displays a more evenly spread electrostatic surface potential (Figure 7D). The highly basic surface of myostatin may hinder the ability of Fst288 to bind, as this brings together two electropositive surfaces. This may explain the studies finding that the KD of Fst288 for myostatin is higher than that for activin A (Hashimoto et al, 2000; Amthor et al, 2004; Nakatani et al, 2008).
Figure 7.
Myostatin exhibits a unique double-sided electrostatic surface potential. (A, C) Surface representation of the electrostatic potential of Fst288 in complex with myostatin and activin A. Surfaces are coloured by potential on the solvent accessible surface on a scale of −12.5 to 12.5 kbT/ec (red to blue). The heparin-binding site in FSD1 is circled in green. The myostatin:Fst288 complex exhibits a strikingly electropositive surface, especially in comparison to the same surface on the activin A:Fst288 complex. (B, D) Electrostatic surface potential of individual ligands coloured on a scale of −5 to 5 kbT/ec. The myostatin dimer itself actually shows an extremely polar electrostatic surface potential. Activin A is shown for comparison.
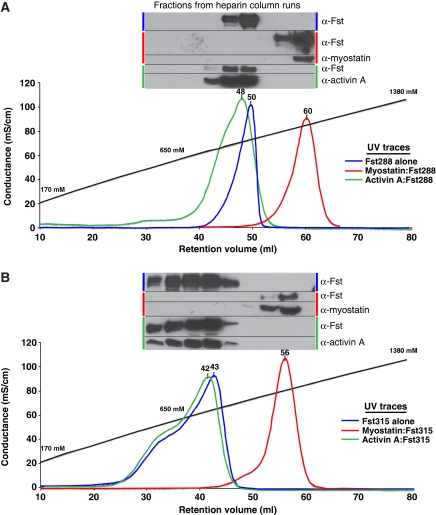
These electrostatic surface potential findings led us to test whether or not the myostatin:Fst288 complex could bind heparin more tightly than either the activin A:Fst288 complex or Fst288 alone. An earlier study validated the use of NaCl elution from a heparin column as a reliable method of determining relative differences in heparin affinity and showed that there is a linear relationship between [NaCl] elution and KD values (Thompson et al, 1994). Individual Fst proteins or preformed ligand:Fst complexes were applied to a heparin column and eluted with an NaCl gradient (Figure 8). Activin A:Fst288 and Fst288 alone bind heparin with similar affinities, eluting at nearly the same concentration of NaCl. However, the affinity of myostatin:Fst288 for heparin is greatly increased. This effect also extends to Fst315. In fact, the myostatin:Fst315 complex has a striking increase in heparin affinity when compared with the activin A:Fst315 complex or Fst315 alone. The electrostatic surface potential of the activin A:Fstl3 complex was also examined, and its corresponding surface is drastically less electropositive (Supplementary Figure 6A). As expected, Fstl3 did not bind the heparin column alone or in complex with either ligand (Supplementary Figure 6B).
Figure 8.
Myostatin greatly increases the affinity of Fst for heparin compared with Fst alone or activin A:Fst complexes. Fst288 (A) and Fst315 (B) alone and in complex with ligands were bound to a heparin column and eluted with a NaCl gradient. Y axis is shown in terms of conductance, and the approximate corresponding NaCl concentration is labelled along the gradient. Peaks traces are in terms of absorbance at 280 nm, and retention volumes are shown above each peak. Western blots were performed on every other fraction collected, and samples are pictured above the peaks they represent.
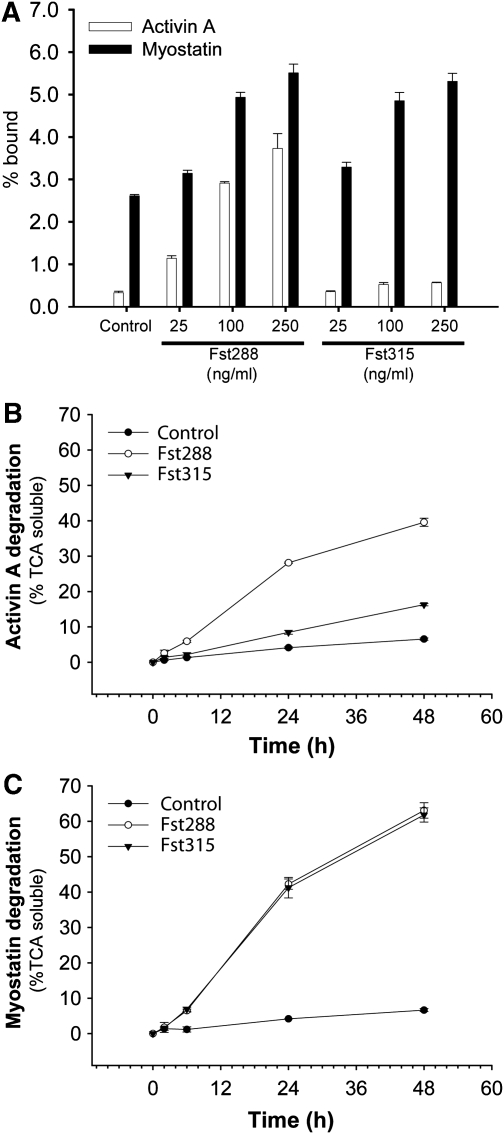
These results, combined with the earlier study on Fst288 facilitating activin A degradation (Hashimoto et al, 1997), led us to investigate whether the increase in complex affinity for heparin has biological implications. We first tested the abilities of free ligands and ligands in the presence of increasing concentrations of either Fst288 or Fst315 to bind to the surface of immortalized gonadotrope (LβT2) cells. As expected, Fst288, but not Fst315, increased cell surface binding of activin A (Figure 9A). In contrast, both Fst288 and Fst315 increased cell surface binding of myostatin, which correlates well with our heparin-binding experiments. Moreover, it seems that myostatin itself may have increased cell surface binding in comparison to activin A. Fst288 binding to activin A leads to ligand degradation through internalization and lysosomal degradation (Hashimoto et al, 1997). Therefore, we assessed the effects of Fst288 and Fst315 binding on myostatin and activin A degradation. As expected Fst288, but not Fst315, substantially increased activin A degradation, whereas both Fst288 and Fst315 increased myostatin degradation (Figure 9B and C). In addition, both Fst288 and Fst315 binding led to both more rapid and more extensive myostatin degradation relative to activin A when bound to Fst288. These findings represent an unrecognized mode of myostatin regulation by Fst and heparin, especially with respect to the serum available form of Fst, Fst315.
Figure 9.
Both Fst288 and Fst315 enhance myostatin cell surface binding and degradation. (A) 2 nM 125I-activin A or 125I-myostatin were incubated in the presence or absence of varying amounts of Fst288 and Fst315 and added to LβT2 cells for 2 h as described. Cell surface binding was calculated as the fraction of radioactive ligand that remained cell-associated after washing. (B, C) The effect of 250 ng/ml Fst288 or Fst315 on degradation of 1 nM radio-labelled activin A (B) or myostatin (C) in LβT2 cells as assessed by the TCA soluble fraction and measured as a function of time.
Discussion
Myostatin is an important negative regulator of muscle growth and an attractive target for the development of therapeutics for the treatment of muscle-wasting disorders (Bradley et al, 2008; Tsuchida, 2008). As all TGF-β family members exhibit a common fold, identifying characteristics unique to myostatin is critical to the rational design of highly specific myostatin inhibitors. The myostatin:Fst288 structure has led us to several discoveries concerning myostatin regulation involving both receptor utilization and antagonist specificity.
The prehelix region is a critical component that permits ligands to discriminate type I receptors (Keller et al, 2004; Nickel et al, 2005; Korupolu et al, 2008; Kotzsch et al, 2009). We discovered that the prehelix region of myostatin is most similar to that of a TGF-β class member and determined that this conformation of the prehelix is likely a structural feature that permits signalling through Alk5. Still, it is somewhat questionable as to how this signalling occurs. Signalling of TGF-β through Alk5 requires the association of Alk5 with the type II receptor, TβRII, as the affinity of Alk5 for TGF-β itself is very low (Groppe et al, 2008). This interaction is facilitated by TβRII binding more distally on the ligand, towards the fingertips, which brings it closer in proximity to the type I receptor (Figure 1B–D). However, myostatin uses the activin type II receptors, which likely bind on the convex surface of the ligand, as has been seen with activin A and BMP receptor structures (Greenwald et al, 2003; Thompson et al, 2003; Weber et al, 2007). This would seemingly preclude the association of Alk5 with the type II receptor on myostatin. One possibility is that the tilt of the wrist helix influences the position of Alk5. The wrist helix of myostatin is tilted more towards the centre of the dimer in comparison to that of TGF-β and may allow Alk5 to form closer contacts with myostatin (Figure 3A; Supplementary Figure 1C). This brings into question whether or not Alk5 actually interacts more strongly with myostatin than it does with TGF-β, possibly through the N-terminus of myostatin, which seems to come in close proximity in the Alk5 superposition (Figure 3A,*). Even a small increase in Alk5 affinity might be sufficient to allow myostatin to signal through a membrane-dependent receptor assembly mechanism, as proposed for activin A signalling through Alk4 (Greenwald et al, 2004; Sebald et al, 2004). It is also possible that intracellular interactions exist between Alk5 and the type II receptor when bound to myostatin, as has been shown for Alk5 and TβRII (Rechtman et al, 2009).
Despite lack of structural data on how activin A or myostatin interact with type I receptors and lack of an Alk4 structure, several speculations can be made on how these ligands may both signal through Alk4, whereas only myostatin signals through Alk5. Mutational analysis has shown that residues in the finger region of activin A are important for Alk4 binding (Harrison et al, 2004; Cook et al, 2005), and these residues are conserved on myostatin. These residues can be correlated with those on Alk4 that, when mutated, decrease activin A signalling (Harrison et al, 2003) (based on modelling of Alk5 into the type I receptor slot of activin A and sequence alignment of Alk4 with Alk5 (Groppe et al, 2008)). Therefore, it is likely that activin A and myostatin share this Alk4 interaction site. As we have shown that altering the prehelix loop of activin A to that of myostatin allows it to signal through Alk5, it is likely that this area influences Alk5 specificity. As it does not seem that there are specific residues in the prehelix loop of myostatin that confer signalling, it is possible that either flexibility or steric hindrance of the prehelix region of activin A prohibits interaction with Alk5. In addition, modelling of Alk5 into the type I receptor site of myostatin shows a loop of Alk5 that points towards the prehelix region of myostatin and could have a function in receptor binding (residues 63–75, shaded pink, Figure 3A). The corresponding region on Alk4 is truncated in comparison (residues 62–71). We propose that the disparity of myostatin and activin A signalling through type I receptors can be explained by the ligands using alternate areas for receptor interaction. The finger regions, which are conserved between activin A and myostatin, may be important for Alk4 binding, whereas the prehelix regions, which are non-conserved, may be an important feature for Alk5 binding.
The structure of the myostatin:Fst288 complex has revealed several unanticipated interactions with Fst288 that have implications towards ligand specificity. Overall, Fst288 antagonizes myostatin similarly to activin A. As this is the first structure of Fst in complex with a ligand other than activin A, we have been able to identify characteristics of Fst288 that allow broad antagonism within the family as well as those that lend specificity to binding. Residues contained within type II receptor-binding sites are conserved between myostatin, activins, and several BMPs, and it seems that Fst288 uses this site similarly between ligands, likely with high affinity. The type I receptor-binding sites on the ligands are significantly more diverse because of the variability of the prehelix regions. Fittingly, this seems to be a site of specificity for the antagonist, as the ND becomes remodelled and forms alternate interactions here. The case is similar for Fst288 and Fstl3 binding to activin A in that FSD1 and FSD2 bind similarly, with differences seen in the NDs (Stamler et al, 2008). In further support, aligning BMP2 or BMP7 with myostatin in the complex structure shows that the prehelix loop of BMP would also likely interfere with ND binding. Data suggests that the ND lacks the ability to bind BMPs, possibly explaining why it is a weak BMP antagonist (Iemura et al, 1998). Overall, it seems that FSD1 and FSD2 are required for high affinity, but that the ND confers specificity to the antagonist.
Myostatin forms further contacts with Fst288 that are not present in the activin A:Fst288 complex, and, with the addition of Sc analysis, evidence points to myostatin forming a more intimate interaction with Fst288 at the ND:type I interface. This is supported by the observation that a truncated Fst containing ND-FSD1-FSD1 binds preferentially to myostatin (Nakatani et al, 2007; Schneyer et al, 2008) and that activin A mutations at the ND interface have little effect on Fst288 binding (Harrison et al, 2006). Therefore, it is unclear as to why Fst has a higher affinity for activin A than myostatin (Hashimoto et al, 2000; Amthor et al, 2004; Nakatani et al, 2008). One explanation is that decreased Fst288 affinity for myostatin is the result of bringing together two electropositive surfaces, which does not occur with activin A. Alternatively, ligand-affinity differences could be explained by an induced fit model in which the conformation of the ND needed to bind myostatin may not be as energetically favourable as that needed to bind activin A and is, thus, induced through myostatin binding. Both models support the observation that Fst association rates are 15-fold slower for myostatin than activin A, whereas the dissociation rates are similar, but fail to explain why the ND-FSD1-FSD1 form binds tightly to myostatin and not activin A (Nakatani et al, 2007).
The effect of myostatin binding on the affinity of Fst for heparin is intriguing. In a study by Hashimoto et al (1997), Fst288 was shown to be a staunch regulator of activin A by binding to heparin on cell surfaces and subsequently causing increased cellular uptake and degradation of the complex (Hashimoto et al, 1997). Here, we observed that both myostatin:Fst288 and Fst315 complexes have greatly increased affinity for the cell surface leading to enhanced ligand degradation. This implies that distant sources of Fst315 production might have a function in regulating myostatin degradation, as it is the prevalent Fst isoform found in serum (Schneider-Kolsky et al, 2000; Schneyer et al, 2004). The increased heparin affinity may explain why myostatin:Fst315 complexes are not observed in the serum, as they would be predicted to interact with cell surfaces once formed (Hill et al, 2002).
Although the Fst residues involved in heparin binding have been identified (Sidis et al, 2005), the mechanism of how large heparin molecules bind Fst in the ligand-bound or unbound state is unclear (Innis and Hyvonen, 2003). On the basis of the myostatin:Fst288 complex structure it seems that a composite heparin-binding site is generated in which a single heparin molecule could bind a continuous electropositive crevice that spans both molecules of Fst. The crevice measures ∼60 Å wide and would fit a heparin molecule ∼14–16 hexoses in length, similar to that observed in the FGF–FGF receptor heparin complex (Schlessinger et al, 2000). To generate this putative composite-binding site, two unfavourable electropositive surfaces are brought together in the complex. It is likely that to maintain the high-affinity interaction with myostatin, the additional contacts that we have identified at the myostatin:Fst288 interface must be formed. Furthermore, heparin binding might actually stabilize the myostatin complex by bridging both Fst molecules concurrently and balancing the electrostatic surface charge. Moreover, heparin-bound Fst may have an increased affinity for myostatin, as the heparin would diminish the putative electrostatic repulsion of myostatin and Fst coming together. This could lead to preferential binding of cell surface-bound Fst to myostatin over activin A. The unexpected observation that myostatin augments heparin binding of Fst suggests that the combination of TGF-β family ligands with other antagonists might also enhance or create novel interfaces with new binding properties for heparin, receptors, or additional antagonists, as expected in the BMP/Chordin/Tsg complex (Oelgeschlager et al, 2000).
Altogether, we have identified several characteristics that make myostatin unique relative to other TGF-β family members. Myostatin and activins share type II receptors, the type I receptor Alk4, and the Fst family of antagonists. On the other hand, myostatin and TGF-βs are similar in that they share the type I receptor Alk5, form latent complexes with their respective propeptides, and interact with the antagonist decorin (Lawrence et al, 1985; Lee and McPherron, 2001; Zhu et al, 2007). Myostatin is an intriguing molecule in that it combines functional and structural characteristics of both the activin and TGF-β classes and is thereby a distinctive family member.
Materials and methods
Protein purification and complex formation
Individual proteins were produced and purified as published earlier with minor alterations (Jiang et al, 2004; Thompson et al, 2005; Lerch et al, 2007; Stamler et al, 2008). Chinese hamster ovary (CHO) cells overexpressing myostatin (McPherron et al, 1997; Lee and McPherron, 2001) were kindly provided by Dr Se-Jin Lee and used to make conditioned media (CM). Additional CM was purchased from Cell Trends (Middletown, MD). CHO cells overexpressing Fst288 and Fst315 were obtained from Dr Shunichi Shimasaki. The Fstl3 plasmid was kindly provided by Dr Henry Keutmann (Stamler et al, 2008). Myostatin complexes were formed by adding myostatin to an excess of Fst or Fstl3 (at least 1:4 molar ratio) and incubating anywhere from 1 h to overnight. Complexes were purified on a Superdex 200 column (Amersham Biosciences). To make activin A complexes, Fst-type proteins were added at molar ratios of 1:2.5 and incubated overnight.
Myostatin:Fst288 complex crystal structure determination
The purified complex of myostatin:Fst288 was concentrated to 3.6 mg/ml and mixed 1:1 in a hanging drop experiment with a well solution containing 125 mM phosphate citrate pH 4.2, 18% ethanol, and 2.5% PEG 1000. Diffraction experiments were performed at the Argonne National Laboratory Advanced Photon Source 22ID beamline. Data were integrated and scaled to 2.15 Å resolution using HKL2000 (Otwinowski et al, 1997). Molecular replacement using PHASER (McCoy et al, 2007) and the activin:Fst288 complex as a search model were used to locate the position of one myostatin:Fst288 complex in the asymmetric unit. The atomic coordinates were refined using REFMAC (Murshudov et al, 1997) along with repeated rounds of model building with COOT (Emsley and Cowtan, 2004). Positional displacement of each chain was described by 7–11 translation/libration/screw (TLS) groups that were identified by the TLSMD server (Painter and Merritt, 2006). Data collection and refinement statistics are shown in Table I, and an example of the electron density map of residues in the prehelix region is shown in Supplementary Figure 2. Structural alignments were performed using CE (Shindyalov and Bourne, 1998), unless otherwise stated. Coordinates and structure factors have been deposited in the PDB with the identifier 3HH2. Buried surface area calculations were done using the PISA webserver (Krissinel and Henrick, 2007). Reported values are the solvent accessible area at the interface and are the average of the areas buried on each of the two ligands. Centre of mass distances were calculated with the CALCOM web server (Costantini et al, 2008). Shape complementarity was calculated using SC (Lawrence and Colman, 1993). Domain motion analysis was done using the DynDom web server (Hayward and Berendsen, 1998)]. Electrostatic surface potentials were calculated using APBS and default parameters (Baker et al, 2001). Ramachandran plot statistics were calculated using MolProbity (Davis et al, 2007) and PROCHECK (Laskowski et al, 1993). Structure figures were rendered using PyMOL (DeLano, 2002).
Table 1.
Data collection and refinements statistics (molecular replacement)
| Native (collected at 100 K) | |
|---|---|
| Data collection | |
| Space group | P212121 |
| Cell dimensions (Å, °) | a=56.6, b=59.5, c=286.1, α=β=γ=90 |
| Resolution (Å) | 50–2.15 (2.23–2.15)a |
| Observations | 471 437 |
| Unique reflections | 53 203 |
| Completeness (%) | 98.7 (88.9) |
| Redundancy | 8.9 (5.5) |
| Rmerge (%) | 9.8 (42.7) |
| Mean I/σI | 16.5 (2.6) |
| Wilson plot B factor (Å2) | 42.2 |
| Model refinement | |
| Reflections (total/free) | 50 306/2702 |
| Rwork/Rfree (%) | 20.8/24.8 |
| Atoms (total/protein) | 6252/5890 |
| Mean B factors (Å2) | |
| All atoms | 25.4 |
| Myostatin (chain A/chain B) | 18.8/20.3 |
| Fst288 (chain C/chain D) | 25.2/28.9 |
| Water (375) | 30.2 |
| Phosphate (3) | 54.7 |
| Citrate (2) | 63.9 |
| r.m.s.d. from ideal | |
| Bonds (Å) | 0.007 |
| Angles (°) | 1.025 |
| Ramachandran plot statistics (number/%)b | |
| Most favoured | 586/89.2 |
| Additionally allowed | 62/9.4 |
| Generously allowed | 7/1.1 |
| Disallowedc | 2/0.3 |
| aValues in parentheses are for highest-resolution shell. | |
| bAs determined for non-gly/pro residues with Procheck. | |
| cAsn138 of both chains of Fst288. | |
Activin A prehelix-switch mutant generation
An activin A construct in the pRK5 vector was a gift of Dr Teresa Woodruff. The prehelix region of activin A (residues 45–58) was replaced with that of myostatin (residues 48–56) using the substitution mutagenesis protocol of Qi and Scholthof (2008).
Luciferase-reporter gene assays
RIB L17 cells (gift of Dr Joan Massagué) were plated in 24-well plates at ∼5 × 104 cells/well. After 24 h, cells were transfected with the CAGA12 luciferase-reporter construct (gift of Dr Anita Roberts (Dennler et al, 1998)) along with empty pRK5 vector, activin A, activin A prehelix-switch, or TGF-β1 (Open Biosystems) constructs using the Mirus LT-1 transfection reagent. A construct containing rat Alk5 or Alk4 in the pRK5 vector was also transfected. Total DNA content was normalized using empty pRK5 vector. Luciferase production was quantified 18 h later using the Luciferase Assay System from Promega.
Binding of activin A:Fst and myostatin:Fst complexes to cultured cells
Activin A and myostatin (R&D Systems) were labelled with Na125I (Perkin Elmer) using the chloramine T (Sigma) method as described earlier (Chapman et al, 2002). Binding of iodinated activin A or myostatin in complex with Fst288 or Fst315 to immortalized murine gonadotrope cells (LβT2) was performed essentially as described in Hashimoto et al (1997) with primary rat pituitary cells. LβT2 cells were plated in 96-well plates (5 × 104 cells/well) in DMEM/10%FBS. After ∼24 h, cells were washed once with growth media and then incubated with 100 μl/well of ligand:Fst complexes for 2 h at 37°C. Complexes were preformed by mixing iodinated activin A or myostatin (0.2 or 2 nM final concentrations) with 0, 25, 100, or 25 ng/ml Fst288 or Fst315 in growth media for 1 h at 37°C before application to cells. Cells were rinsed three times with fresh media and then lysed in 10% SDS (100 μl/well). The amount of cell-bound complex was quantified by γ-counting and is expressed as a percentage bound ((cpm bound/total cpm added) × 100). All treatments were performed in triplicate.
Activin A and myostatin degradation assay
Degradation assays for activin A or myostatin in complex with Fst288 or Fst315 were performed in LβT2 cells essentially as described in Hashimoto et al (1997). Cells were cultured in 96-well plates as described above. Iodinated activin A or myostatin (1 nM) was pre-incubated with Fst288 or Fst315 (0 or 250 ng/ml) in growth media for 1 h at 37°C. Media was aspirated from the wells and replaced with 100 μl/well ligand:Fst complexes. Medium was recovered after 2, 6, 24, or 48 h and mixed with an equal volume of 30% trichloroacetic acid (TCA) solution. After 30 min incubation on ice, samples were centrifuged and radioactivity measured in the total and pelleted fractions by γ-counting. The percentage ligand degradation was calculated ((cpm in TCA soluble fraction/total cpm added per well) × 100). All treatments were performed in triplicate.
Heparin-affinity chromatography
Fst288, Fst315, or Fstl3, alone or in complex with ligand, were applied to a HiTrap 1 ml heparin HP column equilibrated with 50 mM NaHCO3, 150 mM NaCl, pH 7.4, and eluted with a 120-min gradient to 2 M NaCl at 1 ml/min; 2 ml fractions were collected. The following antibodies from R&D Systems were used: anti-Fst (AF669), anti-activin A (AF338), anti-myostatin (AF788), and anti-FLRG (AF1288).
Supplementary Material
Supplementary Figure 1
Supplementary Figure 2
Supplementary Figure 3
Supplementary Figure 4
Supplementary Figure 5
Supplementary Figure 6
Review Process File
Acknowledgments
We thank C Kattamuri and E Angerman for activin A purification, and R Kovall and A Herr for comments on the paper. This work was supported by research grants from the NIHGM R01 (GM084186), AHA (0635097N), and MDA (93887) to TBT. JNC was supported by an AHA predoctoral fellowship. Support was provided by Canadian Institutes of Health Research grant MOP-89991 to DJB who is a Chercheur-boursier of the Fonds de la recherche en santé du Québec. Support was contributed by the Intramural Research Program of the NIDDK, NIH, USA (ACM). CAR was partly supported by a CIHR award to Terry Hébert. Author contributions: JNC performed crystallographic structure determination and analysis, luciferase assays, heparin-binding experiments, and prepared the paper; JNC and TBT designed experiments and discussed results; DJB and CAR performed cell surface binding and degradation assays; ACM provided correspondence and conditioned media from Cell Trends; TBT assisted with data collection, analysis, and paper preparation.
Footnotes
The authors declare that they have no conflict of interest.
References
- Allendorph GP, Isaacs MJ, Kawakami Y, Belmonte JC, Choe S (2007) BMP-3 and BMP-6 structures illuminate the nature of binding specificity with receptors. Biochemistry 46: 12238–12247 [DOI] [PubMed] [Google Scholar]
- Allendorph GP, Vale WW, Choe S (2006) Structure of the ternary signaling complex of a TGF-β superfamily member. Proc Natl Acad Sci USA 103: 7643–7648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amthor H, Nicholas G, McKinnell I, Kemp CF, Sharma M, Kambadur R, Patel K (2004) Follistatin complexes myostatin and antagonises myostatin-mediated inhibition of myogenesis. Dev Biol 270: 19–30 [DOI] [PubMed] [Google Scholar]
- Andersson O, Reissmann E, Ibanez C (2006) Growth differentiation factor 11 signals through the transforming growth factor-β receptor ALK5 to regionalize the anterior-posterior axis. EMBO Rep 7: 831–837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Attisano L, Carcamo J, Ventura F, Weis FM, Massague J, Wrana JL (1993) Identification of human activin and TGF β type I receptors that form heteromeric kinase complexes with type II receptors. Cell 75: 671–680 [DOI] [PubMed] [Google Scholar]
- Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA (2001) Electrostatics of nanosystems: application to microtubules and the ribosome. Proc Natl Acad Sci USA 98: 10037–10041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassing CH, Yingling JM, Howe DJ, Wang T, He WW, Gustafson ML, Shah P, Donahoe PK, Wang XF (1994) A transforming growth factor β type I receptor that signals to activate gene expression. Science 263: 87–89 [DOI] [PubMed] [Google Scholar]
- Bradley L, Yaworsky P, Walsh F (2008) Myostatin as a therapeutic target for musculoskeletal disease. Cell Mol Life Sci 65: 2119–2124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carcamo J, Weis FM, Ventura F, Wieser R, Wrana JL, Attisano L, Massague J (1994) Type I receptors specify growth-inhibitory and transcriptional responses to transforming growth factor β and activin. Mol Cell Biol 14: 3810–3821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman SC, Bernard DJ, Jelen J, Woodruff TK (2002) Properties of inhibin binding to betaglycan, InhBP/p120 and the activin type II receptors. Mol Cell Endocrinol 196: 79–93 [DOI] [PubMed] [Google Scholar]
- Clop A, Marcq F, Takeda H, Pirottin D, Tordoir X, Bibe B, Bouix J, Caiment F, Elsen JM, Eychenne F, Larzul C, Laville E, Meish F, Milenkovic D, Tobin J, Charlier C, Georges M (2006) A mutation creating a potential illegitimate microRNA target site in the myostatin gene affects muscularity in sheep. Nat Genet 38: 813–818 [DOI] [PubMed] [Google Scholar]
- Cook RW, Thompson TB, Kurup SP, Jardetzky TS, Woodruff TK (2005) Structural basis for a functional antagonist in the transforming growth factor beta superfamily. JBC 280: 40177–40186 [DOI] [PubMed] [Google Scholar]
- Costantini S, Paladino A, Facchiano AM (2008) CALCOM: a software for calculating the center of mass of proteins. Bioinformation 2: 271–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daopin S, Piez KA, Ogawa Y, Davies DR (1992) Crystal structure of transforming growth factor-β 2: an unusual fold for the superfamily. Science 257: 369–373 [DOI] [PubMed] [Google Scholar]
- Davis I, Leaver-Fay A, Chen V, Block J, Kapral G, Wang X, Murray L, Arendall W, Snoeyink J, Richardson J, Richardson D (2007) MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res 35: W375–W383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLano WL (2002) The PyMOL Molecular Graphics System. Palo Alto, CA, USA: DeLano Scientific [Google Scholar]
- Dennler S, Itoh S, Vivien D, ten Dijke P, Huet S, Gauthier JM (1998) Direct binding of Smad3 and Smad4 to critical TGF β-inducible elements in the promoter of human plasminogen activator inhibitor-type 1 gene. EMBO J 17: 3091–3100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Cowtan K (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60: 2126–2132 [DOI] [PubMed] [Google Scholar]
- Franzèn P, ten Dijke P, Ichijo H, Yamashita H, Schulz P, Heldin C-H, Miyazono K (1993) Cloning of a TGFβ type I receptor that forms a heteromeric complex with the TGFβ type II receptor. Cell 75: 681–692 [DOI] [PubMed] [Google Scholar]
- Greenwald J, Groppe J, Gray P, Wiater E, Kwiatkowski W, Vale W, Choe S (2003) The BMP7/ActRII extracellular domain complex provides new insights into the cooperative nature of receptor assembly. Mol Cell 11: 605–617 [DOI] [PubMed] [Google Scholar]
- Greenwald J, Vega ME, Allendorph GP, Fischer WH, Vale W, Choe S (2004) A flexible activin explains the membrane-dependent cooperative assembly of TGF-β family receptors. Mol Cell 15: 485–489 [DOI] [PubMed] [Google Scholar]
- Grobet L, Martin LJ, Poncelet D, Pirottin D, Brouwers B, Riquet J, Schoeberlein A, Dunner S, Menissier F, Massabanda J, Fries R, Hanset R, Georges M (1997) A deletion in the bovine myostatin gene causes the double-muscled phenotype in cattle. Nat Genet 17: 71–74 [DOI] [PubMed] [Google Scholar]
- Groppe J, Greenwald J, Wiater E, Rodriguez-Leon J, Economides AN, Kwiatkowski W, Affolter M, Vale WW, Belmonte JC, Choe S (2002) Structural basis of BMP signalling inhibition by the cystine knot protein Noggin. Nature 420: 636–642 [DOI] [PubMed] [Google Scholar]
- Groppe J, Hinck CS, Samavarchi-Tehrani P, Zubieta C, Schuermann JP, Taylor AB, Schwarz PM, Wrana JL, Hinck AP (2008) Cooperative assembly of TGF-β superfamily signaling complexes is mediated by two disparate mechanisms and distinct modes of receptor binding. Mol Cell 29: 157–168 [DOI] [PubMed] [Google Scholar]
- Harrington AE, Morris-Triggs SA, Ruotolo BT, Robinson CV, Ohnuma S, Hyvonen M (2006) Structural basis for the inhibition of activin signalling by follistatin. EMBO J 25: 1035–1045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison CA, Chan KL, Robertson DM (2006) Activin-A binds follistatin and type II receptors through overlapping binding sites: generation of mutants with isolated binding activities. Endocrinology 147: 2744–2753 [DOI] [PubMed] [Google Scholar]
- Harrison CA, Gray PC, Fischer WH, Donaldson C, Choe S, Vale W (2004) An activin mutant with disrupted ALK4 binding blocks signaling via type II receptors. J Biol Chem 279: 28036–28044 [DOI] [PubMed] [Google Scholar]
- Harrison CA, Gray PC, Koerber SC, Fischer W, Vale W (2003) Identification of a functional binding site for activin on the type I receptor ALK4. J Biol Chem 278: 21129–21135 [DOI] [PubMed] [Google Scholar]
- Hart PJ, Deep S, Taylor AB, Shu Z, Hinck CS, Hinck AP (2002) Crystal structure of the human TβR2 ectodomain—TGF-β3 complex. Nat Struct Biol 9: 203–208 [DOI] [PubMed] [Google Scholar]
- Hashimoto O, Kawasaki N, Tsuchida K, Shimasaki S, Hayakawa T, Sugino H (2000) Difference between follistatin isoforms in the inhibition of activin signalling: activin neutralizing activity of follistatin isoforms is dependent on their affinity for activin. Cell Signal 12: 565–571 [DOI] [PubMed] [Google Scholar]
- Hashimoto O, Nakamura T, Shoji H, Shimasaki S, Hayashi Y, Sugino H (1997) A novel role of follistatin, an activin-binding protein, in the inhibition of activin action in rat pituitary cells. Endocytotic degradation of activin and its acceleration by follistatin associated with cell-surface heparan sulfate. J Biol Chem 272: 13835–13842 [DOI] [PubMed] [Google Scholar]
- Hayward S, Berendsen HJ (1998) Systematic analysis of domain motions in proteins from conformational change: new results on citrate synthase and T4 lysozyme. Proteins 30: 144–154 [PubMed] [Google Scholar]
- Hill JJ, Davies MV, Pearson AA, Wang JH, Hewick RM, Wolfman NM, Qiu Y (2002) The myostatin propeptide and the follistatin-related gene are inhibitory binding proteins of myostatin in normal serum. J Biol Chem 277: 40735–40741 [DOI] [PubMed] [Google Scholar]
- Hill JJ, Qiu Y, Hewick RM, Wolfman NM (2003) Regulation of myostatin in vivo by growth and differentiation factor-associated serum protein-1: a novel protein with protease inhibitor and follistatin domains. Mol Endocrinol 17: 1144–1154 [DOI] [PubMed] [Google Scholar]
- Iemura S, Yamamoto TS, Takagi C, Uchiyama H, Natsume T, Shimasaki S, Sugino H, Ueno N (1998) Direct binding of follistatin to a complex of bone-morphogenetic protein and its receptor inhibits ventral and epidermal cell fates in early Xenopus embryo. Proc Natl Acad Sci USA 95: 9337–9342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Innis CA, Hyvonen M (2003) Crystal structures of the heparan sulfate-binding domain of follistatin. Insights into ligand binding. J Biol Chem 278: 39969–39977 [DOI] [PubMed] [Google Scholar]
- Innis CA, Shi J, Blundell TL (2000) Evolutionary trace analysis of TGF-β and related growth factors: implications for site-directed mutagenesis. Protein Eng 13: 839–847 [DOI] [PubMed] [Google Scholar]
- Jiang MS, Liang LF, Wang S, Ratovitski T, Holmstrom J, Barker C, Stotish R (2004) Characterization and identification of the inhibitory domain of GDF-8 propeptide. Biochem Biophys Res Commun 315: 525–531 [DOI] [PubMed] [Google Scholar]
- Kambadur R, Sharma M, Smith TP, Bass JJ (1997) Mutations in myostatin (GDF8) in double-muscled Belgian Blue and Piedmontese cattle. Genome Res 7: 910–916 [DOI] [PubMed] [Google Scholar]
- Keller S, Nickel J, Zhang JL, Sebald W, Mueller TD (2004) Molecular recognition of BMP-2 and BMP receptor IA. Nat Struct Mol Biol 11: 481–488 [DOI] [PubMed] [Google Scholar]
- Keutmann HT, Schneyer AL, Sidis Y (2004) The role of follistatin domains in follistatin biological action. Mol Endocrinol 18: 228–240 [DOI] [PubMed] [Google Scholar]
- Kirsch T, Sebald W, Dreyer MK (2000) Crystal structure of the BMP-2-BRIA ectodomain complex. Nat Struct Biol 7: 492–496 [DOI] [PubMed] [Google Scholar]
- Korupolu RV, Muenster U, Read JD, Vale W, Fischer WH (2008) Activin A/bone morphogenetic protein (BMP) chimeras exhibit BMP-like activity and antagonize activin and myostatin. J Biol Chem 283: 3782–3790 [DOI] [PubMed] [Google Scholar]
- Kotzsch A, Nickel J, Seher A, Sebald W, Muller TD (2009) Crystal structure analysis reveals a spring-loaded latch as molecular mechanism for GDF-5-type I receptor specificity. EMBO J 28: 937–947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krissinel E, Henrick K (2007) Inference of macromolecular assemblies from crystalline state. J Mol Biol 372: 774–797 [DOI] [PubMed] [Google Scholar]
- Laskowski RA, MacArthur MW, Moss DS, Thornton JM (1993) PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Crystallogr 26: 283–291 [Google Scholar]
- Lavery K, Swain P, Falb D, Alaoui-Ismaili MH (2008) BMP-2/4 and BMP-6/7 differentially utilize cell surface receptors to induce osteoblastic differentiation of human bone marrow-derived mesenchymal stem cells. J Biol Chem 283: 20948–20958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence DA, Pircher R, Jullien P (1985) Conversion of a high molecular weight latent β-TGF from chicken embryo fibroblasts into a low molecular weight active β-TGF under acidic conditions. Biochem Biophys Res Commun 133: 1026–1034 [DOI] [PubMed] [Google Scholar]
- Lawrence MC, Colman PM (1993) Shape complementarity at protein/protein interfaces. J Mol Biol 234: 946–950 [DOI] [PubMed] [Google Scholar]
- Lee SJ (2007) Quadrupling muscle mass in mice by targeting TGF-β signaling pathways. PLoS One 2: e789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SJ, McPherron AC (2001) Regulation of myostatin activity and muscle growth. Proc Natl Acad Sci USA 98: 9306–9311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SJ, Reed LA, Davies MV, Girgenrath S, Goad ME, Tomkinson KN, Wright JF, Barker C, Ehrmantraut G, Holmstrom J, Trowell B, Gertz B, Jiang MS, Sebald SM, Matzuk M, Li E, Liang LF, Quattlebaum E, Stotish RL, Wolfman NM (2005) Regulation of muscle growth by multiple ligands signaling through activin type II receptors. Proc Natl Acad Sci USA 102: 18117–18122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerch TF, Shimasaki S, Woodruff TK, Jardetzky TS (2007) Structural and biophysical coupling of heparin and activin binding to follistatin isoform functions. J Biol Chem 282: 15930–15939 [DOI] [PubMed] [Google Scholar]
- Ling N, Ying SY, Ueno N, Shimasaki S, Esch F, Hotta M, Guillemin R (1986) Pituitary FSH is released by a heterodimer of the β-subunits from the two forms of inhibin. Nature 321: 779–782 [DOI] [PubMed] [Google Scholar]
- Little SC, Mullins MC (2009) Bone morphogenetic protein heterodimers assemble heteromeric type I receptor complexes to pattern the dorsoventral axis. Nat Cell Biol 11: 637–643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy A, Grosse-Kunstleve R, Adams P, Winn M, Storoni L, Read R (2007) Phasercrystallographic software. J Appl Crystallogr 40: 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McPherron AC, Lawler AM, Lee SJ (1997) Regulation of skeletal muscle mass in mice by a new TGF-β superfamily member. Nature 387: 83–90 [DOI] [PubMed] [Google Scholar]
- McPherron AC, Lee SJ (1997) Double muscling in cattle due to mutations in the myostatin gene. Proc Natl Acad Sci USA 94: 12457–12461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellor SL, Ball EM, O'Connor AE, Ethier JF, Cranfield M, Schmitt JF, Phillips DJ, Groome NP, Risbridger GP (2003) Activin βC-subunit heterodimers provide a new mechanism of regulating activin levels in the prostate. Endocrinology 144: 4410–4419 [DOI] [PubMed] [Google Scholar]
- Mosher DS, Quignon P, Bustamante CD, Sutter NB, Mellersh CS, Parker HG, Ostrander EA (2007) A mutation in the myostatin gene increases muscle mass and enhances racing performance in heterozygote dogs. PLoS Genet 3: e79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Dodson EJ (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr 53: 240–255 [DOI] [PubMed] [Google Scholar]
- Nakatani M, Takehara Y, Sugino H, Matsumoto M, Hashimoto O, Hasegawa Y, Murakami T, Uezumi A, Takeda S, Noji S, Sunada Y, Tsuchida K (2007) Transgenic expression of a myostatin inhibitor derived from follistatin increases skeletal muscle mass and ameliorates dystrophic pathology in mdx mice. Faseb J 22: 477–487 [DOI] [PubMed] [Google Scholar]
- Nakatani M, Takehara Y, Sugino H, Matsumoto M, Hashimoto O, Hasegawa Y, Murakami T, Uezumi A, Takeda S, Noji S, Sunada Y, Tsuchida K (2008) Transgenic expression of a myostatin inhibitor derived from follistatin increases skeletal muscle mass and ameliorates dystrophic pathology in mdx mice. FASEB J 22: 477–487 [DOI] [PubMed] [Google Scholar]
- Nickel J, Kotzsch A, Sebald W, Mueller TD (2005) A single residue of GDF-5 defines binding specificity to BMP receptor IB. J Mol Biol 349: 933–947 [DOI] [PubMed] [Google Scholar]
- Oelgeschlager M, Larrain J, Geissert D, De Robertis EM (2000) The evolutionarily conserved BMP-binding protein twisted gastrulation promotes BMP signalling. Nature 405: 757–763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W, Charles Wcarter Jr (1997) [20] Processing of X-ray diffraction data collected in oscillation mode. In Methods in Enzymology, Vol. 276, Macromolecular Crystallography, part A, pp 307–326. Academic Press [DOI] [PubMed] [Google Scholar]
- Painter J, Merritt EA (2006) TLSMD web server for the generation of multi-group TLS models. J Appl Cryst 39: 109–111 [Google Scholar]
- Qi D, Scholthof KB (2008) A one-step PCR-based method for rapid and efficient site-directed fragment deletion, insertion, and substitution mutagenesis. J Virol Methods 149: 85–90 [DOI] [PubMed] [Google Scholar]
- Rebbapragada A, Benchabane H, Wrana JL, Celeste AJ, Attisano L (2003) Myostatin signals through a transforming growth factor β-like signaling pathway to block adipogenesis. Mol Cell Biol 23: 7230–7242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rechtman MM, Nakaryakov A, Shapira K, Ehrlich M, Henis Y (2009) Different domains regulate homomeric and heteromeric complex formation among type I and type II TGF-β receptors. J Biol Chem 284: 7843–7852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saremba S, Nickel J, Seher A, Kotzsch A, Sebald W, Mueller TD (2008) Type I receptor binding of bone morphogenetic protein 6 is dependent on N-glycosylation of the ligand. FEBS J 275: 172–183 [DOI] [PubMed] [Google Scholar]
- Schlessinger J, Plotnikov AN, Ibrahimi OA, Eliseenkova AV, Yeh BK, Yayon A, Linhardt RJ, Mohammadi M (2000) Crystal structure of a ternary FGF-FGFR-heparin complex reveals a dual role for heparin in FGFR binding and dimerization. Mol Cell 6: 743–750 [DOI] [PubMed] [Google Scholar]
- Schmierer B, Hill CS (2005) Kinetic analysis of Smad nucleocytoplasmic shuttling reveals a mechanism for transforming growth factor β-dependent nuclear accumulation of Smads. Mol Cell Biol 25: 9845–9858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider-Kolsky M, D'Antona D, Evans LW, Taylor N, O'Connor A, Groome NP, de Kretser D, Wallace EM (2000) Maternal serum total activin A and follistatin in pregnancy and parturition. BJOG 107: 995–1000 [DOI] [PubMed] [Google Scholar]
- Schneyer AL, Sidis Y, Gulati A, Sun JL, Keutmann H, Krasney PA (2008) Differential antagonism of activin, myostatin and growth and differentiation factor 11 by wild-type and mutant follistatin. Endocrinology 149: 4589–4595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneyer AL, Wang Q, Sidis Y, Sluss PM (2004) Differential distribution of follistatin isoforms: application of a new FS315-specific immunoassay. J Clin Endocrinol Metab 89: 5067–5075 [DOI] [PubMed] [Google Scholar]
- Schuelke M, Wagner KR, Stolz LE, Hubner C, Riebel T, Komen W, Braun T, Tobin JF, Lee SJ (2004) Myostatin mutation associated with gross muscle hypertrophy in a child. N Engl J Med 350: 2682–2688 [DOI] [PubMed] [Google Scholar]
- Sebald W, Nickel J, Zhang JL, Mueller TD (2004) Molecular recognition in bone morphogenetic protein (BMP)/receptor interaction. Biol Chem 385: 697–710 [DOI] [PubMed] [Google Scholar]
- Shimmi O, Umulis D, Othmer H, O'Connor MB (2005) Facilitated transport of a Dpp/Scw heterodimer by Sog/Tsg leads to robust patterning of the Drosophila blastoderm embryo. Cell 120: 873–886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shindyalov IN, Bourne PE (1998) Protein structure alignment by incremental combinatorial extension (CE) of the optimal path. Protein Eng 11: 739–747 [DOI] [PubMed] [Google Scholar]
- Sidis Y, Mukherjee A, Keutmann H, Delbaere A, Sadatsuki M, Schneyer A (2006) Biological activity of follistatin isoforms and follistatin-like-3 is dependent on differential cell surface binding and specificity for activin, myostatin, and bone morphogenetic proteins. Endocrinology 147: 3586–3597 [DOI] [PubMed] [Google Scholar]
- Sidis Y, Schneyer AL, Keutmann HT (2005) Heparin and activin-binding determinants in follistatin and FSTL3. Endocrinology 146: 130–136 [DOI] [PubMed] [Google Scholar]
- Sidis Y, Schneyer AL, Sluss PM, Johnson LN, Keutmann HT (2001) Follistatin: essential role for the N-terminal domain in activin binding and neutralization. J Biol Chem 276: 17718–17726 [DOI] [PubMed] [Google Scholar]
- Stamler R, Keutmann HT, Sidis Y, Kattamuri C, Schneyer A, Thompson TB (2008) The structure of FSTL3.activin A complex. Differential binding of N-terminal domains influences follistatin-type antagonist specificity. J Biol Chem 283: 32831–32838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugino K, Kurosawa N, Nakamura T, Takio K, Shimasaki S, Ling N, Titani K, Sugino H (1993) Molecular heterogeneity of follistatin, an activin-binding protein. Higher affinity of the carboxyl-terminal truncated forms for heparan sulfate proteoglycans on the ovarian granulosa cell. J Biol Chem 268: 15579–15587 [PubMed] [Google Scholar]
- ten Dijke P, Yamashita H, Ichijo H, Franzen P, Laiho M, Miyazono K, Heldin CH (1994) Characterization of type I receptors for transforming growth factor-β and activin. Science 264: 101–104 [DOI] [PubMed] [Google Scholar]
- Thies RS, Chen T, Davies MV, Tomkinson KN, Pearson AA, Shakey QA, Wolfman NM (2001) GDF-8 propeptide binds to GDF-8 and antagonizes biological activity by inhibiting GDF-8 receptor binding. Growth Factors 18: 251–259 [DOI] [PubMed] [Google Scholar]
- Thompson L, Pantoliano M, Springer B (1994) Energetic characterization of the basic fibroblast growth factor-heparin interaction: identification of the heparin binding domain. Biochemistry 33: 3831–3840 [DOI] [PubMed] [Google Scholar]
- Thompson TB, Lerch TF, Cook RW, Woodruff TK, Jardetzky TS (2005) The structure of the follistatin:activin complex reveals antagonism of both type I and type II receptor binding. Dev Cell 9: 535–543 [DOI] [PubMed] [Google Scholar]
- Thompson TB, Woodruff TK, Jardetzky TS (2003) Structures of an ActRIIB:activin A complex reveal a novel binding mode for TGF-β ligand:receptor interactions. EMBO J 22: 1555–1566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchida K (2008) Targeting myostatin for therapies against muscle-wasting disorders. Curr Opin Drug Discov Devel 11: 487–494 [PubMed] [Google Scholar]
- Ueno N, Ling N, Ying SY, Esch F, Shimasaki S, Guillemin R (1987) Isolation and partial characterization of follistatin: a single-chain Mr 35,000 monomeric protein that inhibits the release of follicle-stimulating hormone. Proc Natl Acad Sci USA 84: 8282–8286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber D, Kotzsch A, Nickel J, Harth S, Seher A, Mueller U, Sebald W, Mueller TD (2007) A silent H-bond can be mutationally activated for high-affinity interaction of BMP-2 and activin type IIB receptor. BMC Struct Biol 7: 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita H, ten Dijke P, Huylebroeck D, Sampath TK, Andries M, Smith JC, Heldin CH, Miyazono K (1995) Osteogenic protein-1 binds to activin type II receptors and induces certain activin-like effects. J Cell Biol 130: 217–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Li Y, Shen W, Qiao C, Ambrosio F, Lavasani M, Nozaki M, Branca MF, Huard J (2007) Relationships between transforming growth factor-β1, myostatin, and decorin: implications for skeletal muscle fibrosis. J Biol Chem 282: 25852–25863 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1
Supplementary Figure 2
Supplementary Figure 3
Supplementary Figure 4
Supplementary Figure 5
Supplementary Figure 6
Review Process File