Abstract
Magnetic resonance spectroscopy (MRS) has been shown to have great clinical potential as a supplement to magnetic resonance imaging in the detection of prostate cancer (CaP). MRS provides functional information in the form of changes in the relative concentration of specific metabolites including choline, creatine, and citrate which can be used to identify potential areas of CaP. With a view to assisting radiologists in interpretation and analysis of MRS data, some researchers have begun to develop computer-aided detection (CAD) schemes for CaP identification from spectroscopy. Most of these schemes have been centered on identifying and integrating the area under metabolite peaks which is then used to compute relative metabolite ratios. However, manual identification of metabolite peaks on the MR spectra, and especially via CAD, is a challenging problem due to low signal-to-noise ratio, baseline irregularity, peak overlap, and peak distortion. In this article the authors present a novel CAD scheme that integrates nonlinear dimensionality reduction (NLDR) with an unsupervised hierarchical clustering algorithm to automatically identify suspicious regions on the prostate using MRS and hence avoids the need to explicitly identify metabolite peaks. The methodology comprises two stages. In stage 1, a hierarchical spectral clustering algorithm is used to distinguish between extracapsular and prostatic spectra in order to localize the region of interest (ROI) corresponding to the prostate. Once the prostate ROI is localized, in stage 2, a NLDR scheme, in conjunction with a replicated clustering algorithm, is used to automatically discriminate between three classes of spectra (normal appearing, suspicious appearing, and indeterminate). The methodology was quantitatively and qualitatively evaluated on a total of 18 1.5 T in vivo prostate T2-weighted (w) and MRS studies obtained from the multisite, multi-institutional American College of Radiology (ACRIN) trial. In the absence of the precise ground truth for CaP extent on the MR imaging for most of the ACRIN studies, probabilistic quantitative metrics were defined based on partial knowledge on the quadrant location and size of the tumor. The scheme, when evaluated against this partial ground truth, was found to have a CaP detection sensitivity of 89.33% and specificity of 79.79%. The results obtained from randomized threefold and fivefold cross validation suggest that the NLDR based clustering scheme has a higher CaP detection accuracy compared to such commonly used MRS analysis schemes as z score and PCA. In addition, the scheme was found to be robust to changes in system parameters. For 6 of the 18 studies an expert radiologist laboriously labeled each of the individual spectra according to a five point scale, with 1∕2 representing spectra that the expert considered normal and 3∕4∕5 being spectra the expert deemed suspicious. When evaluated on these expert annotated datasets, the CAD system yielded an average sensitivity (cluster corresponding to suspicious spectra being identified as the CaP class) and specificity of 81.39% and 64.71%, respectively.
Keywords: magnetic resonance spectroscopy, computer-aided diagnosis, prostate cancer, nonlinear dimensionality reduction, hierarchical clustering, unsupervised classification, k-means, replicated clustering, principal component analysis, locally linear embedding, graph embedding, spectral clustering, z score
INTRODUCTION
Prostatic adenocarcinoma (CaP) is the most commonly occurring malignancy among men with 186 320 new cases and 28 660 deaths estimated to occur in the United States in 2008 (American Cancer Society, 2008). Early detection of CaP offers the best hope of curing it; however, early prostate cancer is usually asymptomatic.1 Screening of CaP is based on digital rectal examination and monitoring elevated levels of the blood serum prostate specific antigen (PSA). Definitive diagnosis of CaP involves histological examination of biopsy specimens obtained via a blinded sextant transrectal ultrasound (TRUS) directed biopsy for patients with elevated PSA levels. Since prostate ultrasound is limited in its ability to identify CaP, biopsy locations are chosen at random within the prostate sextants. Consequently, the CaP detection accuracy associated with TRUS is only 20%–25% in patients with elevated PSA levels (4–10 μg∕ml).2 Recently, in vivo endorectal T2-weighted (w) structural magnetic resonance (MR) imaging (MRI) of the prostate has allowed for greater discrimination between benign and cancerous prostatic structures as compared to TRUS.3 However, structural T2-w MRI by itself has been shown to be limited in its ability to detect small foci of carcinoma contributing to a relatively low detection specificity.3
Over the past few years, MR spectroscopic (MRS) imaging (MRSI) has emerged as a useful complement to structural MRI for potential screening of CaP.4, 5 MRSI is a noninvasive technique used to obtain the metabolic concentrations of specific molecular markers and biochemicals in the prostate including citrate, creatine, and choline, and changes in the ratio [choline∕citrate or (choline+creatine)∕citrate] of which have been shown to be linked to presence of CaP.6, 7, 8, 9 The spectra are obtained at either single or multiple locations from a rectangular spectral grid placed on a corresponding T2-w MR image. It has been demonstrated previously that the relative concentrations of choline, citrate, and creatine are significantly different in CaP and normal regions within the prostate.6, 10
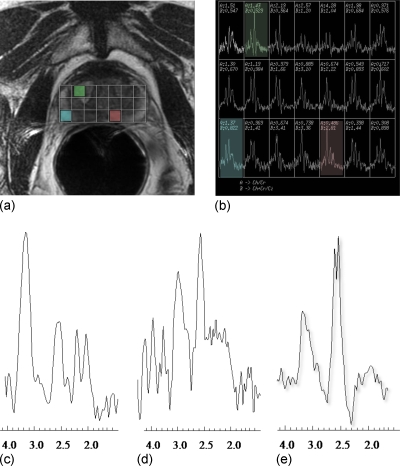
The relative concentrations of choline, creatine, and citrate are obtained by calculating the area under the peak for these metabolites to assess the presence of CaP at a specific prostate location on the T2-w MRI. Identification of precise location of specific metabolites on the MR spectra is a difficult task for radiologists due to (a) a low signal to noise ratio and (b) the presence of biomedical signal artifacts associated with MR spectra such as peak overlap and peak and baseline distortion. Figure 1a shows a MRS grid superimposed on the corresponding T2-w MRI slice, while Fig. 1b shows the MR spectra acquired from individual MRS voxels from the grid shown in Fig. 1a. Figures 1c, 1d, 1e correspond to representative CaP, noisy and normal spectra obtained from three different locations (shown in red, blue, and green, respectively) within the spectral grid [Fig. 1b]. Note the amount of noise in the MRS in Fig. 1d which, in some cases, can severely limit the ability of a radiologist to accurately identify individual metabolite peaks and quantitate corresponding peak areas. Thus the usefulness of MRSI as an adjunct to MRI as a means of detecting, localizing, and characterizing CaP is highly dependent on the quality of the spectral examinations obtained, owing to the challenges in visually identifying spectroscopic CaP signatures (through the identification of abnormal metabolite peak area ratios) with poor data quality.11
Figure 1.
(a) shows a T2-w MRI slice of the prostate with a user-selected 3×7 voxel grid overlaid on the prostate; the MRS spectral grid corresponding to the T2 slice is shown in (b). An abnormal appearing spectra [red voxel in (a) and (b)] is shown in (c), while (e) shows the normal appearing spectra [green voxel in (a) and (b)] (d) shows additional spectra [blue voxel in (a) and (b)] with poor signal to noise ratio and with the baseline affected by a tail of broad upfield lipid resonance (2.0–2.5 ppm). Note that lipid contamination in edge voxels is relatively common in prostate MRSI and can affect the accuracy of the calculated metabolite ratios.
Computer-aided diagnosis (CAD) in medical imaging serves as an adjunctive method for image or data analysis, aiding the radiologist in identifying areas of disease or abnormality.12 The low detection accuracy associated with current TRUS prostate biopsies points to a need for developing an image guided decision support system to direct needle placement in the prostate to increase CaP detection accuracy. While our group13, 14 and others15 have begun to develop CAD systems for CaP detection from structural and functional MR imaging, corresponding developments in MRS have not been as forthcoming. This is in spite of evidence to suggest that integration of structural and metabolic imaging7, 16 could boost diagnostic yield over that which could be obtained from any individual modality. To date, computer-based approaches for prostate MRSI analysis have focused largely on the use of semiautomated peak area integration to determine metabolite ratios, although a few researchers17, 18, 19, 20, 21 explored the use of more sophisticated techniques for automated peak finding.
Previous attempts at computerized analysis of MRS can be classified into two broad categories: (a) signal quantification (model dependent) and (b) pattern recognition (model independent) approaches. Commonly used quantification methods include VARPRO,17AMARES,18 and QUEST,19 which are software utilities where the objective is to minimize the squared distance between the acquired data and a model basis function built on prior knowledge about the metabolic profiles of a typical MR spectrum. However, the performance of these quantification models is usually dependent on (i) the choice of correct number of model components, (ii) optimal choice of prior knowledge (model function), (iii) presence of noise and contributions from nonprostate spectra, (iv) peak overlap owing to contributions from multiple metabolites, and (v) baseline distortion and line broadening.22 In order to avoid the limitations of model and peak detection based approaches for MRS, recently some researchers have begun to explore domain independent techniques such as z score and principal component analysis (PCA). An excellent comprehensive comparison of quantification and pattern recognition schemes used for MRS analysis is provided in Ref. 22.
z score is a statistical quantity obtained as the ratio of the difference in each individual sample’s score and the population mean to the population variance, z score analysis20, 23 aims to quantify the totality of contributions of all metabolites in the spectral vector. In Refs. 20, 23, z score was employed for MRS based detection of glioma and CaP, respectively. Another statistical technique, canonical correlation analysis (CCA),24 based on calculating the canonical coefficients to obtain correlated linear relationships between two multidimensional variables was shown to be useful in successfully classifying prostate MRSI datasets into four classes: Aggressive tumor, tumor, mixed tissue, and healthy tissue.
Devos et al.25 used linear discriminant analysis (LDA) and a least squares support vector machine (SVM) classifier to discriminate between different tumor classes on brain MRS. Ma and Sun26 and Simonetti et al.27 explored other linear dimensionality reduction methods such as independent component analysis and PCA, in conjunction with support vector machine (SVM) classifier, to differentiate between brain tissue classes via MRS.
Dimensionality reduction (DR) refers to the projection of high dimensional data into a reduced dimensional feature space without a significant loss in class discriminatory information. The low dimensional representation of the data is easier to visualize and DR algorithms aim to preserve object relationships so that objects that are close to one another in the high dimensional ambient space are mapped to adjacent locations in the resulting low dimensional embedding space. However, linear DR schemes such as PCA assume the original high dimensional data to be inherently linear and hence employ linear projection methods to reduce data dimensionality. Recently several nonlinear dimensionality reduction (NLDR) algorithms have been proposed for the analysis and visualization of nonlinear data.29, 30, 31 The objective behind NLDR methods is to nonlinearly map objects, c, d belonging to the same object class and characterized by M-dimensional feature vectors F(c), F(d), to adjacent locations S(c), S(d) in the low dimensional embedding; S(c), S(d) representing the m-dimensional dominant eigenvectors corresponding to c, d (m⪡M). Unlike linear DR schemes that typically employ the Euclidean distance measure to estimate object distances, most NLDR schemes aim at preserving geodesic distances between objects while computing the embedding. We have previously demonstrated that the use of NLDR methods for representation of high dimensional gene- and protein-expression data, results in better classification compared to linear DR based representation (PCA and LDA).28
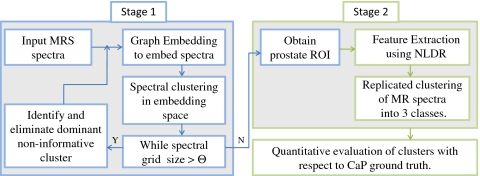
In this paper, we present a fully automated CAD system for detecting abnormal∕suspicious regions on the prostate using 1.5 T prostate MRSI data. Figure 2 illustrates the organization of our CAD scheme. Our methodology comprises of two stages. In stage 1, a novel hierarchical classification scheme is employed to recursively distinguish prostatic from extracapsular (noninformative) spectra via graph embedding (GE),31, 32 a well known NLDR scheme, in order to hone in the region of interest (ROI) corresponding to the prostate. In a typical field of view (FOV) of an in vivo endorectal prostate T2-w MR image, the prostate occupies a small percentage [approximately 10% in Fig. 1a] of the total volume within a prostate MRI scene. Since the extracapsular spectra are most populous, the largest cluster is identified at each iteration as being noninformative and eliminated. This process is repeated until the number of MR spectra remaining is approximately equal to the number usually contained in the prostate (Θ) [Fig. 1a]. The removal of extracapsular spectra in stage 1 makes it easier to discriminate between suspicious appearing and normal appearing spectra within the prostate. In stage 2, a NLDR scheme is applied to nonlinearly embed the informative spectra into a reduced dimensional space. The individual prostate spectra now characterized by their low dimensional embedding coordinates are clustered into distinct classes via a “replicated clustering” scheme. All spectra are aggregated into three classes based on the assumption that they correspond to normal, CaP, and indeterminate classes.
Figure 2.
Flowchart showing various system components and methodological overview of the CAD scheme. Hierarchical spectral clustering is performed in stage 1 to automatically obtain the prostate grid, followed in stage 2 by the identification of different tissue classes via NLDR and replicated clustering.
The rest of this paper is organized as follows: In Sec. 2 we provide a detailed description of the feature extraction and clustering schemes employed in this paper. In Sec. 3 we provide the details of our CAD methodology for prostate MRS classification. In Sec. 4 we explain the evaluation scheme employed in this work followed in Sec. 5 by results of qualitative and quantitative evaluation of our scheme on a total of 18 prostate MRI-MRS studies. Concluding remarks and future research directions are presented in Sec. 6.
DESCRIPTION OF FEATURE EXTRACTION AND REPLICATED CLUSTERING METHODS
Feature extraction methods
Linear dimensionality reduction scheme: PCA
PCA is a linear DR method widely used to visualize high dimensional data and discern object relationships in the data by finding orthogonal axes that contain the greatest amount of variance in the data.33 These orthogonal eigenvectors corresponding to the largest eigenvalues are called “principal components.” To obtain these principal components, each data point c in set C is first centered by subtracting the mean of all the features for each observation c from its original M dimensional feature value fu(c), u∊{1,…,M} as shown by
| (2.1) |
From feature values for each c∊C, a new ∣C∣×M matrix Y is constructed, where ∣C∣ is the cardinality of set C. The matrix Y is then decomposed into corresponding singular values as shown by
| (2.2) |
where via singular value decomposition a ∣C∣×∣C∣ diagonal matrix WPCA containing the eigenvalues of the principal components, a m×∣C∣ left singular matrix U, and a M×∣C∣ matrix V are obtained. The eigenvalues in WPCA represent the amount of variance for each eigenvector , v∊{1,2,…,m}, in matrix VT and are used to rank the corresponding eigenvectors in the order of greatest variance. Thus the first m eigenvectors that represent a prespecified percentage of the variance in the data are extracted, while the remaining Eigen vectors are discarded. Thus each data sample c∊C is now described by an m-dimensional embedding vector SPCA(c). In spite of the fact that PCA assumes that the data lie on a linear manifold, it allows for specification of the number of eigenvectors required to explain a prespecified percentage of the variance in the data.
z score
For a set of objects, CV⊂C, c∊CV, a mean vector and the corresponding standard deviation vector , where and are obtained. At each c∊C, z score (Sz(c)) is defined as
| (2.3) |
where F(c) corresponds to the feature vector at each c and Sz(c) reflects the degree to which the value of an object deviates from the normal based on a statistical linear model. A predefined threshold θz is then used to classify each c∊C into one class or the other based on whether Sz(c)⩾θz.
Nonlinear dimensionality reduction methods
In this work we consider two popular NLDR schemes, locally linear embedding (LLE) (Ref. 30) and GE (Ref. 32) for MRS analysis. We aim to demonstrate the use of NLDR schemes for representation of MRS data results in superior discrimination between CaP and non-CaP spectra compared to the use of linear DR schemes such as PCA.
Graph embedding. The aim of graph embedding32 is to find an embedding vector SGE(ci), ∀ci∊C, i∊{1,…,∣C∣}, such that the relative ordering of the distances between objects in high dimensional space is maximally preserved in the lower dimensional space. Thus, if locations ci,cj∊C,i,j∊{1,…,∣C∣} are adjacent in the high dimensional feature space, then ‖SGE(ci)−SGE(cj)‖2 should be small, where ‖⋅‖2 represents the Euclidean norm. This will only be true if the distances between all ci, cj∊C are preserved in the low dimensional mapping of the data. To compute the optimal embedding, an adjacency matrix WGE∊R∣C∣×∣C∣ is first defined as
| (2.4) |
SGE(ci) is then obtained via maximization of the following objective function:
| (2.5) |
where XGE=[SGE(c1),SGE(c2),…,SGE(cn)], n=∣C∣, and γ=∣C∣−1. Additionally, D is a diagonal matrix where ∀ci∊C, i∊{1,…,∣C∣}, the diagonal element is defined as D(i,i)=∑jWGE(i,j). The embedding space is defined by the eigenvectors corresponding to the smallest m eigenvalues of (D−WGE) XGE=λDXGE. The matrix XGE∊R∣C∣×m of the first m eigenvectors is constructed, and ∀ci∊C, SGE(ci) is defined as row i of XGE.
Locally linear embedding (LLE). LLE (Ref. 30) operates by assuming that objects within a local neighborhood in a high dimensional feature space are linearly related. Consider the set of high dimensional feature vectors F={F(c1),F(c2),…,F(cn)}, ∀ci∊C, i∊{1,…,n}. LLE aims to map the set F to the corresponding set XLLE={SLLE(c1),SLLE(c2),…,SLLE(cn)} of embedding coordinates. Let d(1),…,d(K) be the K nearest neighbors of ci and let ηK(ci) be the indices of the location of the K-nearest neighbors (K-NN) of ci∊C. The feature vector F(ci) and its K-NN’s {F(d(1)),F(d(2)),…,F(d(K))} are assumed to lie on a patch of the manifold that is locally linear, allowing us to use the Euclidean metric to determine distance between neighbors. Each F(ci) can then be approximated by a weighted sum of its K-NN. The optimal reconstruction weights are given by the sparse matrix WLLE (subject to the constraint ∑jWLLE(i,j)=1) that minimizes
| (2.6) |
Having determined the weighting matrix WLLE, the next step is to find a low dimensional representation of the points in F that preserves this weighting. Thus, for each F(ci) approximated as the weighted combination of its K-NN, its projection SLLE(ci) will be the weighted combination of the projections of these same K-NN. The optimal XLLE in the least squares sense minimizes
| (2.7) |
where XLLE=[SLLE(c1),SLLE(c2),…,SLLE(cn)], , and I is the identity matrix. The minimization of Eq. 2.7 subject to the constraint (a normalization constraint that prevents the solution XLLE=0) is an eigenvalue problem whose solutions are the eigenvectors of the Laplacian matrix L. Since the rank of L is n−1, the first eigenvector is ignored and the second smallest eigenvector represents the best one-dimensional projection of all the samples. The best two-dimensional projection is given by the eigenvectors with the second and third smallest eigenvalues, and so forth.
Replicated k-means clustering in the reduced feature space
For the DR schemes, ϕ∊{PCA,LLE,GE}, unsupervised replicated clustering is used to classify all objects c∊C into one of the k classes based on Sϕ(c), the low dimensional representation of F(c). Replicated clustering is a variant of the popular k-means34 clustering scheme. The k-means algorithm is initialized by randomly partitioning the data into k clusters and computing the cluster center for each partition. The distance of each point from each of the k centroids is computed and each object is reassigned to the closest cluster centroid to minimize the intraclass variance. This random initialization may lead to local minima leading, in turn, to different clustering results. The motivation behind replicated clustering is to make the final aggregation results from k-means more deterministic. In replicated clustering, multiple weak clusterings of the data are generated. The optimal clustering solution is then chosen among the various weak clusterings as the one with the least intracluster variance. Below we briefly describe the various steps involved in this algorithm.
Step 1. At each of T iterations, k-means is applied to cluster all objects c∊C into one of the k classes , t∊{1,…,T}, where each c∊C is characterized by a high dimensional feature vector F(c). For each , q∊{1,2,…,k}, t∊{1,2,…,T} the centroid of each cluster is determined as
| (2.8) |
Step 2. At each iteration t∊{1,…,T}, the average Euclidean distance between each and corresponding cluster center , t∊{1,…,T}, q∊{1,2,…,k}, is then determined as
| (2.9) |
The average intracluster distance over all k clusters is then obtained as
| (2.10) |
Step 3. Finally, the clusterings , q∊{1,2,…,k} within a specific iteration t∊{1,…,T} are identified as the stable clustering result for which is minimum over all t.
Note that replicated clustering identifies stable clusterings as those that minimize intraclass variance. Note further that while we are not explicitly seeking to increase intercluster distance, our empirical results suggest that replicated k-means clustering tends to also push the cluster centers farther apart.
METHODOLOGY
In Sec. 3A we provide a brief description of the notation employed in this paper. In Sec. 3B we provide a brief description of the data sets considered in this study. Details concerning determination of the approximate ground truth for spatial extent of CaP on MRI are provided in Sec. 3C. Methodological details regarding the two-stage CAD system for CaP detection on MRS are detailed in Secs. 3D, 3E, respectively.
Notation
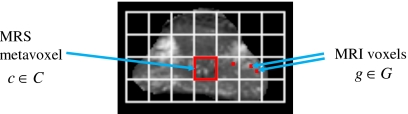
We represent the 3D prostate T2-w MRI scene by G=(G,f), where G is a 3D grid of voxels g∊G and f(g) is a function that assigns an intensity value to every g∊G. We also define a spectral scene C=(C,F) where C is a 3D grid of metavoxels, c∊C. Each metavoxel c is associated with a corresponding M-dimensional spectral vector F(c)=[fu(c)∣u∊{1,…,M}], where fu(c) represents the MRS signal intensity at each c along the frequency domain. Figure 3 shows the spatial relationship between the MR spectral metavoxel c∊C and T2-w MRI voxel g∊G. Note that the distance between any two adjacent metavoxels ci,cj∊C, ‖ci−cj‖2, where ‖⋅‖2 denotes the L2 norm, i,j∊{1,…,∣C∣}, and ∣C∣ is the cardinality of C, is roughly 13 times the distance between any two adjacent MRI voxels gi,gj∊G, where i,j∊{1,…,∣G∣}. A list of commonly used notations and symbols in this paper is given in Table 1.
Figure 3.
Illustration of the spatial relationship between MRS metavoxels c∊C and T2-w MRI voxels g∊G. The spectral grid C comprising of 28 metavoxels has been overlaid on a T2-w MRI prostate slice and is shown in white. Note the region outlined in red on C corresponds to the area occupied by a metavoxel, but may contain multiple MRI voxels (highlighted in red).
Table 1.
List of commonly used notation and symbols in this paper.
| Symbol | Description |
|---|---|
| G | 3D MRI scene |
| G | 3D grid of MRI voxels |
| g | Voxel location in G,g∊G |
| fu(c) | MR signal intensity at c |
| f(g) | MR intensity value at g |
| M | Number of original high dimensions |
| ϕ | DR method, ϕ∊{PCA,LLE,GE} |
| i,j | Two adjacent spatial locations on an image |
| R | Maximum CaP diameter |
| Cp | Potential cancer space, Cp⊂C |
| Stable clusters obtained for q∊{1,2,…,k} | |
| ΔX,ΔY | Size of metavoxel in X and Y dimensions |
| α | Threshold parameter for z score, α∊[0,1] |
| Cs | Set of precise spatial locations of CaP |
| Ca,o | Spatial locations of Ca outside CP |
| Sensitivity value associated with CaP class | |
| Wϕ | Distance matrix for ϕ∊{GE,LLE,PCA} |
| κ | Locally linear parameter for ϕ∊{LLE} |
| C | 3D MR spectral scene |
| C | 3D grid of metavoxels |
| c | A metavoxel in C,c∊C |
| u | Frequency index |
| F(c) | Vector of spectral content at c |
| m | Number of reduced dimensions, m⪡M |
| Sϕ(c) | Low dimensional embedding vector at c |
| Sz(c) | z scores at each |
| Ks | Contiguous slices with CaP presence |
| Ng | Number of CaP voxels in CP,Ng∊CP |
| VT | Prostate grid |
| Θ | Threshold for prostate size |
| θz | z score threshold |
| Set of spatial locations for clusters | |
| Ca,i | Spatial locations of Ca inside CP |
| Specificity value associated with CaP class | |
| η | Confidence estimate associated with SN and SP |
| u | Dimension parameter for ϕ∊{LLE,GE,PCA} |
MRSI data description
A total of 18 deidentified and anonymized prostate cancer MRI∕MRS datasets from the ACRIN trial was chosen randomly for this pilot study. All exams were performed on a 1.5 T magnet (GE Medical Systems, Milwaukee, WI). Oblique axial T2-w spectra (TR 4000–6000 ms, TE 90–120 ms, slice thickness of 3 mm, acquisition matrix of 256×192, and FOV of 12–14 cm) were first obtained. Using the PROSE (Prostate spectroscopy and imaging examination) software package (www.gehealthcare.com∕usen∕mr∕applications∕products∕prose.html), MRSI (TR 1000 ms, TE 130 ms, spectral width of 1000 Hz, and number of points is 512) was then prescribed from the oblique axial T2-w images, with outer voxel suppression and oblique suppression planes manually set to exclude the majority of fat about the prostate capsule. A FOV of 11 cm was used for the 16×16 MRSI grid (voxel dimension of 6.75×6.75×3 mm3). MRS was obtained as a PRESS (point resolved spectroscopy sequence) sequence.35 The MRS spectral grid was contained in DICOM image sets, from which the 16×16 grid containing 256 complex spectra per slice was obtained using IDL 6.4 (ITT Visual Information Systems).
Determining approximate ground truth for spatial extent of CaP on MRI-MRS
Following radical prostatectomy, the gland was fixed in formalin and sectioned per institutional routine (whole mount or standard sections). Sections were then embedded in formalin, and duplicate slides for each block were prepared for central review. Slides were stained with Hemtoxylin and Eosin and reviewed by a single central pathologist for areas of cancer. MRI-pathology correlation was established through a joint review session of trial imagers and pathologists, who determined slice-by-slice histology-MRI concordance. In order to ensure uniform spectral and histologic sextant assignment, sextant boundaries (i.e., between apex and midgland, and between midgland and base) were based on the MRI slice assignments that had previously been determined by each site radiologist. Using these sextant boundaries, and the best approximation of MRI-histologic concordance, the presence and the diameter of CaP in each sextant were established with maximum tumor diameter, denoted as R, recorded for all positive sextants.
Potential cancer space
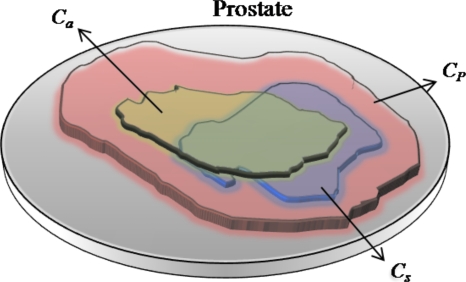
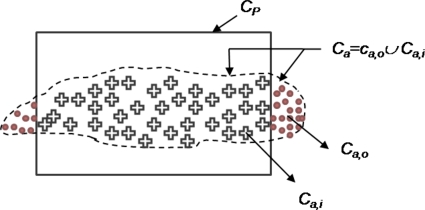
In order to quantitatively evaluate CAD performance in terms of performance metrics such as sensitivity and specificity, the precise spatial location of the target class within C is required. Unfortunately, this information was not readily available for the studies considered in this project. Hence, we define a probabilistic ground truth for CaP that involves first defining a potential cancer space representing a spatial region (CP) within C within which the tumor is embedded. To appreciate the need for CP, let us assume the ideal case scenario (Fig. 4) where the precise spatial location of CaP is known a priori and is denoted by the set Cs. If Ca denotes the set of spatial locations corresponding to CaP identified by the CAD system, the true positive (TP) area could be calculated as ∣Cs∩Ca∣. Similarly, the false positive (FP) area for the CAD system is ∣Ca−(Cs∩Ca)∣ and false negative (FN) area is ∣Cs−(Cs∩Ca)∣. For the problem we are considering, Cs is unavailable and hence the need for CP which is defined as the set of spatial locations within which a total number of Ng CaP locations are contained. The true number of metavoxels c within CP that represent CaP can be calculated as
| (3.1) |
where Ks represents the number of contiguous MR sections containing CaP, ⌈ ⌉ refers to the ceiling operation, and ΔX and ΔY refer to the size of the metavoxel c in the X and Y dimensions. Thus for a MRS scene C, with known cancer in left midgland (LM), the prostate being contained in a 3×6 grid and the prostate midgland region extending over two contiguous slices, the total number of CaP metavoxels ∣CP∣ is 18 (3×3×2). The 3×6 prostate grid is divided into two equal right and left halves. Given that the tumor has a maximum diameter of 13.75 mm in LM, with ΔX, ΔY=6.875, Ng=8 metavoxels corresponding to CaP within CP. Both CP and Ng are integral to defining probabilistic estimates of CAD sensitivity, specificity, and positive predictive value (PPV), details of which are provided in Sec. 4.
Figure 4.
An illustration of the precise ground truth location (Cs) on the prostate and spatial location of the class (Ca) identified as CaP by a CAD system. Note that in this case sensitivity, specificity of CaP detection via CAD can be determined precisely since Cs is known exactly. CP represents the potential cancer space that needs to be defined when Cs is not available and contains within it Ng CaP metavoxels.
Expert annotations
We also endeavored to obtain a more precise estimate of CaP ground truth for a subset of the studies via expert annotation of individual MR spectra. For 6 of the 18 ACRIN studies, an expert radiologist laboriously annotated each individual spectra with labels 1–5; with 1 and 2 being spectra that appeared to be normal and spectra 3–5 being suspicious appearing. For this expert annotated dataset, our CAD system attempted to discriminate all spectra as being either normal or suspicious appearing (i.e., distinguish between spectras labeled as 1∕2 and those labeled as 3∕4∕5).
Stage 1: Localization of prostate using hierarchical spectral clustering of MRS
For the FOV for studies considered in this work, a majority of the spectra lay outside the prostate. The motivation behind stage 1 of our algorithm is to exploit the differences in spectral characteristics of spectra inside and outside the prostate to hone in the prostate ROI automatically. This is done by identifying and eliminating the dominant cluster at every iteration until a cluster size threshold is attained. The nonlinear DR scheme graph embedding32 is employed to project all spectra into a reduced dimensional embedding SGE(c), followed by replicated k-means clustering to aggregate all c∊C into two clusters , corresponding to prostatic and extracapsular spectral classes. At each iteration t∊{1,…,T}, a subset of voxels in C is obtained by eliminating the noninformative extracapsular spectra identified as the dominant cluster . The approximate number of prostate spectra (Θ) of a MRS grid is learned during the offline training phase. The automatic cascaded scheme stops when the number of remaining spectra in the MRS grid is approximately equal to Θ. The result of the HierarclustMRS algorithm is a spectral grid containing all the prostate spectra.
Algorithm.
HierarclustMRS
| Input: F(c) for all c∊C, Θ, C. |
| Output: . |
| begin |
| 0. Initialize , t=0; |
| 1. whiledo |
| 2. Apply Graph Embedding (Ref. 32), to F(c), for all to obtain ; |
| 3. Apply replicated k-means clustering on to obtain two stable clusters ; |
| 4. Identify larger cluster , where w∊{1,2}; |
| 5. Create set by eliminating all from ; |
| 6. t=t+1; |
| 7. endwhile; |
| 8. ; |
| end |
Note that in general the algorithm is terminated when the total number of spectra is marginally greater than (or equal to) Θ, which usually occurs within two to three iterations.
Stage 2: CaP identification on MRS via NLDR
Following stage 1, we attempt to apply more sophisticated analysis to the spectra in to be able to discriminate between different tissue classes in the prostate. Apart from NLDR schemes (LLE and graph embedding) that were considered, two other feature extraction schemes (z score and PCA) were also evaluated in terms of their ability to discriminate between different classes of the prostate spectra (abnormal appearing, normal appearing and indeterminate). Following feature extraction, replicated k-means clustering for the DR schemes (LLE, graph embedding, and PCA) and thresholding for z score was applied to obtain hard classification of the spectra into CaP and non-CaP categories. For z score evaluation, a predefined threshold θz is used to classify each as abnormal or normal appearing based on whether Sz(c)>θz or Sz(c)<θz. Three commonly employed DR methods (explained previously in Sec. 3) are applied to the MR spectra in so that for any , the high dimensional ambient feature vector F(c) is mapped to Sϕ(c), where ϕ∊{PCA,LLE,GE}. Replicated clustering is then employed to cluster each , into one of the three possible classes, corresponding to abnormal appearing, normal appearing, or an indeterminate class, with the intermediate class corresponding to undefined, equivocal, or noisy spectra. For the manually annotated datasets, replicated clustering was used to identify all analyzable spectra as being either normal or suspicious appearing (two clusters).
EVALUATION METHODS
Identification of cancer cluster
As mentioned previously in Sec. 3C1, a potential CaP space CP is defined within which the number of CaP locations Ng is determined. Following replicated clustering of Sϕ(c), , we need to identify which of the k clusters correspond to the CaP class. We represent the corresponding sets of spatial locations for clusters as , respectively. With respect to CP (as illustrated in Fig. 5), some part of may be within or outside CP. Thus , q∊{1,2,…,k}. The TP, FP, true negative (TN), and FN ratios associated with each class , q∊{1,2,…,k} with respect to CP and Ng are then obtained. The following heuristic algorithm is then used to identify which of , q∊{1,2,…,k} represents the CaP class .
Algorithm.
IdentifyCaPCluster
| Input: , q∊{1,2,..,k}, CP, Ng, C. |
| Output: |
| begin |
| 0. forq=1 tokdo; |
| 1. ; |
| 2. ; |
| 3. Ifthen |
| TPq=Ng, , FNq=0, ; |
| 4. else, , , ; |
| 5. endif; |
| 6. endfor; |
| 7. ; |
| end |
Figure 5.
Illustration of the potential ground truth space (CP) containing Ng metavoxels corresponding to CaP. Ca represents the CaP segmentation obtained by the MRS CAD analysis scheme and Ca,o and Ca,i represent those regions of Ca that lie outside and within CP respectively.
Thus Algorithm IdentifyCaPCluster identifies the CaP cluster as the one that maximizes true positive fraction while simultaneously minimizing the false positive fraction.
Performance evaluation metrics
Having identified , the corresponding CaP detection sensitivity and specificity values are determined as
| (4.1) |
| (4.2) |
Given that and are estimates of the sensitivity and specificity (given the probabilistic estimates of the CaP ground truth extent), we also compute a confidence estimate (η) associated with the performance measures as a function of ∣CP∣ and Ng. η is determined as
| (4.3) |
Thus for each study for which we only have CP and Ng available as surrogates of CaP ground truth, we report η along with and for those studies.
RESULTS
Qualitative results
Stage 1: Qualitative evaluation of the hierarchical clustering scheme
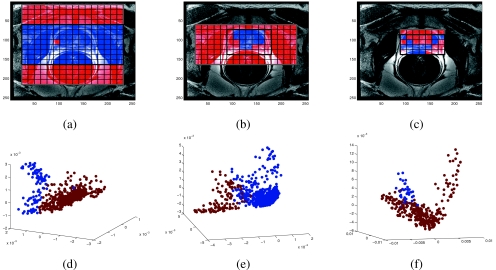
Figure 6 shows the qualitative results of the hierarchical cascade scheme for distinguishing prostatic from extracapsular spectra. Figure 6a represents spatial maps of the spectral grid (16×16 spectral voxels) superimposed on the corresponding T2-w MRI scene for one patient study. Every in Fig. 6a is assigned one of two colors (blue and red) corresponding to spectra identified by the algorithm as prostatic or extracapsular. Note that the dominant cluster [spatial locations in red in Fig. 6a] has been eliminated in the second iteration [ (16×8 spectral voxels)] [Fig. 6b]. The final spectral grid [ in Fig. 6c] is obtained after elimination of extracapsular spectra (red locations) during the third iteration of the cascade. Figures 6d, 6e, 6f represent the embedding plots [where each original spectral vector F(c), c∊C is plotted in 3D eigenvector space using the three dominant embedding values as coordinates] from (16×16 spectral voxels) (d) to (7×4 spectral voxels) (f) for one study at three different levels of the cascade. Note that at the end of the third iteration, the prostate ROI has been accurately identified and the spectral grid accurately overlaid on the prostate. Further note that in Figs. 6a, 6b, 6c, the spectral grid with the pronounced boundary indicates the ROI during the current iteration.
Figure 6.
Spectral grids for a single 2D slice of a T2-w MRI scene for a patient at (a) the first cascade level , (b) second cascade level , and (c) third cascade level . Note that the size of the grid reduces from 16×16 metavoxels in (a) to 7×4 in (c) by elimination of extracapsular spectra in the dominant cluster (red). The corresponding clustered embedding plots at each of the cascade levels are also shown in (d)–(f), corresponding, in turn, to the metavoxel grids shown in (a)–(c).
Stage 2: Evaluation of feature extraction schemes for CaP detection
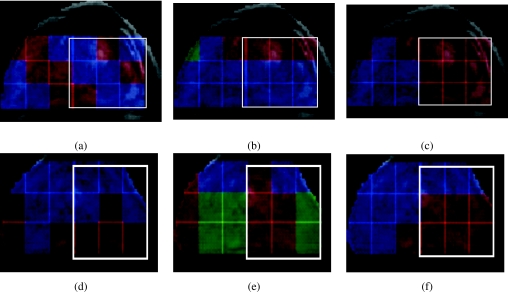
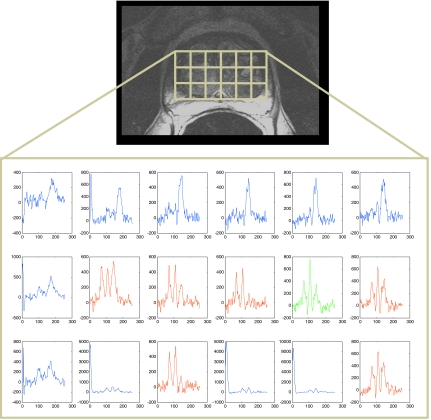
The identification of the prostate grid in stage 1 allows for the resolvability of the three MR spectral classes (abnormal appearing, normal appearing, and indeterminate). The differences between these three spectral classes within the prostate spectra cluster at the higher levels in the cascade become discriminable only after the removal of extracapsular spectra. Note that replicated clustering was used to identify only two clusters (suspicious and normal appearing spectra) for the six manually annotated studies. Figure 7 shows the qualitative results of the four feature extraction schemes employed in this work for CaP detection for two different patient studies, each row in Fig. 7 corresponding to a different study. The three colors assigned to the spectral voxels in Fig. 7 correspond to the three clusters obtained via replicated clustering on the reduced dimensional spectra Sϕ(c), ϕ∊{PCA,LLE,GE} for c∊C. For the z score scheme, each metavoxel was classified as belonging to one of the two classes [red and blue in Figs. 7a, 7d], reflecting abnormal and normal appearing spectra. The white box superposed in Figs. 7a, 7b, 7c, 7d, 7e, 7f shows the potential cancer space (CP) for corresponding slices. In each of Figs. 7a, 7b, 7c, 7d, 7e, 7f, the red cluster was identified as comprising abnormal using the IdentifyCaPCluster algorithm (sec. 4) and following feature extraction and replicated clustering. Figures 7a, 7d show the results obtained via z score; while Figs. 7b, 7e show the results of PCA in identifying abnormal appearing, normal appearing, and indeterminate spectra. Figures 7c, 7f show the corresponding results for graph embedding and LLE, respectively. For the first study (first row in Fig. 7), graph embedding [Fig. 7c] appears to yield a near perfect CaP detection in terms of sensitivity and specificity as only CaP voxels are identified within the white cancer grid (CP). The corresponding results for z score [Fig. 7a] and PCA [Fig. 7b] both yield poor detection sensitivity and specificity. Similarly, for the second study shown in Fig. 7, LLE [Fig. 7f] appears to yield higher CaP detection sensitivity and specificity compared to z score [Fig. 7d], and PCA [Fig. 7e]. Figure 8 shows an example of the MR spectral grid with classification labels obtained from graph embedding and replicated clustering plotted back on the individual spectra; the spectra in red corresponding to those identified as abnormal appearing, blue spectra corresponding to normal appearing, and green spectra corresponding to indeterminate.
Figure 7.
Qualitative comparison of prostate MRS analysis via four different feature extraction schemes employed for two different studies; each row corresponding to a different study. (a) and (d) represent the suspicious appearing (red voxels) and benign appearing (blue voxels) spectra identified via z score, and (b) and (e) illustrate the clustering results obtained via PCA [red (suspicious appearing), blue (benign appearing), green (intermediate)]. Results for the NLDR schemes (c) GE and (f) LLE are also shown [the red, blue, and green colors having the same meaning as for PCA in (b) and (e)]. The white box superposed on (a)–(f) shows the locations of the potential cancer space (CP). In each of (a)–(f) the cluster with the red metavoxels was the one identified as the CaP class based on the IdentifyCaPCluster algorithm (Sec. 5A).
Figure 8.
MRS spectral grid plotted with the classification labels (three colors correspond to three different clusters) obtained from graph embedding and replicated clustering. The red spectra correspond to those identified as abnormal appearing, the blue correspond to normal appearing, and the green correspond to indeterminate spectra (possibly noise in this case).
Quantitative results
Quantitative evaluation of stage 1: Hierarchical clustering
At the end of stage 1, the largest rectangular box that contains all prostate spectra is then overlaid on the T2-w image. Note that in quantitative evaluation of stage 1, the precise spatial extent of the prostate is all that is needed. This ground truth is ascertained by manual placement of a spectral grid on the prostate by an expert radiologist. Table 2 shows the average sensitivity, specificity, and PPV in automated identification of the prostate grid with respect to and averaged over 18 studies.
Table 2.
Average sensitivity, specificity, PPV values for automated identification of prostate grid using hierarchical spectral clustering averaged over 18 studies.
| Sensitivity | Specifity | PPV |
|---|---|---|
| 97.66% | 98.87% | 89.29% |
Quantitative evaluation of stage 2: Identifying suspicious appearing spectra
Evaluation ofzscore via receiver operating characteristic (ROC) curve analysis. In order to define the optimal threshold θz for performing a z score-based classification of each c∊C as normal or suspicious, a set of cancerous voxels obtained via annotation by an expert radiologist, CV⊂C, was defined during an offline training phase. For each c∊CV, a corresponding Sz(c) was obtained which was then used to define the average μz and standard deviation σz of z score values for CaP. The threshold θz was then defined as μz±ασz, where α∊[0,1]. The value of α was uniformly varied between [0,1] and a corresponding values for θz obtained. At each value of θz, each metavoxel is identified as belonging to CaP if Sz(c)>θz, normal otherwise. Thus at each θz, the corresponding sensitivity and specificity of CaP detection via z score is computed [Eqs. (4.12) and (4.13)] with respect to CP and Ng. A curve is then fit to the sensitivity, specificity values to obtain the ROC curve. The optimal threshold was determined as the operating point on the ROC curve; the location on the ROC curve closest to 100% sensitivity, specificity. Average sensitivity and specificity at the operating point of ROC curve were found to be 72.85% and 65.45%, respectively.
Evaluation of DR methods via cross validation using probabilistic ground truth for CaP extent. The ability of a classifier to distinguish between object classes in embedding spaces obtained via DR methods is dependent on to the choice of the number of dimensions (v) of the embedding space in which the data are represented. For the NLDR methods, the low dimensional data representations obtained via LLE (Ref. 30) is also a function of κ, the parameter controlling the size of the local neighborhood within which linearity is assumed. In order to evaluate the parameter sensitivity of different DR methods, the robustness of the DR methods over different values of κ and v was quantitatively evaluated. LLE was evaluated by varying κ∊{6,7,…,15} and v∊{3,4,…,10}, a total of 80 different combinations of parameter values. PCA and graph embedding were evaluated for eight different values of v∊{3,4,…,10}.
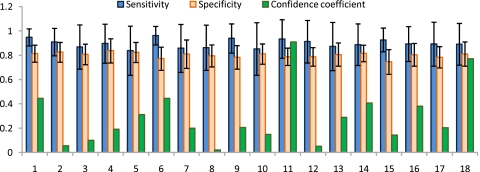
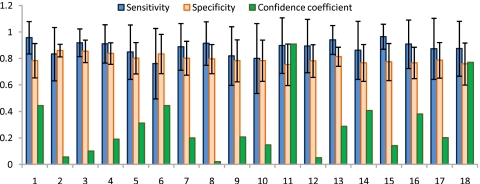
For each of ϕ∊{LLE,GE,PCA}, threefold and fivefold cross validation averaged over 20 iterations were also performed to obtain average sensitivity and specificity in terms of CaP detection performance for all 18 studies. Threefold cross validation was performed by randomly choosing three datasets and calculating average CaP detection sensitivity and specificity across three studies on the 80-dimensional parameter space. The parameter set (vmax for PCA, GE and vmax, κmax for LLE) with maximum sensitivity and specificity in this space were then identified as optimal values and used for the CAD scheme on the remaining 15 studies. The average CaP detection sensitivity and specificity on these 15 studies was then recorded. On the next trial, 3 random training studies from 18 were again selected and used to optimize the parameters and evaluation again was done on the remaining 15 studies. This entire process was repeated for a total of 20 times. The mean μϕ and standard deviation σϕ in CaP detection sensitivity and specificity across these 20 iterations were reported in Table 3(a) for ϕ∊{LLE,GE,PCA}. A similar routine was employed when performing fivefold cross validation. The corresponding results are reported in Table 3(b). All NLDR schemes employed in this work were found to have higher sensitivity of CaP detection compared to PCA; graph embedding performing the best with a sensitivity of almost 90%. In Figs. 910 barplots are shown representing CaP detection sensitivity and specificity (μϕ,σϕ) for each of the 18 studies considered in this work via threefold and fivefold cross validation, respectively, over 20 iterations for GE. The confidence estimates (η) associated with the sensitivity and specificity measurements of each study are also shown. Note that while η is low for a majority of the studies, for studies 11 and 18, the confidence associated with CaP detection sensitivity and specificity for graph embedding were almost 90% and 80%, respectively.
Table 3.
This shows the average and standard deviation in CaP detection sensitivity and specificity for different feature extraction methods over κ∊{5,…,15} and v∊{3,…,10} for 18 different studies via (a) threefold cross validation, and (b) fivefold cross validation.
| Method | Sensitivity | Specificity |
|---|---|---|
| (a) | ||
| GE | 89.33±1.87 | 79.79±2.24 |
| LLE | 83.70±4.50 | 81.04±4.60 |
| PCA | 78.53±3.20 | 83.97±2.81 |
| (b) | ||
| GE | 89.09±2.10 | 79.22±2.10 |
| LLE | 84.93±4.42 | 81.32±4.05 |
| PCA | 78.78±4.31 | 84.11±2.46 |
Figure 9.
Barplot showing average and standard deviation in CaP detection sensitivity and specificity for the individual 18 studies averaged over 20 iterations of threefold cross validation via graph embedding. The coefficient of confidence (η) associated with the estimation of the performance measures is also shown in green.
Figure 10.
Barplot showing the average and standard deviation in CaP detection sensitivity and specificity for the individual 18 studies averaged over 20 iterations for fivefold cross validation via graph embedding. The coefficient of confidence (η) associated with the estimation of the performance measures is also shown in green.
Evaluation of all feature extraction methods against expert annotated spectra. Table 4 shows the average sensitivity and specificity results obtained for ϕ∊{LLE,GE,PCA,z} for six studies for which each spectra had been labeled according to the five point scale36 by an expert radiologist. For all methods, we made the assumption that spectra labeled as 1∕2 were all normal appearing and the spectra labeled 3∕4∕5 were suspicious appearing. For each of ϕ∊{LLE,GE,PCA,z}, the IdentifyCaPCluster algorithm was employed to identify the cluster most likely to correspond to CaP spectra. Since we only had six studies available, the mean and standard deviation in CaP detection sensitivity and specificity across all the expert annotated datasets were determined (based on previously determined vmax and κmax) and shown in Table 4.
Table 4.
This shows the average and standard deviation in CaP detection sensitivity and specificity for different feature extraction methods obtained via comparison with expert annotations for six ACRIN studies.
| Method | Sensitivity | Specificity |
|---|---|---|
| GE | 79.74±13.18 | 65.49±7.58 |
| LLE | 81.36±5.69 | 64.71±9.84 |
| PCA | 76.68±14.07 | 48.26±12.95 |
| z score | 62.70±18.67 | 50.73±7.39 |
CONCLUDING REMARKS
In this paper we have presented a novel application of nonlinear dimensionality reduction and hierarchical clustering for automated identification of (a) the prostate ROI based on the classification of MR spectral data alone and (b) suspicious appearing spectra within the prostate ROI. A total of 18 MRI∕MRS studies from the ACRIN trial was considered for the evaluation of four different feature extraction algorithms (PCA, z score, LLE, GE) in conjunction with replicated clustering in terms of their ability to identify suspicious appearing spectra. Owing to the fact that only limited knowledge regarding precise spatial extent of CaP was available for the studies considered in this work, we defined a probabilistic ground truth estimate for CaP and a confidence coefficient to assess the degree of certainty associated with the CAD performance measures reported. The high confidence estimates associated with two of the studies (study 11 and 18) seem to suggest that the consistently high CaP detection sensitivity and specificity measurements for the other studies are not erroneous. For a subset (6) of the 18 studies, a radiologist laboriously annotated each of the spectra according to the five point scale. For these studies, we assumed that the spectra corresponding to 3∕4∕5 were cancerous. In comparing the four feature extraction schemes on the 18 datasets with partial CaP ground truth estimates, as well as the six studies for which expert annotations was available, the NLDR schemes (GE and LLE) consistently outperformed PCA and z score in terms of both CaP detection sensitivity and specificity. In addition, the nonlinear DR schemes were found to be relatively robust to change in the value of the system parameters (κ,v). The use of the replicated clustering scheme helped overcome the instability associated with k-means clustering, yielding consistently stable clusters. Our scheme is also highly efficient with stage 1 only requiring an average of 11.24 s to analyze a 256×256×8 MRI∕MRS 3D 1.5 T scene on a Pentium IV, 2 Gbyte RAM Intel processor machine. Stage 2 took an average of 5.23 s for automated classification of MR spectra as being suspicious, normal appearing, or indeterminate.
Future work will involve integration of our automated CAD MRS scheme with T2-w MRI to incorporate both structural and functional information for more accurate identification of CaP. We also aim to perform more rigorous analysis of the scheme on a larger cohort of data. The availability of more precise knowledge of spatial location of CaP on the MR imagery will help to further confirm and validate the efficacy of our methods.
While replicated clustering is more stable than k-means, it still requires specification of the value of k. In this work we assumed k=3 based on the assumption that all the spectra were either normal appearing, suspicious appearing, or indeterminate. While Jung et al.,36 identified five distinct classes of prostate MR spectra (five point scale), in our experiments (owing perhaps to the quality of data) we were unable to find five unique clusters. A future avenue of exploration in the future will be to look at fully unsupervised clustering schemes (e.g., mean shift) which do not require prespecification of the number of data clusters.
ACKNOWLEDGMENTS
This work was made possible via grants from the Wallace H. Coulter Foundation, New Jersey Commission on Cancer Research, National Cancer Institute (Grant Nos. R01CA136535-01, ARRA-NCl-3 R21CA127186–02S1, R21CA127186–01, and R03CA128081-01), the Society for Imaging Informatics in Medicine (SIIM), The Cancer Institute of New Jersey, and the Life Science Commercialization Award from Rutgers University.
References
- Catalona W., Smith D., Ratliff T., Dodds K., Coplen D., Yuan J., Petros J., and Andriole G., “Measurement of prostate-specific antigen in serum as a screening test for prostate cancer,” N. Engl. J. Med. 324, 1156–1161 (1991). [DOI] [PubMed] [Google Scholar]
- Borboroglu P., Comer S., Riffenburgh R., and Amling C., “Extensive repeat transrectal ultrasound guided prostate biopsy in patients with previous benign sextant biopsies,” J. Urol. (Paris) 163(1), 158–162 (2000). [PubMed] [Google Scholar]
- Yu Kyle K., and Hricak H., “Imaging prostate cancer,” Radiol. Clin. North Am. 38(1), 59–85 (2000). 10.1016/S0033-8389(05)70150-0 [DOI] [PubMed] [Google Scholar]
- Scheidler J., Hricak H., Vigneron D., Yu K., Sokolov D., Huang R., Zaloudek C., Nelson S., Carroll P., and Kurhanewicz J., “Prostate cancer: Localization with three-dimensional proton MR spectroscopic imaging-clinicopathologic study,” Radiology 213, 473–480 (1999). [DOI] [PubMed] [Google Scholar]
- Carroll P., Coakley F., and Kurhanewicz J., “Magnetic resonance imaging and spectroscopy of prostate cancer,” Rev. Neurol. 8(1), S4–S10 (2006). [PMC free article] [PubMed] [Google Scholar]
- Ullrich G., Mueller-Lisse , Swanson M., Vigneron D., Hricak H., Bessette A., Males R., Wood P., Noworolski S., Nelson S., Barken I., Carroll P., and Kurhanewicz J., “Time-dependent effects of hormone-deprivation therapy on prostate metabolism as detected by combined magnetic resonance imaging and 3D magnetic resonance spectroscopic imaging,” Magn. Reson. Med. 46, 49–57 (2001). 10.1002/mrm.1159 [DOI] [PubMed] [Google Scholar]
- Kurhanewicz J., Swanson M., Nelson S., and Vigneron D., “Combined magnetic resonance imaging and spectroscopic imaging approach to molecular imaging of prostate cancer,” J. Magn. Reson Imaging 16, 451–463 (2002). 10.1002/jmri.10172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heerschap A., Jager G. J., van der Graaf M., Barentsz J. O., Rosetten J., De La Oosterhof G., Ruijter E., and Ruijs J., “In vivo proton MR spectroscopy reveals altered metabolite content in malignant prostate tissue,” Anticancer Res. 17, 1455–1460 (1997). [PubMed] [Google Scholar]
- Kurhanewicz J., Vigneron D. B., Hricak H., Narayan P., Carroll P., and Nelson S. J., “Three-dimensional H-1 MR spectroscopic imaging of the in situ human prostate with high (0.24-0.7-cm3) spatial resolution,” Radiology 198(3), 795–805 (1996). [DOI] [PubMed] [Google Scholar]
- Zakian K. L., Eberhardt S., Hricak H., Shukla-Dave A., Kleinman S., Muruganandham M., Sircar K., Kattan M. W., Reuter V. E., Scardino P. T., and Koutcher J. A., “Transition zone prostate cancer: Metabolic characteristics at H MR spectroscopic imaging—initial results,” Radiology 229, 241–247 (2003). [DOI] [PubMed] [Google Scholar]
- Wetter A., Engl T., Nadjmabadi D., Fliessbach K., Lehnert T., Gurung J., Beecken W., and Vogl J., “Combined MRI and MR spectroscopy of the prostate before radical prostatectomy,” AJR, Am. J. Roentgenol. 187, 724–730 (2006). [DOI] [PubMed] [Google Scholar]
- Madabhushi A. and Metaxas D., “Ultrasound techniques in digital mammography and their application in breast cancer diagnosis,” Medical Imaging Systems Technology: Analysis and Computational Methods (World Scientific, Singapore, 2005), pp. 119–150. [Google Scholar]
- Madabhushi A., Feldman M., Metaxas D., Tomaszeweski J., and Chute D., “Automated detection of prostatic adenocarcinoma from high-resolution ex vivo MRI,” IEEE Trans. Med. Imaging 24(12), 1611–1625 (2005). 10.1109/TMI.2005.859208 [DOI] [PubMed] [Google Scholar]
- Viswanath S., Rosen M., and Madabhushi A., “A consensus embedding approach for segmentation of high resolution in vivo prostate magnetic resonance imagery,” Society of Photo-Optical Instrumentation Engineers Medical Imaging Conference (SPIE 2008), San Diego (SPIE, 2008), Vol. 6915, 69150U.
- Chan I., Wells W., Mulkern R., Haker S., Zhang J., Zou K., Maier S., and Tempany C., “Detection of prostate cancer by integration of line-scan diffusion, T2-mapping and T2-weighted magnetic resonance imaging: A multichannel statistical classifier,” Med. Phys. 30(9), 2390–2398 (2003). 10.1118/1.1593633 [DOI] [PubMed] [Google Scholar]
- Kurhanewicz J., Vigneron D. B., Males R. G., Swanson M. G., Yu K. K., and Hricak H., “The prostate: MR imaging and spectroscopy. Present and future,” Radiology 38(1), 115–138 (2000). [DOI] [PubMed] [Google Scholar]
- Van der Veen J. W. C., De Beer R., Luyten P. R., and Van Ormondt D., “Accurate quantification of in vivo 31P NMR signals using the variable projection method and prior knowledge,” Magn. Reson. Med. 6(1), 92–98 (1988). 10.1002/mrm.1910060111 [DOI] [PubMed] [Google Scholar]
- Vanhamme L., Boogaart A., and Huffel S., “Improved method for accurate and efficient quantification of MRS data with use of prior knowledge,” J. Magn. Reson. 129(1), 35–43 (1998). 10.1006/jmre.1997.1244 [DOI] [PubMed] [Google Scholar]
- Ratiney H., Sdika M., Coenradie Y., Cavassila S., van Ormondt D., and Graveron-Demilly D., “Time-domain semiparametric estimation based on a metabolite basis set,” Pediatr. Nephrol. 18(1), 1–13 (2005). [DOI] [PubMed] [Google Scholar]
- Zakian K., Sircar K., Hricak H., Chen H., Dave A., Eberhardt S., Muruganandham M., Ebora L., Kattan M., Reuter V., Scardino P., and Koutcher J., “Correlation of proton MR spectroscopic imaging with gleason score based on sept-section pathologic analysis after radical prostatectomy,” Radiology 234, 804–814 (2005). 10.1148/radiol.2343040363 [DOI] [PubMed] [Google Scholar]
- Pels P., Ozturk-Isik E., Swanson M., Vanhamme L., Kurhanewicz J., Nelson S., and Huffel S., “Quantification of prostate MRSI data by model-based time domain fitting and frequency domain analysis,”Pediatr. Nephrol. 19, 188–197 (2006). [DOI] [PubMed] [Google Scholar]
- Kelm M., Menze B., Zechmann C., Baudendistel K., and Hanprecht F., “Automated estimation of tumor probability in prostate MRSI: Pattern recognition vs. quantification,” Magn. Reson. Med. 57, 150–159 (2006). 10.1002/mrm.21112 [DOI] [PubMed] [Google Scholar]
- Quan H. and Bao S., “z score analysis for H-MRSI data of glioma and prostate cancer,” Asia-Oceania Federation of Organizations for Medical Physics, 2004.
- Laudadio T., Pels P., Lathauwer L., Hecke P., and Huffel S., “Tissue segmentation and classification of MRSI data using canonical correlation analysis,” Magn. Reson. Med. 54, 1519–1529 (2005). 10.1002/mrm.20710 [DOI] [PubMed] [Google Scholar]
- Devos A., Lukas L., Suykens J. A. K., Vanhamme L., Tate A. R., Howe F. A., Majs C., Moreno-Torres A., van der Graaf M., Ars C., and Van Huffel S., “Classification of brain tumours using short echo time 1H MR spectra,” J. Magn. Reson Imaging 170, 164–175 (2004). [DOI] [PubMed] [Google Scholar]
- Ma J. and Sun J., “MRS classification based on independent component analysis and support vector machines,” IEEE Fifth International Conference on Hybrid Intelligent Systems (IEEE, Nov. 2005), pp. 81–84.
- Simonetti A. W., Melssen W. J., Szabo de Edelenyi F., van Asten J. J., Heerschap A., and Buydens L. M., “Combination of feature-reduced MR spectroscopic and MR imaging data for improved brain tumor classification,” NMR Biomed. 18, 34–43 (2005). 10.1002/nbm.919 [DOI] [PubMed] [Google Scholar]
- Lee G., Madabhushi A., and Rodriguez C., “Investigating the efficacy of nonlinear dimensionality reduction schemes in classifying gene and protein expression studies,” IEEE/ACM Trans. Comput. Biol. Bioinf. 5, 368–384 (2008). 10.1109/TCBB.2008.36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenenbaum J., de Silva V., and Langford J. C., “A global geometric framework for nonlinear dimensionality reduction,” Science 290, 2319–2322 (2000). 10.1126/science.290.5500.2319 [DOI] [PubMed] [Google Scholar]
- Roweis S. and Saul L., “Nonlinear dimensionality reduction by locally linear embedding,” Science 290, 2323–2326 (2000). 10.1126/science.290.5500.2323 [DOI] [PubMed] [Google Scholar]
- Madabhushi A., Shi J., Rosen M., Tomasezweski J., and Feldman M., “Graph embedding to improve supervised classification: Detecting prostate cancer,” Medical Image Computing and Computer-Assisted Intervention (MICCAI 2005), LNCS, 2005, Vol. 3749, pp. 729–737. [DOI] [PubMed]
- Shi J. and Malik J., “Normalized cuts and image segmentation,” IEEE Trans. Pattern Anal. Mach. Intell. 22(8), 888–905 (2000). 10.1109/34.868688 [DOI] [Google Scholar]
- Hotelling H., “Analysis of a complex of statistical variables into principal components,” J. Educ. Psychol. 24, 417–441 (1933). 10.1037/h0071325 [DOI] [Google Scholar]
- MacQueen J. B., “Some methods for classification and analysis of multivariate observations,” Proceedings of the Fifth Berkeley Symposium on Mathematical Statistics and Probability (University of California Press, Berkeley, 1967), Vol. 1, pp. 281–297.
- Bottomley P. A., “Spatial localization in NMR spectroscopy in vivo,” Ann. N.Y. Acad. Sci. 508, 333–348 (1997). 10.1111/j.1749-6632.1987.tb32915.x [DOI] [PubMed] [Google Scholar]
- Jung J., Coakley F., Vigneron D., Swanson M., Qayyum A., Weinberg V., Jones K., Carroll P., and Kurhanewicz J., “Prostate depiction at endorectal MR spectroscopic imaging: investigation of a standardized evaluation system,” Radiology 233, 701–708 (2004). 10.1148/radiol.2333030672 [DOI] [PubMed] [Google Scholar]