Abstract
The last few years have seen significant advances in our understanding of the molecular mechanisms of stem-cell-fate specification. New and emerging high-throughput techniques, as well as increasingly accurate loss-of-function perturbation techniques, are allowing us to dissect the interplay among genetic, epigenetic, proteomic, and signaling mechanisms in stem-cell-fate determination with ever-increasing fidelity (Boyer et al. 2005, 2006; Ivanova et al. 2006; Loh et al. 2006; Cole et al. 2008; Jiang et al. 2008; Johnson et al. 2008; Kim et al. 2008; Liu et al. 2008; Marson et al. 2008; Mathur et al. 2008). Taken together, recent reports using these new techniques demonstrate that stem-cell-fate specification is an extremely complex process, regulated by multiple mutually interacting molecular mechanisms involving multiple regulatory feedback loops. Given this complexity and the sensitive dependence of stem cell differentiation on signaling cues from the extracellular environment, how are we best to develop a coherent quantitative understanding of stem cell fate at the systems level? One approach that we and other researchers have begun to investigate is the application of techniques derived in the computational disciplines (mathematics, physics, computer science, etc.) to problems in stem cell biology. Here, we briefly sketch a few pertinent results from the literature in this area and discuss future potential applications of computational techniques to stem cell systems biology.
FROM MOLECULES TO NETWORKS
Modern stem cell studies now typically use a variety of different high-throughput techniques to deconstruct the molecular basis of cell-fate specification. Nevertheless, individual studies inevitably only focus on one chosen aspect of stem cell self-renewal, fate specification, or reprogramming. Consequently, although they typically produce a wealth of data, each individual study still only represents a small aspect of our collective knowledge of stem cell behavior. Therefore, it is useful for information from a large number of individual studies to be collated and cataloged into structured meta-data sets representing the collective knowledge about the molecular regulatory mechanisms that control stem cell self-renewal and differentiation. However, the task of constructing and maintaining such collective knowledge data sets is computationally and biologically challenging because different experimental studies consider different types of stems cells, under different culture conditions, using different experimental techniques that may naturally produce biased results due to inherent limitations of experimental techniques. For example, proteomic experiments are known to enrich for highly abundant proteins, whereas gene expression microarrays are noisy and mRNA levels often only partially correlate with protein expression and function. To tackle the data integration and knowledge accumulation challenge, applications of techniques from the mathematical field of graph theory (Ma’ayan 2009) have been particularly successful. The realization that complex biological systems can be conceptually represented as networks (also known as graphs in the mathematical literature) has revolutionized our approach to exploring complex biochemical systems. To construct a biological regulatory network, elements such as genes, proteins, mRNAs, microRNAs (miRNAs), or any other kind of molecular species are represented as nodes, whereas the biochemical interactions between species, for example, protein–protein interactions or transcription factor regulation of gene expression, are represented as edges or links. Because a variety of different types of regulatory mechanisms can be represented as networks (Ma’ayan et al. 2005a), representing complex biochemical systems as networks allows the merging of different types of experimental data into a single conceptual framework (Ma’ayan 2008). An example of the successful application of graph-theoretic techniques to data integration in stem cell biology was recently given by Franz-Josef Müller, Jeanne Loring, and coworkers (Müller et al. 2008). They first classified different types of human stem cells on the basis of their genome-wide mRNA expression signatures (Müller et al. 2008) and identified a set of genes that are specifically up-regulated across a variety of different types of stem cells. Then, using available mammalian protein–protein interaction databases, they “connected” their identified stem cell gene set into a network of protein interactions, naming this integrated network PluriNet. To build PluriNet, the authors made use of a graph-theoretic algorithm and software package called Matisse (Ulitsky and Shamir 2007) to identify modules in gene expression data using background knowledge about known protein–protein interactions. In general, algorithms such as Matisse can be used to identify functional modules in complex data sets (Berger et al. 2007), whereas statistical tools can be used to characterize the functional theme of such modules (Subramanian et al. 2005). Alternatively, protein–protein interaction networks can be readily reconstructed experimentally using proteomic techniques such as immunoprecipitation-based “pull-downs” followed by mass spectrometry (IP-MS) (Gygi and Aebersold 2000). For example, a protein–protein interaction network centered around the transcription factor Nanog was recently constructed by Jianlong Wang, Stuart Orkin, and coworkers using a set of serial IP-MS experiments in which they pulled down different components of the Nanog interaction complex one at a time (Wang et al. 2006). Resources such as the PluriNet and empirically constructed interaction networks are useful because they can be used as a reference upon which to “project” future data and interpret new findings within the context of known biology.
Another source of data for building regulatory networks comes from high-throughput chromatin immunoprecipitation (ChIP)-chip (Kidder et al. 2008), ChIP-seq (Chen et al. 2008), and ChIP-PET (Loh et al. 2006) experiments. These techniques are commonly used to identify transcription factor—DNA interactions and thereby connect transcription factors to the putative sets of genes that they regulate. These techniques can also be used to identify a broad range of epigenetic chromatin modifications such as methylation/acetylation status of histone proteins. Several studies have used these techniques to identify targets of a number of the core pluripotency transcription factors. To clarify the nature of the observed binding events, high-throughput transcription-factor-binding studies are often coupled to loss-of-function experiments and genome-wide mRNA expression profiling to assess the functionality of any identified putative regulatory interactions (e.g., whether observed transcriptional binding induces activation or repression of the target gene). In addition to transcriptional regulation of stem cell fate, accumulating evidence, first in Drosophila (Hatfield et al. 2005) and more recently in mammalian stem cells (Houbaviy et al. 2003; Tay et al. 2008), suggests that miRNAs are also intimately involved in the regulation of stem-cell-fate decisions (Gangaraju and Lin 2009). For example, it has recently been shown that mir-21 suppresses a set of core pluripotency genes and is itself transcriptionally suppressed by the pluripotency factor Rest (Singh et al. 2008). Consequently, databases and network analyses deconstructing the place of miRNAs in the regulation of mammalian cells are also rapidly emerging (Altuvia et al. 2005; Griffiths-Jones et al. 2006). Although transcriptional regulation of stem cell fate is now being dissected with increasing detail, the complex signaling network (Ma’ayan et al. 2005b) upstream of the transcriptional network is less well understood. Even less information is available that sheds light on how signaling networks converge on events that occur in the nucleus. However, we anticipate that emerging high-throughput phosphoproteomics and RNA interference experiments will provide insights into the structure and function of stem cell signaling networks and their relationship to the core transcriptional network. Indeed, some progress has already been made toward this end (Chen et al. 2008).
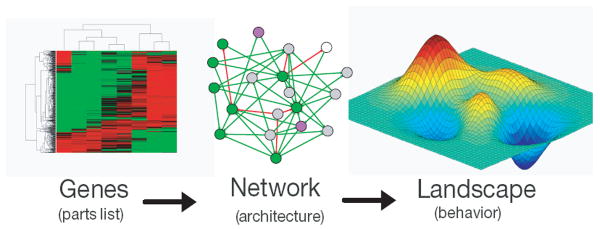
Taken together, these reports suggest that stem-cell-fate determination is an intrinsically complex process, regulated by a dynamic interplay among genetic, proteomic, miRNA, and epigenetic mechanisms. To begin to make sense of this complexity, it is useful to cast this multiplicity of biochemical interactions in the form of networks that encode the architecture of the regulatory mechanisms of stem-cell-fate specification at the system level (Fig. 1). However, this approach inevitably leads to a paradox: As our understanding of the molecular basis of cell fate becomes more detailed, the networks that arise from these integrative studies become correspondingly more complex and difficult to interpret. For this reason, there is now a pressing need to generate new tools to make sense of these complex networks to understand how internal molecular circuitry defines cell fate at the systems level. In a sense, new tools are needed to “see the forest and not just the trees.” In the following section, we discuss ways in which mathematical models may be fruitfully used to make sense of this complexity.
Figure 1.
Understanding stem cells at the systems level: From genes to networks and landscapes.
FROM NETWORKS TO ATTRACTORS
A mentioned above, a number of recent reports have begun to reconstruct the transcriptional circuitry underpinning the maintenance of stem cell pluripotency and self-renewal (Boyer et al. 2005; Ivanova et al. 2006; Kim et al. 2008). Taken together, these studies report a complex transcriptional regulatory circuit centered around a set of core pluripotency factors (including Oct4, Sox2, Nanog, Esrrb, Tbx3, Tcl1, Dppa4, Tcf3, and others) connected to an extended set of lineage-specifying factors. Crucially, this extended circuit appears to have a highly enriched feedback loop structure, in which the core pluripotency factors regulate the expression of their target genes in a highly combinatorial manner and are themselves regulated in a coordinated way. The multiplicity of positive and negative feedback loops present in this core circuit makes determination and prediction of cell behavior from regulatory architecture intrinsically difficult. To tackle this problem, it is conceptually convenient to take a physical approach and think of transcriptional activation and inhibition as forces that “push” and “pull” the cell’s internal transcriptional state in different directions: some synergistically, pushing the cell in the same genetic direction; others competitively, pushing the cell in divergent directions. Within this framework, cell-fate determination may be seen as resulting from the sum of the internal forces that the cell experiences in response to environmental signaling cues, and cell “types” as equilibrium states in which the core transcription factors are expressed at a level that balances the system. Within the mathematical literature, such balanced configurations are referred to as attractors because if perturbed away, the system is attracted back to the balanced state over time. A useful analogy is that of a marble perturbed from the bottom of a bowl that tracks out a transient trajectory around the sides of the bowl only to eventually return to rest again at its bottom. A system that supports the existence of multiple different attractor states is said to exhibit multistability.
The notion that different cell types may result from multistability of an underlying dynamical system was first suggested by the Nobel-Prize-winning physicist Max Delbrück in the late 1940s (Delbrück 1949; Thomas 1998) and has been developed extensively in a theoretical context by Stuart Kauffman and other researchers since the late 1960s (Kauffman 1969, 1993; Thomas 1998; MacArthur et al. 2008). However, although this notion has received much attention in the theoretical literature, experimental evidence that distinct mammalian cell fates may correspond to attractors of underlying high-dimensional regulatory networks has only recently been provided by Sui Huang, Donald Ingber, and coworkers (Huang et al. 2005). To do so, these authors made use of the experimental observation that similar in vitro cellular responses can often be induced by disparate chemical stimuli. In particular, they used the fact that human promyelocytic HL60 cells may be triggered to neutrophil differentiation in vitro, either by treatment with retinoic acid (RA) or by treatment with dimethylsulfoxide (DMSO). By taking time courses of microarrays during differentiation, they found that RA and DMSO initially triggered widely divergent patterns of gene expression; however, although genome-scale patterns of expression were initially divergent, they found that, over time, the patterns of gene expression induced by RA and DMSO ultimately converged to a common end point. The fact that alternative perturbations affect a common response through divergent routes is characteristic of an attracting state, and their results therefore suggest that the HL60 neutrophil state is an attractor of a (as yet undefined) complex regulatory network. Since this initial report, further evidence that other mammalian cell types may be high-dimensional attractors has been provided (Chang et al. 2006, 2008; Ying et al. 2008). For instance, by blocking fibroblast growth factor (FGF) receptor and extracellular signal-regulated kinase (ERK) signaling, Qi-Long Ying, Austin Smith, and coworkers demonstrated that, if protected from external inductive differentiation stimuli, mouse embryonic stem cells may be maintained in culture in a self-renewing state without the need for the additional culture stimuli usually required for their maintenance. This result suggests that in the mouse, the core self-renewing pluripotent state is internally stable and self-sustaining, indicating that it may correspond to an attractor of the complex pluripotency circuit. Within this context, recent reports that fully differentiated cell types may be reprogrammed to a primitive pluripotent state by a variety of different means (Yu et al. 2007; Nakagawa et al. 2008; Feng et al. 2009) are also indicative of the presence of a core pluripotent attracting state.
Taken together, these experimental reports are consistent with the notion that cell fates, including the primitive pluripotent self-renewing state, may correspond to different high-dimensional attractors of the cell’s internal regulatory circuitry. However, current reports have generally only provided indirect evidence of attractors or evidence for cellular attractors at the RNA level. Because cell fate is controlled by complex feedback among genetic, epigenetic, and proteomic mechanisms, a current challenge in stem cell systems biology is to extend these initial reports, to map not only the genetic profile of cellular attractors, but also the proteomic and epigenetic profiles of cellular attractors. For example, from a biological point of view, it is usual to think of cell types as characterized by fixed molecular signatures (Ivanova et al. 2002); however, from a mathematical point of view it is also natural to suspect that the complex circuitry at the core of cell-fate specification may allow not just static “fixed-point” attractors, but also stable self-sustaining oscillatory states, in which transcriptional forces balance in a dynamic manner. Oscillators are ubiquitous in complex systems containing feedback loops, and many biochemical oscillators have correspondingly been described (Winfree 2001). In the context of stem cell differentiation, recent data indicating that Nanog expression fluctuates in murine embryonic stem cells (Chambers et al. 2007) are possibly indicative of a dynamic, rather than static, attracting state. The notion of dynamic stem cell attractors is intuitively appealing because, if present, they may allow individual cells to be dynamically primed: At the Nanog high-expression phase, cells are resistant to inductive stimuli, whereas at the Nanog low-expression phase, cells are sensitive to inductive differentiation stimuli. Evidence for dynamic stem cell attractors is currently lacking; however, we anticipate that this may be a fruitful area for future stem cell systems biology research.
FROM ATTRACTORS TO LANDSCAPES
In an attempt to understand the robustness of cellular differentiation, Conrad Waddington suggested his now famous epigenetic landscape (Waddington 1957). His view was that development occurs rather like a marble rolling down a tilted, funneled landscape containing multiple “hills” and “valleys”: As differentiation progresses, the cell adopts a more and more specific state, corresponding to a deeper valley in the landscape, and is barred from spontaneous movement between states by the hills that split the landscape into discrete valleys. Crucially, within Waddington’s view, cell types are not terminally fixed, but rather, they are maintained by “epigenetic” barriers that can, given sufficient perturbation, be overcome. Recent demonstrations that cells can be reprogrammed from one type to another (Jaenisch and Young 2008; Zhou et al. 2008) suggest that this is indeed the case, and these reports have correspondingly led to a revived interest in Waddington’s ideas (Goldberg et al. 2007). The notion that cell fate is guided by an underlying regulatory landscape is also appealing from a theoretical point of view because, for many complex systems, attractors may be directly associated in a precise way with local minima of an appropriately defined potential energy (or energy-like). This observation has led other authors to conjecture that Waddington’s epigenetic landscape may, in fact, correspond to the “energy” landscape of a cell’s underlying regulatory architecture (Huang and Ingber 2007). Energy landscapes have proven to be successful in helping to understand many other complex phenomena (such as the protein folding problem, for example [Wales 2003; Janke 2007]), and we therefore anticipate that applications of energy landscape theory will be useful in addressing the relationship between internal regulatory circuitry and cell-fate determination. In particular, by determining the topology of cellular “energy” landscapes, it may be possible to understand not just the nature of individual cellular attractors, but also the ways in which individual attractors relate to one another (e.g., the heights of the barriers separating them). In the context of cellular reprogramming, such information would be particularly useful because it would provide a means to determine how efficiently different cell types may be reprogrammed, either to the pluripotent state or to alternative differentiated or multipotent states.
CONCLUSIONS
These are exciting times for stem cell biology. New and emerging high-throughput technologies are allowing us to deconstruct the mechanisms of cell-fate determination with ever-increasing detail. By representing the multiplicity of regulatory interactions underpinning stem cell fate as networks, we are beginning to dissect stem-cell-fate specification at the systems level. However, it is becoming clear that cell-fate specification is a fundamentally complex process and this complexity makes it intrinsically difficult to determine and predict cell behavior from regulatory network architecture. One potential way to connect cell fate to regulatory circuitry is by using regulatory architecture to define a cellular “energy” landscape—in which valleys are associated with different cell types and hills are associated with the barriers between them—and computationally explore the topology of this landscape. This approach is conceptually reminiscent of Waddington’s epigenetic landscape but has, until recently, been hampered by lack of data. However, with the advent of high-resolution high-throughput techniques, we are now beginning to accumulate sufficient data at multiple molecular and biochemical levels to make Waddington’s vision quantitative. Doing so will require interdisciplinary collaboration among experimentalists, mathematicians, physicists, and computer scientists. Thus, this is not only an outstanding problem in stem cell systems biology, but also an area rich in collaborative opportunities between experimentalists and theoreticians. Consequently, developing a rigorous understanding of stem-cell-fate determination at the systems level is a significant challenge as well as a great opportunity.
References
- Altuvia Y, Landgraf P, Lithwick G, Elefant N, Pfeffer S, Aravin A, Brownstein MJ, Tuschl T, Margalit H. Clustering and conservation patterns of human microRNAs. Nucleic Acids Res. 2005;33:2697–2706. doi: 10.1093/nar/gki567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger S, Posner J, Ma’ayan A. Genes2Networks: Connecting lists of gene symbols using mammalian protein interactions databases. BMC Bioinformatics. 2007;8:372. doi: 10.1186/1471-2105-8-372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer LA, Lee TI, Cole MF, Johnstone SE, Levine SS, Zucker JP, Guenther MG, Kumar RM, Murray HL, Jenner RG, et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer LA, Plath K, Zeitlinger J, Brambrink T, Medeiros LA, Lee TI, Levine SS, Wernig M, Tajonar A, Ray MK, et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441:349–353. doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- Chambers I, Silva J, Colby D, Nichols J, Nijmeijer B, Robertson M, Vrana J, Jones K, Grotewold L, Smith A. Nanog safeguards pluripotency and mediates germline development. Nature. 2007;450:1230–1234. doi: 10.1038/nature06403. [DOI] [PubMed] [Google Scholar]
- Chang H, Oh P, Ingber D, Huang S. Multistable and multistep dynamics in neutrophil differentiation. BMC Cell Biol. 2006;7:11. doi: 10.1186/1471-2121-7-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang HH, Hemberg M, Barahona M, Ingber DE, Huang S. Transcriptome-wide noise controls lineage choice in mammalian progenitor cells. Nature. 2008;453:544–547. doi: 10.1038/nature06965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Xu H, Yuan P, Fang F, Huss M, Vega VB, Wong E, Orlov YL, Zhang W, Jiang J, et al. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell. 2008;133:1106–1117. doi: 10.1016/j.cell.2008.04.043. [DOI] [PubMed] [Google Scholar]
- Cole MF, Johnstone SE, Newman JJ, Kagey MH, Young RA. Tcf3 is an integral component of the core regulatory circuitry of embryonic stem cells. Genes Dev. 2008;22:746–755. doi: 10.1101/gad.1642408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delbrück M. Unités Biologiques Douées de Continuité Génétique. Editions du Centre National de la Recherche Scientifique; Paris: 1949. Discussion; pp. 33–35. [Google Scholar]
- Feng B, Jiang J, Kraus P, Ng J-H, Heng J-CD, Chan Y-S, Yaw L-P, Zhang W, Loh T-H, Han J, et al. Reprogramming of fibroblasts into induced pluripotent stem cells with orphan nuclear receptor Esrrb. Nat Cell Biol. 2009;11:197–203. doi: 10.1038/ncb1827. [DOI] [PubMed] [Google Scholar]
- Gangaraju VK, Lin H. microRNAs: Key regulators of stem cells. Nat Rev Mol Cell Biol. 2009;10:116–125. doi: 10.1038/nrm2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg AD, Allis CD, Bernstein E. Epigenetics: A landscape takes shape. Cell. 2007;128:635–638. doi: 10.1016/j.cell.2007.02.006. [DOI] [PubMed] [Google Scholar]
- Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34:D140–D144. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gygi SP, Aebersold R. Mass spectrometry and proteomics. Curr Opin Chem Biol. 2000;4:489–494. doi: 10.1016/s1367-5931(00)00121-6. [DOI] [PubMed] [Google Scholar]
- Hatfield SD, Shcherbata HR, Fischer KA, Nakahara K, Carthew RW, Ruohola-Baker H. Stem cell division is regulated by the microRNA pathway. Nature. 2005;435:974–978. doi: 10.1038/nature03816. [DOI] [PubMed] [Google Scholar]
- Houbaviy HB, Murray MF, Sharp PA. Embryonic stem cell-specific microRNAs. Dev Cell. 2003;5:351–358. doi: 10.1016/s1534-5807(03)00227-2. [DOI] [PubMed] [Google Scholar]
- Huang S, Ingber DE. A non-genetic basis for cancer progression and metastasis: Self-organizing attractors in cell regulatory networks. Breast Dis. 2007;26:27–54. doi: 10.3233/bd-2007-26104. [DOI] [PubMed] [Google Scholar]
- Huang S, Eichler G, Bar-Yam Y, Ingber DE. Cell fates as high-dimensional attractor states of a complex gene regulatory network. Phys Rev Lett. 2005;94:128701-1–128701-4. doi: 10.1103/PhysRevLett.94.128701. [DOI] [PubMed] [Google Scholar]
- Ivanova N, Dobrin R, Lu R, Kotenko I, Levorse J, DeCoste C, Schafer X, Lun Y, Lemischka IR. Dissecting self-renewal in stem cells with RNA interference. Nature. 2006;442:533–538. doi: 10.1038/nature04915. [DOI] [PubMed] [Google Scholar]
- Ivanova NB, Dimos JT, Schaniel C, Hackney JA, Moore KA, Lemischka IR. A stem cell molecular signature. Science. 2002;298:601–604. doi: 10.1126/science.1073823. [DOI] [PubMed] [Google Scholar]
- Jaenisch R, Young R. Stem cells, the molecular circuitry of pluripotency and nuclear reprogramming. Cell. 2008;132:567–582. doi: 10.1016/j.cell.2008.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janke W, editor. Rugged free energy landscapes: Common computational approaches to spin glasses, structural glasses and biological macromolecules. Springer; New York: 2007. [Google Scholar]
- Jiang J, Chan YS, Loh YH, Cai J, Tong GQ, Lim CA, Robson P, Zhong S, Ng HH. A core Klf circuitry regulates self-renewal of embryonic stem cells. Nat Cell Biol. 2008;10:353–360. doi: 10.1038/ncb1698. [DOI] [PubMed] [Google Scholar]
- Johnson R, Teh CH, Kunarso G, Wong KY, Srinivasan G, Cooper ML, Volta M, Chan SS, Lipovich L, Pollard SM, et al. REST regulates distinct transcriptional networks in embryonic and neural stem cells. PLoS Biol. 2008;6:e256. doi: 10.1371/journal.pbio.0060256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauffman S. Homeostasis and differentiation in random genetic control networks. Nature. 1969;224:177–178. doi: 10.1038/224177a0. [DOI] [PubMed] [Google Scholar]
- Kauffman S. The origins of order: Self-organization and selection in evolution. Oxford University Press; New York: 1993. [Google Scholar]
- Kidder BL, Yang J, Palmer S. Stat3 and c-Myc genome-wide promoter occupancy in embryonic stem cells. PLoS ONE. 2008;3:e3932. doi: 10.1371/journal.pone.0003932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Chu J, Shen X, Wang J, Orkin SH. An extended transcriptional network for pluripotency of embryonic stem cells. Cell. 2008;132:1049–1061. doi: 10.1016/j.cell.2008.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Huang J, Chen T, Wang Y, Xin S, Li J, Pei G, Kang J. Yamanaka factors critically regulate the developmental signaling network in mouse embryonic stem cells. Cell Res. 2008;18:1177–1189. doi: 10.1038/cr.2008.309. [DOI] [PubMed] [Google Scholar]
- Loh Y-H, Wu Q, Chew JL, Vega VB, Zhang W, Chen X, Bourque G, George J, Leong B, Liu J, et al. The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat Genet. 2006;38:431–440. doi: 10.1038/ng1760. [DOI] [PubMed] [Google Scholar]
- Ma’ayan A. Network integration and graph analysis in mammalian molecular systems biology. IET Syst Biol. 2008;2:206–221. doi: 10.1049/iet-syb:20070075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma’ayan A. Insights into the organization of biochemical regulatory networks using graph theory analyses. J Biol Chem. 2009;284:5451–5455. doi: 10.1074/jbc.R800056200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma’ayan A, Blitzer RD, Iyengar R. Toward predictive models of mammalian cells. Annu Rev Biophys Biomol Struct. 2005a;34:319–349. doi: 10.1146/annurev.biophys.34.040204.144415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma’ayan A, Jenkins SL, Neves S, Hasseldine A, Grace E, Dubin-Thaler B, Eungdamrong NJ, Weng G, Ram PT, Rice JJ, et al. Formation of regulatory patterns during signal propagation in a mammalian cellular network. Science. 2005b;309:1078–1083. doi: 10.1126/science.1108876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacArthur BD, Please CP, Oreffo ROC. Stochasticity and the molecular mechanisms of induced pluripotency. PLoS ONE. 2008;3:e3086. doi: 10.1371/journal.pone.0003086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marson A, Levine SS, Cole MF, Frampton GM, Brambrink T, Johnstone S, Guenther MG, Johnston WK, Wernig M, Newman J, et al. Connecting microRNA genes to the core transcriptional regulatory circuitry of embryonic stem cells. 2008;134:521–533. doi: 10.1016/j.cell.2008.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathur D, Danford TW, Boyer LA, Young RA, Gifford DK, Jaenisch R. Analysis of the mouse embryonic stem cell regulatory networks obtained by ChIP-chip and ChIP-PET. Genome Biol. 2008;9:R126. doi: 10.1186/gb-2008-9-8-r126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller F-J, Laurent LC, Kostka D, Ulitsky I, Williams R, Lu C, Park IH, Rao MS, Shamir R, Schwartz PH, et al. Regulatory networks define phenotypic classes of human stem cell lines. Nature. 2008;455:401–405. doi: 10.1038/nature07213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa M, Koyanagi M, Tanabe K, Takahashi K, Ichisaka T, Aoi T, Okita K, Mochiduki Y, Takizawa N, Yamanaka S. Generation of induced pluripotent stem cells without Myc from mouse and human fibroblasts. Nat Biotechnol. 2008;26:101–106. doi: 10.1038/nbt1374. [DOI] [PubMed] [Google Scholar]
- Singh SK, Kagalwala MN, Parker-Thornburg J, Adams H, Majumder S. REST maintains self-renewal and pluripotency of embryonic stem cells. Nature. 2008;453:223–227. doi: 10.1038/nature06863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tay Y, Zhang J, Thomson AM, Lim B, Rigoutsos I. microRNAs to Nanog, Oct4 and Sox2 coding regions modulate embryonic stem cell differentiation. Nature. 2008;455:1124–1128. doi: 10.1038/nature07299. [DOI] [PubMed] [Google Scholar]
- Thomas R. Laws for the dynamics of regulatory networks. Int J Dev Biol. 1998;42:479–485. [PubMed] [Google Scholar]
- Ulitsky I, Shamir R. Identification of functional modules using network topology and high-throughput data. BMC Systems Biol. 2007;1:8. doi: 10.1186/1752-0509-1-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waddington CH. The strategy of the genes. Allen and Unwin; London: 1957. [Google Scholar]
- Wales DJ. Energy landscapes: Applications to clusters, biomolecules and glasses. Cambridge University Press; Cambridge: 2003. [Google Scholar]
- Wang J, Rao S, Chu J, Shen X, Levasseur DN, Theunissen TW, Orkin SH. A protein interaction network for pluripotency of embryonic stem cells. Nature. 2006;444:364–368. doi: 10.1038/nature05284. [DOI] [PubMed] [Google Scholar]
- Winfree A. The geometry of biological time. 2. Springer; New York: 2001. [Google Scholar]
- Ying Q-L, Wray J, Nichols J, Batlle-Morera, Doble B, Woodgett J, Cohen P, Smith A. The ground state of embryonic stem cell self-renewal. Nature. 2008;453:519–523. doi: 10.1038/nature06968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, Stewart R, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- Zhou Q, Brown J, Kanarek A, Rajagopal J, Melton DA. In vivo reprogramming of adult pancreatic exocrine cells to β-cells. Nature. 2008;455:627–632. doi: 10.1038/nature07314. [DOI] [PMC free article] [PubMed] [Google Scholar]