Abstract
G proteins are key molecular switches in the regulation of membrane protein function and signal transduction. The prokaryotic membrane protein FeoB is involved in G protein coupled Fe2+ transport, and is unique in that the G protein is directly tethered to the membrane domain. Here, we report the structure of the soluble domain of FeoB, including the G protein domain, and its assembly into an unexpected trimer. Comparisons between nucleotide free and liganded structures reveal the closed and open state of a central cytoplasmic pore, respectively. In addition, these data provide the first observation of a conformational switch in the nucleotide-binding G5 motif, defining the structural basis for GDP release. From these results, structural parallels are drawn to eukaryotic G protein coupled membrane processes.
Keywords: crystallography, GDP release, GTPase, iron transport
Introduction
The nucleotide GTP and its cognate GTPases (G proteins) are critical for normal physiology in all organisms by acting as molecular ‘on' (GTP bound) or ‘off' (GDP bound) switches for fundamental cellular processes, such as protein synthesis and sorting, cell growth, and regulation of ion channels (Cachero et al, 1998; Finlin et al, 2006). As such, their aberrant functions have resulted in a number of pathophysiological disorders, such as asthma and cancer (Karnoub and Weinberg, 2008; Schaafsma et al, 2008). Structural studies have anatomically characterized G proteins, revealing a highly conserved GTP-binding domain of roughly 160 residues distinguished by five short amino-acid motifs, termed G1–G5, that are critical in the binding of both a magnesium (Mg2+) ion and the guanine nucleotide (Sprang, 1997; Vetter and Wittinghofer, 2001). Activation of G proteins requires the initial dissociation of GDP, leading to an intermediate and transient nucleotide-free (apo) state. In all G proteins, the subsequent binding of GTP turns the G protein ‘on' and causes two regions, defined as ‘Switch I' and ‘Switch II', to change conformation, which allows specific ‘effector' proteins to interact and convey the signalling pathway. Hydrolysis of GTP to GDP eventuates in an ‘off' state, reversing the structural changes with attenuation of downstream signalling, completing the G protein cycle (Sprang, 1997; Vetter and Wittinghofer, 2001).
Central to understanding G protein coupled signal transduction is the mechanism by which the dissociation of GDP is catalysed. For the heterotrimeric Gαβγ proteins, the dissociation of GDP from the Gα subunit is catalysed by activated membrane-bound G protein coupled receptors (GPCRs), which associate with the GDP-bound Gαβγ and act as a guanine exchange factor (GEF) (Oldham et al, 2007; Oldham and Hamm, 2008). Recent studies have indicated that the GEF activity conferred by GPCRs is achieved by the modulation of structural changes in the G5 motif of the Gα subunit, leading to a reduced affinity for GDP (Hamm et al, 1988; Johnston and Siderovski, 2007a, 2007b; Scheerer et al, 2008). Further indication of the role played by the G5 motif is a Gαs mutant (Ala366Ser), clinically characterized by testotoxicosis (precocious puberty) in males due to an accelerated GDP release (Iiri et al, 1994). This mutation was mapped to the G5 motif and, in effect, it was proposed that the mutation mimics the accelerated dissociation of GDP normally induced by the agonist-activated GPCR. In structural studies of G proteins with bound GDP and GTP, the G5 motif has been shown to coordinate to the nucleotide base through a conserved hydrogen-bonding network (Sprang, 1997). However, a full understanding of the involvement of the G5 motif in GDP release requires structural characterization of the nucleotide-free transition state, a challenging feat due to its inherent instability in these systems (Sprang, 1997; Johnston and Siderovski, 2007a, 2007b; Oldham and Hamm, 2008).
Prokaryotic uptake of ferrous iron (Fe2+) has recently emerged as a G protein coupled process involving the membrane protein FeoB, which has possible evolutionary links to eukaryotic G protein coupled membrane processes (Marlovits et al, 2002; Hantke, 2003). FeoB is unique in that it includes an intracellular G protein directly fused to a polytopic membrane transport protein, separated by a short linker domain of unknown function (Marlovits et al, 2002; Daley et al, 2005) (Supplementary Figure S1). Mutational studies and sequence similarity between the G protein domain and the human oncogene p21-Ras and Gα proteins have identified the G1–G4 signature motifs in FeoB, although the critical G5 loop could not be assigned (Marlovits et al, 2002). Crucially, as with its eukaryotic counterparts, critical aspects regarding the regulation and mechanism of nucleotide exchange within FeoB remain unexplained. Furthermore, the nature of the coupling between the G protein and the membrane domain is still undetermined (Marlovits et al, 2002; Cartron et al, 2006; Eng et al, 2008). In nucleotide-coupled transporters (e.g. ABC transporters), the coupling occurs through structural changes catalysed by the nucleotide hydrolysis, whereas in channels (e.g. the cyclic nucleotide-gated ion channels), it is the binding of nucleotide that is associated with structural changes (Clayton et al, 2004; Hollenstein et al, 2007; Bocquet et al, 2009; Rees et al, 2009). Functional characterization has shown that FeoB has intrinsically slow GTP hydrolysis, which suggests that FeoB functions as a nucleotide-gated Fe2+ channel. However, previous studies have revealed intrinsically high GDP release rates, which would yield a constitutive ‘on' (GTP bound) state in the Fe2+ influx-machinery. To reconcile this apparent conundrum, FeoB would require stringent regulation of GDP dissociation.
With a tethered G protein, reminiscent of mammalian Ras and Gαβγ, FeoB presents a unique opportunity to study the regulation of nucleotide exchange and the coupling between a G protein and its cognate membrane component. To address this, we have determined crystal structures of both the apo and GTP-bound form of the entire soluble N-terminal domain of FeoB. We have discovered that FeoB forms a trimer around a central cytoplasmic pore, which is closed in the apo structure and open in the GTP-bound structure. The linker domain of FeoB is proposed to act as an intrinsic effector, coupling GTP binding with pore opening. On the basis of these results, a gating model for G protein coupled Fe2+ transport is discussed. Importantly, unambiguous identification and structural characterization of the G5 signature motif reveals it to play a deciding role in GDP release.
Results
FeoB1−270 assembles into a trimeric complex
On the basis of sequence and hydropathy analysis, we generated a construct composed of the entire hydrophilic domain (residues 1–270) of FeoB (FeoB1−270) from Escherichia coli. With this construct, we pursued structural studies and initially obtained crystals of the apo (nucleotide free) FeoB1−270. The model was refined to a resolution of 2.2 Å with R and Rfree values of 22.5 and 28.7%. In addition, the non-hydrolyzable GTP analogue 5′[β,γ-imido]triposphate derivative mant-GMPPNP (mGTP) was used to form a stable FeoB1−270–mGTP complex, which crystallized in the spacegroup P21. The structure was refined to 2.74 Å with R and Rfree of 22.7 and 29.4%, respectively. There was no clear density for the C-terminal nine amino acids (residues 262–270), which have been omitted from all models. Data collection and refinement statistics are presented in Table I (see Supplementary Figure S2 for representative electron density map).
Table 1.
Data collection, phasing and refinement statistics for the Pb derivative, Native, mGTP-FeoB1−270 and apo-FeoB1−270 structures
| Pb derivative | Native | apo-FeoB1−270 | mGTP-FeoB1−270 | |
|---|---|---|---|---|
| Data collection | ||||
| Space group | P212121 | P212121 | C2 | P21 |
| Cell dimensions | ||||
| a, b, c (Å) | 47.7, 122.9, 134.5 | 47.4, 122.4, 134.7 | 146.1, 84.4, 66.2 | 76.0, 46.4, 120.1 |
| α, β, γ (deg) | 90, 90, 90 | 90, 90, 90 | 90, 108, 90 | 90, 95, 90 |
| Wavelength (Å) | 1.5418 | 0.9753 | 0.9793 | 0.9793 |
| Resolution (Å)a | 20–2.90 (3.06–2.90) | 60–2.4 (2.53–2.4) | 70–2.2 (2.32–2.2) | 50–2.72 (2.82–2.72) |
| Rmergea | 0.093 (0.270) | 0.066 (0.248) | 0.176 (0.459) | 0.088 (0.421) |
| I/σIa | 7.0 (2.8) | 8.6 (3.0) | 3.3 (1.8) | 14.3 (2.5) |
| Completeness (%)a | 96.8 (83.4) | 99.1 (99.4) | 99.9 (99.9) | 96.7 (82.0) |
| Redundancy | 5.6 | 4.0 | 7.3 | 3.2 |
| apo-FeoB1−270 | mGTP-FeoB1−270 | |||
| Refinement | ||||
| PDB Code | 3HYR | 3HYT | ||
| Resolution (Å)a | 72–2.20 (2.26–2.20) | 45–2.74 (2.81–2.74) | ||
| No. reflections | 36896 | 20674 | ||
| Rwork/Rfreea | 0.225/0.287 (0.322/0.361) | 0.227/0.294 (0.316/0.423) | ||
| Residues included in the final model | A/B/C2-64, A/B/C71-260 | A2-30, A40-64, A71-261, B2-30, B40-64, B71-260, C2-30, C40-63, C74-261 | ||
| No. atoms | ||||
| Protein | 5841 | 5722 | ||
| Ligand/ion | 0 | 129 | ||
| Water | 480 | 102 | ||
| B-factors | ||||
| Protein | 21.2 | 39.8 | ||
| Ligand/ion | 38.4 | |||
| Water | 21.3 | 25.3 | ||
| r.m.s.d. | ||||
| Bond lengths (Å) | 0.013 | 0.006 | ||
| Bond angles (deg) | 1.486 | 1.000 | ||
| Ramachandran analysisb | ||||
| Favoured regions (%) | 98.0 | 96.6 | ||
| Allowed regions (%) | 100.0 | 100.0 | ||
| PDB accession code | ||||
| aValues in parentheses are for the highest-resolution shell. | ||||
| bAs assessed by MOLPROBITY ([10]). | ||||
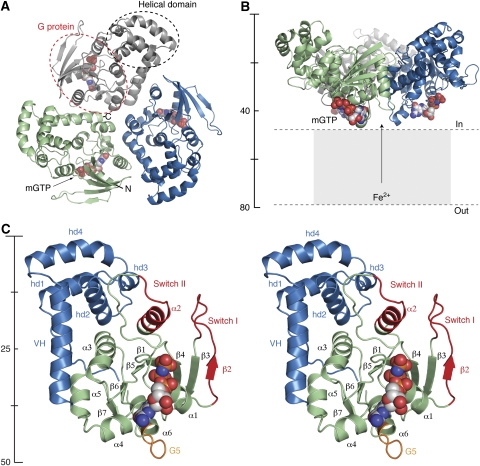
FeoB1−270 crystallized in four space groups (C2 and P21 present work, P212121 and P31 data not shown). In all crystal forms, the overall structure of FeoB1−270 resembles a funnel generated by a non-crystallographic trimeric assembly, with approximate three-fold symmetry between protomers (Figure 1). The protomer to protomer interface (buried surface of ∼1280 Å2) is mainly stabilized by hydrophobic interactions, although hydrogen bonds and salt bridges between residues in neighbouring molecules further stabilize the trimer (see Supplementary Figure S1). To test the biological relevance of the oligomer, we designed a double cysteine mutant (Ala158Cys and Ala254Cys) to form an intersubunit disulphide bond linking subunits together according to Yernool et al (2004) (Supplementary Figure S3). The double Cys mutant, when treated with Cu(II) (1,10-phenantroline)3, forms a stable 88 kDa trimer, illustrating the same oligomeric state of FeoB1−270 in solution (Supplementary Figure S3 and Materials and methods). In addition, purified wild-type protein was cross-linked with glutaraldehyde, which clearly illustrated formation of a trimer in solution (Supplementary Figure S3). As all previous GTPases reside as either monomer or hetero trimers, FeoB1−270 represents the first homo trimer constellation structurally defined. The orientation of the trimer in relation to the inner membrane is predicted from the location of the C-termini (on convex side) and membrane topology analyses (Daley et al, 2005) to be such that the concave side faces the cytoplasm (Figure 1B; Supplementary Figure S4). The surface facing the membrane has a charge distribution resembling that of the soluble domain of CorA (Eshaghi et al, 2006) and the K+ channel KirBac1.1 (Kuo et al, 2003), with a negatively charged centre and a positively charged rim (Supplementary Figure S4).
Figure 1.
Overall structure of FeoB1−270. (A) Ribbon representation of the trimer viewed from the intracellular space. Individual protomers are coloured in grey, green, and blue. Dotted lines indicate the spatial organization of the G protein and helical domain. (B) Side view of the trimer, with colours as in (A). The mGTP molecule is shown as space-fill. (C) Stereo view of one protomer. The N-terminal G protein and helical domain are illustrated in green and blue, respectively. The Switch regions are shown in red, and the G5 motif is depicted in orange. All structural figures were made using PyMOL (http://pymol.sourceforge.net).
Structure of (apo) FeoB1−270
The overall fold of the FeoB1−270 monomer is divided into two distinct domains connected through a flexible linker sequence; an N-terminal GTP-binding domain (G protein, residues 1–166), and a C-terminal helical domain (residues 178–261; Figure 1C). The C-terminal domain forms a helical bundle followed by an extended α-helix, here designated as the ‘valve helix' (residues 241–261; see Figure 1C). The valve helix terminates close to the centre of the trimer and is in an ideal position to regulate the Fe2+ translocation process. The entire helical domain is a new structural feature, not observed in other G proteins, and a search using DALI revealed no significant structural homologues, whereas BLAST indicated low-sequence conservation in this domain between FeoB proteins from other prokaryotic species (see Supplementary Figure S1). Previous biochemical data have indicated this domain to be functioning as a guanine dissociation inhibitor (Eng et al, 2008), although our structure indicates that the domain is situated too far away from the nucleotide-binding site for a direct involvement in inhibition of nucleotide release. The N-terminal G protein, on the other hand, exhibits a polypeptide fold similar to the core of other prokaryotic and eukaryotic GTPases, consisting of a seven-stranded β-sheet (β1–β7) surrounded by five α-helices (α1–α5; Figure 1C). The structural similarity between the G protein domain of FeoB1−270 and other G proteins is illustrated by the r.m.s.d in Cα positions from their superimpositions; 1.83 Å for p21-Ras (114 Cα atoms, PDB code 5P21) and 1.63 Å for Gαi1 (112 Cα atoms, PDB code 2G83; Supplementary Figure S5).
The four consensus elements, G1–G4, which are involved in GTP and Mg2+ binding in all G proteins (Bourne et al, 1991), are also present in FeoB (Supplementary Figure S1). In addition, the effector-binding regions recognized as Switch I (residues 25–40) and Switch II (residues 70–84) are structurally analogous to those of other GTPases (Sprang, 1997). In the apo FeoB1−270 structure, the Switch I region forms an extension to the central canonical β-sheet (β2), similar to the GDP-liganded structure of Ran, another G protein in the Ras superfamily (Scheffzek et al, 1995).
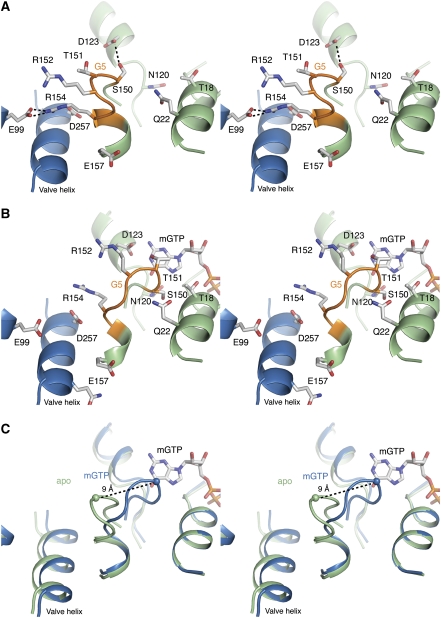
The G5 motif is, despite low sequence conservation, attributed to critical guanine base coordination (Sprang, 1997) and believed to be key to the GPCR catalysed nucleotide exchange in Gα subunits (Oldham et al, 2006; Oldham and Hamm, 2007, 2008). Previous attempts to clearly identify this motif in FeoB1−270 by sequence alignments and mutational analysis were unsuccessful (Marlovits et al, 2002; Hantke, 2003; Eng et al, 2008). In our structure, the G5 motif can unambiguously be identified as residues (150)STRGRG(155) by superimposition with Gα and Ras. This reveals the G5 motif to be in a different position in the amino-acid sequence than suggested earlier (Marlovits et al, 2002; Hantke, 2003). Structural characterization of the G5 motif in the intermediate apo state is rare due to the inherent instability of this state (Sprang, 1997; Johnston and Siderovski, 2007a, 2007b; Oldham and Hamm, 2008). However, in the apo structure of FeoB1−270, the G5 motif is stabilized through a hydrogen bond between Ser150 and Asp123, and through a salt bridge formed between the G5 residue Arg154 and the conserved Glu99 in the neighbouring protomer (Figure 2A). The hydrogen bond between Ser150 and Asp123 is noteworthy, as Asp123 has been shown to be essential for GDP/GTP binding (Marlovits et al, 2002).
Figure 2.
Stereo views of the G5 motif and mGTP-binding site. (A) Apo state. The G protein is shown in green, with the G5 motif highlighted in orange. The neighbouring protomer is shown in blue. An interprotomeric salt bridge between Arg154 and Glu99, and an intraprotomeric hydrogen bond between Asp123 and Ser150 stabilizes the G5 ‘out' state. (B) mGTP state. Contraction of the G protein after GTP binding leads to a high affinity G5 ‘in' state. In the transition, the salt bridge between Arg154 and Glu99, and hydrogen bond between Asp123 and Ser150, are broken. (C) Superimposition reveals a structural shift of ∼9 Å in the Cα position of Thr151 between the ‘in' and ‘out' conformation.
The structure of mGTP complex reveals conformational changes
To confirm the GTP binding properties of FeoB, we determined the structure of FeoB1−270 bound to the non-hydrolysable GTP analogue mGTP. The structure of mGTP-FeoB1−270 was refined to 2.74 Å resolution. Electron density clearly identified the position of a single mGTP molecule per protomer, and a putative Mg2+ ion bound adjacent to the γ-phosphate of mGTP (Supplementary Figure S2). The Mg2+-binding site and mGTP coordination pattern mirror those found in other G proteins, including p21-Ras and Gα subunits (Wall et al, 1995; Sprang, 1997). The phosphate moieties of mGTP interact with the P-loop (G1; residues 10–18), particularly through Asn13, the hydroxyl group of Thr18, the amino group of Lys16, and backbone carbonyl oxygen and amine groups. The hydrophobic guanine group interacts with highly conserved residues in the G4 region, including Asn120 and Asp123, which form hydrogen bonds with the O6/N7, and N1/N2 atoms of mGTP, respectively (Figure 2B).
In relation to the apo structure, one significant observation is a large structural change in the newly identified G5 motif. In the mGTP-FeoB1−270 structure, this motif has shifted ∼9 Å closer to the GTP-binding site, breaking the salt bridge between Arg154 and Glu99 and the hydrogen bond between Asp123 and Ser150 (Figure 2C). This ‘in' conformation of the G5 motif is stabilized by hydrogen bonds between Thr151 (G5) and the N1 atom of mGTP, and between Ser150 (G5) and Gln22/Asn120. In addition, the ‘in' state in the mGTP-liganded structure is likely further stabilized and dependent on the minor structural changes taking place in other motifs, including Switch I/Switch II, following mGTP binding.
Stopped-flow measurements corroborate the role of the G5 motif in GDP release
Comparison of the apo and mGTP-bound structures indicates that the conformational transition in the G5 motif has an important function in GDP release. To corroborate this hypothesis, we performed a structural comparison with the Gαi1 subunit mutant (Ala326Ser; PDB Entry code 1BH2), which is the corresponding mutant to the precocious puberty causing Gαs Ala366Ser. Native FeoB1−270 has a polypeptide sequence that mimics the Gα mutants with a Serine residue occupying the structurally equivalent position in the G5 motif (Gαi1 Ala326Ser versus FeoB1−270 Ser150). As the Gαs mutant has been characterized with an 80-fold acceleration in its rate constant of GDP release (koff,GDP 0.23 s−1, compared with koff,GDP 0,003 s−1 in wt protein) (Iiri et al, 1994), we wanted to explore whether we could decelerate the GDP release in FeoB1−270 by inversely mutating Ser150 to an Alanine residue. Consistent with the structural parallels, stopped-flow measurements confirmed native FeoB1−270 to have an extremely high GDP release rate constant (koff,GDP 145 (±9) s−1, 20°C; Table II) (Marlovits et al, 2002; Eng et al, 2008). Conversely, the Ser150Ala mutant showed drastically altered release rate constant (koff,GDP 10 (±0.1) s−1, 20°C), consistent with our hypothesis that the G5 motif is involved in the rapid GDP release in native FeoB1−270. To further investigate the relevance that the G5 ‘out' hydrogen-bonding network has on the GDP release rate, we also mutated Arg154 (Arg154Ala). As with the Ser150Ala mutant, the Arg154Ala protein shows a drastically decreased release rate constant (koff,GDP 19 (±0.5) s−1, 20°C; Table II).
Table 2.
Rate constants for the association (kobs) and dissociation (koff) of mant-nucleotides with FeoB1−270
| Protein | Concentration (μM) | mGMPPNP kobs s−1 (20°C) | mGMPPNP koff s−1 (20°C) | mGDP koff,GDP s−1 (20°C) |
|---|---|---|---|---|
| FeoB1−270 | 2.5 | 1.63 | ||
| 5 | 1.93 | 1.85 | 145 (±9) | |
| 10 | 2.68 | |||
| 20 | 4.01 | |||
| 40 | 6.71 | |||
| S150A | 5 | 3.87 | 1.77 | 10 (±0.1) |
| R154A | 5 | 2.14 | 1.89 | 19 (±0.5) |
mGTP binding leads to the opening of a central cytoplasmic pore
All G proteins present an effector site on GTP binding; for Ras type G proteins and Gα subunits, the effector association is through the active conformation of Switch I and Switch II (Spoerner et al, 2001; Tesmer et al, 2005; Sprang et al, 2007). In FeoB, the helical domain is situated in proximity to Switch II (Figure 1C), and structural changes in Switch II following mGTP binding are accompanied by observed disorder in the helical domain, indicating that the helical domain is acting as an intrinsic effector. This structural change is transmitted to the valve helix, which in relation to the apo structure has shifted 1–2 Å away from the centre of the trimer.
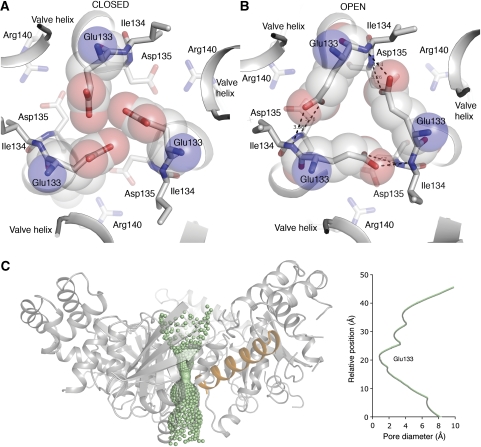
Following this, a striking aspect of the trimeric mGTP-bound structure is that it forms a central cytoplasmic pore, about 20 Å in length and with a diameter of ∼1.2 Å at the narrowest point (Figure 3). The cytoplasmic pore could facilitate gating and passage of un-hydrated Fe2+ ions (ionic radius of Fe2+ ∼0.76 × 10−10 m) delivered from the transmembrane domain. In the apo structure (both C2 and P212121 data), Glu133 is in a conformation that effectively blocks the pore (Figure 3A). However, the structural adaptation occurring on mGTP binding is accompanied by a change in the hydrogen-bonding pattern in the centre of the trimer interface. In the FeoB1−270-mGTP structure, the Glu133 residue has a different conformation, which is stabilized by hydrogen bonds to the backbone amino group of Ile134 in the neighbouring molecule (Figure 3B). The driving force for the conformational change likely originates from mGTP binding, followed by small changes in the structure of the helical domain and the valve helix, which are ideally positioned to regulate the pore (Figure 3C).
Figure 3.
Details of the cytoplasmic gate. (A) Apo structure. View from the membrane side down the centre of the trimer. The trimer is stabilized through a salt bridge formed between Asp135 and Arg140 in neighbouring protomers. In the apo state, Glu133 is pointing to the centre, effectively closing the pore. (B) In the mGTP-bound structure, structural changes leads to a shift in the position of Cα atoms surrounding the centre of the trimer. This enables Glu133 to change conformation and form hydrogen bonds with mainchain N-atoms of Ile134. The new conformation leads to opening of a central cytoplasmic pore. (C) The pore diameter along the proposed cytoplasmic gate in the open state. The main site of constriction and gating (residue Glu133) is depicted. The valve helix from one protomer is shown in orange to illustrate its position in relation to the pore. The cytoplasmic pore was generated using HOLE (Smart et al, 1993).
Discussion
To delineate the complex structure/function relationships of channels or transporter proteins, the individual structural and functional domains are often studied in isolation. To this end, we solved the structures of the apo and mGTP-bound states of the soluble domain of FeoB. Our structural data provides, for the first time, anatomical information of a G protein, which is directly involved in the gating of membrane ion transport processes. A comparative analysis between the apo and mGTP-bound structures along with measured and known biochemical data, begin to reveal the mechanism for GDP release and gating of G protein coupled Fe2+ translocation.
Mechanism of GDP release
The structural changes recognized when the apo- and mGTP-FeoB1−270 structures are compared have not been observed in other systems. In particular, the lack of an equivalent nucleotide-free (apo) structure for the heterotrimeric Gαβγ protein has left the mechanism of GDP release open to debate. As we have determined the structures of both apo and liganded states of FeoB1−270, we can depict the structural transitions driving GDP release. A rapid GDP release rate for FeoB1−270 indicates that the G5 motif has an inherent propensity to switch to a stable ‘out' state, a state in which the affinity for GDP is nominal. This propensity is likely due to the ability of residues in the G5 motif to participate in stabilizing interactions (salt bridges and hydrogen bonds) when in the ‘out' conformation. This was corroborated by our stopped-flow experiments, which revealed altered GDP release rates when the residues involved in the stabilizing interactions were mutated. Despite the reduced rate of the GDP release in the mutated protein, it remains high when compared with Ras type G proteins and Gαβγ proteins. This intrinsically elevated GDP release rate could be due to other residues in the G5 loop, such as Gly153 or Gly155, which could act as ‘hinge' residues and allow for a highly flexible structure. In addition, the movement of the G5 loop could be regulated by an as-yet-unidentified factor to slow down GDP release.
From this we propose that Gαβγ proteins could harbour a similar structural flexibility as observed in FeoB1−270 and that the elusive apo state in the activated GPCR/Gαβγ complex includes an ‘out' conformation of the G5 motif. In support of the hypothesis that Gα subunits have a conformational two-way G5 motif, are previous studies of the GPCR/Gαβγ interactions that have indicated that conformational changes in the G5 motif are key for GDP release (Hamm et al, 1988; Oldham and Hamm, 2008; Scheerer et al, 2008). In addition, recent NMR analysis of a receptor-associated (apo) Gα revealed the nucleotide-free form to represent a structurally dynamic intermediate state (Abdulaev et al, 2006).
Mechanism of G protein coupled gating
It is unclear from the structures presented here whether the pore in the mGTP-bound state of FeoB1−270 has reached maximum aperture, or whether the structure represents a transition state. However, the structural data presented here in concert with published functional data facilitates a novel and simple mechanism for G protein coupled gating of Fe2+ membrane translocation; (i) binding of GTP-Mg2+ is coupled to structural transitions in Switch I and Switch II; (ii) the ‘active' conformation of the Switch regions causes the helical domain, which acts as an intrinsic effector, to move towards the presented effector site (Switch II); (iii) the shift in the helical domain drives a lateral movement of the valve helix, which leads to the opening of a central cytoplasmic pore, allowing Fe2+ to enter the cell through the transmembrane domain (see Figure 3; Supplementary Figure S6); (iv) hydrolysis of GTP to GDP leads to a concerted relaxation of the Switch regions, release of the helical domain from the effector site, and closing of the pore.
Materials and methods
Expression and purification of FeoB1−270
FeoB1−270 was sub-cloned from a full-length E. coli FeoB construct (Daley et al, 2005) and inserted into a pGEX4-T1 vector (GE Healthcare Life Sciences) and expressed as a fusion protein with glutathion S-transferase (GST). Native and mutant protein was purified using GST affinity chromatography according to the manufacturer's protocol. The GST moiety was removed by thrombin cleavage and the protein was polished by size exclusion chromatography (Superdex 75; GE Healthcare Life Sciences). Purified protein was buffer exchanged to 20 mM Tris pH 8.0 and concentrated to 20 mg ml−1.
Crystallization and X-ray data collection
Crystals of native FeoB1−270 (∼20 mg ml−1) were grown by mixing protein and reservoir solution (30% PEG 400, 100 mM MgCl2, 0.1 M MES pH 6.5) in a 2:1 ratio using the hanging-drop technique. Crystals appeared after 48 h at 20°C and grew to maximal dimensions of 100 × 100 × 70 μm within a week. Crystals of native FeoB1−270 grew in space group P212121, whereas crystals of Se-Met labelled protein (apo-FeoB1−270) crystallized in C2. A nucleotide-bound protein was obtained by co-crystallizing the native FeoB1−270 protein in the presence of 1 mM mGTP. Crystals appeared in the same reservoir solution as the native protein, although crystals grew in spacegroup P21. The analogue GMPPMP was also used for co-crystallization trials, although the mGTP provided the highest resolution structure. A Pb-derivative crystal was prepared by soaking a crystal of the native protein in a solution containing trimethyllead acetate (5 mM) for 1 h, before data collection.
X-ray data were collected from native, mGTP-FeoB1−270, and Se-Met labelled apo-FeoB1−270 crystals using a MARmosaic 300 detector at beamline ID23 B/D at the Advanced Photon Source (APS, Chicago). Data from heavy atom soaked crystals (Pb-derivative) were collected on a Rigaku MicroMax 007HF Cu rotating anode X-ray generator with Osmic VariMax HF focusing optics and a MAR345 Image Plate detector. All image data were collected at 100 K and processed using Denzo & Scalepack (Otwinowski et al, 1997) or Mosflm (Leslie, 1992) and Scala (Collaborative Computational Project, Number 4, 1994). Cell dimensions and data collection statistics are presented in Table I and Supplementary data.
Structure determination and refinement of apo and liganded FeoB1−180
The structure of FeoB1−270 was determined by SIRAS phasing, using data from Pb-derivatized and native crystals. Heavy atom sites were determined with SHELXD (Sheldrick, 2008) and refined with SHARP (Bricogne et al, 2003). SIRAS phases to 2.9 Å were applied to the native data set and extended to 2.2 Å using three-fold averaging, solvent fattening and histogram mapping in DM (Collaborative Computational Project, Number 4, 1994). The atomic model was built using the programs O (Jones et al, 1991) and COOT (Emsley and Cowtan, 2004). During the model building process, it was rapidly recognized that significant regions of the FeoB1−270 structure were disordered in the native crystals. The coordinates of the preliminary model were therefore used as a molecular replacement model to determine the structure of FeoB1−270 in crystals grown from the Se-Met labelled protein (space group C2), using PHASER. Model building was completed using this data and refinement was carried out using REFMAC5 (Murshudov et al, 1997) (with TLS) and PHENIX (Zwart et al, 2008). As the data from the Se-Met labelled protein produced the most complete model, this structure will hereafter be referred to as apo-FeoB1−270. The coordinates of the resulting apo-FeoB1−270 model were used as a molecular replacement model to determine the structure of mGTP-FeoB1−270 using PHASER (McCoy et al, 2007). The coordinates of both the apo-FeoB1−270 and mGTP-FeoB1−270 structures have been deposited in the Protein Data Bank with accession codes 3HYR and 3HYT, respectively.
GTPase activity measurements
GTP hydrolysis assay was performed using a Malachite Green Phosphate Assay (BioAssay Systems) that provides colorimetric detection of inorganic phosphate. FeoB1−270 (2 μM) in Tris buffer (20 mM Tris pH 8.0, 100 mM NaCl) was incubated for 8–20 h with GTP (20 μM) at 25°C. Reactions were terminated on addition of malachite green reagent (40 μl) and A620 was measured after 30 min to monitor hydrolysis. Appropriate controls were performed to ensure that there was no background interference from the reaction components.
Mutagenesis
The plasmid pGEX-4T1/FeoB1−270 was used to generate expression vectors with single amino-acid mutations (S150A and R154A) and one double mutant (A158C/A254C), by using Stratagene's QuickChange site-directed mutagenesis kit. Mutants were expressed and purified as the wt protein.
Cross-linking
Purified FeoB1−270 double mutant, A158C/A254C, was concentrated to ∼4 mg ml−1 and left at 4°C for 3 days to allow for assembly of oligomer. For the cross-linking reaction, Cu(II) (1,10-phenantroline)3 was prepared by adding fresh mixture of equal volumes of 0.5 mM CuSO4 and 1.5 mM 1,10-phenantroline prepared in ethanol and diluted 1 in 10 with 1 M sodium citrate (pH 5.6). 1 mM of Cu(II) (1,10-phenantroline)3 was added to gel-filtration fractions of FeoB1−270 trimer and reactions were carried out at room temperature for 1 h. The samples were analysed by SDS–polyacrylamide gel electrophoresis (PAGE) under non-reducing conditions. The protein bands were identified and molecular mass of protein in solution was determined by mass spectrometry (SUPRU, University of Sydney).
Purified wild-type FeoB1−270 was also cross-linked with glutaraldehyde (0.01 mM) at 25°C for 30 min and reactions were quenched using 150 mM Tris pH 7.5. Cross-linked protein was subsequently evaluated by SDS–PAGE.
Stopped-flow fluorescence assay
A stopped-flow based assay was performed to determine binding and release of fluorescent nucleotides in wt FeoB1−270 and the S150A/Arg154Ala mutants as described (Marlovits et al, 2002). Briefly, to determine release rate constants, 10 mM of native FeoB1−270 or mutant protein was incubated with 0.5 mM of mGDP in stopped-flow buffer (20 mM MES, pH 6.5, 100 mM NaCl, 10 mM MgCl2,) for 30 min at room temperature. Equal volumes of the protein-mGDP mix and 1 mM GTP in stopped-flow buffer were rapidly mixed into a 22.5 μl optical cell of a pneumatically driven stopped-flow apparatus (SF-61, Hi-Tech Scientific Ltd., Salisbury, UK). The mant group was excited using the 355 nm line of a 100 W short arc mercury lamp (Osram, Berlin, Germany) and the fluorescence was monitored at right angles to the incident light beam using an R928 multialkali side-on photomultiplier. The fluorescence was collected at wavelengths ⩾400 nm by using a GG400 glass cut-off filter (Schott, Mainz, Germany) in front of the photomultiplier. Similarly, observed binding rate constants were determined by rapidly mixing 2.5–40 mM FeoB1−270 or 5 mM mutant protein with 0.5 mM mGMPPNP in the stopped-flow apparatus. All data reported are averages of 7–10 independent experimental traces performed under identical conditions. Reactions were performed at 20°C. At this temperature the rate constants kobs,mGDP>1000 s−1 were obtained, indicating that GDP binding and dissociation occurred on a time scale faster than what can be reliably measured by stopped-flow.
Supplementary Material
Supplementary Figure S1
Supplementary Figure S2
Supplementary Figure S3
Supplementary Figure S4
Supplementary Figure S5
Supplementary Figure S6
Supplementary Figure Legends
Supplementary Information
Review Process File
Acknowledgments
This study was supported by the Australian Research Council (ARC; DP0666970 to MJ). MR was a recipient of the Australian Research Council Linkage Fellowship (LX0881956) and supported by Stiftelsen Bengt Lundqvist Minne. MJM was supported by a Cancer Institute of NSW Career Development Fellowship. Data collection was done at the Advanced Photon Source (APS). We acknowledge the support of S Corcoran, N Venugopolan, and M Becker at GM/CA-CAT, for scientific support and assistance with data collection. GM/CA-CAT beamline (ID23) is supported by the US National Cancer Institute and the US National Institute of General Medical Science. Visits to APS were supported by the Australian Nuclear Science Technology Organization (ANSTO). This research was facilitated by access to the Sydney University Proteome Research Unit, established under the Australian Government's Major National Research Facilities program and supported by the University of Sydney. G von Heijne is acknowledged for supplying the FeoB-GFP vector and B Crossett for mass spectrometry. We thank Jeff Abramson and Gunnar von Heijne for critical assessment of the paper.
Footnotes
The authors declare that they have no conflict of interest.
References
- Abdulaev NG, Ngo T, Ramon E, Brabazon DM, Marino JP, Ridge KD (2006) The receptor-bound ‘empty pocket' state of the heterotrimeric G-protein alpha-subunit is conformationally dynamic. Biochemistry 45: 12986–12997 [DOI] [PubMed] [Google Scholar]
- Bocquet N, Nury H, Baaden M, Le Poupon C, Changeux JP, Delarue M, Corringer PJ (2009) X-ray structure of a pentameric ligand-gated ion channel in an apparently open conformation. Nature 457: 111–114 [DOI] [PubMed] [Google Scholar]
- Bourne HR, Sanders DA, McCormick F (1991) The GTPase superfamily: conserved structure and molecular mechanism. Nature 349: 117–127 [DOI] [PubMed] [Google Scholar]
- Bricogne G, Vonrhein C, Flensburg C, Schiltz M, Paciorek W (2003) Generation, representation and flow of phase information in structure determination: recent developments in and around SHARP 2.0. Acta Crystallogr D Biol Crystallogr 59: 2023–2030 [DOI] [PubMed] [Google Scholar]
- Cachero TG, Morielli AD, Peralta EG (1998) The small GTP-binding protein RhoA regulates a delayed rectifier potassium channel. Cell 93: 1077–1085 [DOI] [PubMed] [Google Scholar]
- Cartron M, Maddocks S, Gillingham P, Craven C, Andrews S (2006) Feo—transport of ferrous iron into bacteria. BioMetals 19: 143–157 [DOI] [PubMed] [Google Scholar]
- Clayton GM, Silverman WR, Heginbotham L, Morais-Cabral JH (2004) Structural basis of ligand activation in a cyclic nucleotide regulated potassium channel. Cell 119: 615–627 [DOI] [PubMed] [Google Scholar]
- Collaborative Computational Project, Number 4 (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr 50: 760–763 [DOI] [PubMed] [Google Scholar]
- Daley DO, Rapp M, Granseth E, Melen K, Drew D, von Heijne G (2005) Global topology analysis of the Escherichia coli inner membrane proteome. Science 308: 1321–1323 [DOI] [PubMed] [Google Scholar]
- Davis IW, Leaver-Fay A, Chen VB, Block JN, Kapral GJ, Wang X, Murray LW, Arendall WB, Snoeyink J, Richardson JS, Richardson DC (2007) MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res 35: W375–W383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Cowtan K (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60: 2126–2132 [DOI] [PubMed] [Google Scholar]
- Eng ET, Jalilian AR, Spasov KA, Unger VM (2008) Characterization of a novel prokaryotic GDP dissociation inhibitor domain from the G protein coupled membrane protein FeoB. J Mol Biol 375: 1086–1097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eshaghi S, Niegowski D, Kohl A, Molina DM, Lesley SA, Nordlund P (2006) Crystal structure of a divalent metal ion transporter CorA at 2.9 Angstrom resolution. Science 313: 354–357 [DOI] [PubMed] [Google Scholar]
- Finlin BS, Correll RN, Pang C, Crump SM, Satin J, Andres DA (2006) Analysis of the complex between Ca2+ channel beta-subunit and the Rem GTPase. J Biol Chem 281: 23557–23566 [DOI] [PubMed] [Google Scholar]
- Hamm HE, Deretic D, Arendt A, Hargrave PA, Koenig B, Hofmann KP (1988) Site of G protein binding to rhodopsin mapped with synthetic peptides from the alpha subunit. Science 241: 832–835 [DOI] [PubMed] [Google Scholar]
- Hantke K (2003) Is the bacterial ferrous iron transporter FeoB a living fossil? Trends Microbiol 11: 192–195 [DOI] [PubMed] [Google Scholar]
- Hollenstein K, Dawson RJ, Locher KP (2007) Structure and mechanism of ABC transporter proteins. Curr Opin Struct Biol 17: 412–418 [DOI] [PubMed] [Google Scholar]
- Iiri T, Herzmark P, Nakamoto JM, van Dop C, Bourne HR (1994) Rapid GDP release from Gs alpha in patients with gain and loss of endocrine function. Nature 371: 164–168 [DOI] [PubMed] [Google Scholar]
- Johnston CA, Siderovski DP (2007a) Receptor-mediated activation of heterotrimeric G-proteins: current structural insights. Mol Pharmacol 72: 219–230 [DOI] [PubMed] [Google Scholar]
- Johnston CA, Siderovski DP (2007b) Structural basis for nucleotide exchange on Gαi subunits and receptor coupling specificity. Proc Natl Acad Sci 104: 2001–2006 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Jones TA, Zou JY, Cowan SW, Kjeldgaard M (1991) Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A 47 (Pt 2): 110–119 [DOI] [PubMed] [Google Scholar]
- Karnoub AE, Weinberg RA (2008) Ras oncogenes: split personalities. Nat Rev Mol Cell Biol 9: 517–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo A, Gulbis JM, Antcliff JF, Rahman T, Lowe ED, Zimmer J, Cuthbertson J, Ashcroft FM, Ezaki T, Doyle DA (2003) Crystal structure of the potassium channel KirBac1.1 in the closed state. Science 300: 1922–1926 [DOI] [PubMed] [Google Scholar]
- Leslie AGW (1992) Recent changes to the MOSFLM package for processing film and image plate data. Joint CCP4+ ESF-EAMCB Newslett Protein Crystallogr 26: 11–20 [Google Scholar]
- Marlovits TC, Haase W, Herrmann C, Aller SG, Unger VM (2002) The membrane protein FeoB contains an intramolecular G protein essential for Fe(II) uptake in bacteria. Proc Natl Acad Sci USA 99: 16243–16248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ (2007) Phaser crystallographic software. J Appl Crystallogr 40: 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Dodson EJ (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr 53: 240–255 [DOI] [PubMed] [Google Scholar]
- Oldham WM, Hamm HE (2007) How do receptors activate G proteins? In Mechanisms and Pathways of Heterotrimeric G Protein Signaling, Sprang SR (ed) pp 67–93. San Diego: Academic Press [Google Scholar]
- Oldham WM, Hamm HE (2008) Heterotrimeric G protein activation by G-protein-coupled receptors. Nat Rev Mol Cell Biol 9: 60–71 [DOI] [PubMed] [Google Scholar]
- Oldham WM, Van Eps N, Preininger AM, Hubbell WL, Hamm HE (2006) Mechanism of the receptor-catalyzed activation of heterotrimeric G proteins. Nat Struct Mol Biol 13: 772–777 [DOI] [PubMed] [Google Scholar]
- Oldham WM, Van Eps N, Preininger AM, Hubbell WL, Hamm HE (2007) Mapping allosteric connections from the receptor to the nucleotide-binding pocket of heterotrimeric G proteins. Proc Natl Acad Sci USA 104: 7927–7932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W, Charles W, Carter J (1997) Processing of X-ray diffraction data collected in oscillation mode. In Methods in Enzymology Macromolecular Crystallography Part A, pp 307–326. San Diego: Academic Press [DOI] [PubMed] [Google Scholar]
- Rees DC, Johnson E, Lewinson O (2009) ABC transporters: the power to change. Nat Rev Mol Cell Biol 10: 218–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaafsma D, Roscioni SS, Meurs H, Schmidt M (2008) Monomeric G-proteins as signal transducers in airway physiology and pathophysiology. Cell Signal 20: 1705–1714 [DOI] [PubMed] [Google Scholar]
- Scheerer P, Park JH, Hildebrand PW, Kim YJ, Krausz N, Choe H, Hofmann KP, Ernst OP (2008) Crystal structure of opsin in its G-protein-interacting conformation. Nature 455: 497–502 [DOI] [PubMed] [Google Scholar]
- Scheffzek K, Klebe C, Fritz-Wolf K, Kabsch W, Wittinghofer A (1995) Crystal structure of the nuclear Ras-related protein Ran in its GDP-bound form. Nature 374: 378–381 [DOI] [PubMed] [Google Scholar]
- Sheldrick GM (2008) A short history of SHELX. Acta Crystallogr A 64: 112–122 [DOI] [PubMed] [Google Scholar]
- Smart OS, Goodfellow JM, Wallace BA (1993) The pore dimensions of gramicidin A. Biophys J 65: 2455–2460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spoerner M, Herrmann C, Vetter IR, Kalbitzer HR, Wittinghofer A (2001) Dynamic properties of the Ras switch I region and its importance for binding to effectors. Proc Natl Acad Sci USA 98: 4944–4949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprang SR (1997) G protein mechanisms: insights from structural analysis. Annu Rev Biochem 66: 639–678 [DOI] [PubMed] [Google Scholar]
- Sprang SR, Chen Z, Du X (2007) Structural basis of effector regulation and signal termination in heterotrimeric G[alpha] proteins. In Mechanisms and Pathways of Heterotrimeric G Protein Signaling, Sprang SR (ed) pp 1–65. San Diego: Academic Press [DOI] [PubMed] [Google Scholar]
- Tesmer VM, Kawano T, Shankaranarayanan A, Kozasa T, Tesmer JJG (2005) Snapshot of activated G proteins at the membrane: the Galphaq-GRK2-Gbetagamma complex. Science 310: 1686–1690 [DOI] [PubMed] [Google Scholar]
- Vetter IR, Wittinghofer A (2001) The guanine nucleotide-binding switch in three dimensions. Science 294: 1299–1304 [DOI] [PubMed] [Google Scholar]
- Wall MA, Coleman DE, Lee E, Iñiguez-Lluhi JA, Posner BA, Gilman AG, Sprang SR (1995) The structure of the G protein heterotrimer Gi[alpha]1[beta]1[gamma]2. Cell 83: 1047–1058 [DOI] [PubMed] [Google Scholar]
- Yernool D, Boudker O, Jin Y, Gouaux E (2004) Structure of a glutamate transporter homologue from Pyrococcus horikoshii. Nature 431: 811–818 [DOI] [PubMed] [Google Scholar]
- Zwart PH, Afonine PV, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, McKee E, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Storoni LC, Terwilliger TC, Adams PD (2008) Automated structure solution with the PHENIX suite. Methods Mol Biol 426: 419–435 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure S1
Supplementary Figure S2
Supplementary Figure S3
Supplementary Figure S4
Supplementary Figure S5
Supplementary Figure S6
Supplementary Figure Legends
Supplementary Information
Review Process File