Abstract
If replication forks are perturbed, a multifaceted response including several DNA repair and cell cycle checkpoint pathways is activated to ensure faithful DNA replication. Here, we show that poly(ADP-ribose) polymerase 1 (PARP1) binds to and is activated by stalled replication forks that contain small gaps. PARP1 collaborates with Mre11 to promote replication fork restart after release from replication blocks, most likely by recruiting Mre11 to the replication fork to promote resection of DNA. Both PARP1 and PARP2 are required for hydroxyurea-induced homologous recombination to promote cell survival after replication blocks. Together, our data suggest that PARP1 and PARP2 detect disrupted replication forks and attract Mre11 for end processing that is required for subsequent recombination repair and restart of replication forks.
Keywords: homologous recombination, Mre11, poly(ADP-ribose) polymerase, replication restart, stalled replication forks
Introduction
Poly(ADP-ribose) polymerase 1 (PARP1) is an abundant nuclear protein that is activated by DNA-strand breaks; activation of PARP1 leads to automodification and modification of other acceptor proteins with poly(ADP-ribose) (PAR) polymers (Satoh and Lindahl, 1992). PARP1 protects DNA breaks and chromatin structure and recruits DNA repair and checkpoint proteins to sites of damage (Allinson et al, 2003; Ahel et al, 2008). Inhibition of PARP1 is synthetic lethal with defects in homologous recombination (HR) and is currently being tested as a monotherapy for heritable breast and ovarian cancers deficient in the BRCA1 or BRCA2 genes (Bryant et al, 2005; Farmer et al, 2005; Helleday et al, 2008). PARP2, another nuclear member of the PARP family with largely unknown function, shares homology with PARP1 and is also activated by DNA breaks (Ame et al, 1999). Embryonic knockout of either PARP1 or PARP2 is well tolerated; however, double knockout is embryonic lethal (Menissier de Murcia et al, 2003) suggesting that PARP1 and PARP2 can compensate for some of each other's functions.
PARP activity has been found to be enhanced in replicating cells (Lehmann et al, 1974), in the vicinity of replication forks (Jump et al, 1979) and in newly replicated chromatin (Anachkova et al, 1989). In addition, PARP1 has been shown to interact with several DNA replication proteins, many of which were poly(ADP-ribosyl)ated (Simbulan et al, 1993; Simbulan-Rosenthal et al, 1996; Dantzer et al, 1998). It has, therefore, been postulated that PARP1 might be involved in mammalian DNA replication. After treatment with hydroxyurea (HU), which stalls replication forks by depleting dNTP pools, PARP-1−/− cells have been shown to display delayed progress from S into G2/M phase (Yang et al, 2004), and although the underlying molecular mechanisms are not yet clear, a role for PARP1 in the response to replication fork stalling has been suggested. The PARP proteins are unique to higher eukaryotes and there is no evidence of PARP activity in Saccharomyces cerevisiae or prokaryotes.
Regulation of DNA replication has been recognised as an important mechanism for preventing carcinogenesis, as impaired replication fork progression and increased replication-dependent DNA damage were observed in early stages of tumour development (Bartkova et al, 2006; Di Micco et al, 2006). In particular, the efficient reactivation of stalled replication forks is considered essential to maintain faithful replication and genomic stability. In Escherichia coli, stalled or collapsed replication forks are reactivated by recombination-dependent or -independent pathways, catalysed by the RuvABC or PriA and PriC proteins, respectively (Heller and Marians, 2006). These proteins are not conserved in eukaryotes, and eukaryotic mechanisms of replication fork reactivation are not well characterised. In yeast, HU-induced fork stalling is reversible and forks only collapse in certain backgrounds, for example rad53 mutants (Lopes et al, 2001); it is only in those backgrounds that HR is triggered for repair (Meister et al, 2005). This is in contrast to higher eukaryotes, in which HU triggers HR directly in wild-type mammalian cells (Arnaudeau et al, 2001; Saintigny et al, 2001; Lundin et al, 2002), suggesting that recombination-dependent replication restart mechanisms might be important in higher eukaryotes. Stalled forks are likely to need processing before HR medicated restart can take place, and this is supported by the fact that the end-processing Mre11 protein relocates to stalled replication forks after replication inhibition (Robison et al, 2004; Hanada et al, 2007). Here, we show that PARP1 binds to and is activated at stalled replication forks to mediate recruitment of Mre11 to initiate the end processing required for replication restart and HR.
Results
PARP is activated on replication stalling and required for survival
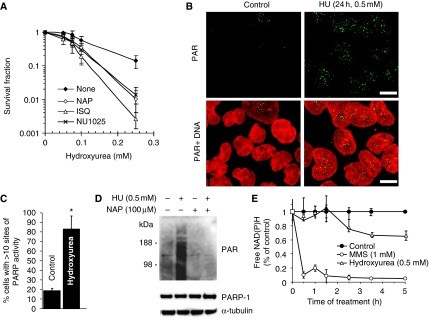
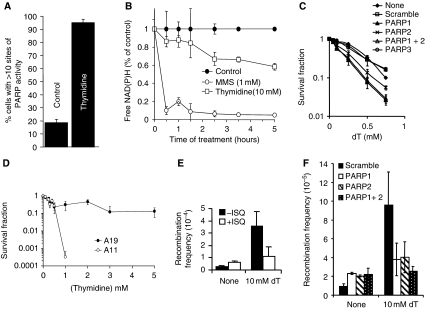
Several lines of evidence suggest that PARP is associated with replication forks (Lehmann et al, 1974; Jump et al, 1979; Anachkova et al, 1989; Simbulan et al, 1993; Simbulan-Rosenthal et al, 1996; Dantzer et al, 1998; Yang et al, 2004). Here, we wanted to test whether PARP1 itself is involved in the response to stalled replication forks. We treated PARP1−/− mouse embryonic fibroblasts (MEFs) or PARP-inhibited cells with HU, which depletes dNTP pools and stalls replication forks (Bianchi et al, 1986). We found that both PARP-inhibited and PARP1−/− cells are sensitive to increasing doses of HU as compared with wild-type cells (Figure 1A; Supplementary Figure S1), showing that PARP is required for survival of replication fork stalling and confirming the HU sensitivity in PARP1−/− MEFs reported earlier (Yang et al, 2004). To further understand the role of PARP in promoting survival to HU, we tested whether PARP activity is also triggered by replication fork stalling, which would indicate an active role for PARP in the response to stalled forks. We found that the products of PARP activity, PAR polymers, are formed in cells treated with HU (Figure 1B–D). It is well established that PARP is activated by DNA single-strand breaks (SSBs), produced directly or as a base excision repair intermediate (Satoh and Lindahl, 1992), and that this attracts SSB repair proteins for repair (El-Khamisy et al, 2003). When investigating the kinetics of PARP activation, we found that HU treatment triggers a much slower PARP response than treatment with the alkylating agent methylmethane sulphonate (MMS) (Figure 1E) and, this is likely to be because of the low number of active forks that can be stalled by HU treatment in an asynchronous cell population.
Figure 1.
PARP is activated at stalled replication forks and required for survival of HU-induced replication stalling. (A) Surviving fraction of AA8 hamster cells treated for 10 days with increasing doses of HU in the presence or absence of PARP inhibitors NU1025 (100 nM), 1,5-dihydroxyisoquinoline (ISQ; 0.6 mM) or 4-amino-1,8-NAP (100 μM). (B) Immunofluorescence staining for PAR in AA8 hamster cells treated for 24 h with or without 0.5 mM HU. DNA was counterstained with TO-PRO-3 iodide. Bar 10 μm. (C) Quantification of immunofluorescence staining above. Percentage of AA8 cells containing sites of PARP activity induced by a 24-h treatment with 0.5 mM HU. Differences are statistically significant (Student's t-test, P<0.05). (D) Western blot analysis of PAR (top), PARP1 (middle) and α-tubulin (bottom) in myc-PARP-expressing U2OS cells treated with combinations of 0.5 mM HU and 100 μM NAP. (E) PARP activity measured by the decrease of free NAD(P)H over time during incubation with 0.5 mM HU or 1 mM MMS.
PARP binds to and is activated by stalled replication fork structures in vitro
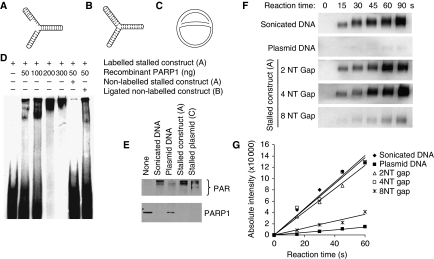
It is well established that the PARP1 zinc-finger domains bind with high affinity to DNA double-strand ends as well as to other DNA structures (D'Silva et al, 1999). It is likely that PARP1 can bind to double-strand breaks (DSBs) formed when replication forks collapse (Saintigny et al, 2001; Lundin et al, 2002). However, we wanted to test whether PARP1, in addition to its ability to bind DSBs, can bind to and is activated by stalled replication forks without DSBs. To test whether PARP1 has the ability to bind a fork structure, an artificial stalled fork substrate was designed and PARP1 binding determined in an electrophoretic mobility shift assay (Figure 2A; Supplementary Figure S2). The substrate contained sealed ends to exclude the binding of PARP1 to double-stranded DNA ends, which has been reported (D'Amours et al, 1999). We found that the PARP1 protein binds to the stalled fork substrate in a concentration-dependent manner (Figure 2D). PARP1 binds specifically to the gap in the replication fork-like region, as the binding was not competed for by a similar substrate ligated to form a closed DNA structure (Figure 2B). We also found that recombinant PARP1 is activated by the same replication fork substrate (Figure 2E). To further understand the DNA structure activating PARP1 at stalled forks, we tested how different gap sizes influence PARP1 activation. We found that a gap size of four nucleotides activated PARP1 equally effectively as the positive control (sonicated DNA) and increased the activity 10-fold as compared with the plasmid control suggesting that PARP1 is fully activated by a short gap (Figure 2F and G). Increasing the gap size to eight nucleotides dramatically decreased PARP1 activation, suggesting that PARP1 only recognises fork structures without extensive single-stranded DNA (ssDNA) regions.
Figure 2.
PARP1 binds to and is activated by DNA fork structures in vitro. (A) Biotin-labelled stalled fork construct and (B) ligated construct, containing sealed DNA ends. (C) Early replication intermediate of the plasmid pBROTB535, containing replication forks stalled in vitro by omission of topoisomerase from the replication reaction. (D) Electrophoretic mobility shift assay using biotin-labelled artificial stalled fork substrate and increasing concentrations of purified PARP1 protein with or without a 10-fold excess of non-labelled competitor stalled fork substrate or ligated construct. (E) Western blot analysis of PARP1 (bottom) and PAR (top) after incubation of 50 ng purified PARP protein with 50 ng of different DNA substrates. Automodification reduces the electrophoretic mobility of PARP1, accounting for the decreased amounts of unmodified PARP1 protein detectable at its expected molecular size in these samples. (F) PARP1 activation by increasing length of gap within the stalled fork structure (A). Recombinant human PARP1 (5 nM) was incubated with DNA constructs and biotinylated NAD+ for the times indicated, and blots were probed with anti-biotin antibody. Sonicated DNA was used as positive control and plasmid DNA as negative control. (G) Quantification of PARP1 activation as in (F).
We wanted to know whether naturally stalled replication forks could also activate PARP1. To create such a replication intermediate, a standard oriC DNA replication mixture was used to initiate replication of the DNA plasmid pBROTB535 (Hiasa and Marians, 1994); however, topoisomerase was omitted resulting in prevention of fork progression by positively supercoiled DNA and accumulation of an early replication intermediate (McGlynn et al, 2001) (Figure 2C). It has been shown earlier that such stalled replication forks can reverse into a chicken foot structure that includes a Holliday Junction (Postow et al, 2001). We found that PARP1 is five-fold more activated by the stalled plasmid as compared with plasmid control (Figure 2E), suggesting that stalled replication forks are substrates for PARP1, at least in vitro.
In conclusion, we believe that PARP1 may bind to and be activated in the absence of DSBs at stalled replication forks, which contain short ssDNA regions.
PARP is activated at sites of stalled forks in mammalian cells
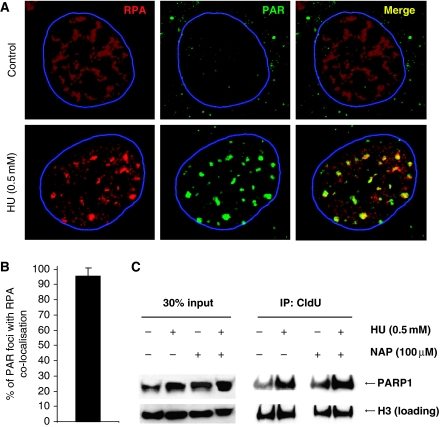
It has been shown that ssDNA regions form at replication forks stalled by HU and that this ssDNA is rapidly coated with Replication Protein A (RPA) to activate the ATR kinase (Zou and Elledge, 2003; Robison et al, 2004). In line with earlier reports, we found that the RPA relocates into nuclear foci after HU treatment (Robison et al, 2004). Here, we found that the HU-induced RPA foci co-localise with HU-induced PAR polymers, supporting the hypothesis that PARP is activated at sites of stalled and collapsed replication forks in mammalian cells (Figure 3A and B).
Figure 3.
HU induces RPA foci that co-localise with sites of activated PARP. (A) HU-induced RPA foci in wild-type MEFs, representing ssDNA, co-localise with PAR polymers formed in response to 0.5 mM HU treatment for 24 h. DNA was counterstained with TO-PRO-3 iodide. The nuclei borders are marked with blue. (B) Percentages of PAR foci co-localising with RPA. Cells with at least 10 PAR foci with a diameter over 1 μm in an optical section of 1.5 μm taken in the middle of the cell were analysed for co-localisation. The mean and standard deviation from 10 cells are depicted. (C) PARP1 associates with stalled replication forks independently of PARP activity. Forks were isolated by CldU co-immunoprecipitation (co-IP) after a 3-h treatment with 0.5 mM HU and/or inhibition of PARP using 100 μM NAP. The level of histone H3 was used as loading control.
The PARP1 protein is highly abundant and does not form foci on HU treatment (Schultz et al, 2003, data not shown). Nevertheless, we wanted to investigate whether the PARP1 protein is present at stalled replication forks. For this experiment, we labelled newly replicated DNA with chlorodeoxyuridine (CldU) after HU stalling and investigated whether PARP1 co-immunoprecipitated (co-IP) with CldU (i.e. restarted replication forks). We found that PARP1 did co-IP with restarted replication forks (Figure 3C). The amount of PARP1 present in the chromatin (DNA) fraction and co-IP with CldU is increased after HU treatment and was not affected by PARP inhibition, suggesting that PARP1 is present at sites of stalled replication forks and that this association with replication forks is independent of PARP activity (Figure 3C). This is in line with earlier data showing that PARP1 binding to DNA is independent of the enzyme activity (Satoh and Lindahl, 1992). Interestingly, PARP1 co-IP with newly replicated DNA in untreated as well as HU-treated cells, suggesting that PARP is always bound to DNA or that it is involved in overcoming spontaneously stalled replication forks.
PARP1 is required for replication restart of stalled replication forks
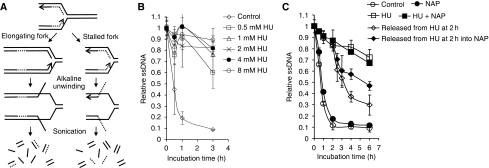
As PARP1 binds to and is activated at stalled replication forks and promotes survival after stalling, we wanted to investigate how PARP influences the reactivation and repair of stalled replication forks. To test this, we used a novel method based on the principle that each replication fork provides a pair of single-stranded DNA ends that serve as starting points for DNA unwinding in alkaline solution (Johansson et al, 2004). The cells are pulse labelled for 30 min with 3H-thymidine and the speed of replication fork elongation is monitored as the time required for the labelled DNA to be progressed into the double-stranded DNA fraction after alkaline unwinding (Figure 4A). This procedure only measures elongation of replication forks present at the time of labelling.
Figure 4.
PARP activity is required for restart of stalled replication forks. (A) The alkaline DNA unwinding technique releases 3H-thymidine-labelled DNA onto the ssDNA fraction when replication elongation is inhibited. The speed of replication fork elongation is measured as the time required for 3H-thymidine-labelled DNA not to be released into the ssDNA fraction (Johansson et al, 2004). (B) Dose-dependent replication elongation inhibition in AA8 hamster cells after addition of HU. (C) Time course of replication fork progression in AA8 cells during HU or after HU treatment with/without PARP inhibitor (NAP, 50 μM). The means and standard errors (bars) of three independent experiments are shown.
We found that addition of HU directly after the pulse label inhibits replication elongation and inhibition of replication elongation is saturated at 2 mM (Figure 4B). We then investigated replication progression when PARP is inhibited. A co-treatment with 2 mM HU and PARP inhibitor did not affect the HU-induced slowing of replication elongation (Figure 4C). These data show that HU-induced stalling of replication elongation is independent of PARP activity. We also wanted to test the possibility that PARP is involved in repair and reactivation of stalled replication forks. In this experiment, we arrested replication elongation with HU for 2 h and then released the cells in fresh media with or without inhibition of PARP. We found that cells released from HU resumed replication elongation quickly after the HU treatment. In contrast, cells released into media containing PARP inhibitor showed a replication elongation delay (Figure 4C). These data suggest that PARP activity is required to resume replication elongation at stalled forks.
Our replication elongation assay is unable to determine whether the reduced replication elongation after PARP inhibition is due to a fraction of the replication forks being stalled or whether the speed of replication elongation is globally reduced. To address this, we used the DNA fibre assay (Petermann et al, 2008); we incorporated 5-CldU, blocked with HU and released into 5-iododeoxyuridine (IdU) in the presence or absence of PARP inhibition; DNA spreads were then analysed by immunofluorescence (Figure 5A). We found that rather than globally slowing replication, PARP inhibition increases the number of forks that do not resume replication after release from HU (Figure 5B; see Supplementary Figure S3 for a more detailed analysis). The same results were observed after depletion of PARP1 using siRNA (Figure 5E; Supplementary Figure S4), showing that it is not inactive PARP protein trapped on DNA that prevents replication restart. An alternative siRNA directed against a different target sequence of PARP1 had the same effect (Figure 7). This shows that PARP activity and PARP1 protein are required to reactivate stalled replication forks. PARP inhibition or depletion did not, however, significantly alter normal replication elongation rates, showing that PARP is not required for replication elongation per se (Figure 5C and F).
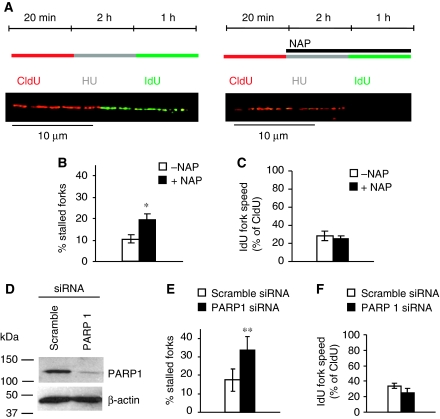
Figure 5.
PARP1 is required for replication restart as determined using the DNA fibre assay. DNA fibre analysis of replication fork restart in U2OS cells treated with PARP inhibitor NAP or depleted of PARP1. (A) Labelling protocols for DNA fibre analysis of replication forks. U2OS cells were pulse labelled with CldU, treated with HU for 2 h, and released into IdU. Example images of replication forks are shown. (B) Fork restart in the presence or absence of 100 μM NAP (left). Stalled replication forks are shown as percentage of CldU-labelled tracks. (C) Speed of restarting forks in the presence or absence of 100 μM NAP (right). IdU fork speeds are shown as percentage of CldU fork speeds. (D) Protein levels of PARP1 and β-actin (control) in U2OS cells after 48 h depletion with siRNA. (E) Fork restart in PARP1-depleted cells, as above (left). (F) Speed of restarting forks in PARP1-depleted cells, as above (right). The means and s.d. (bars) of three independent experiments are shown. Values marked with asterisks are significantly different from control (*P<0.05 or **P<0.01).
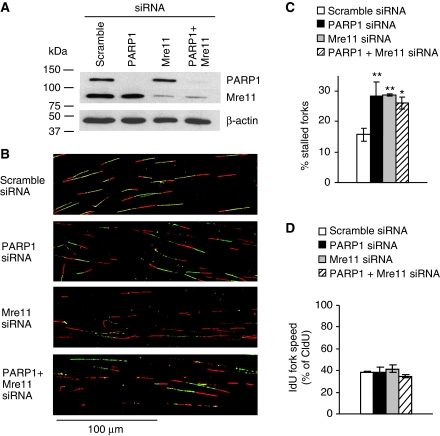
Figure 7.
PARP1 exerts an effect in the same pathway as Mre11 to restart stalled replication forks. DNA fibre analysis of replication fork restart in U2OS cells depleted of PARP1, Mre11 or co-depleted of PARP1 and Mre11. Cells were labelled to analyse fork restart after 2 h HU as in Figure 5. (A) Western blot analysis of PARP1, Mre11 and β-actin in siRNA-treated U2OS cells. (B) Representative images of DNA fibre tracks. (C) Fork restart in PARP1- and/or Mre11-depleted cells. (D) Speed of restarting forks in PARP1- and/or Mre11-depleted cells. IdU fork speeds are shown as percentage of CldU fork speeds. Means and s.d. of three independent experiments are shown. Values marked with asterisks are significantly different from control (*P<0.05 or **P<0.01).
PARP1 is required for Mre11 foci formation and efficient ssDNA formation at stalled replication forks
Little is known about the mechanism for replication restart in mammalian cells, but processing of DNA structures and HR are likely to have a function. It has been shown earlier that Mre11, part of the Mre11/RAD50/Nbs1 complex and involved in DNA resection to promote HR (Sartori et al, 2007; Buis et al, 2008), has a function in the restart of damaged replication forks (Trenz et al, 2006).
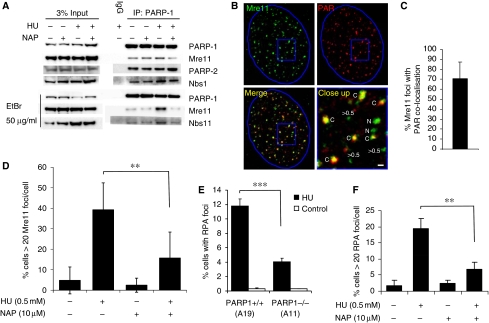
Here, we confirmed an interaction of PARP1 with Nbs1 and Mre11 (Haince et al, 2008) that is resistant to ethidium bromide, showing that the interaction is direct and not DNA dependent (Figure 6A). The amount of co-IP Mre11 was reduced when PARP was inhibited, suggesting that PARP activity is required to promote the interaction. Treatment with HU causes Mre11 to re-localise into discrete foci. We found that a portion of the Mre11 foci formed in response to HU treatment co-localise with PAR polymers (Figure 6B and C), reminiscent of the observation that the localisation of Mre11 to sites of DNA damage is dependent on PARP (Haince et al, 2008). In addition, the number of cells containing >20 HU-induced Mre11 foci was reduced when PARP was inhibited (Figure 6D). Mre11 is involved in resecting DNA ends (Williams et al, 2008) and is critical for repair of collapsed replication forks (Dolganov et al, 1996; Costanzo et al, 2001); this resection is thought to be essential because it allows HR-induced restart of forks. Our data, therefore, suggest that PARP exerts an effect to attract or retain Mre11 at sites of stalling, thus promoting resection, which could in turn allow for HR-mediated restart. The portion of Mre11 foci not overlapping with PAR polymers may reflect sites of non-homologous end joining, which can also repair HU-induced DSBs (Saintigny et al, 2001; Lundin et al, 2002).
Figure 6.
PARP is required for Mre11 localisation and resection at stalled replication forks. (A) Co-immunoprecipitation in U2OS cells showing proteins interacting with PARP1 in presence and absence of 0.5 mM HU and 100 μM PARP inhibitor NAP. (B) Immunofluorescence staining for PAR polymers and Mre11 protein in U2OS cells treated with 0.5 mM HU for 24 h. DNA was counterstained with TO-PRO-3 iodide. The nuclei borders are marked with blue. The close up panel shows Mre11 foci that co-localise with PAR (labelled C) those that do not co-localise (labelled N) and Mre11 foci with a diameter <0.5 μm. Bar is 0.5 μm. (C) Percentages of Mre11 foci co-localising with PAR. Cells with at least 10 Mre11 foci with a diameter over 0.5 μm in an optical section of 1.5 μm taken in the middle of the cell were analysed for co-localisation. The mean and standard deviation from 22 cells are depicted. (D) Quantification of Mre11 foci in U2OS cells induced by 0.5 mM HU in the presence or absence of PARP inhibitor. The means and s.d. (bars) of three experiments are shown. Values marked with asterisks are significantly different (Student's t-test, P<0.01). (E) Quantification of large RPA foci induced by 0.5 mM HU in PARP+/+ (A19) and PARP−/− (A11) MEFs. The means and s.d. (bars) of five experiments are shown. Values marked with asterisks are significantly different (Student's t-test, P<0.001). (F) Quantification of RPA foci in U2OS cells induced by 0.5 mM HU in the presence or absence of PARP inhibitor. The means and s.d. (bars) of three experiments are shown. Values marked with asterisks are significantly different (Student's t-test, P<0.01).
Using RPA foci as a marker of the amount of ssDNA produced, we tested whether resection of DNA is dependent on PARP. We found that fewer HU-induced RPA foci form in PARP1−/− as compared with PARP1+/+ MEFs and that PARP inhibitor reduced the formation of HU-induced RPA foci in wild-type cells (Figure 6E and F), suggesting that PARP1 activation is required for efficient formation of ssDNA regions at stalled replication forks; this is consistent with a role for PARP in recruitment of Mre11 for resection of DNA.
PARP1 and Mre11 work in the same pathway for restart of stalled replication forks
Although PARP1 facilitates Mre11 recruitment to stalled forks and promotes ssDNA formation, it is not clear whether this is related to the role of PARP in replication restart. To investigate the interplay between PARP1 and Mre11 during the reactivation of stalled replication forks, we depleted Mre11 using siRNA (Figure 7A) and found that Mre11-depleted cells showed a similar defect in replication restart as PARP1-depleted cells (Figure 7B and C; see Supplementary Figure S5 for a more detailed analysis). Moreover, we found that co-depletion of both PARP1 and Mre11-inhibited replication restart to the same extent as depletion of either of the two proteins alone (Figure 7C), suggesting that PARP1 and Mre11 exert an effect in the same pathway to restart replication forks. PARP1 and/or Mre11 knockdown had no influence on the speeds of restarting forks (Figure 7D).
PARP1 and PARP2 are required for HR induced at stalled replication forks
It is well established that HR is involved in replication restart in bacteria (Heller and Marians, 2006) and that agents that stall or collapse replication forks such as HU, thymidine (dT) and camptothecin (CPT) strongly induce HR in mammalian cells (Arnaudeau et al, 2001; Lundin et al, 2002), indicating that this method of restart may also be preserved in mammalian cells. Mre11 resection is required for HR (Buis et al, 2008), suggesting that PARP1 may promote recombination induced at stalled replication forks through recruitment of Mre11. Furthermore, the genetic (Menissier de Murcia et al, 2003) and physical (Figure 6A) interaction between PARP1 and PARP2 suggests that PARP2 may also mediate replication restart. To test this directly, we measured HR between two non-functional neoR genes in the human cell line SW480SN.3 (Saleh-Gohari and Helleday, 2004). We found that siRNA depletion of either PARP1 and/or PARP2 abrogates HU-induced recombination (Figure 8A), showing that both these proteins collaborate to activate recombination at stalled replication forks. These results are in contrast to recombination induced by a replication-independent DSB that does not require PARP1 for completion (Schultz et al, 2003; Yang et al, 2004).
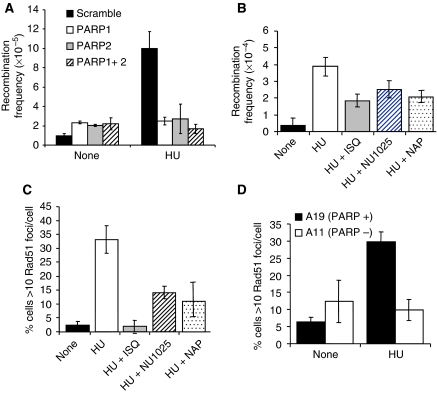
Figure 8.
PARP1 and PARP2 are required for RAD51 foci formation and HR induced at stalled replication forks. (A) Recombination frequency in the reporter construct SCneo in PARP1 and/or PARP2 depleted SW480SN.3 cells treated with 0.5 mM HU for 24 h. Means and s.d. of three independent experiments are shown. Values marked with asterisks are significantly different from control (*P<0.05 or **P<0.01). (B) Recombination frequency measured in the hprt gene in SPD8 hamster cells after a 24-h treatment with 0.5 mM HU with/without PARP inhibitors NU1025 (100 nM), 1,5-dihydroxyisoquinoline (ISQ-0.6 mM) or 4-amino-1,8 NAP (100 μM). (C) Rad51 foci formation in AA8 hamster cells induced by a 24-h HU treatment (0.5 mM) with/without PARP inhibitors NU1025 (100 nM), ISQ (0.6 mM) or NAP (100 μM). (D) Rad51 foci formation induced by 0.5 mM HU treatment in PARP+/+ and PARP−/− MEFs. The means (symbols) and standard deviations (error bars) from at least three independent experiments are depicted.
To further consolidate the role of PARP in HR, we made use of the SPD8 cell line that carries an endogenous recombination substrate for HR in the hprt gene (Helleday et al, 1998). Using this recombination system, we found that HU-induced recombination is decreased by inhibition of PARP activity with the PARP inhibitors ISQ, 4-amino-1,8 napthalimide or NU1025 (Figure 8B), showing that PARP activity is required to activate recombination repair at stalled replication forks.
In agreement with these results, we found that depletion of either PARP1 or PARP2 sensitises cells to HU, whereas depletion of PARP3 had no effect (Supplementary Figure S6). Depletion of PARP1 and PARP2 together did not further enhance HU sensitivity, suggesting that they collaborate to promote survival after HU treatment. We also found that fewer cells contain HU-induced RAD51 foci when PARP is inhibited (Figure 8C) or absent in the PARP−/− MEFs (Figure 8D), further supporting a requirement for PARP in the activation of recombination at stalled replication forks.
Replication forks stalled with dT also trigger PAR formation and require PARP1 and PARP2 for survival and HR
Several DNA lesions are suggested to form at stalled replication forks; the lesion formed is likely to depend on the agent used for stalling, the length of time the fork is stalled for and the sequence in which stalling occurs (Helleday, 2003). The data presented here suggest that PARP is potentially activated at replication forks directly or after collapse of replication forks into DSBs. To test whether a different type of stalled replication fork is sufficient to trigger PARP activity and repair, we investigated the response to excess dT. dT depletes cells only of dCTP and as a result slows down replication fork progression substantially, but unlike HU, it does not completely stop fork progression (Bjursell and Reichard, 1973). Replication forks do not collapse after dT treatments, and although fewer than four DSBs per cell are formed, even after prolonged dT treatments, HR is activated (Lundin et al, 2002; Bolderson et al, 2004). Here, we find increased PARP activity after dT treatment as indicated by the increased amount of PAR foci formation (Figure 9A). Furthermore, dT activates PARP with similar kinetics as HU (Figure 9B), suggesting that the activating substrate is the same as after HU treatments. Survival assays using PARP−/− MEFs show that PARP1 is required for survival of dT treatment (Figure 9D). siRNA depletion of PARP1 and PARP2 also showed that both proteins are required for survival after dT treatment. There is no additional effect observed when both PARP1 and PARP2 are depleted (Figure 9C), suggesting that as is the case for HU treatment, PARP1 and PARP2 collaborate for survival after dT treatment. Furthermore both PARP1 and PARP2 are required to activate HR in the SCneo reporter construct after dT treatments (Figure 9F), which was also confirmed using the hprt recombination assay using PARP inhibitors as detailed above (Figure 9E). Altogether, these results suggest that PARP is activated at stalled replication forks, although we cannot exclude the possibility that PARP is also activated at collapsed replication forks that include a DSB.
Figure 9.
PARP is activated and required for repair of replication forks stalled after dT treatments. (A) Percentage of AA8 Chinese hamster cells containing sites of PARP activity induced by a 24-h dT (2 mM) treatment. (B) PARP activity measured by the decrease of free NAD(P)H over time during incubation with 10 mM dT or 1 mM MMS. (C) Survival fraction of SW480SN.3 cells depleted of various PARP proteins after treatment for 24 h with increasing doses of the replication inhibitor dT. P<0.001 for PARP1, PARP2 and PARP1+2 as compared with scramble control. (D) Survival fraction of A11 (PARP−/−) and A19 (PARP+/+) after continuous exposure to increasing doses of dT. (E) Recombination frequency measured in the hprt gene in SPD8 hamster cells induced by a 24-h treatment with 10 mM dT with/without PARP inhibitor 1,5-dihydroxyisoquinoline (ISQ-0.6 mM). (F) Recombination frequency in the reporter construct SCneo within SW480SN.3 cells after 24 h treatment with 0.5 mM HU, depleted with PARP1 and/or PARP2 siRNAs. The means (symbol) and standard deviations (error bar) from at least three experiments are depicted.
Discussion
PARP1 facilitates DNA repair
Although PARP1-inhibited or knockout cells have a defect in SSB repair, the PARP1 protein is not essential for the SSB repair process itself. Instead, it coordinates and increases the speed of repair, probably by detecting the lesions. This is clear, as a complete defect in repairing the approximate 5 × 104 SSBs occurring in a human cell every day would not be compatible with the survival seen in PARP1−/− mice. Here, we show that PARP has a role at a subset of stalled replication forks that likely do not include extensive ssDNA to facilitate replication restart and HR. In analogy with the role of PARP1 in SSB repair, we suggest that it increases the speed of repair, by detecting stalled forks, rather than participating in the replication restart process itself.
PARP has a role in detecting stalled replication forks
Although there are numerous observations supporting a role for PARP at replication forks (Lehmann et al, 1974; Jump et al, 1979; Simbulan et al, 1993; Simbulan-Rosenthal et al, 1996; Dantzer et al, 1998; Yang et al, 2004), there is no consensus as to its function. Here, we show for the first time, using three different types of assay and two different types of replication stress, that PARP1 is activated after replication fork stress in mammalian cells (Figures 1 and 9). PARP1 efficiently binds DSBs (D'Silva et al, 1999). Hence, it is easy to envisage that PARP1 binds DSBs formed at collapsed replication forks. However, it is possible that PARP1 also binds stalled replication forks. Here, we show that PARP1 can bind stalled replication fork structures in vitro that contain short gaps (Figure 2), which may be a consequence after ssDNA annealing to stabilise stalled forks. We also find that PARP1 is present at stalled replication forks in cells (Figure 3). Furthermore, we show that PARP is also efficiently activated after dT treatments that are not associated with DSBs (Figure 9) (Lundin et al, 2002; Bolderson et al, 2004). Together these data suggest that in addition to DSBs, PARP1 is able to bind stalled replication forks in mammalian cells. The molecular structures activating PARP at stalled forks are likely to contain short gaps of ssDNA, which is likely only a subset of stalled replication forks.
PARP attracts Mre11 to stalled replication forks and promotes resection and replication restart
Mre11 is involved in the resection of DNA ends (Sartori et al, 2007; Buis et al, 2008) and it has been shown that HU induces Mre11 foci that co-localise with RPA (Robison et al, 2004). It was also shown that IR-induced RPA foci are dependent on Mre11 function (Buis et al, 2008). We find a clear link between PARP1 and Mre11 in reactivating stalled replication forks. HU-induced Mre11 foci are dependent on PARP activity and co-localise with PAR foci (Figure 6). Furthermore, Mre11 resection seems dependent on PARP as the amount of ssDNA (as measured by RPA foci) is lower in PARP1−/− cells. It is, therefore, likely that PARP serves to recruit Mre11 to sites of stalling in which it resects the DNA causing RPA foci to form. These results are consistent with earlier reports suggesting that localisation of Mre11 to laser-induced damage is dependent on PARP activity (Haince et al, 2008). We find epistasis between Mre11 and PARP1 in reactivating stalled replication forks, suggesting that the two proteins exert an effect in the same replication restart pathway. Interestingly, 30% of Mre11 foci did not co-localise with PAR foci (Figure 6C) and approximately one-third of Mre11 still relocated to foci in presence of a PARP inhibitor (Figure 6D). This suggests that there is a portion of Mre11 that is brought to stalled forks independently of PARP1. Consistent with this, a portion of the RPA foci formed in response to HU is independent of PARP1 (Figure 6E and F), suggesting that some ssDNA formation at stalled forks is independent of PARP activity. These ssDNA regions may represent the uncoupling of replicative polymerase and helicase, which occurs at stalled forks and also generates ssDNA (Walter and Newport, 2000; Byun et al, 2005), these may be longer stretches of ssDNA, which we show do not activate PARP and may, therefore, be sensed in some other way.
Earlier, it has been shown that PARP1−/− cells display delayed progress from S into G2/M phase after HU arrest (Yang et al, 2004), but a molecular explanation for these observations has been missing. Using two separate methods, we show here that PARP1 is directly involved in facilitating restart of HU stalled replication forks (Figures 4 and 5). A simple explanation for the results could be that inhibited PARP is stuck on stalled replication forks preventing access of the restart machinery. However, we see the same replication restart defect in PARP1 siRNA-depleted cells (Figure 5E), showing that PARP1 has an active role in replication restart.
It is clear that only a portion of stalled replication forks require PARP activity for efficient restart (Figure 4C). This is likely to be because in the absence of dNTPs after HU treatment, some of the forks only stop temporarily and that they can restart immediately when the dNTP supply is restored. Other structural changes may occur in those forks requiring PARP for reactivation.
The role of PARP in HR
The overall role of PARP1 in HR is complex. PARP-inhibited or knockout cells show an increase in spontaneous SCE levels (Oikawa et al, 1980), and increased spontaneous HR (Wang et al, 1997; Simbulan-Rosenthal et al, 1999; Bryant and Helleday, 2006). In contrast to this, the authors and others earlier reported that the rate of gene conversion induced by the rare-cutting restriction endonuclease I-SceI is independent of PARP1 (Schultz et al, 2003; Yang et al, 2004). The reason for PARP-inhibited or -defective cells showing an elevated level of spontaneous HR is their defect in SSB repair. The increased amount of SSBs results in increased replication fork collapse and conversion to DSBs. These one-ended DSBs are then substrates for HR repair that results in SCE. Consequently, increased SCE and recombination is a common phenotype for cells defective in SSB repair (Oikawa et al, 1980; Thompson et al, 1982; Saleh-Gohari et al, 2005). In contrast, recombination induced by the restriction endonuclease I-SceI produces a two-ended DSB, which is repaired by synthesis-dependent-strand annealing and results in gene conversion (Johnson and Jasin, 2000; Saleh-Gohari et al, 2005). In this case, recombination is not induced by the increased number of SSBs and PARP1 is itself not essential for the gene conversion event repairing these two-ended DSBs (Schultz et al, 2003; Yang et al, 2004).
Here, we show, using two different HR assays and by investigating RAD51 foci formation, that both PARP1 and PARP2 are required for HR induced at stalled replication forks by HU or dT (Figures 8 and 9). Our model is that PARP1 and PARP2 are required to recruit Mre11 for resection and ssDNA formation at stalled forks, this then allows RAD51 loading and subsequent HR (Figure 10).
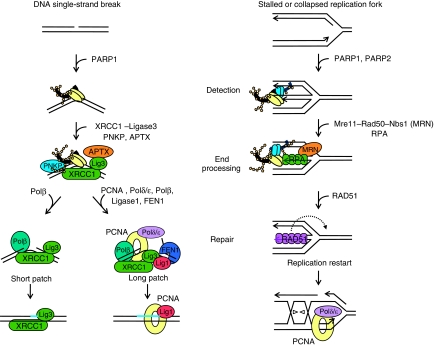
Figure 10.
Model for PARP-mediated repair of DNA SSBs and stalled replication forks. PARP1 rapidly binds to nicked, gapped or broken DNA regions, for example DNA SSB or stalled or collapsed replication forks, which activates the enzyme. The detection of stalled or collapsed replication forks is likely to involve activation of both PARP1 and PARP2. The PNKP protein and Mre11–RAD50–Nbs1 (MRN) complex are recruited to SSBs and stalled replication forks, respectively, to initiate end processing. Subsequent repair of SSBs includes XRCC1-ligase3, polymerase β, aprataxin (APTX) and the additional factors PCNA, polymerases δ/ɛ, FEN1 and ligase 1 in the case of long patch repair (Caldecott, 2008). The machinery involved in repair and replication restart in mammalian cells is poorly investigated, but likely to include the RAD51 protein and HR to reactivate normal PCNA-mediated replication by polymerases δ/ɛ.
Concluding remarks
It is well established that PARP1 has an early role in detecting SSBs and attracting proteins involved in the end-processing part of the SSB repair machinery, which is then followed by repair, using the intact DNA strand as a template (Figure 10) (Caldecott, 2008). Similarly, we suggest that PARP1 and PARP2 are important in detecting stalled or collapsed replication forks (with or without a DSB intermediate) to attract the Mre11–Rad50–Nbs1 complex, which is required for end processing (Figure 10). Subsequently, the resected DNA will be coated with RPA, which is then replaced by RAD51, to initiate HR to restart stalled replication forks (Figure 10). To support this model here, we show that inhibition or loss of PARP impairs all downstream steps in the model, that is Mre11 localisation to stalled forks, RPA and RAD51 foci formation, HR and replication restart.
Materials and methods
Cell culture
The AA8, irs1SF and CXR3 cell lines were provided by Larry Thompson (Livermore, CA). Malgorzata Z. Zdzienicka generously provided the VC8, VC8#13 and VC8+B2 cell lines. The SW480SN.3 cell line contains a stably integrated single copy of the recombination reporter vector SCneo (Mohindra et al, 2002; Saleh-Gohari et al, 2005). The A11 and A19 cell lines were a kind gift from Zhao-Qi Wang (Lyon, FRANCE), and U2OS cells were obtained from ATCC. All cell lines in this study were grown in Dulbecco's modified Eagle's Medium (DMEM) with 10% foetal bovine serum and penicillin (100 U/ml) and streptomycin sulphate (100 μg/ml) at 37°C under an atmosphere containing 5% CO2. SW480.SN3 media was supplemented with hygromycin (0.05 mM) to maintain the SCneo vector. The SPD8 cell line was grown in the presence of 5 μg/ml 6-thioguanine to suppress spontaneous recombination. All cells were routinely checked for mycoplasma infection.
Immunofluorescence
Cells were plated onto coverslips and grown for 24 h in the presence or absence of treatments as indicated. Coverslips were rinsed in phosphate-buffered saline (PBS) at 37°C and fixed in 3% paraformaldehyde, 0.1% Triton X-100 for 20 min at room temperature. Coverslips were extensively washed (PBS, 0.1–0.3% Triton X-100, 0.15% BSA) before incubation with primary antibody for 16 h at 4°C. The coverslips were washed as above followed by 1 h incubation at room temperature with the appropriate secondary antibody and washed again as above. Coverslips were washed in PBS, DNA stained with 1 μg/ml To Pro (Molecular Probes) and mounted in SlowFade Antifade (Molecular Probes).
Primary antibodies used were rabbit polyclonal antibodies against PAR (Trevigen), Mre11 (Cell Signaling) and Rad51 (H-92, Santa Cruz), mouse monoclonal anti-PAR (H10, BD Pharmingen), goat polyclonal anti-Rad51 (C-20, Santa Cruz) and rat monoclonal anti-RPA32 (4E4, Cell Signaling). The secondary antibodies were Cy-3-conjugated goat anti-rabbit IgG antibody (Zymed), AlexaFluor 555 goat anti-rabbit F(ab′)2 IgG, AlexaFluor 546 donkey anti-goat IgG and AlexaFluor 488 donkey anti-rabbit IgG (all Molecular Probes). Antibodies were diluted in PBS containing 3% BSA. Images were obtained with a Zeiss LSM 510 inverted confocal microscope using a planapochromat 63X/NA 1.4 oil immersion objective. Through focus maximum projection images were acquired from optical sections 0.50 μm apart and with a section thickness of 1.0 μm. Images were processed using Adobe PhotoShop (Abacus Inc). The frequencies of cells containing foci were determined in at least two separate experiments. At least 300 nuclei were counted on each slide.
In all co-localisation studies, cells were treated with 0.5 mM of HU for 24 h and subsequently stained as described above. For co-localisation studies, between Mre11 and PAR foci U2OS cells with >10 Mre11 foci larger than 0.5 μm in diameter in a single optical section of 1.5 μm from the middle of the cell were tracked in the microscope. The frequency of Mre11 foci that co-localised with PAR foci were then determined in 22 cells from two separate experiments. For co-localisation studies, between PAR and RPA foci A19 cells with >10 PAR foci larger than 1.0 μm in diameter in a single optical section of 1.5 μm from the middle of the cell were tracked in the microscope. The frequency of PAR foci that co-localised with RPA foci were then determined in 10 cells from two separate experiments.
Measurements of NAD(P)H levels
A water-soluble tetrazolium salt (5 mM WST-8) was used to monitor the amount of NAD(P)H through its reduction to a yellow-coloured formazan dye (Nakamura et al, 2003). A total of 5000 cells were plated at least in triplicate into wells of a 96-well plate and cultured in 100 μl normal growth media for 4 h at 37°C. CK8 buffer (Dojindo Molecular Technology), containing WST-8, was then added either with or without treatment with DNA damaging agents at concentrations indicated. Reduction of WST-8 in the presence of NAD(P)H was determined by measuring visible absorbance (OD450) every 30 min. Between measurements, cells were incubated at 37°C. A medium blank was also prepared containing just media and CK8 buffer. Changes in NAD(P)H levels were calculated by comparing the absorbance of wells containing cells treated with DNA damaging agents to those treated with DMSO only. Alternately, relative levels of NAD(P)H in different cells lines were calculated after 4 h incubation in CK8 buffer.
Inhibitors
PARP inhibitors used were NU1025 and 1,8-naphthalimide, which were generous gift from Dr Nicola Curtin (Newcastle, UK), and 1,5-dihydroxyisoquinoline and 4-amino-1,8-NAP, which were purchased from Sigma.
siRNA treatment
siRNA against PARP1, PARP2 or PARP3 were designed in house as described earlier (Bryant et al, 2005). Alternative PARP1 siRNA directed against a different PARP1 target sequence is described in Fisher et al (2007) and was used for the experiments in Figure 7. siRNA against Mre11 was designed as described in Myers and Cortez (2006). Scrambled siRNA was purchased from Dharmacon and Qiagen. Cells were transfected with 100 nM scrambled siRNA, 50 nM targeting +50 nM scrambled siRNA or 50 nM of each targeting siRNA for co-depletion experiments. For toxicity or recombination assays, 10 000 cells grown in 6-well plates overnight were transfected with 100 nM siRNA using Oligofectamine Reagent (Invitrogen) according to manufacturer's instructions. Cells were then cultured in normal growth medium for 48 h before trypsinisation and replating. Depletion was confirmed by RT–PCR as described earlier (Bryant et al, 2005). For the DNA fibre assay, 5000 cells grown in 6-well plates overnight were transfected using Dharmafect 1 reagent (Dharmacon) according to manufacturer's instructions for HeLa cells. Cells were then cultured in normal growth media for 48 or 72 h before DNA labelling and fibre preparation. Depletion was confirmed by western blotting.
Toxicity assay
A total of 500 cells were plated in triplicate onto 100 mm dishes or 200 cells were plated in triplicate into 6-well plates 4 h before the addition of PARP inhibitors and increasing doses of HU as indicated. Treatment was for 24 h, after which cells were washed in PBS and fresh media added, or continuous, in which case the original medium was left on cells; 7–12 days later, when colonies could be observed, they were fixed and stained with methylene blue in methanol (4 g/l). Colonies consisting of >50 cells were subsequently counted. Each colony was assumed to represent one cell surviving from the original 500/200 and the surviving fraction for each dose was calculated. When siRNA-depleted cells were used, they were transfected as above for 48 h and, then replated in the presence or absence of increasing doses of HU for 24 h.
Electrophoretic mobility shift assay
Oligonucleotides: Construct A 5′-CGATAGAAGAACTATCGCAATGTATCGAGTCAAGCCGACTCGATACTGGA CTGGAACAACCAGTCCA-3′ with or without 3′ biotin modification.
Construct B 5′-CGATAGAAGAACTATCGCAATGTATCGAGTCAAGCCGACTCGATACTG GACTGGAACAACCAGTCCAATTG-3′ with 5′ phosphate modification; this oligo was self-ligated using T4 ligase.
Before use, constructs were heated to 80°C and then allowed to cool slowly on the bench; 2.5 ng of labelled probe was then incubated with/without increasing amounts of recombinant PARP1 in the presence or absence of various competitor DNAs in EMSA buffer (40 mM Tris, 1 mM EDTA, 20 mM NaCl, 20 μg/ml BSA, 8% glycerol) for 30 min on ice. Reactions were then separated on a 6% non-denaturing polyacylamide gel, transferred to nylon membrane and bands detected by using the ‘Lightshift Chemiluminescent EMSA kit' (Pierce Biotechnology, UK).
In vitro PARP activity assay
Stalled plasmid was produced as described earlier (McGlynn et al, 2001). A measure of 50 ng of each DNA substrate DNA and 50 ng recombinant PARP (Alexis, UK) were incubated together for 10 min at room temperature in a final reaction volume of 25 μl in activity buffer (50 mM Tris, 4 mM MgCl, 200 mM DTT, 0.1 mg/ml BSA and 0.4 mM NAD). The reaction mixture was then loaded on an 8% SDS–PAGE gel, transferred to nitrocellulose and western blotted for PARP and PAR.
Auto-ADP ribosylation of PARP1 was assayed using biotinylated NAD+, 6-biotin-17-NAD+ (Bio-NAD+, Trevigen); 5 nM of recombinant human PARP1 (Alexis) was incubated in PARP reaction buffer (50 mM Tris–HCl buffer, 2 mM MgCl2) with 12.5 μM Bio-NAD+ and 37.5 μM NAD in the presence of 200 ng of various DNA constructs. The reaction was terminated at indicated time points by adding LDS sample buffer. The samples were separated on 4–12% Bis–Tris NuPAGE Novex gels (Invitrogen) under denaturing conditions, and the gels were blotted onto nitrocellulose membranes (GE Healthcare). The blots were probed with anti-biotin antibody (Santa Cruz, USA) followed by incubation with horseradish peroxidase-conjugated secondary antibody (Thermo Scientific), and the bands were visualised using SuperSignal West Femto chemiluminescence substrate (Thermo Scientific). Quantification of PARP1 ADP ribosylation was carried out using Adobe Photoshop CS2.
Recombination in SW480SN.3 cells
After siRNA treatments, cells were rinsed in PBS and left in fresh non-selective media for 48 h. The cell plates were then rinsed in 2 × PBS, trypsinised and reseeded for cloning (2 plates with 500 cells/plate), and for selection of recombinants in G418 (1 mg/ml; 25 cells/mm2). After 12–14 days, the colonies obtained were stained with methylene blue in methanol (4 g/l).
Recombination in SPD8 cells
A total of 1.5 × 106 cells were inoculated into 100 mm dishes in medium 4 h before a 24-h treatment with drugs as indicated. After treatments, the cells were rinsed three times with PBS and 10 ml medium added before allowing the cells to recover for 48 h. After recovery, cells were released by trypsinisation and counted. HPRT+ revertants were selected by plating 3 × 105 treated cells per dish in the presence of HAsT (50 μM hypoxanthine, 10 μM L-azaserine and 5 μM dT). To determine cloning efficiency, two dishes were plated with 500 cells each. The colonies obtained were stained with methylene blue in methanol (4 g/l), after 7 (in the case of cloning efficiency) or 10 (for reversion) days of incubation.
Western blotting
Cells were lysed in RIPA buffer in the presence of 1 × protease and phosphatase inhibitor cocktails (Sigma). An aliquot of 50 μg total protein was run on an SDS–PAGE gel and transferred to Hybond ECL membrane (Amersham Pharmacia). This membrane was immunoblotted with rabbit polyclonal antibodies against PARP2 (Yuc Alexis Biochemicals), Mre11 (Cell Signaling), goat anti-NbsI (C-19, Santa Cruz Biotechnology) or mouse monoclonal antibodies against PARP1 (F2 Santa Cruz Biotechnology), PAR (10H, Trevigen) and β-actin (Sigma) in 5% milk overnight. Immunoreactive protein was visualised using ECL reagents (Amersham Pharmacia) following manufacturer's instructions.
PARP1 co-immunoprecipitation
The cDNA encoding myc-tagged PARP1 was cloned into the pcDNA3.1 expression vector as described (Hassa et al, 2001). The resulting myc-tagged PARP1 expression vector was transfected into U2OS cells and a stably transfected cell line was selected by growing the cells in the presence of 1 mg/ml Geneticin (GIBCO). The myc-PARP1 overexpressing cells were treated with 0.5 mM HU, 10 μM NAP or 0.5 mM HU and 10 μM NAP or left untreated. After a 24-h treatment, the cells were washed twice with ice cold PBS. The cells were lysed on ice for 10 min using a whole-cell lysis buffer (HEPES 50 mM, NaCl 150 mM, EDTA 1 mM, EGTA 1 mM, Glycerol 10%, Triton X-100 1%, complete protease inhibitor complex (Roche), phospatase inhibitor complex 1 and phosphatase inhibitor complex 2 (SIGMA)) and the cell lysate was collected. The cell lysate was cleared by centrifugation at 16 000 r.p.m. for 20 min. The protein concentration of the lysates was determined by using the Coomassie Plus—The Better Bradford Assay (PIERCE).
A total of 1.0 mg whole-cell lysate was used for each co-immunoprecipitation reaction. The lysate was precleared by incubating the lysates with normal mouse IgG agarose conjugate (Santa Cruz Biotechnology). The precleared lysates were collected and incubated with mouse anti-myc agarose conjugate (9E10, Santa Cruz Biotechnology) or normal mouse IgG agarose conjugate using top over end rotation over night at 4°C. To check whether the interaction was dependent on chromatin, ethidium bromide (50 μg/ml) was added to the lysate. The co-immunoprecipitation reaction was washed four times with lysis buffer containing 0.3 M NaCl and the immunoprecipitated proteins were eluted by boiling in SDS–PAGE reducing loading buffer. For SDS–PAGE followed by western blot, 25% of the eluted material was loaded.
CldU co-immunoprecipitation of proteins present at stalled replication forks
A total of 2 × 106 cells were treated with 1 mM HU and/or 10 μM NAP for 3 h. HU was washed away and cells were labelled with CldU (100 μM) for 40 min. Cells were cross-linked in 1% formaldehyde for 15 min R/T and treated with 0.125 M glycine for 15 min RT. Cells were harvested by scraping in cold PBS. Cytoplasmic proteins were removed by incubation in hypotonic buffer (10 mM HEPES pH7, 50 mM NaCl, 0.3 M sucrose, 0.5% TX-100, protease inhibitors (cocktail, Roche)) for 10 min on ice and centrifugation at 1500 g for 5 min. Nuclear-soluble fraction was removed by incubation with nuclear buffer (10 mM HEPES pH 7, 200 mM NaCl, 1 mM EDTA, 0.5% NP-40, protease inhibitors (cocktail, Roche)) for 10 min on ice and centrifugation at 13 000 r.p.m. for 2 min. Pellets were resuspended in lysis buffer (10 mM HEPES pH 7, 500 mM NaCl, 1 mM EDTA, 1% NP-40, protease inhibitors (cocktail, Roche)), sonicated, centrifuged for 30 s at 13 000 r.p.m. and the supernatant collected. Protein concentration was measured by Bradford assay.
A measure of 150 μg of total protein (from above fraction) was used for IP reaction with 2 μg of anti-CldU antibody (rat-anti-CldU, AbD Serotec) and 20 μl of protein A/G-PLUS agarose (Santa Cruz Biotechnology). The IP reaction was washed 2 times with nuclear buffer, 2 times with washing buffer (10 mM HEPES, 0.1 mM EDTA protease inhibitors (cocktail, Roche)), incubated in 2x sample loading buffer (100 mM Tris–HCl pH 6.8, 100 mM DTT, 4% SDS, 0.2% bromphenol blue, 20% glycerol) for 30 min at 90°C and used for western blot as described above.
Elongation at replication forks
Cells were seeded onto 24-well plates at a density of 2 × 105 per well and grown for 24 h before pulse labelling of replication forks with 3H-TdR (37 kBq/ml) (for a period of 30 min in DMEM at 37°C under 5% CO2). HU and/or NAP, at the doses shown, were added to the medium during the post-labelling period. In certain cases, after a 2-h exposure to HU, cells were washed twice with PBS and fresh medium with/without adding NAP. Inhibition of fork progression by DNA lesions results in an elevated amount of labelled, ssDNA as detected by alkaline unwinding (Johansson et al, 2004).
DNA fibre assay
Exponential cell cultures of U2OS cells were pulse labelled with 25 μM CldU for 20 min, washed once with medium and pulse labelled with 250 μM IdU with 4-amino-1,8 napthalimide (100 μM) or equal volumes of DMSO for another 20 min. For HU treatment, cells were pulse labelled with 25 μM CldU for 20 min, washed thrice with medium, and incubated in 2 mM HU with 4-amino-1,8 napthalimide or equal volumes of DMSO for 2 h. Afterwards, cells were washed thrice with medium and pulse labelled with 250 μM IdU for 1 h with 4-amino-1,8 napthalimide or equal volumes of DMSO. For RNAi experiments, U2OS cells were treated with scrambled siRNA or siRNA directed against PARP1, Mre11 or both for 48 or 72 h, then pulse labelled with CldU, treated with HU and released from HU into IdU as above. Labelled cells were harvested and DNA fibre spreads prepared as described earlier (Henry-Mowatt et al, 2003). For immunodetection of CldU-labelled tracts, acid-treated fibre spreads were incubated with rat anti-BrdU monoclonal antibody that recognises CldU, but not IdU AbD Serotec for 1 h at room temperature. Slides were fixed with 4% paraformaldehyde and incubated with an AlexaFluor 594-conjugated goat anti-rat IgG (Molecular Probes) for 1.5 h at room temperature. IdU-labelled patches were detected using a mouse anti-BrdU monoclonal antibody that recognises IdU, but not CldU (Becton Dickinson) over night at 4°C, followed by an AlexaFluor 488-conjugated goat anti-mouse F(ab′)2 fragment (Molecular Probes) for 1.5 h at room temperature. Fibres were examined using a Leica SP2 confocal microscope using a × 63 (1.3 NA) glycerol immersion objective. The lengths of red (AF 594) or green (AF 488)-labelled patches were measured using the ImageJ software (National Institutes of Health) and arbitrary length values were converted into μm using the scale bars created by the microscope. At least 100 replication tracks were measured per experiment as described earlier (Merrick et al, 2004). For quantification of replication structures, at least 250 structures were counted per experiment.
Supplementary Material
Supplementary Online Material
Review Process File
Acknowledgments
We thank Drs Zhao-Qi Wang and Larry Thompson for materials and The Swedish Cancer Society, the Swedish Children's Cancer Foundation, the Swedish Research Council, the Swedish Pain Relief Foundation, Cancer Research UK, the Medical Research Council and Yorkshire Cancer Research for supportng this work financially.
Footnotes
The authors declare that they have no conflict of interest.
References
- Ahel I, Ahel D, Matsusaka T, Clark AJ, Pines J, Boulton SJ, West SC (2008) Poly(ADP-ribose)-binding zinc finger motifs in DNA repair/checkpoint proteins. Nature 451: 81–85 [DOI] [PubMed] [Google Scholar]
- Allinson SL, Dianova II, Dianov GL (2003) Poly(ADP-ribose) polymerase in base excision repair: always engaged, but not essential for DNA damage processing. Acta Biochim Pol 50: 169–179 [PubMed] [Google Scholar]
- Ame JC, Rolli V, Schreiber V, Niedergang C, Apiou F, Decker P, Muller S, Hoger T, Menissier-de Murcia J, de Murcia G (1999) PARP-2, a novel mammalian DNA damage-dependent poly(ADP-ribose) polymerase. J Biol Chem 274: 17860–17868 [DOI] [PubMed] [Google Scholar]
- Anachkova B, Russev G, Poirier GG (1989) DNA replication and poly(ADP-ribosyl)ation of chromatin. Cytobios 58: 19–28 [PubMed] [Google Scholar]
- Arnaudeau C, Lundin C, Helleday T (2001) DNA double-strand breaks associated with replication forks are predominantly repaired by homologous recombination involving an exchange mechanism in mammalian cells. J Mol Biol 307: 1235–1245 [DOI] [PubMed] [Google Scholar]
- Bartkova J, Rezaei N, Liontos M, Karakaidos P, Kletsas D, Issaeva N, Vassiliou LV, Kolettas E, Niforou K, Zoumpourlis VC, Takaoka M, Nakagawa H, Tort F, Fugger K, Johansson F, Sehested M, Andersen CL, Dyrskjot L, Orntoft T, Lukas J et al. (2006) Oncogene-induced senescence is part of the tumorigenesis barrier imposed by DNA damage checkpoints. Nature 444: 633–637 [DOI] [PubMed] [Google Scholar]
- Bianchi V, Pontis E, Reichard P (1986) Changes of deoxyribonucleoside triphosphate pools induced by hydroxyurea and their relation to DNA synthesis. J Biol Chem 261: 16037–16042 [PubMed] [Google Scholar]
- Bjursell G, Reichard P (1973) Effects of thymidine on deoxyribonucleoside triphosphate pools and deoxyribonucleic acid synthesis in Chinese hamster ovary cells. J Biol Chem 248: 3904–3909 [PubMed] [Google Scholar]
- Bolderson E, Scorah J, Helleday T, Smythe C, Meuth M (2004) ATM is required for the cellular response to thymidine induced replication fork stress. Hum Mol Genet 31: 2937–2945 [DOI] [PubMed] [Google Scholar]
- Bryant HE, Helleday T (2006) Inhibition of poly (ADP-ribose) polymerase activates ATM which is required for subsequent homologous recombination repair. Nucleic Acids Res 34: 1685–1691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant HE, Schultz N, Thomas HD, Parker KM, Flower D, Lopez E, Kyle S, Meuth M, Curtin NJ, Helleday T (2005) Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose)polymerase. Nature 434: 913–917 [DOI] [PubMed] [Google Scholar]
- Buis J, Wu Y, Deng Y, Leddon J, Westfield G, Eckersdorff M, Sekiguchi JM, Chang S, Ferguson DO (2008) Mre11 nuclease activity has essential roles in DNA repair and genomic stability distinct from ATM activation. Cell 135: 85–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byun TS, Pacek M, Yee MC, Walter JC, Cimprich KA (2005) Functional uncoupling of MCM helicase and DNA polymerase activities activates the ATR-dependent checkpoint. Genes Dev 19: 1040–1052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldecott KW (2008) Single-strand break repair and genetic disease. Nat Rev Genet 9: 619–631 [DOI] [PubMed] [Google Scholar]
- Costanzo V, Robertson K, Bibikova M, Kim E, Grieco D, Gottesman M, Carroll D, Gautier J (2001) Mre11 protein complex prevents double-strand break accumulation during chromosomal DNA replication. Mol Cell 8: 137–147 [DOI] [PubMed] [Google Scholar]
- D'Amours D, Desnoyers S, D'Silva I, Poirier GG (1999) Poly(ADP-ribosyl)ation reactions in the regulation of nuclear functions. Biochem J 342 (Part 2): 249–268 [PMC free article] [PubMed] [Google Scholar]
- D'Silva I, Pelletier JD, Lagueux J, D'Amours D, Chaudhry MA, Weinfeld M, Lees-Miller SP, Poirier GG (1999) Relative affinities of poly(ADP-ribose) polymerase and DNA-dependent protein kinase for DNA strand interruptions. Biochim Biophys Acta 1430: 119–126 [DOI] [PubMed] [Google Scholar]
- Dantzer F, Nasheuer HP, Vonesch JL, de Murcia G, Menissier-de Murcia J (1998) Functional association of poly(ADP-ribose) polymerase with DNA polymerase alpha-primase complex: a link between DNA strand break detection and DNA replication. Nucleic Acids Res 26: 1891–1898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Micco R, Fumagalli M, Cicalese A, Piccinin S, Gasparini P, Luise C, Schurra C, Garre M, Nuciforo PG, Bensimon A, Maestro R, Pelicci PG, d'Adda di Fagagna F (2006) Oncogene-induced senescence is a DNA damage response triggered by DNA hyper-replication. Nature 444: 638–642 [DOI] [PubMed] [Google Scholar]
- Dolganov GM, Maser RS, Novikov A, Tosto L, Chong S, Bressan DA, Petrini JH (1996) Human Rad50 is physically associated with human Mre11: identification of a conserved multiprotein complex implicated in recombinational DNA repair. Mol Cell Biol 16: 4832–4841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Khamisy SF, Masutani M, Suzuki H, Caldecott KW (2003) A requirement for PARP-1 for the assembly or stability of XRCC1 nuclear foci at sites of oxidative DNA damage. Nucleic Acids Res 31: 5526–5533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farmer H, McCabe N, Lord CJ, Tutt AN, Johnson DA, Richardson TB, Santarosa M, Dillon KJ, Hickson I, Knights C, Martin NM, Jackson SP, Smith GC, Ashworth A (2005) Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature 434: 917–921 [DOI] [PubMed] [Google Scholar]
- Fisher A, Hochegger H, Takeda S, Caldecott KW (2007) Poly (ADP-ribose) polymerase-1 accelerates single-strand break repair in concert with poly (ADP-ribose) glycohydrolase. Mol Cell Biol 27: 5597–5605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haince JF, McDonald D, Rodrigue A, Dery U, Masson JY, Hendzel MJ, Poirier GG (2008) PARP1-dependent kinetics of recruitment of MRE11 and NBS1 proteins to multiple DNA damage sites. J Biol Chem 283: 1197–1208 [DOI] [PubMed] [Google Scholar]
- Hanada K, Budzowska M, Davies SL, van Drunen E, Onizawa H, Beverloo HB, Maas A, Essers J, Hickson ID, Kanaar R (2007) The structure-specific endonuclease Mus81 contributes to replication restart by generating double-strand DNA breaks. Nat Struct Mol Biol 14: 1096–1104 [DOI] [PubMed] [Google Scholar]
- Hassa PO, Covic M, Hasan S, Imhof R, Hottiger MO (2001) The enzymatic and DNA binding activity of PARP-1 are not required for NF-kappa B coactivator function. J Biol Chem 276: 45588–45597 [DOI] [PubMed] [Google Scholar]
- Helleday T (2003) Pathways for mitotic homologous recombination in mammalian cells. Mutat Res 532: 103–115 [DOI] [PubMed] [Google Scholar]
- Helleday T, Arnaudeau C, Jenssen D (1998) A partial hprt gene duplication generated by non-homologous recombination in V79 Chinese hamster cells is eliminated by homologous recombination. J Mol Biol 279: 687–694 [DOI] [PubMed] [Google Scholar]
- Helleday T, Petermann E, Lundin C, Hodgson B, Sharma RA (2008) DNA repair pathways as targets for cancer therapy. Nat Rev Cancer 8: 193–204 [DOI] [PubMed] [Google Scholar]
- Heller RC, Marians KJ (2006) Replisome assembly and the direct restart of stalled replication forks. Nat Rev Mol Cell Biol 7: 932–943 [DOI] [PubMed] [Google Scholar]
- Henry-Mowatt J, Jackson D, Masson JY, Johnson PA, Clements PM, Benson FE, Thompson LH, Takeda S, West SC, Caldecott KW (2003) XRCC3 and Rad51 modulate replication fork progression on damaged vertebrate chromosomes. Mol Cell 11: 1109–1117 [DOI] [PubMed] [Google Scholar]
- Hiasa H, Marians KJ (1994) Primase couples leading- and lagging-strand DNA synthesis from oriC. J Biol Chem 269: 6058–6063 [PubMed] [Google Scholar]
- Johansson F, Lagerqvist A, Erixon K, Jenssen D (2004) A method to monitor replication fork progression in mammalian cells: nucleotide excision repair enhances and homologous recombination delays elongation along damaged DNA. Nucleic Acids Res 32: e157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson RD, Jasin M (2000) Sister chromatid gene conversion is a prominent double-strand break repair pathway in mammalian cells. EMBO J 19: 3398–3407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jump DB, Butt TR, Smulson M (1979) Nuclear protein modification and chromatin substructure. 3. Relationship between poly(adenosine diphosphate) ribosylation and different functional forms of chromatin. Biochemistry 18: 983–990 [DOI] [PubMed] [Google Scholar]
- Lehmann AR, Kirk-Bell S, Shall S, Whish WJ (1974) The relationship between cell growth, macromolecular synthesis and poly ADP-ribose polymerase in lymphoid cells. Exp Cell Res 83: 63–72 [DOI] [PubMed] [Google Scholar]
- Lopes M, Cotta-Ramusino C, Pellicioli A, Liberi G, Plevani P, Muzi-Falconi M, Newlon CS, Foiani M (2001) The DNA replication checkpoint response stabilizes stalled replication forks. Nature 412: 557–561 [DOI] [PubMed] [Google Scholar]
- Lundin C, Erixon K, Arnaudeau C, Schultz N, Jenssen D, Meuth M, Helleday T (2002) Different roles for nonhomologous end joining and homologous recombination following replication arrest in mammalian cells. Mol Cell Biol 22: 5869–5878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGlynn P, Lloyd RG, Marians KJ (2001) Formation of Holliday junctions by regression of nascent DNA in intermediates containing stalled replication forks: RecG stimulates regression even when the DNA is negatively supercoiled. Proc Natl Acad Sci USA 98: 8235–8240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meister P, Taddei A, Vernis L, Poidevin M, Gasser SM, Baldacci G (2005) Temporal separation of replication and recombination requires the intra-S checkpoint. J Cell Biol 168: 537–544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menissier de Murcia J, Ricoul M, Tartier L, Niedergang C, Huber A, Dantzer F, Schreiber V, Ame JC, Dierich A, LeMeur M, Sabatier L, Chambon P, de Murcia G (2003) Functional interaction between PARP-1 and PARP-2 in chromosome stability and embryonic development in mouse. EMBO J 22: 2255–2263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrick CJ, Jackson D, Diffley JF (2004) Visualization of altered replication dynamics after DNA damage in human cells. J Biol Chem 279: 20067–20075 [DOI] [PubMed] [Google Scholar]
- Mohindra A, Hays LE, Phillips EN, Preston BD, Helleday T, Meuth M (2002) Defects in homologous recombination repair in mismatch-repair-deficient tumour cell lines. Hum Mol Genet 11: 2189–2200 [DOI] [PubMed] [Google Scholar]
- Myers JS, Cortez D (2006) Rapid activation of ATR by ionizing radiation requires ATM and Mre11. J Biol Chem 281: 9346–9350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura J, Asakura S, Hester SD, de Murcia G, Caldecott KW, Swenberg JA (2003) Quantitation of intracellular NAD(P)H can monitor an imbalance of DNA single strand break repair in base excision repair deficient cells in real time. Nucleic Acids Res 31: e104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oikawa A, Tohda H, Kanai M, Miwa M, Sugimura T (1980) Inhibitors of poly(adenosine diphosphate ribose) polymerase induce sister chromatid exchanges. Biochem Biophys Res Commun 97: 1311–1316 [DOI] [PubMed] [Google Scholar]
- Petermann E, Helleday T, Caldecott KW (2008) Claspin promotes normal replication fork rates in human cells. Mol Biol Cell 19: 2373–2378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postow L, Ullsperger C, Keller RW, Bustamante C, Vologodskii AV, Cozzarelli NR (2001) Positive torsional strain causes the formation of a four-way junction at replication forks. J Biol Chem 276: 2790–2796 [DOI] [PubMed] [Google Scholar]
- Robison JG, Elliott J, Dixon K, Oakley GG (2004) Replication protein A and the Mre11.Rad50.Nbs1 complex co-localize and interact at sites of stalled replication forks. J Biol Chem 279: 34802–34810 [DOI] [PubMed] [Google Scholar]
- Saintigny Y, Delacote F, Vares G, Petitot F, Lambert S, Averbeck D, Lopez BS (2001) Characterization of homologous recombination induced by replication inhibition in mammalian cells. EMBO J 20: 3861–3870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saleh-Gohari N, Bryant HE, Schultz N, Parker KM, Cassel TN, Helleday T (2005) Spontaneous homologous recombination is induced by collapsed replication forks that are caused by endogenous DNA single-strand breaks. Mol Cell Biol 25: 7158–7169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saleh-Gohari N, Helleday T (2004) Conservative homologous recombination preferentially repairs DNA double-strand breaks in the S phase of the cell cycle in human cells. Nucleic Acids Res 32: 3683–3688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sartori AA, Lukas C, Coates J, Mistrik M, Fu S, Bartek J, Baer R, Lukas J, Jackson SP (2007) Human CtIP promotes DNA end resection. Nature 450: 509–514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh MS, Lindahl T (1992) Role of poly(ADP-ribose) formation in DNA repair. Nature 356: 356–358 [DOI] [PubMed] [Google Scholar]
- Schultz N, Lopez E, Saleh-Gohari N, Helleday T (2003) Poly(ADP-ribose) polymerase (PARP-1) has a controlling role in homologous recombination. Nucleic Acids Res 31: 4959–4964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simbulan CM, Suzuki M, Izuta S, Sakurai T, Savoysky E, Kojima K, Miyahara K, Shizuta Y, Yoshida S (1993) Poly(ADP-ribose) polymerase stimulates DNA polymerase alpha by physical association. J Biol Chem 268: 93–99 [PubMed] [Google Scholar]
- Simbulan-Rosenthal CM, Haddad BR, Rosenthal DS, Weaver Z, Coleman A, Luo R, Young HM, Wang ZQ, Ried T, Smulson ME (1999) Chromosomal aberrations in PARP(−/−) mice: genome stabilization in immortalized cells by reintroduction of poly(ADP-ribose) polymerase cDNA. Proc Natl Acad Sci USA 96: 13191–13196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simbulan-Rosenthal CM, Rosenthal DS, Hilz H, Hickey R, Malkas L, Applegren N, Wu Y, Bers G, Smulson ME (1996) The expression of poly(ADP-ribose) polymerase during differentiation-linked DNA replication reveals that it is a component of the multiprotein DNA replication complex. Biochemistry 35: 11622–11633 [DOI] [PubMed] [Google Scholar]
- Thompson LH, Brookman KW, Dillehay LE, Carrano AV, Mazrimas JA, Mooney CL, Minkler JL (1982) A CHO-cell strain having hypersensitivity to mutagens, a defect in DNA strand-break repair, and an extraordinary baseline frequency of sister-chromatid exchange. Mutat Res 95: 427–440 [DOI] [PubMed] [Google Scholar]
- Trenz K, Smith E, Smith S, Costanzo V (2006) ATM and ATR promote Mre11 dependent restart of collapsed replication forks and prevent accumulation of DNA breaks. EMBO J 25: 1764–1774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter J, Newport J (2000) Initiation of eukaryotic DNA replication: origin unwinding and sequential chromatin association of Cdc45, RPA, and DNA polymerase alpha. Mol Cell 5: 617–627 [DOI] [PubMed] [Google Scholar]
- Wang ZQ, Stingl L, Morrison C, Jantsch M, Los M, Schulze-Osthoff K, Wagner EF (1997) PARP is important for genomic stability but dispensable in apoptosis. Genes Dev 11: 2347–2358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams RS, Moncalian G, Williams JS, Yamada Y, Limbo O, Shin DS, Groocock LM, Cahill D, Hitomi C, Guenther G, Moiani D, Carney JP, Russell P, Tainer JA (2008) Mre11 dimers coordinate DNA end bridging and nuclease processing in double-strand-break repair. Cell 135: 97–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang YG, Cortes U, Patnaik S, Jasin M, Wang ZQ (2004) Ablation of PARP-1 does not interfere with the repair of DNA double-strand breaks, but compromises the reactivation of stalled replication forks. Oncogene 23: 3872–3882 [DOI] [PubMed] [Google Scholar]
- Zou L, Elledge SJ (2003) Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science 300: 1542–1548 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Online Material
Review Process File