Abstract
The application of protein biomarkers as an aid for the detection and treatment of diseases has been subject to intensified interest in recent years. The quantitative assaying of protein biomarkers in easily obtainable biological fluids such as serum and urine offers the opportunity to improve patient care via earlier and more accurate diagnoses in a convenient, non-invasive manner as well as providing a potential route towards more individually targeted treatment. Essential to achieving progress in biomarker technology is the ability to screen large numbers of proteins simultaneously in a single experiment with high sensitivity and selectivity. In this article, we highlight recent progress in the use of microarrays for high-throughput biomarker profiling and discuss some of the challenges associated with these efforts.
I Introduction
The discovery of protein biomarkers whose change in expression level or state correlates with the progression of a disease is becoming increasingly important. Once validated, proposed biomarkers can be involved in achieving an earlier diagnosis, differentiating between disease types with greater accuracy, and assessing response to treatment. Prominent examples of biomarkers include proteins that are associated with a variety of cancers,1-4 as well as heart,5,6 renal,7,8 andAlzheimer's9,10 diseases.Most biomarker studies reported to date have focused on serum-, plasmaand urine-based samples. A number of other possibilities include saliva, tears, breath condensates, cerebrospinal fluid and tissue lysates extracted from biopsy samples. There are several excellent reviews detailing biomarker discovery and validation.11-15 The potential benefits of biomarkers have greatly motivated both academic and industry researchers to apply new proteomic technologies for biomarker discovery and to develop quantitative analytical methodologies for rapid and sensitive biomarker detection. Many clinically relevant biomarkers reside in blood at picomolar concentrations and lower, which is five to seven orders of magnitude lower than the most abundant plasma proteins. Therefore, protein detection with high specificity and sensitivity is required; unfortunately, no universal ultrasensitive enzymatic amplification method (such as PCR for the case of nucleic acid detection) exists for proteins. Additionally, it is desirable to screen multiple proteins simultaneously in a single sample. Multiplexed measurements are attractive not only for economic reasons but also for identifying characteristic signal patterns associated with the relative changes in entire sets of proteins as this will provide much more insight and diagnostic accuracy than individual biomarker measurements.
The most widely used techniques for the discovery and simultaneous profiling of multiple protein biomarkers are mass spectrometry,16-18 2-D western blotting,19,20 2-D gel electrophoresis,21,22 and immunological assays such as enzyme-linked immunosorbent assay (ELISA).2,20,23 Each of these detection platforms plays an important role in the various steps involved in establishing new biomarkers; however, none of these techniques are well-suited for the comparative analysis of large numbers of samples or the multiplexed detection of many targets within an individual sample. An excellent and efficient alternative is microarray-based technologies.14,19,24-30 Although microarray methods are wellestablished for high-throughput nucleic acid studies, their application for protein detection has been limited by issues such as the surface immobilization of protein capture probes without loss in bioactivity as well as the availability of highly specific probes suitable for use in complex biological samples where there is a high risk of assay cross-reactivity. Herein, we discuss some of the latest developments aimed at expanding the applicability of microarray biosensors for the detection of biomarkers for disease analysis.
II Application of antibody microarrays for disease analysis
Antibodies, being natural binders of proteins, are by far the most widely applied type of capture probe agent used for the detection and profiling of biomarkers in biological fluids. However, despite the widespread use of DNA microarray technology for the analysis of genetic materials, the feasibility of creating large-scale (i.e. >100 binding pairs) antibody microarrays for high-throughput proteomics was only first successfully demonstrated in 2001 by Haab et al.31 Since then, antibody microarray technology has progressed rapidly by addressing the multiple challenges associated with the surface immobilization of antibodies in a stable, high-density and reproducible manner without subsequent loss of bioactivity.27 Conventional methods of attaching antibodies to surfaces include adsorption to planar glass slides coated with, for example, nitrocellulose and poly-L-lysine polymers; covalent coupling through a cross-linker via thiol, maleimide or amino groups to functionalized surfaces; as well as the use of affinity binding such as biotin/streptavidin and fusion tagged proteins (e.g. histidine-tag, glutathione Stransferase) to obtain a more controlled antibody surface orientation. Alternatives to glass substrates that are less welldeveloped include nylon membranes, gelbased 3-D structures, plastic microwells and suspension arrays of beads. Each approach has a number of relative advantages and disadvantages that have been previously discussed in several recent reviews.27,32-34 At present, the best choice of surface attachment chemistry is not yet firmly established and also depends on additional factors such as the variety of protein types to be immobilized, the analyte sample medium and the detection method.
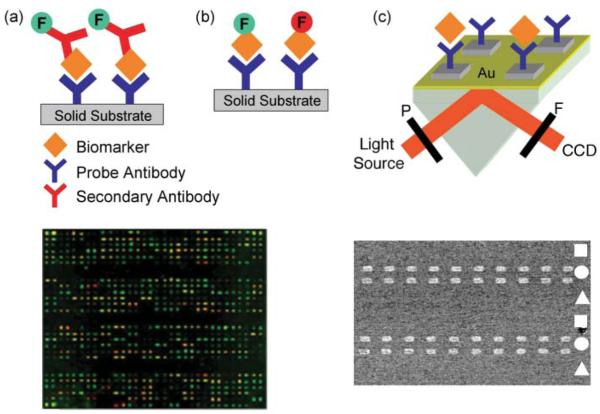
Various signal generation strategies have been used in antibody arrays and typically involve either directly labeling the target itself along with every other protein in the sample or, alternatively, using a labeled secondary binding probe which has a high specificity towards the target antigen. Awide variety of labeling species exist which enable detection by conventional methods such as radioactivity, colorimetry, chemiluminescence, light scattering and fluorescence with the latter by far the most widely applied.35 In a sandwich assay [see Fig. 1(a)], immobilized antibodies capture unlabeled proteins followed by the binding of a second fluorescently labeled antibody specific to a different site (epitope) on the target protein. The use of two antibodies helps to improve specificity and reduce background noise since a detection signal is only obtained when the target biomarker is simultaneously bound to both probes. However, sandwich assays are difficult to develop for largescale multiplexed measurements (i.e. >30 antigens) due to requiring two complete sets of antibodies for each target and the increasing likelihood of cross-reactivity between detection antibodies and nonspecific array elements as the number of targets increases. On the other hand, label-based assays [Fig. 1(b)] are limited more by the availability of single antibody– target pairs and the spatial resolution and size of the array. The direct (fluorophore) and indirect (biotin or digoxigenin) labeling of complex protein mixtures such as serum is readily achieved using various commercially available kits based on Nhydroxysuccinimide or universal linkage system chemistries. Although less sensitive than the sandwich format as well as a risk of epitope impairment upon labeling, the majority of disease-related applications involving antibody microarrays reported to date are based on the direct sample labeling. For example, the fluorescence image in the bottom left of Fig. 1 represents a proteomic signature pattern associated with directly labeled sera samples from cystic fibrosis patients. 36 Currently, a number of companies including Gentel Biosurfaces, Whatman and Clontech offer microarrays containing from around 50 up to several hundred antibody probes for the label-based analysis of human cancerrelated proteins.
Figure 1.
Simplified scheme for the detection of biomarker binding onto an antibody microarray through (a) a fluorescently labeled secondary antibody in a sandwich assay, (b) a fluorescence label (F) directly attached to the biomarker, and (c) label-free measurements on a thin gold film using SPRI. Here, the output from a collimated white light source is passed through a polarizer (P), onto a prism/microarray assembly and collected through a filter (F) onto a CCD camera. On the bottom left is a representative fluorescence image based on a two-color analysis of directly labeled sera from cystic fibrosis patients. This image is compared to images acquired from normal patient samples: proteins that are elevated in one source compared to the other either fluoresce green or red, while proteins with similar levels in both samples appear in yellow. Adapted with permission from ref. 36. The bottom right is an SPRI difference image obtained by subtracting images acquired before and after 10 nM β2-microglobulin binding onto a three-component antibody array composed of anti-cysC (square), anti-β2m (circle) and anti-SEB (triangle). Adapted with permission from ref. 39.
An alternative detection method for array-based measurements that does not require the use of a protein tag or label is surface plasmon resonance imaging [SPRI, see Fig. 1(c)]. SPRI is a surfacesensitive optical technique that quantitatively measures bioaffinity interactions (e.g. DNA–DNA, antibody–protein, RNA–protein) at microarrays created on a thin gold film by detecting changes in the local index of refraction upon surface binding.37,38 This technique is particularly advantageous for the analysis of biological samples containing multiple unknown biomarkers and where labeling is undesirable and secondary antibodies might not be available. The SPRI image in the bottom right of Fig. 1 is from a study on the detection of low-molecular weight renal disease biomarkers, β2-microglobulin (β2m) and cystatin C (cysC), using antibody microarrays created on planar gold surfaces.39 For both biomarkers the SPRI detection limit was 1 nM, which is approx. 100-fold less sensitive than comparable fluorescence imaging measurements. Neither detection method is capable of biomarker affinity monitoring at concentrations of around 10 pM and lower without some form of signal amplification. Methods to significantly improve the sensitivity of both SPRI and fluorescencebased measurements are discussed in a later section.
The maturation of antibody microarray fabrication and detection methods has enabled researchers to begin exploring a number of ways how this versatile technology can be applied to provide insight into cancer diagnosis and therapeutics. The earliest and most numerous applications have involved applying the highthroughput capabilities of arrays to measure the relative abundances of sets of proteins in various biological fluids, mostly serum, to identify associations with disease and new candidate markers for cancer diagnosis. Haab and colleagues have used direct labeling methods to profile proteins in the serum of patients with prostate,40 lung,29 pancreatic41 and bladder4 cancers. In the pancreatic study,41 using arrays of 92 antibodies and samples from 142 patients revealed multiple proteins associated with both malignant and benign disease, some of which were previously unknown to be associated with pancreatic cancer. Particularly important is that antibody arrays could be used to differentiate between disease states with high reproducibility and accuracy. This helps establish the principle that analysis of distinctive serum profiling patterns can significantly improve diagnostic accuracy compared to monoplexed studies of individual markers.
In addition to the analysis of proteins in body fluids, other novel applications of antibody microarrays recently demonstrated include the study of tissue extracts and whole cancer cells. For example, comparative fluorescence assays of tumor tissues removed by laser capture microdissection have been reported for breast cancer studies42 with distinctive protein profiles obtained for diseased tissue compared to respective healthy counterparts. Antibody arrays have also been used to quantify the expression levels of surface molecules on colorectal cancer cells.43 Suspensions of cells were incubated on an antibody array targeting various membrane-specific antigens with further distinguishing of sub-populations of cells present in the tumor sample obtained using fluorescentlabeled antibodies. Certain antigens were clearly up-regulated specifically on the tumor cell surfaces suggesting a potential method for defining potential drug targets and assessing treatment effectiveness.
Despite the rapid progress being made in the design and application of antibody microarrays, there are still multiple challenges remaining. These include improving array surface chemistries to achieve a better understanding of antibody immobilization as well as preventing nonspecific adsorption and obtaining better chip reproducibility. Amajor obstacle that becomes more prominent with increasing numbers of probe elements and targets is assay cross-reactivity. This problem is exacerbated by the fact that most monoclonal antibodies have been selected for use in single immunoassays. Therefore, large sets of highly specific antibody probes need to be created whose performance has been validated for multiplexed analyses in complex samples. Developing pattern recognition algorithms23,41,44 alongside the introduction of controls and calibrators will also significantly improve confidence in protein identification and quantitative data analysis. In the remainder of this article we focus on some of the latest developments aimed at circumventing difficulties associated with antibody microarrays through the use of alternative biomolecular probes as well as describing novel signal amplification technologies that are compatible with array-based measurements.
III Emergingmicroarray methods for protein biomarker detection
While most research efforts have centered on the development of antibody microarray-based applications for the high-throughput detection and discovery of biomarkers, some of the technical challenges described above have encouraged researchers to explore alternative biomolecular probes whose proteinbinding properties are similar to or better than those of antibodies. In this section, we discuss some recent highlights in the creation and application of nucleic acid, lysate, peptide and small molecule microarrays for biomarker analysis.
III.1 Nucleic acid aptamer microarrays
Aptamers are short nucleic acid sequences (single-stranded RNA or DNA) that selectively bind with high specificity and affinity to non-nucleic targets such as proteins as well as a large variety of other targets that include ions, toxins, drug molecules, cells and tissues45-48 (see ref. 49 for a comprehensive aptamer database). Binding occurs not through sequence hybridization but via interaction of the target with particular 3-D stemloop and internal loop structures formed by the nucleic acids. Typically, aptamers are isolated from large combinatorial libraries using a reiterative selection and amplification process known as SELEX (systematic evolution of ligands by exponential enrichment).50,51 This process is especially attractive as, unlike antibody production, it is not in vivo but rather in vitro, and does not require a host organism. Once a particular sequence is obtained from the selection process, well-established methods for high-throughput nucleic acid synthesis can then be applied. Other advantages of nucleic acids compared to the use of antibodies for bioanalytical applications include less susceptibility to denaturation as well as being more amenable to chemical modification. This provides opportunities to introduce additional functionalities that can be used for covalent surface attachment and integration into novel sensing platforms. In addition, nucleic acid surfaces tend to be much less susceptible to non-specific adsorption than protein-modified surfaces.
Aptamers have been generated against a range of targets and adapted for numerous chemical or biological sensing systems with the vast majority of work focused on the sensitive detection of single aptamer–protein and aptamer–drug complexes.46,52-55 Consequently, the merging of aptamer and microarray technologies to create bioaffinity sensors for the simultaneous detection of multiple protein or biomarker targets from a biological sample is still at a very early stage.
DNA aptamer microarrays
There have been a few recent proof-of-principle reports exploring the applicability of DNA aptamers as capture agents arrayed on a solid surface for specific protein detection.56-59 Lindner and co-workers58 immobilized 5 - amino-modified DNA aptamers via glutaraldehyde linkage on amino-silanized glass substrates and compared the binding affinity of aptamer microarrays with antibody microarrays which are both selective towards the same fluorescently-labeled thrombin target. It was clearly demonstrated that aptamer-based analyte recognition was at least as sensitive as antibody-based detection. In addition, the specificity of the aptamer arrays was superior to that of the antibody array, which exhibited a much higher level of non-specific protein adsorption that resulted in a higher background signal.
Currently, one of the most promising approaches for multiplexed protein detection is the development of photoaptamer technology, which is designed to increase the binding affinity of photoaptamers to protein targets via a photoinitiated cross-linking reaction.60,61 This is achieved by substituting thymine bases with 5-bromodeoxyuridine in the DNA aptamer sequence with the resulting photoaptamer selected for a specific protein biomarker via the photochemical SELEX process;61 this technology is under commercial development by Somalogic, Inc. High-density photoaptamer arrays are created by synthesizing selected photoaptamers with an amine on the 5′-terminus to enable immobilization onto a modified glass surface. Cross-linkage between photoaptamers and specific sites on their target proteins is achieved by irradiating at 308 nm. The photo-linked proteins at each aptamer array element can then be quantified with a fluorescent-based universal protein stain. Petach's group60 demonstrated this approach by creating a 17-plex photoaptamer array exhibiting detection limits of 10 fM for several biomarkers including interleukin-16, vascular endothelial growth factor (VEGF), and end ostatin as well as being able to detect proteins in 10% serum samples. One drawback is that this technology is currently limited to DNA photoaptamers; further development work on other photoactive modified nucleotides should allow the introduction of RNA photoaptamers.
RNA aptamer microarrays
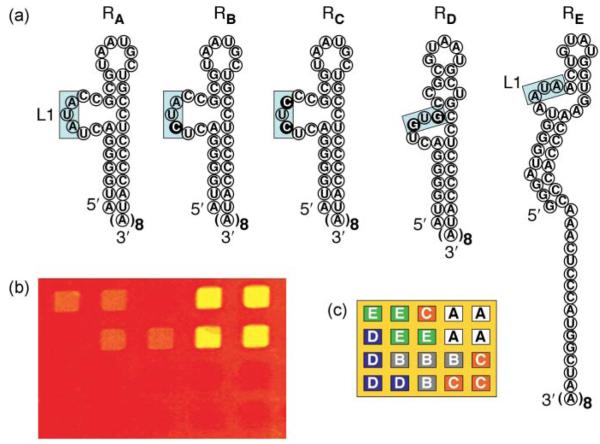
In contrast to thewealth of information available involving DNA microarrays, there are only a handful of reports describing the fabrication of RNA microarrays.52,56,62-68 This is due to the difficulty in tethering RNA molecules to a surface without loss of functionality. Most initial efforts at RNA microarray fabrication have involved modified RNA sequences such as biotinylated RNA56,64,65 or thiol-modified RNA.67 For example, Ellington and coworkers56 created an aptamer microarray by printing four different (two DNA and two RNA) biotin-modified aptamers onto streptavidin-coated glass slides for the quantitative and simultaneous detection of multiple protein targets. Recently, we developed a novel methodology to create RNA aptamer microarrays on gold surfaces and applied these arrays to detect various biomarker proteins including thrombin, factor protein IXa (fIXa) and VEGF.62,63 By using a surface DNA– RNA enzymatic ligation reaction, robust and bioactive RNA microarrays could be prepared using unmodified RNA and thus circumventing several costly and time-consuming preparative steps that often lead to RNA degradation. Fig. 2 shows the specific binding of fIXa onto a multi-component RNA aptamer microarray created via the surface ligation of unmodified probe RNA aptamer sequences onto universal DNA microarrays. The array was composed of five RNA aptamer variants (labeled RA–RE) obtained from SELEX experiments. On exposure of the array to a 100 nM solution of fIXa, increases in the SPRI detection signal identified two aptamer sequences (RA and RE) that specifically bind to fIXa, with RA having the strongest binding affinity. The lowest concentration detected was reported as 5 nM with no crosstalk.63
Figure 2.
(a) Schematic displaying the structures of five different fIXa aptamer variants (RA–RE) used to create a single-stranded RNA microarray. Both RA and RE share a conserved AUA sequence that is boxed in Loop 1 (L1), but RE differs significantly elsewhere. The sequences RB, RC and RD are modified RA aptamers with single or double base mutations indicated using the dark circles. (b) A representative SPRI difference image obtained for the differential binding of 100 nM fIXa onto the five-component array, whose pattern is shown in (c). A prominent increase in percent reflectivity was observed at the RA and RE array elements: the RA aptamer has a very strong binding affinity (2.9% increase in ΔR), while the RE aptamer has a weaker affinity for fIXa (1.1% increase in ΔR). The RB, RC and RD aptamers do not have a quantifiable binding affinity to fIXa. Adapted from ref. 63.
III.2 Lysate microarrays
An alternative approach to the sandwichand label-based antibody detection formats described in Section II is instead to immobilize protein lysates taken from multiple clinical samples in an array format.69-73 These antigen, or reversephase microarrays are incubated with a labeled antibody specific for a particular protein, therefore allowing the simultaneous quantification of a single protein present in many samples; this is in contrast to antibody array experiments where many proteins in a single sample are profiled. Demonstrations of this technology for cancer studies include measuring the phosphorylation of signaling proteins and associating changes between malignant and normal prostate tissuewith disease progression,73 and the screening of compounds for anticancer activity.72 One particular area of research significantly benefiting from lysate microarray development is the characterization of autoantibodies secreted into blood and tissue fluids as part of the humoral immune response. Autoantibodies raised against a number of different cancers74-77 can be probed at each array element with a labeled anti-immunoglobulin (e.g. anti- IgE). Recent advances also include the use of quantum dots for signal enhancement78 and combining phage-display and protein microarray technologies to improve the earlier detection of prostate77 and head and neck74cancers.
III.3 Peptide and small molecule microarrays
The last few years have seen tremendous progress in the development of new technologies that combine combinatorial chemistry and array fabrication methods for the site-specific immobilization of up to several thousand peptides and small molecules on a single solid substrate (see refs 79 and 80 for reviews). The most widely investigated applications of small molecule microarrays have been for the identifying of protein-binding ligands and function inhibitors, while peptide arrays have been mostly utilized for screening the substrate preferences of various proteinmodifying enzymes. In either case, there has been to date only a limited focus on the application of small molecules and peptides as affinity and diagnostic reagents useful for bioanalytical research. Reddy andKodadek81 have prepared microarrays comprised of several thousand (around 8000) peptide-like molecules, called pep toids, for the quantitative identification of proteins present in biological solutions. Incubating the array with an unlabeled protein solution resulted in a distinctive pattern or fingerprint that is visualized by the secondary binding of a fluorescently-labeled antibody selective towards a particular protein of interest. The protein fingerprint could be discerned with high reproducibility in a solution containing a large excess of bacterial proteins, indicating the potential of this technology for biomarker profiling in serum and other sample types. In a separate example, Hupp and co-workers82 demonstrated that peptide combinatorial libraries could be used to rapidly acquire a high-affinity peptide aptamer that can selectively bind to a target protein that is overexpressed in human cancers.
IV Advanced signal amplification methods for ultrasensitive microarray detection
One of the fundamental difficulties impeding the further development of microarray-based technologies for biomarker profiling and disease diagnostics is achieving sufficient sensitivity to reliably detect low-abundant proteins. This problem is further compounded by the need to operate in complex biological solutions where there may be a huge excess (as much as 107) of more abundant proteins and if the binding affinity between the microarray probe and target is weak (in the μM to mM range). Conventional microarray methods involving the single fluorophore labeling of either the protein target or a secondary probe molecule (as described in Fig. 1) typically have a detection limit in the low- to mid-picomolar range, which is comparable in sensitivity to traditional solution-based ELISA measurements. The ability to routinely measure protein targets at femtomolar concentrations and lower would be extremely valuable for early disease diagnosis. In this section, we highlight recent efforts aimed at novel signal amplification strategies based on (i) surface enzyme reactions, and (ii) functionalized nanomaterials that can both significantly improve sensitivity and be applied in an array format.
IV.1 Enzymatic amplification
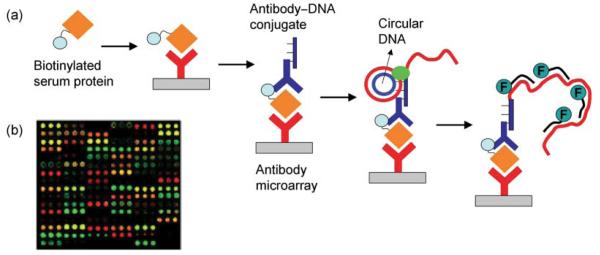
One of the most promising approaches recently developed for the on-chip amplification of detection signals associated with the binding of serum proteins onto an antibody microarray is a surface-localized rolling circle amplification (RCA) reaction. RCA has been more commonly applied to DNA quantitation and mutation detection,83,84 and in order to adapt it for protein detection, antibodies covalently conjugated to a DNA primer sequence were designed.85-87 Fig. 3(a) outlines the RCA mechanism for the amplified detection of target proteins that are first directly labeled with biotin in serum solution before incubating on the antibody array. Following this step, primer DNA conjugated antibodies (i.e. anti-biotin) that recognize the biotin tag are selectively bound to the surface-immobilized protein target. A circular DNA sequence with a portion complementary to the primer sequence is then hybridized to the surface probe/target/antibody conjugate complex. The enzyme DNA polymerase significantly extends the primer sequence by repeatedly traveling around the circular DNA template. Next, multiple labeled oligomers are hybridized to the extended primer resulting in a significantly amplified fluorescent signal (>30-fold compared to single fluorophore-labeled assays) that is also proportional to the amount of target protein in the original sample. A two-color RCA method was applied by Haab and co-workers for the multiplexed profiling of serum proteins associated with pancreatic41 and lung29 cancers. The fluorescence image in Fig. 3(b) is a distinctive serum protein profile obtained by first separately labeling the test and reference samples with biotin and digoxigenin respectively before co-incubation on an 84-component antibody microarray. 29 Two RCA reactions are performed simultaneously using Cy3 to detect the biotin-labeled proteins and Cy5 to detect the digoxigenin-labeled proteins bound to the microarray. Distinct expression profiles could be reproducibly established involving proteins at femtomolar concentrations, which was not previously achievable using other detection methods.
Figure 3.
(a) Schematic showing enzymatic amplification of biomarker detection signals via RCA. First, target proteins are labeled with biotin in serum solution and incubated on the antibody array. Primer DNA-conjugated antibodies (i.e. anti-biotin) are then bound to the surface-immobilized biotinylated protein target. Following the binding of circular DNA, a polymerase reaction significantly extends the primer sequence using the circle as a template. This enables the hybridization of multiple labeled oligonucleotides resulting in an amplified fluorescent signal. Adapted from ref. 85. (b) Fluorescence image of an antibody microarray analyzing sera from lung cancer and healthy samples using two-color RCA. Adapted from ref. 29.
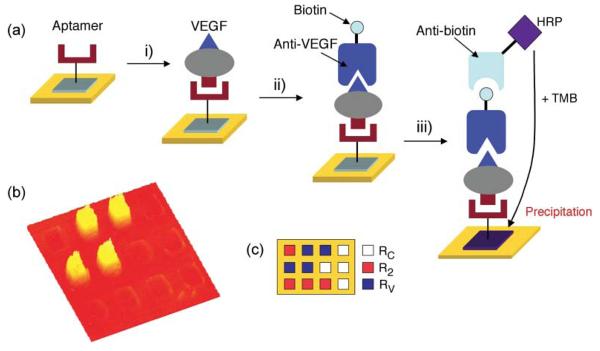
Recently, we demonstrated a different microarray-based enzymatic amplifi- cation approach involving an aptamer– antibody sandwich assay where the secondary antibody is conjugated to the enzyme horseradish peroxidase (HRP).62 HRP-conjugated antibodies are widely used in ELISA microwell or membrane assays.88,89 However, these solution-based measurements have limited spatial res olution and cannot be used in an array format. Fig. 4(a) outlines a scheme where the enzyme substrate 3,3′,5,5′-tetramethylbenzidine (TMB) is used instead to form a localized surface precipitation pattern with high spatial resolution that can be detected optically using SPRI.62 First, the target protein binds with high affinity to a single-stranded RNA aptamer array element followed by the introduction of biotin-labeled secondary antibody that recognizes a different epitope on the target to create a surface aptamer–protein–antibody sandwich structure. Next, anti-biotin conjugated to the enzyme HRP is incubated on the microarray. In a final step, the surface is exposed to a solution of TMB, which reacts with HRP to form a precipitate on the gold surface array element containing the target protein. This results in a significantly amplified SPRI response as demonstrated in Fig. 4(b) obtained for the detection of a 1 pM concentration of VEGF.62
Figure 4.
(a) Schematic of the enzymatically amplified detection of VEGF using an aptamer– protein–antibody sandwich assay. A three-component aptamer microarray is exposed sequentially to i) 1 pM VEGF, ii) 10 nM biotinylated VEGF antibody, and iii) 50 nM anti-biotinconjugated HRP. (b) The SPRI difference image shows that an increase in Δ%R was observed only at the RV array elements, which is a known RNA aptamer specific to VEGF. R2 and RC are negative control sequences. (c) Pattern of the three-component RNA microarray. Adapted with permission from ref. 62.
IV.2 Nanomaterial-enhanced detection
The exploitation of the unique optical, electronic and magnetic properties exhibited by nanomaterials such as metal and semiconductor nanoparticles, nanotubes and nanowires is opening up many new avenues for ultrasensitive protein detection.90,91 Compared with the enzymatic amplification methods described above, the use of nanomaterials for the labeling and detection of biomolecules offers the advantages of reducing the cost and number of detection steps as well as potentially improving reliability, sensitivity and accuracy. Most studies applying nanomaterials for protein detection have been limited to single target-based assays.91-93 This is not surprising considering that the issues described earlier such as chemical stability, cross-reactivity between different probes and targets as well as non-specific adsorption are also equally applicable to nanomaterial enhanced biosensing.
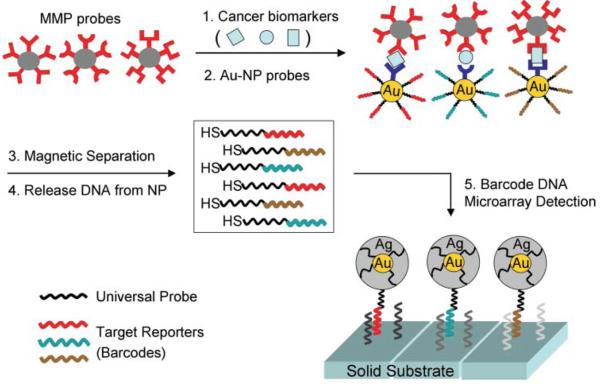
A recent exception is the development of a multiplexed version of the biobarcode amplification method established by Mirkin and co-workers94 for the ultrasensitive detection of three established cancer biomarkers present in either serum or buffer. As outlined in Fig. 5, three Au nanoparticle (NP) probes are cofunctionalized with both an antibody that recognizes the target protein and a barcode oligonucleotide.94 Half of the barcode is composed of a target reporting sequence, while the remaining half is a universal sequence common to all the barcodes. In addition, three magnetic microparticle (MMP) probes are conjugated to a monoclonal antibody specific to an epitope on the target protein different from the one recognized by the Au- NP probe. The target is captured in a MMP–biomarker–Au-NP sandwich complex, which is separated from unreacted Au-NP probes via an applied electric field. Next, the barcode strands from nanoparticles bound to the biomarker are released into solution by a ligand exchange process and subsequently detected and quantified with the chip-based scanometric method.95 This involves measuring the light scattered from array spots complementary to each barcode strand following the surface binding of Au-NPs and a subsequent silver amplification step. This allowed the amplified detection of prostratespecific antigen (prostrate cancer), human chorionic gonadotropin (testicular cancer marker), and afetoprotein (hepatocellular carcinoma marker) at femtomolar concentrations. One possible limitation of the biobarcode assay is the need to prepare both gold and magnetic probe particles specific for each target. Consequently, the potential of this approach to achieve the high-throughput capacity of conventional microarray techniques remains to be proven.
Figure 5.
Schematic of the biobarcode assay for the multiplexed detection of protein biomarkers. For each of the three protein targets, a corresponding set of Au-NP and MMP probes are prepared. The Au-NPs are conjugated to both antibodies that recognize the target protein and ‘reporter’ barcode oligonucleotides, while the MMPs are functionalized with antibodies specific to a different epitope on the target. Each MMP and Au-NP bind in a sandwich assay with the resulting complexes isolated under an applied magnetic field and the barcode strands released by a ligand exchange process. Half of each released barcode sequence is unique to a particular target while the remaining half is universal to all the barcodes. The released DNA is quantified via hybridization onto a complementary microarray element followed by hybridization of a second Au-NP probe that is modified with DNA complementary to the universal component of each barcode. A silver deposition step is then used to further amplify the light scattering detection signal. Adapted from ref. 94.
The large signal amplification possible using Au nanoparticle-labeled secondary antibodies and electroless silver amplifi- cation was also demonstrated by Bailey et al.96 as part of a new method termed DNA-encoded antibody libraries (DEAL) that enables the multiplexed detection of both nucleic acids and proteins on the same DNA microarray platform. In this approach, antibody probes are conjugated with a unique set of single-stranded DNA oligomers that are complementary to a particular DNA array element. DEAL antibodies are first introduced to the biological fluid containing the protein target along with a secondary-labeled antibody in a sandwich assay format. The entire complex (DEAL antibody–protein target–secondary antibody) is then captured on a particular array element by DNAhybridization. The simultaneous detection of multiple proteins and nucleic acids in a single microfluidic channel was demonstrated with the nanoparticle signal amplification enabling a protein detection limit of 10 fM.
Another emerging technology with the potential for high-throughput, ultrasensitive biomarker detection is the application of silicon nanowires to create field effect transistor (FET) devices where binding of charged molecules onto the functionalized nanowire surface causes a change in conductance that can be measured quantitatively in real-time.97 Pioneering work by the Lieber research group98 demonstrated that multiple cancer biomarkers could be detected simultaneously on a single nanowire array with each individually addressable nanowire functionalized with a particular capture antibody. Particularly impressive is that detection limits in the low femtomolar range were reported without additional labeling of the target or the use of a secondary nanoparticle-labeled binding probe. This is around 105 times better than equivalent label-free measurements using conventional SPR techniques. Furthermore, these electronic nanodevices can be readily integrated into miniaturized analytical systems for target delivery which, in combination with continual advances in the assembly of larger and more complex nanowire sensor arrays should create a powerful sensing platform that will be of benefit to many areas of biological research.
V Concluding remarks
The ability to perform rapid, multiplexed protein biomarker analyses in complex biological fluids will undoubtedly lead to new opportunities for the understanding and treatment of disease. The versatility of microarray-based detection platforms for the high-throughput screening of nucleic acids is now well-established; however, developing microarray technologies for the highly specific and ultrasensitive detection of low-abundant protein biomarkers has proven to be considerably more challenging. The biggest hurdle remaining in the use of microarrays for large-scale proteomic studies is the low availability of suitable capture probes. The predictive value of a single biomarker is uncertain in many studies and there is a growing consensus that multiplexed measurements examining the relative abundance ofmany types of proteins will provide far more insight. In addition, new developments utilizing aptamers, peptides and small molecules will provide complementary options to antibodies, and the effective integration of microarrays with other proteomics techniques should lead to faster methods for biomarker discovery. The next generation of microarrays capable of ultrasensitive, high-throughput, lowcost biomarker analysis will likely involve a combination of surface enzyme reactions, nanotechnology, microfluidic networks and advanced data analysis tools. However, care will be required for signal amplification strategies that associate multiple fluorophores or nanoparticles with a single biomarker in order to ensure that the amplification mechanism remains controlled and quantifiable over several orders of magnitude. In general, we expect microarray technologies to continue to increase in scope and effectiveness that will in turn accelerate markedly the rate of biomarker discovery and the characterization of disease-specific pathways.
Acknowledgements
The authors acknowledge funding support from the National Institutes of Health (2RO1 GM059622-08) and the National Science Foundation (CHE-0551935). H. J. L. thanks the Kyungpook National University Research Fund 2008 for their support.
References
- 1.Basil CF, Zhao Y, Zavaglia K, Jin P, Panelli MC, Voiculescu S, Mandruzzato S, Lee HM, Seliger B, Freedman RS, Taylor PR, Hu N, Zanovello P, Marincola FM, Wang E. Cancer Res. 2006;66:2953–2961. doi: 10.1158/0008-5472.CAN-05-3433. [DOI] [PubMed] [Google Scholar]
- 2.Zangar RC, Daly DS, White AM. Expert Rev. Proteomics. 2006;3:37–44. doi: 10.1586/14789450.3.1.37. [DOI] [PubMed] [Google Scholar]
- 3.Srivastava S. Mol. Diagn. Ther. 2006;10:221–230. doi: 10.1007/BF03256460. [DOI] [PubMed] [Google Scholar]
- 4.Sanchez-Carbayo M, Socci ND, Lozano JJ, Haab BB, Cordon-Cardo C. Am. J. Pathol. 2006;168:93–103. doi: 10.2353/ajpath.2006.050601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Masson S, Latini R, Anand IS, Vago T, Angelici L, Barlera S, Missov ED, Clerico A, Tognoni G, Cohn JN. Clin. Chem. (Washington, D. C.) 2006;52:1528–1538. doi: 10.1373/clinchem.2006.069575. [DOI] [PubMed] [Google Scholar]
- 6.Felker GM, Petersen JW, Mark DB. Can. Med. Assoc. J. 2006;175:611–617. doi: 10.1503/cmaj.060236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bonventre JV. Contrib. Nephrol. 2007;156:213–219. doi: 10.1159/000102086. [DOI] [PubMed] [Google Scholar]
- 8.Hewitt SM, Dear J, Star RA. J. Am. Soc. Nephrol. 2004;15:1677–1689. doi: 10.1097/01.asn.0000129114.92265.32. [DOI] [PubMed] [Google Scholar]
- 9.Keating CD. Proc. Natl. Acad. Sci. U. S. A. 2005;102:2263–2264. doi: 10.1073/pnas.0500024102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Georganopoulou DG, Chang L, Nam J-M, Thaxton CS, Mufson EJ, Klein WL, Mirkin CA. Proc. Natl. Acad. Sci. U. S. A. 2005;102:2273–2276. doi: 10.1073/pnas.0409336102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gunawardana CG, Diamandis EP. Cancer Lett. 2007;249:110–119. doi: 10.1016/j.canlet.2007.01.002. [DOI] [PubMed] [Google Scholar]
- 12.Rifai N, Gillette MA, Carr SA. Nat. Biotechnol. 2006;24:971–983. doi: 10.1038/nbt1235. [DOI] [PubMed] [Google Scholar]
- 13.Danna EA, Nolan GP. Curr. Opin. Chem. Biol. 2006;10:20–27. doi: 10.1016/j.cbpa.2005.12.021. [DOI] [PubMed] [Google Scholar]
- 14.Mattoon D, Michaud G, Merkel J, Schweitzer B. Expert Rev. Proteomics. 2005;2:879–889. doi: 10.1586/14789450.2.6.879. [DOI] [PubMed] [Google Scholar]
- 15.Reynolds MA, Kirchick HJ, Dahlen JR, Anderberg JM, McPherson PH, Nakamura KK, Laskowitz DT, Valkirs GE, Buechler KF. Clin. Chem. (Washington, D. C.) 2003;49:1733–1739. doi: 10.1373/49.10.1733. [DOI] [PubMed] [Google Scholar]
- 16.Ackermann BL, Hale JE, Duffin KL. Curr.DrugMetab. 2006;7:525–539. doi: 10.2174/138920006777697918. [DOI] [PubMed] [Google Scholar]
- 17.Kolch W, Neususs C, Pelzing M, Mischak H. Mass Spectrom. Rev. 2005;24:959–977. doi: 10.1002/mas.20051. [DOI] [PubMed] [Google Scholar]
- 18.Zangar RC, Varnum SM, Covington CY, Smith RD. DiseaseMarkers. 2004;20:135–148. doi: 10.1155/2004/754640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sanchez-Carbayoa M. Clin. Chem. (Washington, D. C.) 2006;52:1651–1659. doi: 10.1373/clinchem.2005.059592. [DOI] [PubMed] [Google Scholar]
- 20.Imafuku Y, Omenn GS, Hanash S. Disease Markers. 2004;20:149–153. doi: 10.1155/2004/829450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Choe LH, Werner BG, Lee KH. NeuroRx. 2006;3:327–335. doi: 10.1016/j.nurx.2006.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen J-H, Chang Y-W, Yao C-W, Chiueh T-S, Huang S-C, Chien K-Y, Chen A, Chang F-Y, Wong C-H, Chen Y-J. Proc. Natl. Acad. Sci. U. S. A. 2004;101:17039–17044. doi: 10.1073/pnas.0407992101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mor G, Visintin I, Lai Y, Zhao H, Schwartz P, Rutherford T, Yue L, Bray-Ward P, Ward DC. Proc. Natl. Acad. Sci. U. S. A. 2005;102:7677–7682. doi: 10.1073/pnas.0502178102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nettikadan S, Radke K, Johnson J, Xu J, Lynch M, Mosher C, Henderson E. Mol. Cell. Proteomics. 2006;5:895–901. doi: 10.1074/mcp.M500350-MCP200. [DOI] [PubMed] [Google Scholar]
- 25.Eisenstein M. Nature. 2006;444:959–962. doi: 10.1038/444959a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yu X, Xu D, Cheng Q. Proteomics. 2006;6:5493–5503. doi: 10.1002/pmic.200600216. [DOI] [PubMed] [Google Scholar]
- 27.Angenendt P. Drug Discovery Today. 2005;10:503–511. doi: 10.1016/S1359-6446(05)03392-1. [DOI] [PubMed] [Google Scholar]
- 28.Haab BB. Mol. Cell. Proteomics. 2005;4:377–383. doi: 10.1074/mcp.M500010-MCP200. [DOI] [PubMed] [Google Scholar]
- 29.Gao W, Kuick R, Orchekowski RP, Misek DE, Qiu J, Greenberg AK, Rom WN, Brenner DE, Omenn GSS, Haab BB, Hanash SM. BMC Cancer. 2005;5:110. doi: 10.1186/1471-2407-5-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gl̈okler J, Angenendt P. J. Chromatogr., B. 2003;797:229–240. doi: 10.1016/j.jchromb.2003.08.034. [DOI] [PubMed] [Google Scholar]
- 31.Haab BB, Dunham MJ, Brown PO. Genome Biol. 2001;2 doi: 10.1186/gb-2001-2-2-research0004. research0004.1–0004.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen GYJ, Uttamchandani M, Lue RYP, Lesaicherre M-L, Yao SQ. Curr. Top. Med. Chem. 2003;3:705–724. doi: 10.2174/1568026033452375. [DOI] [PubMed] [Google Scholar]
- 33.Taussig MJ, Landegren U. Targets. 2003;2:169–176. [Google Scholar]
- 34.Zhu H, Snyder M. Curr. Opin. Chem. Biol. 2003;7:55–63. doi: 10.1016/s1367-5931(02)00005-4. [DOI] [PubMed] [Google Scholar]
- 35.Kingsmore SF. Nat. Rev. Drug Discovery. 2006;5:310–321. doi: 10.1038/nrd2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Srivastava M, Eidelman O, Jozwik C, Paweletz C, Huang W, Zeitlin PL, Pollard HB. Mol. Genet. Metab. 2006;87:303–310. doi: 10.1016/j.ymgme.2005.10.021. [DOI] [PubMed] [Google Scholar]
- 37.Lee HJ, Wark AW, Corn RM. Langmuir. 2006;22:5241–5250. doi: 10.1021/la060223o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Smith EA, Corn RM. Appl. Spectrosc. 2003;57:320A–332A. doi: 10.1366/000370203322554446. [DOI] [PubMed] [Google Scholar]
- 39.Lee HJ, Nedelkov D, Corn RM. Anal. Chem. 2006;78:6504–6510. doi: 10.1021/ac060881d. [DOI] [PubMed] [Google Scholar]
- 40.Miller JC, Zhou H, Kwekel J, Cavallo R, Burke J, Butler EB, Teh BS, Haab BB. Proteomics. 2003;3:56–63. doi: 10.1002/pmic.200390009. [DOI] [PubMed] [Google Scholar]
- 41.Orchekowski R, Hamelinck D, Li L, Gliwa E, VanBrocklin M, Marrero JA, Vande Woude GF, Feng Z, Brand R, Haab BB. Cancer Res. 2005;65:11193–11202. doi: 10.1158/0008-5472.CAN-05-1436. [DOI] [PubMed] [Google Scholar]
- 42.Hudelist G, Pacher-Zavisin M, Singer CF, Holper T, Kubista E, Schreiber M, Manavi M, Bilban M, Czerwenka K. Breast Cancer Res. Treat. 2004;86:281–291. doi: 10.1023/b:brea.0000036901.16346.83. [DOI] [PubMed] [Google Scholar]
- 43.Ellmark P, Belov L, Huang P, Lee CS, Solomon MJ, Morgan DK, Christopherson RI. Proteomics. 2006;6:1791–1802. doi: 10.1002/pmic.200500468. [DOI] [PubMed] [Google Scholar]
- 44.Binder SR, Hixson C, Glossenger J. Autoimmunity Rev. 2006;5:234–241. doi: 10.1016/j.autrev.2005.07.007. [DOI] [PubMed] [Google Scholar]
- 45.Fischer NO, Tarasow TM, Tok JB-H. Curr. Opin. Chem. Biol. 2007;11:316–328. doi: 10.1016/j.cbpa.2007.05.017. [DOI] [PubMed] [Google Scholar]
- 46.Bunka DHJ, Stockley PG. Nat. Rev. Microbiol. 2006;4:588–596. doi: 10.1038/nrmicro1458. [DOI] [PubMed] [Google Scholar]
- 47.Proske D, Blank M, Buhmann R, Resch A. Appl. Microbiol. Biotechnol. 2005;69:367–374. doi: 10.1007/s00253-005-0193-5. [DOI] [PubMed] [Google Scholar]
- 48.Stojanovic MN, Kolpashchikov DM. J. Am. Chem. Soc. 2004;126:9266–9270. doi: 10.1021/ja032013t. [DOI] [PubMed] [Google Scholar]
- 49.Lee JF, Hesselberth JR, Meyers LA, Ellington AD. Nucleic AcidsRes. 2004;32:D95–D100. doi: 10.1093/nar/gkh094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ellington AD, Szostak JW. Nature. 1990;346:818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- 51.Tuerk C, Gold L. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 52.Lee JF, Stovall GM, Ellington AD. Curr. Opin. Chem. Biol. 2006;10:282–289. doi: 10.1016/j.cbpa.2006.03.015. [DOI] [PubMed] [Google Scholar]
- 53.Akiyama H, Kachi S, Silva RL, Umeda N, Hackett SF, McCauley D, McCauley T, Zoltoski A, Epstein DM, Campochiaro PA. J. Cell. Physiol. 2006;207:407–412. doi: 10.1002/jcp.20583. [DOI] [PubMed] [Google Scholar]
- 54.Di Giusto DA, Wlassoff WA, Gooding JJ, M. B. A., King GCC. Nucleic Acids Res. 2005;33:e64. doi: 10.1093/nar/gni063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rusconi CP, Scardino E, Layzer J, Pitoc GA, Ortel TL, Monroe D, Sullenger BA. Nature. 2002;419:90–94. doi: 10.1038/nature00963. [DOI] [PubMed] [Google Scholar]
- 56.Cho EJ, Collett JR, Szafranska AE, Ellington AD. Anal.Chim. Acta. 2006;564:82–90. doi: 10.1016/j.aca.2005.12.038. [DOI] [PubMed] [Google Scholar]
- 57.Lin C, Katilius E, Liu Y, Zhang J, Yan H. Angew. Chem., Int. Ed. 2006;45:5296–5301. doi: 10.1002/anie.200600438. [DOI] [PubMed] [Google Scholar]
- 58.Stadtherr K, Wolf H, Lindner P. Anal. Chem. 2005;77:3437–3443. doi: 10.1021/ac0483421. [DOI] [PubMed] [Google Scholar]
- 59.Lee M, Walt DR. Anal. Biochem. 2000;282:142–146. doi: 10.1006/abio.2000.4595. [DOI] [PubMed] [Google Scholar]
- 60.Bock C, Coleman M, Collins BD, Davis J, Foulds G, Gold L, Greef C, Heil J, Heilig JS, Hicke B, Hurst MN, Miller D, Ostroff R, Petach H, Schneider D, Vant-Hull B, Waugh S, Weiss A, Wilcox SK, Zichi D. Proteomics. 2004;4:609–618. doi: 10.1002/pmic.200300631. [DOI] [PubMed] [Google Scholar]
- 61.Golden MC, Collins BD, Willis MC, Koch TH. J. Biotechnol. 2000;81:167–178. doi: 10.1016/s0168-1656(00)00290-x. [DOI] [PubMed] [Google Scholar]
- 62.Li Y, Lee HJ, Corn RM. Anal. Chem. 2007;79:1082–1088. doi: 10.1021/ac061849m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li Y, Lee HJ, Corn RM. Nucleic Acids Res. 2006;34:6416–6424. doi: 10.1093/nar/gkl738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Collett JR, Cho EJ, Ellington AD. Methods. 2005;37:4–15. doi: 10.1016/j.ymeth.2005.05.009. [DOI] [PubMed] [Google Scholar]
- 65.Collett JR, Cho EJ, Lee JF, Levy M, Hood AJ, Wan C, Ellington AD. Anal. Biochem. 2005;338:113–123. doi: 10.1016/j.ab.2004.11.027. [DOI] [PubMed] [Google Scholar]
- 66.Kirby R, Cho EJ, Gehrke B, Bayer T, Park YS, Neikirk D, McDevitt J, Ellington AD. Anal. Chem. 2004;76:4066–4075. doi: 10.1021/ac049858n. [DOI] [PubMed] [Google Scholar]
- 67.Goodrich TT, Lee HJ, Corn RM. J. Am. Chem. Soc. 2004;126:4086–4087. doi: 10.1021/ja039823p. [DOI] [PubMed] [Google Scholar]
- 68.McCauley TG, Hamaguchi N, Stanton M. Anal. Biochem. 2003;319:244–250. doi: 10.1016/s0003-2697(03)00297-5. [DOI] [PubMed] [Google Scholar]
- 69.Mendes KN, Nicorici D, Cogdell D, Tabus I, Yli-Harja O, Guerra R, Hamilton SR, Zhang W. J. Proteome Res. 2007;6:2753–2767. doi: 10.1021/pr070184h. [DOI] [PubMed] [Google Scholar]
- 70.Romeo MJ, Wunderlich J, Ngo L, Rosenberg SA, Steinberg SM, Berman DM. Clin. Cancer Res. 2006;12:2463–2467. doi: 10.1158/1078-0432.CCR-05-1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ramaswamy A, Lin E, Chen I, Mitra R, Morrisett J, Coombes K, Ju Z, Kapoor M. Proteome Sci. 2005;3:9. doi: 10.1186/1477-5956-3-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Nishizuka S, Charboneau L, Young L, Major S, Reinhold WC, Waltham M, Kouros-Mehr H, Bussey KJ, Lee JK, Espina V, Munson PJ, Petricoin E, III, Liotta LA, Weinstein JN. Proc. Natl. Acad. Sci. U. S. A. 2003;100:14229–14234. doi: 10.1073/pnas.2331323100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Paweletz CP, Charboneau L, Bichsel VE, Chen SNLT, Gillespie JW, Emmert-Buck MR, Roth MJ, Petricoin IE, Liotta LA. Oncogene. 2001;20:1981–1989. doi: 10.1038/sj.onc.1204265. [DOI] [PubMed] [Google Scholar]
- 74.Lin H-S, Talwar HS, Tarca AL, Ionan A, Chatterjee M, Ye B, Wojciechowski J, Mohapatra S, Basson MD, Yoo GH, Peshek B, Lonardo F, Pan C-JG, Folbe AJ, Draghici S, Abrams J, Tainsky MA. Cancer Epidemiol. Biomarkers Prev. 2007;16:2396–2405. doi: 10.1158/1055-9965.EPI-07-0318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chatterjee M, Mohapatra S, Ionan A, Bawa G, Ali-Fehmi R, Wang X, Nowak J, Ye B, Nahhas FA, Lu K, Witkin SS, Fishman D, Munkarah A, Morris R, Levin NK, Shirley NN, Tromp G, Abrams J, Draghici S, Tainsky MA. Cancer Res. 2006;66:1181–1190. doi: 10.1158/0008-5472.CAN-04-2962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhong L, Coe SP, Stromberg AJ, Khattar NH, Jett JR, Hirschowitz EA. J. Thorac. Oncol. 2006;1:512–519. [PubMed] [Google Scholar]
- 77.Wang X, Yu J, Sreekumar A, Varambally S, Shen R, Giacherio D, Mehra R, Montie JE, Pienta KJ, Sanda MG, Kantoff PW, Rubin MA, Wei JT, Ghosh D, Chinnaiya AM. New Engl. J. Med. 2005;353:1224–1235. doi: 10.1056/NEJMoa051931. [DOI] [PubMed] [Google Scholar]
- 78.Geho D, Lahar N, Gurnani P, Huebschman M, Herrmann P, Espina V, Shi A, Wulfkuhle J, Garner H, Petricoin E, III, Liotta LA, Rosenblatt KP. Bioconjugate Chem. 2005;16:559–566. doi: 10.1021/bc0497113. [DOI] [PubMed] [Google Scholar]
- 79.Uttamchandani M, Walsh DP, Yao SQ, Chang Y-T. Curr. Opin. Chem. Biol. 2005;9:4–13. doi: 10.1016/j.cbpa.2004.12.005. [DOI] [PubMed] [Google Scholar]
- 80.Panicker RC, Huang X, Yao SQ. Comb. Chem. High Throughput Screening. 2004;7:547–556. doi: 10.2174/1386207043328517. [DOI] [PubMed] [Google Scholar]
- 81.Reddy MM, Kodadek T. Proc. Natl. Acad. Sci. U. S. A. 2005;102:12672–12677. doi: 10.1073/pnas.0501208102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Murray E, McKenna EO, Burch LR, Dillon J, Langridge-Smith P, Kolch W, Pitt A, Hupp TR. Biochemistry. 2007;46:13742–13751. doi: 10.1021/bi7008739. [DOI] [PubMed] [Google Scholar]
- 83.Nallur G, Luo C, Fang L, Cooley S, Dave V, Lambert J, Kukanskis K, Kingsmore S, Lasken R, Schweitzer B. Nucleic Acids Res. 2001;29:e118. doi: 10.1093/nar/29.23.e118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lizardi PM, Huang X, Zhu Z, Bray- Ward P, Thomas DC, Ward DC. Nat. Genet. 1998;19:225–232. doi: 10.1038/898. [DOI] [PubMed] [Google Scholar]
- 85.Zhou H, Bouwman K, Schotanus M, Verweij C, Marrero JA, Dillon D, Costa J, Lizardi P, Haab BB. Genome Biol. 2004;5:R28. doi: 10.1186/gb-2004-5-4-r28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Schweitzer B, Roberts S, Grimwade B, Shao W, Wang M, Fu Q, Shu Q, Laroche I, Zhou Z, Tchernev VT, Christiansen J, Velleca M, Kingsmore SF. Nat. Biotechnol. 2002;20:359–365. doi: 10.1038/nbt0402-359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Schweitzer B, Wiltshire S, Lambert J, O'Malley S, Kukanskis K, Zhu Z, Kingsmore SF, Lizardi PM, Ward DC. Proc. Natl. Acad. Sci. U. S. A. 2000;97:10113–10119. doi: 10.1073/pnas.170237197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Roda A, Mirasoli M, Guardigli M, Michelini E, Simoni P, Magliulo M. Anal. Bioanal. Chem. 2006;384:1269–1275. doi: 10.1007/s00216-006-0308-6. [DOI] [PubMed] [Google Scholar]
- 89.Huang RP. J. Immunol. Methods. 2001;255:1–13. doi: 10.1016/s0022-1759(01)00394-5. [DOI] [PubMed] [Google Scholar]
- 90.Geho DH, Jones CD, Petricoin EF, Liotta LA. Curr. Opin. Chem. Biol. 2006;10:56–61. doi: 10.1016/j.cbpa.2006.01.003. [DOI] [PubMed] [Google Scholar]
- 91.Zhang H, Zhao Q, Li X-F, Le XC. Analyst. 2007;132:724–737. doi: 10.1039/b704256f. [DOI] [PubMed] [Google Scholar]
- 92.Haes AJ, Chang L, Klein WL, Van Duyne RP. J. Am. Chem. Soc. 2005;127:2264–2271. doi: 10.1021/ja044087q. [DOI] [PubMed] [Google Scholar]
- 93.Grubisha DS, Lipert RJ, Park H-Y, Driskell J, Porter MD. Anal. Chem. 2003;75:5936–5943. doi: 10.1021/ac034356f. [DOI] [PubMed] [Google Scholar]
- 94.Stoeva SI, Lee J-S, Smith JE, Rosen ST, Mirkin CA. J. Am. Chem. Soc. 2006;128:8378–8379. doi: 10.1021/ja0613106. [DOI] [PubMed] [Google Scholar]
- 95.Nam J-M, Thaxton CS, Mirkin CA. Science. 2003;301:1884–1886. doi: 10.1126/science.1088755. [DOI] [PubMed] [Google Scholar]
- 96.Bailey RC, Kwong GA, Radu CG, Witte ON, Heath JR. J. Am. Chem. Soc. 2007;129:1959–1967. doi: 10.1021/ja065930i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Patolsky F, Zheng G, Lieber CM. Anal. Chem. 2006;78:4260–4269. doi: 10.1021/ac069419j. [DOI] [PubMed] [Google Scholar]
- 98.Zheng G, Patolsky F, Cui Y, Wang WU, Lieber CM. Nat. Biotechnol. 2005;23:1294–1301. doi: 10.1038/nbt1138. [DOI] [PubMed] [Google Scholar]