Abstract
The concept of using stem cells as self-renewing sources of healthy cells in regenerative medicine has existed for decades, but most applications have yet to achieve clinical success. A main reason for the lack of successful stem cell therapies is the difficulty in fully recreating the maintenance and control of the native stem cell niche. Improving the performance of transplanted stem cells therefore requires a better understanding of the cellular mechanisms guiding stem cell behavior in both native and engineered three-dimensional (3D) microenvironments. Most techniques, however, for uncovering mechanisms controlling cell behavior in vitro have been developed using 2D cell cultures and are of limited use in 3D environments such as engineered tissue constructs. Deciphering the mechanisms controlling stem cell fate in native and engineered 3D environments, therefore, requires rigorous quantitative techniques that permit mechanistic, hypothesis-driven studies of cell–microenvironment interactions. Here, we review the current understanding of 2D and 3D stem cell control mechanisms and propose an approach to uncovering the mechanisms that govern stem cell behavior in 3D.
Challenges with the Therapeutic Application of Stem Cells
Stem cells are defined by their ability to both self-renew and differentiate by undergoing asymmetric cell divisions. This unique behavior is made possible by the specialized, three-dimensional (3D) microenvironment that immediately surrounds them in native tissue. These niches provide the structural, biochemical, mechanical, and stimulatory cues necessary for the appropriate functioning of stem cells during homeostasis and in response to physiological change.1,2 In this way, stem cells are defined by their 3D anatomical location, and when we remove them from it, we create derivatives that have altered functionality, phenotype, and responsiveness to environmental cues. Even given these challenges, stem cells have been used in both preclinical and clinical studies, and there is substantial promise in their use for the purpose of regenerating diseased or injured tissues.3–8 However, there are many potential dangers in the clinical application of stem cells when the appropriate characterization, manipulation, sourcing, and quality control are not adequately performed.9–11
In 2005, a 13-year-old boy with ataxia telangiectasia, a hereditary neurodegenerative disease, presented with recurrent headaches after receiving three stem cell transplantations in Moscow, at the ages of 7, 10, and 12. Investigation into the cause of the headaches by magnetic resonance imaging revealed a multifocal brain tumor.12 The tumor was of nonhost origin and therefore from the implanted cells that were derived from at least two different donors.12 The cells that were injected into the patient were described as “fetal neural stem cells”; however, the exact nature of the cells was unknown, and it is not clear whether appropriate quality control and safety and effectiveness studies had been performed.
The devastating result of this stem cell procedure highlights the gap that still exists between our understanding of how a stem cell behaves in vivo in the niche and its unpredictability outside of that niche. The International Society for Stem Cell Research responded to this case with a call to emphasize the importance of rigorous and consistent preclinical trials on the safety and effectiveness of stem cell treatments.13 Mimicking the control present in the native stem cell niche is necessary to achieve consistent and effective application of stem cells in the clinic; however, the spatial and temporal mechanisms that specifically govern stem cell fate determination within these environments remain unclear.
Embryonic stem cells are commonly regarded as the gold standard for regenerative therapies because they are pluripotent: there are no totipotent stem cells, as none have demonstrated replication of the placenta. However, the sheer number of environmental cues and mechanisms required for directing specific fate determination has made them difficult to control in vitro and in vivo. Further, their potential introduces complexity in defining and testing these mechanisms. Multipotent mesenchymal stem cells (MSC), on the other hand, have restricted potential as compared to embryonic stem cells, lending simplicity to their differentiation decision. Additionally, the variety of sources available, including bone marrow,14 cartilage,15 fat,16 lung,17 ligament,18 brain, spleen, liver, kidney, muscle, thymus, pancreas, and more,19 provides multiple starting points for unique applications in tissue engineering. Further, induced pluripotent cells are cells genetically reprogrammed to regenerate pluripotency from various adult tissue cell types. Using this technology, patient-specific induced pluripotent cells can be derived from mature cell types and show potential in drug screening and for the development of disease models.20
The key challenge for all these cell types, however, remains the same, providing the proper environmental cues to regulate the balance between differentiation and self-renewal. Functional tissue engineering will require an understanding of how the niche controls stem cell function, application of those principles to synthetically mimic that niche in 3D, and novel methods of data acquisition and analysis to feed descriptive and predictive models of tissue function. Execution of these aims will require an increasingly interdisciplinary approach to tissue biology and engineering.
Directing Stem Cell Differentiation and Self-Renewal
Stem cells derive their behavior from cues that lie in their extracellular environment. These cues operate on different spatial and temporal scales to pattern specific cellular behaviors that drive tissue morphogenesis and differentiation. A wealth of knowledge, gathered in the last several decades, characterizes stem cell phenotype and function as well as presents methods to direct their differentiation along specific lineages in response to these extracellular cues. Within this context, the extracellular matrix (ECM), soluble growth factors, cell–cell interactions, and mechanical stimuli combine to coordinate the in vivo tissue response on multiple scales creating overlapping influences that are differentiated by the scope of their impact. In vitro, these distinct stimuli can be employed in a controlled fashion to both direct and mechanistically understand stem cell fate determination.
Extracellular matrix
The ECM not only serves to provide structural and organizational guides for tissue development but also defines and maintains cellular phenotype and drives cell fate decisions. Cells are surrounded by a complex architecture of proteins, polysaccharides, and proteoglycans that constantly undergo dynamic change as a result of assembly, remodeling, and degradation events. The flexibility inherent in this structure allows the microenvironment to be tuned on a cell-by-cell basis in response to unique patterns of protein secretion and modification. Adhesion to the specific components of the ECM via integrins, cadherins, and discoidin domain receptors activate signaling programs sensitive to the composition and orientation of that ECM.21 In vitro, MSC produce and use applied ECM to promote their own expansion and inhibit spontaneous and uncontrolled differentiation.22 The ECM also plays a critical role in defining cell shape, which in turn affects cell survival,23 proliferation,24 and differentiation,25 and these roles have also been confirmed in vivo. For example, integrins not only help to position germ stem cells within their niche but also provide feedback between those cells and the extracellular environment. Loss of integrin function in the germ stem cells niche of adult Drosophila melanogaster testis prevents the proper deposition, organization, and composition of the ECM.26
Soluble cues
In vivo, soluble factors, such as peptide growth factors and hormones, play a significant role in directing stem cell fate. These factors diffuse through and are sequestered by the ECM and bind to cell surface receptors activating cellular function. Interactions with the ECM restrict the effect soluble factors can have on the cell population creating overlapping spheres of influence that are ultimately controlled by the cell through tuning of the ECM. Additionally, the acute but transient nature of growth factor signaling is directly contrasted to the slow but sustained signals received from the ECM. For example, when induced to differentiate in restrictive ECM environments, adhesive, flattened cells preferentially adopt an osteogenic phenotype and round cells tend toward adipogenesis.25 Additionally, restricting the dynamics of the actin cytoskeleton, which coordinates intracellular signal transduction with cues in the ECM, promotes osteogenesis irrespective of soluble adipogenic induction. In this way, the ECM primes cells to bias their interpretation of extracellular soluble factors into internal signals.
Cell–cell communication and positioning
Juxtacrine cell–cell communication, propagated by the physical association of two cells, provides a persistent morphogenic cue, while the diffusion of soluble signals from neighboring cells, paracrine signaling, transiently affects proximal populations of cells. The method of association between neighboring cell populations represents a significant difference in commitment to cooperate. These interactions between cell populations influence a range of stem cell behavior, including the induction of programs of differentiation27,28 and promotion of expansion and self-renewal properties.29–31 Mature cell populations physically associate, through adherens junctions, with stem cells in the native niche. For example, osteoblasts interact with hematopoietic stem cells in the bone marrow, endothelium regulates neural stem cell differentiation in the brain, and MSC promote hematopoietic stem cells' self-renewal.32–35 Additionally, positioning within the stem cell niche, achieved by both cell–cell contacts and the spatial distribution of the ECM, physically restricts stem cell self-renewal and differentiation behavior by directing the position of the mitotic axis in asymmetric division.36,37
Mechanical stimulation
Overlaid on all of these mechanisms is the mechanical environment that governs ECM interactions, changes sensitivities to soluble cues, and physically distorts cell populations within tissues. As a global cue, the mechanical environment influences all aspects of tissue behavior, since essentially all cells, with the exception of circulating blood cells, are anchorage dependent and require interaction with the ECM to maintain viability. When adherent, cells are able to exert contractile forces on their environment and sense compliancy to induce appropriate cellular behavior.38 Even cells within soft tissues exhibit these characteristics and the patterning of mechanical forces throughout development guides morphogenesis.39 In particular, matrix elasticity directs stem cell differentiation where 2D substrates, modeled to closely replicate in vivo tissue compliance, are more efficient in promoting their specific lineage commitment.40 Similarly, in clusters of MSC, commitment is location specific, with cells in areas of high stress (edges) becoming osteogenic and those under low stress (interior) assuming an adipogenic profile.41 In addition, stem cells respond to fluid flow–induced shear stresses,42,43 compressive and tensional strains,44,45 cyclical stretching,46–49 and hydrostatic pressures.50
The Limitations of 2D Culture
Most of the above work has been performed in 2D culture systems, and while these systems present useful and powerful methods for probing cellular behavior, many of these studies are difficult, if not impossible to confirm in vivo. These elegant 2D systems allow for the precise control of cue presentation; however, our tendency to simplify the cellular environment detracts from the richness of the data we generate. Even grown in monolayer (2D), cells are still 3D structures and are influenced by the entirety of their environment. Embracing the complexity of these data sets can only be achieved through advances in the generation of quantitative models to operate on and represent the data, but there also exists a point where 2D culture is limited in its ability to ask questions about the complex spatial and temporal patterning of these cues like that seen in vivo. In addition, it is clear that the three-dimensionality of tissue is essential for the maintenance of cellular function and the development of physiologically relevant structures. Three-dimensional matrices in vitro and in vivo define the structural, mechanical, and biochemical make-up of the cellular microenvironment and are crucial for allowing a bidirectional interplay to exist between the cell and tissue during development.51 While cells on rigid, planar surfaces can respond to the mechanical nature of the culture system, they have little capacity to manipulate the composition and mechanical properties of the ECM itself.52 Mature cells cultured in 3D matrices exhibit altered phenotypes inhibiting their proliferative nature and enhancing their ability to form higher order structures.53 This 3D context also enhances stem cell potential by permitting the dynamic interfacing of the differentiating stem cell with its matrix.54
Promising Leads in 3D Biomaterial Engineering
Approaching the problem of controlling cell and tissue function from a different mindset, biomaterials scientists and bioengineers generate constructs that mimic the structure and function of the end tissue product in 3D. This alternative approach critically links 2D mechanistic studies to the in vivo application; however, the mechanisms of cellular interaction and subcellular functioning within these environments are not well known. Using both natural (protein and polysaccharide) and synthetic polymers, material scientists can engineer patterns of adhesion,55,56 composition,57,58 growth factor59–61 and mechanical gradients,62,63 cell positioning,64–68 degradation rates,69–72 and geometry73 to direct tissue morphogenesis.74–77 The application of these materials with a cellular component has been well reviewed recently for a range of tissue targets, including, but not limited to, bone,78–80 cardiac,81,82 tendon,83,84 nerve,85,86 cartilage,87,88 and fat.89–91
Within these artificial environments engineers demonstrate elegant control over cellular function; however, the rationale for their approach has largely been based upon sophisticated trial and error, which lacks efficiency in surveying a range of microenvironments. This biomaterial toolkit, however, provides a powerful mechanism through which one can translate sophisticated 2D experimental design into complex 3D microenvironments. Using engineering biomaterials with tunable physical, chemical, and biological properties as control signals allows the uncoupling of these signals to reduce the complexity of the 3D system. This opens a door for engineers to take a step back from clinical application and use their tools to advance the understanding of tissue biology and disease progression and provide tools for biologists to begin testing mechanistic hypotheses in 3D culture. In this way, we begin to bridge the disconnect that exists between 2D signaling studies and tissue engineering and engraftment.
Translating Mechanism into 3D
The rigorous and reductionist signal transduction studies that are performed in 2D and the biomaterial-based application of stem cells in vivo are inherently disconnected by our lack of understanding of the effect dimensionality has on cellular decision making. Tissue engineering is limited by the amount of complexity it can understand and control, and achieving higher levels of understanding requires mechanistic probing of complex models in 3D. Though based upon highly educated guesses, the lack of mechanistic hypotheses that can guide the development of these biomaterials limits their utility in these types of studies. What is fundamentally needed to further this field is a predictive and descriptive model that governs stem cell–material interactions and tissue development. Reaching this point will require a more efficient fusion of rigorous and mechanistic studies and the novel methods of controlling cellular function in 3D. Using this idea, the field of “cell instructive artificial ECM” has emerged to augment biologically inert polymers with specific bioactivity, and thereby define the local microenvironment based upon biological hypotheses.92 For example, bioactive, synthetic hydrogel matrices exploit the activity of vascular endothelial growth factor (VEGF) to induce local, controlled angiogenesis. VEGF tethered to the artificial matrix is restricted by the matrix until cell-mediated enzymatic degradation of the ECM, thereby coupling cell remodeling processes with local concentrations of VEGF.59,93 In this way, tissue components are dynamically related to each other and the active interplay may be more important in guiding development than the quantitative amount of cells and stimuli present within the tissue.
However, there are still significant limitations in the application of rigorous 2D techniques into the study of 3D signal transduction in the traditional sense. For this reason, few groups have successfully extended the well-developed 2D signal transduction field onto 3D scaffolds94–99 and even fewer in the field of stem cell biology.54,100,101 Instead, demonstration of biomaterial performance has primarily relied upon imaging technologies, morphological studies, histology, and bulk material properties. Many of these methods are inherently qualitative and do not readily contribute to achieving higher throughput efficiencies.
Developing complex models that couple mechanical and biochemical cues to achieve a phenotype more closely related to that found in vivo will be a critical component of tissue biology and engineering moving forward. In fact, the field of tissue engineering is additionally turning its attention to the development of physiologically relevant models of diseased tissue.102–105 Many of the tools available for the development of these types of complex environments have been well reviewed elsewhere.51 Engineering consistency and ease of use into these models will facilitate their widespread adoption by cell and molecular biologists as well as bioengineers. The rigorous signal transduction work, done in basic biology labs, and the complexity being introduced by engineers will jumpstart a powerful understanding of the 3D environment and its role in regulating cell function.
Developing these complex microenvironments for both mechanistic studies and clinical application is not trivial. The combinatorial complexity of incorporating various ECM proteins, signaling molecules, mechanical stimuli, and heterogeneous cell populations in different spatial and temporal patterns creates an enormously large search space from which to begin the engineering of new tissue constructs. In addition, the lack of biologically based, mechanistic hypotheses in 3D to inform these choices prevents efficient material development. These problems of scope and throughput require novel methods of analysis that provide quantitative metrics to direct decision processes in early material engineering and decrease this search space.
3D Modeling of Complex Networks and Cell Function
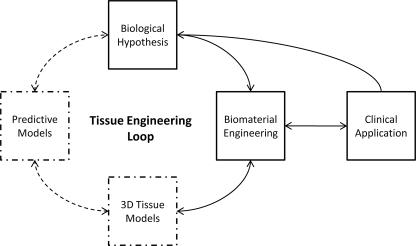
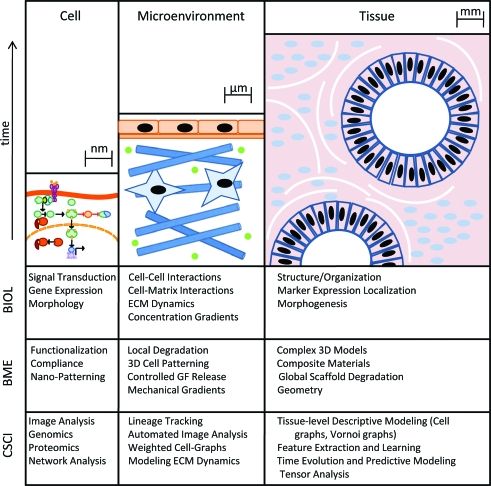
From the single-cell level up to the level of developing tissue and organization dynamics, development of models that quantitatively capture the principles guiding each tier of decision making is an emerging field that has the potential to accelerate tissue engineering efforts. The iterative generation of biological hypotheses and development of new biomaterials based upon those hypotheses is required for effective clinical translation. Developing methods of efficiently closing this “tissue engineering loop” necessitates rigorous quantitative methods for analyzing the spatio-temporal function of cells and tissues (Fig. 1). Quantitative model analysis provides feedback to inform engineering design by testing these hypotheses and improving the throughput of material optimization. There are three levels of tissue function that can be modeled to better understand the dynamic, multiscale interactions: these levels include analysis of the (1) single cell, (2) cells and their neighborhood, and (3) whole tissues (Fig. 2).
FIG. 1.
Engineering efficiency into the tissue engineering loop. Successful clinical application of basic biology and biomedical engineering research is dependent upon the efficient and directed development of new products. Current efforts in the field of biomaterial development are based loosely on biological hypotheses but are largely sophisticated trial-and-error experiments. We can more efficiently move through this process using biomaterials to develop complex three-dimensional (3D) microenvironment models that generate data to feed into descriptive and predictive models. The quantitative metrics that emerge from these models will enable the generation of new mechanistic biological hypotheses based on complex 3D data. Iterating through this loop will create rational approaches to biomaterial development and quantitative metrics that will evaluate material performance and drive further optimization.
FIG. 2.
Multiscale modeling of cell and tissue function. Cells and tissues function across variable time and spatial scales. Capturing quantitative information at each of these stages and modeling the changes observed are critical steps toward closing the tissue engineering loop. Each field encompassed by this work contributes at the spatial scales described above, but as of yet we are unable to fully integrate the information generated at each level to create a cohesive and dynamic picture of the tissue system. Biologists (BIOL) and biomedical engineers (BME) each look across these time scales using a variety of different tools. Bringing these techniques together to reintroduce complexity into 3D cell culture models will aid the development of mechanistic hypotheses that are better able to explain the complexity inherent in physiological structures. Integrating the quantitative tools developed by computer scientists (CSCI) with this 3D data over time will provide quantitative metrics to further inform the design of new materials and culture systems and promote their predictability in the clinical setting. Color images available online at www.liebertonline.com/ten.
Modeling of the single cell involves the complex interaction of signal transduction pathways, cellular differentiation, as well as spatial and morphological information captured by imaging techniques. The cell signaling that leads to changes in cellular behavior and differentiation is a complex and nonlinear network of protein interactions and genetic regulation that, even in 2D, becomes far too complex for traditional methods of data analysis. As relatively few signaling molecules can interact and form thousands and even millions of distinct signaling species, the complexity of these networks requires advanced methods of data mining.106 Computational modeling has presented an approach to understanding network dynamics that defines system function and dysfunction and additionally provides predictive capabilities.107,108 These methods have been well reviewed106,109–111 and have provided insight into the dynamics of specific signaling pathways, interactions between soluble cues, and intracellular interactions.112–120 Additionally, the generation of quantifiable metrics that can define morphological and signaling features at the cell level will facilitate their entry as parameters into predictive and descriptive multiway modeling analyses.
The inner machinery that governs the individual cell exists on a separate temporal and spatial scale as compared to the interactions those cells have with their local microenvironment. Imaging, particularly in 3D, is a powerful tool for cell biologists and engineers and has been used to provide spatial information regarding the location of specific structures within the cellular microenvironment. Critical to the throughput and effectiveness of imaging studies, however, is the development of methods for quantitative and automated analysis of 3D multi-spectral images over time (5D). For example, the use of a series of image sequences to automatically track cell lineage during proliferation and migration provides a wealth of information from positioning, shape, cell–cell contacts, motility, ancestry, and temporal relationships that would be difficult to quantify by hand.121 Analysis of the neural stem cell niche using automated image analysis permits the identification of functionally labeled cells, quantifies cell–cell interactions, and determines cellular proximity to specific structures.122 Inhibition of specific interactions within this model results in the detection of quantitative shifts in cell positioning and proliferation. This work must move toward methods of membrane segmentation and feature definition to extract the interactions between neighboring cells as well as the resources present within the ECM for which they compete. Additionally, tissue engineers employ the use of computational modeling to understand the relationship between cellular behavior and physical forces such as fluid flow, stresses, and strains (i.e., within bioreactors).123
The expansion of the image analysis and quantitative modeling field has, however, allowed us to step even farther out from subcellular localization and microenvironment interactions to global tissue organization and development. The coordination of interaction between cells and their matrix defines tissue structure and function, therefore, the spatio-temporal quantification of these interactions will provide insight into population dynamics and cooperation within developing tissues. Several groups have begun to apply quantitative methods to understanding differentiation, morphogenesis, and remodeling by developing models that predict the sprouting location of branching tubules in 3D type I collagen culture based upon the geometry of the starting structure.124 Another approach based on graph theory and machine learning when applied to tissue structure enables the extraction of parameters that define function and dysfunction in terms of the structural organization of cell nuclei and aids pathology diagnosis.125–128 These cell graphs have also recently been applied to 3D fluorescent images of compacting type I collagen hydrogels, with encapsulated MSC, resulting in the extraction of cellular patterns that govern the spatio-temporal progression of tissue organization in the early stages of MSC culture, matrix remodeling, and differentiation (Lund et al., recently submitted). To further describe the network dynamics of developing tissues, 3D cell-graphs can be enhanced through the quantification of the microenvironment. This method allows for the tracking of ECM dynamics, influenced by MSC, at early time points to identify critical stages of type I collagen remodeling and organization. These stages represent distinct phases with late remodeling events in untreated cells clustering strongly with early phases of remodeling by differentiating cells (Bilgin et al., recently submitted). These methods, taken together, allow us to link engineered tissue structure with tissue maturation and cellular differentiation at multiple scales within a developing tissue and to extract metrics that define these dynamic interactions. Macroscopic tissue spatial distributions are not random. The specific structures present within tissues are developed to support specific function, and therefore understanding the microstructure of developing and diseased tissue will be predictive of function.
The quantitative metrics that emerge from these fields can help to define design targets for efficient engineering of the cell–biomaterial interface and will provide an important and quantifiable link between biological hypotheses and biomaterial performance (Fig. 1). Understanding the early events, both structural and biochemical, that are predictive of long-term functional outputs will increase the throughput, cost-effectiveness, and consistency of biomaterial development by decreasing the search space. Moving forward we must further facilitate the interaction between these different mindsets to establish a new paradigm for the tissue engineering approach.
Moving Forward
There is, however, a data bottleneck in place hindering the implementation of these techniques. Discrepancies exist between the reductionist methodology that reduces complexity by considering fewer data points, and computational techniques that become rigorous only as data volume increases. Biologists have successfully reduced biological complexity to an endless variety of minute parts; however, our ability to translate the minutia of signaling, protein folding, and gene expression into the complexity of development, differentiation, and disease progression depends on understanding their complex multiscale relationships. Computational techniques such as data mining, knowledge discovery, and machine learning can be developed to discover such relationships; however, they require large volumes of multimodal data to be rigorous, which is currently limited by the reductionist paradigm.
By both adapting traditional 2D assays and creating new methods for analysis that capitalize on imaging modalities, biologists and engineers can approach questions of cellular function and dysfunction as a 3D system. However, to do so in a manner that supports the modeling and rigorous statistical standards of computer science we must also engineer methods of high-throughput that can expand biochemical and imaging outputs into large data sets sampled over time and on different scales. In breaking this data barrier there comes an additional practical complexity where the “fast” algorithms developed for current problems will not be fast enough. Linear time algorithms will not be able to handle the quantity and complexity of data that can be generated by advancements in biological and engineering throughput. Genomics data, for example, consume 3–4 mega terabytes of memory and require sublinear (e.g., logarithmic) time algorithms. Another computational challenge that comes from breaking the data bottleneck is the difficulty of representing multimodal, multiscale data in such a way that fast knowledge extraction and data mining algorithms can operate. Techniques for multiway data analysis (e.g., tensor decompositions) can be deployed for such representation and analysis; however, their computational complexity prohibits their applicability. Thus, the challenge in designing fast knowledge extraction and data mining algorithms to operate on and represent this complex data set is to generate novel algorithms that efficiently run through and integrate complex, multiscale data (i.e., tensor analysis). A coordinated interdisciplinary effort will be required to address all the components of this proposed approach and will generate a multiscale analysis of cell and tissue organization and interaction over time. Descriptive and predictive models of this process will lead to improved efficiencies in material optimization for engineered tissue products and increased control over their ultimate structure, function and viability in vivo.
Conclusions
Stem cells present an incredibly flexible technology that has application in tissues as varied as fat, nerve, and bone and disease states from diabetes to cancer and Crohn's disease. However, accelerating their adoption in the clinical setting must be done carefully, to avoid detrimental outcomes that could damage long-term clinical acceptance. The rigorous characterization of stem cell behavior, as a function of their various tissue sources, is necessary to ensure safety and efficacy in vivo. However, the intimate relationship between stem cells and their 3D extracellular environment must be considered in this analysis, and to this point, doing so remains technologically challenging. The biasing of stem cell function, which occurs with traditional methods of 2D culture, leads to unpredictable in vivo results that hamper translation into the clinic. Developing novel methods to test mechanistic hypotheses in engineered 3D microenvironments will guide the translation of the control present in the native stem cell niche to functional tissue engineering products and create more predictable, consistent, and safe technologies.
Acknowledgments
National Institutes of Health Grant #RO1 EB008016 and #RO1 AR053231.
Disclosure Statement
No competing financial interests exist.
References
- 1.Morrison S.J. Spradling A.C. Stem cells and niches: mechanisms that promote stem cell maintenance throughout life. Cell. 2008;132:598. doi: 10.1016/j.cell.2008.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fuchs E. Tumbar T. Guasch G. Socializing with the neighbors: stem cells and their niche. Cell. 2004;116:769. doi: 10.1016/s0092-8674(04)00255-7. [DOI] [PubMed] [Google Scholar]
- 3.Chugh A.R. Zuba-Surma E.K. Dawn B. Bone marrow-derived mesenchymal stems cells and cardiac repair. Minerva Cardioangiol. 2009;57:185. [PubMed] [Google Scholar]
- 4.Dill T. Schachinger V. Rolf A. Mollmann S. Thiele H. Tillmanns H. Assmus B. Dimmeler S. Zeiher A.M. Hamm C. Intracoronary administration of bone marrow-derived progenitor cells improves left ventricular function in patients at risk for adverse remodeling after acute ST-segment elevation myocardial infarction: results of the reinfusion of enriched progenitor cells and infarct remodeling in acute myocardial infarction study (REPAIR-AMI) cardiac magnetic resonance imaging substudy. Am Heart J. 2009;157:541. doi: 10.1016/j.ahj.2008.11.011. [DOI] [PubMed] [Google Scholar]
- 5.Gan Y. Dai K. Zhang P. Tang T. Zhu Z. Lu J. The clinical use of enriched bone marrow stem cells combined with porous beta-tricalcium phosphate in posterior spinal fusion. Biomaterials. 2008;29:3973. doi: 10.1016/j.biomaterials.2008.06.026. [DOI] [PubMed] [Google Scholar]
- 6.Hahn J.Y. Cho H.J. Kang H.J. Kim T.S. Kim M.H. Chung J.H. Bae J.W. Oh B.H. Park Y.B. Kim H.S. Pre-treatment of mesenchymal stem cells with a combination of growth factors enhances gap junction formation, cytoprotective effect on cardiomyocytes, and therapeutic efficacy for myocardial infarction. J Am Coll Cardiol. 2008;51:933. doi: 10.1016/j.jacc.2007.11.040. [DOI] [PubMed] [Google Scholar]
- 7.Perez-Albuerne E.D. Eapen M. Klein J. Gross T.J. Lipton J.M. Baker K.S. Woolfrey A. Kamani N. Outcome of unrelated donor stem cell transplantation for children with severe aplastic anemia. Br J Haematol. 2008;141:216. doi: 10.1111/j.1365-2141.2008.07030.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Duncan C.N. Buonanno M.R. Barry E.V. Myers K. Peritz D. Lehmann L. Bronchiolitis obliterans following pediatric allogeneic hematopoietic stem cell transplantation. Bone Marrow Transplant. 2008;41:971. doi: 10.1038/bmt.2008.19. [DOI] [PubMed] [Google Scholar]
- 9.Bjorklund L.M. Sanchez-Pernaute R. Chung S. Andersson T. Chen I.Y. McNaught K.S. Brownell A.L. Jenkins B.G. Wahlestedt C. Kim K.S. Isacson O. Embryonic stem cells develop into functional dopaminergic neurons after transplantation in a Parkinson rat model. Proc Natl Acad Sci USA. 2002;99:2344. doi: 10.1073/pnas.022438099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Erdo F. Buhrle C. Blunk J. Hoehn M. Xia Y. Fleischmann B. Focking M. Kustermann E. Kolossov E. Hescheler J. Hossmann K.A. Trapp T. Host-dependent tumorigenesis of embryonic stem cell transplantation in experimental stroke. J Cereb Blood Flow Metab. 2003;23:780. doi: 10.1097/01.WCB.0000071886.63724.FB. [DOI] [PubMed] [Google Scholar]
- 11.Reubinoff B.E. Pera M.F. Fong C.Y. Trounson A. Bongso A. Embryonic stem cell lines from human blastocysts: somatic differentiation in vitro. Nat Biotechnol. 2000;18:399. doi: 10.1038/74447. [DOI] [PubMed] [Google Scholar]
- 12.Amariglio N. Hirshberg A. Scheithauer B.W. Cohen Y. Loewenthal R. Trakhtenbrot L. Paz N. Koren-Michowitz M. Waldman D. Leider-Trejo L. Toren A. Constantini S. Rechavi G. Donor-derived brain tumor following neural stem cell transplantation in an ataxia telangiectasia patient. PLoS Med. 2009;6:e1000029. doi: 10.1371/journal.pmed.1000029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Comerford M. (March, 2009) Tumors after attempted stem cell therapy highlight importance of rigorous standards before clinical treatment: ISSCR calls for adherence to recent guidelines. Int Soc Stem Cell Res Press Release. 2009 Apr 11; Retrieved on. [Google Scholar]
- 14.Pittenger M.F. Mackay A.M. Beck S.C. Jaiswal R.K. Douglas R. Mosca J.D. Moorman M.A. Simonetti D.W. Craig S. Marshak D.R. Multilineage potential of adult human mesenchymal stem cells. Science. 1999;284:143. doi: 10.1126/science.284.5411.143. [DOI] [PubMed] [Google Scholar]
- 15.De Bari C. Dell'Accio F. Tylzanowski P. Luyten F.P. Multipotent mesenchymal stem cells from adult human synovial membrane. Arthritis Rheum. 2001;44:1928. doi: 10.1002/1529-0131(200108)44:8<1928::AID-ART331>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 16.Zuk P.A. Zhu M. Mizuno H. Huang J. Futrell J.W. Katz A.J. Benhaim P. Lorenz H.P. Hedrick M.H. Multilineage cells from human adipose tissue: implications for cell-based therapies. Tissue Eng. 2001;7:211. doi: 10.1089/107632701300062859. [DOI] [PubMed] [Google Scholar]
- 17.Sabatini F. Petecchia L. Tavian M. Jodon de Villeroche V. Rossi G.A. Brouty-Boye D. Human bronchial fibroblasts exhibit a mesenchymal stem cell phenotype and multilineage differentiating potentialities. Lab Invest. 2005;85:962. doi: 10.1038/labinvest.3700300. [DOI] [PubMed] [Google Scholar]
- 18.Seo B.M. Miura M. Gronthos S. Bartold P.M. Batouli S. Brahim J. Young M. Robey P.G. Wang C.Y. Shi S. Investigation of multipotent postnatal stem cells from human periodontal ligament. Lancet. 2004;364:149. doi: 10.1016/S0140-6736(04)16627-0. [DOI] [PubMed] [Google Scholar]
- 19.da Silva Meirelles L. Chagastelles P.C. Nardi N.B. Mesenchymal stem cells reside in virtually all post-natal organs and tissues. J Cell Sci. 2006;119:2204. doi: 10.1242/jcs.02932. [DOI] [PubMed] [Google Scholar]
- 20.Yamanaka S. A fresh look at iPS cells. Cell. 2009;137:13. doi: 10.1016/j.cell.2009.03.034. [DOI] [PubMed] [Google Scholar]
- 21.Lukashev M.E. Werb Z. ECM signalling: orchestrating cell behaviour and misbehaviour. Trends Cell Biol. 1998;8:437. doi: 10.1016/s0962-8924(98)01362-2. [DOI] [PubMed] [Google Scholar]
- 22.Chen X.D. Dusevich V. Feng J.Q. Manolagas S.C. Jilka R.L. Extracellular matrix made by bone marrow cells facilitates expansion of marrow-derived mesenchymal progenitor cells and prevents their differentiation into osteoblasts. J Bone Miner Res. 2007;22:1943. doi: 10.1359/jbmr.070725. [DOI] [PubMed] [Google Scholar]
- 23.Huang S. Chen C.S. Ingber D.E. Control of cyclin D1, p27(Kip1), and cell cycle progression in human capillary endothelial cells by cell shape and cytoskeletal tension. Mol Biol Cell. 1998;9:3179. doi: 10.1091/mbc.9.11.3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dike L.E. Chen C.S. Mrksich M. Tien J. Whitesides G.M. Ingber D.E. Geometric control of switching between growth, apoptosis, and differentiation during angiogenesis using micropatterned substrates. In Vitro Cell Dev Biol Anim. 1999;35:441. doi: 10.1007/s11626-999-0050-4. [DOI] [PubMed] [Google Scholar]
- 25.McBeath R. Pirone D.M. Nelson C.M. Bhadriraju K. Chen C.S. Cell shape, cytoskeletal tension, and RhoA regulate stem cell lineage commitment. Dev Cell. 2004;6:483. doi: 10.1016/s1534-5807(04)00075-9. [DOI] [PubMed] [Google Scholar]
- 26.Tanentzapf G. Devenport D. Godt D. Brown N.H. Integrin-dependent anchoring of a stem-cell niche. Nat Cell Biol. 2007;9:1413. doi: 10.1038/ncb1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tsai R.Y. McKay R.D. Cell contact regulates fate choice by cortical stem cells. J Neurosci. 2000;20:3725. doi: 10.1523/JNEUROSCI.20-10-03725.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kiger A.A. White-Cooper H. Fuller M.T. Somatic support cells restrict germline stem cell self-renewal and promote differentiation. Nature. 2000;407:750. doi: 10.1038/35037606. [DOI] [PubMed] [Google Scholar]
- 29.Purpura K.A. Aubin J.E. Zandstra P.W. Sustained in vitro expansion of bone progenitors is cell density dependent. Stem Cells. 2004;22:39. doi: 10.1634/stemcells.22-1-39. [DOI] [PubMed] [Google Scholar]
- 30.Zandstra P.W. Le H.V. Daley G.Q. Griffith L.G. Lauffenburger D.A. Leukemia inhibitory factor (LIF) concentration modulates embryonic stem cell self-renewal and differentiation independently of proliferation. Biotechnol Bioeng. 2000;69:607. [PubMed] [Google Scholar]
- 31.Javazon E.H. Colter D.C. Schwarz E.J. Prockop D.J. Rat marrow stromal cells are more sensitive to plating density and expand more rapidly from single-cell-derived colonies than human marrow stromal cells. Stem Cells. 2001;19:219. doi: 10.1634/stemcells.19-3-219. [DOI] [PubMed] [Google Scholar]
- 32.Calvi L.M. Adams G.B. Weibrecht K.W. Weber J.M. Olson D.P. Knight M.C. Martin R.P. Schipani E. Divieti P. Bringhurst F.R. Milner L.A. Kronenberg H.M. Scadden D.T. Osteoblastic cells regulate the haematopoietic stem cell niche. Nature. 2003;425:841. doi: 10.1038/nature02040. [DOI] [PubMed] [Google Scholar]
- 33.Zhang J. Niu C. Ye L. Huang H. He X. Tong W.G. Ross J. Haug J. Johnson T. Feng J.Q. Harris S. Wiedemann L.M. Mishina Y. Li L. Identification of the haematopoietic stem cell niche and control of the niche size. Nature. 2003;425:836. doi: 10.1038/nature02041. [DOI] [PubMed] [Google Scholar]
- 34.Palmer T.D. Willhoite A.R. Gage F.H. Vascular niche for adult hippocampal neurogenesis. J Comp Neurol. 2000;425:479. doi: 10.1002/1096-9861(20001002)425:4<479::aid-cne2>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 35.Gottschling S. Eckstein V. Saffrich R. Jonas A. Uhrig M. Krause U. Seckinger A. Miesala K. Horsch K. Straub B. K. Ho A.D. Primitive and committed human hematopoietic progenitor cells interact with primary murine neural cells and are induced to undergo self-renewing cell divisions. Exp Hematol. 2007;35:1858. doi: 10.1016/j.exphem.2007.06.013. [DOI] [PubMed] [Google Scholar]
- 36.Yamashita Y.M. Fuller M.T. Asymmetric centrosome behavior and the mechanisms of stem cell division. J Cell Biol. 2008;180:261. doi: 10.1083/jcb.200707083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yamashita Y.M. Fuller M.T. Asymmetric stem cell division and function of the niche in the Drosophila male germ line. Int J Hematol. 2005;82:377. doi: 10.1532/IJH97.05097. [DOI] [PubMed] [Google Scholar]
- 38.Rehfeldt F. Engler A.J. Eckhardt A. Ahmed F. Discher D.E. Cell responses to the mechanochemical microenvironment—implications for regenerative medicine and drug delivery. Adv Drug Deliv Rev. 2007;59:1329. doi: 10.1016/j.addr.2007.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Patwari P. Lee R.T. Mechanical control of tissue morphogenesis. Circ Res. 2008;103:234. doi: 10.1161/CIRCRESAHA.108.175331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Engler A.J. Sen S. Sweeney H.L. Discher D.E. Matrix elasticity directs stem cell lineage specification. Cell. 2006;126:677. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- 41.Ruiz S.A. Chen C.S. Emergence of patterned stem cell differentiation within multicellular structures. Stem Cells. 2008;26:2921. doi: 10.1634/stemcells.2008-0432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Glossop J.R. Cartmell S.H. Effect of fluid flow-induced shear stress on human mesenchymal stem cells: differential gene expression of IL1B and MAP3K8 in MAPK signaling. Gene Expr Patterns. 2009;9:381. doi: 10.1016/j.gep.2009.01.001. [DOI] [PubMed] [Google Scholar]
- 43.Obi S. Yamamoto K. Shimizu N. Kumagaya S. Masumura T. Sokabe T. Asahara T. Ando J. Fluid shear stress induces arterial differentiation of endothelial progenitor cells. J Appl Physiol. 2009;106:203. doi: 10.1152/japplphysiol.00197.2008. [DOI] [PubMed] [Google Scholar]
- 44.Haudenschild A.K. Hsieh A.H. Kapila S. Lotz J.C. Pressure and distortion regulate human mesenchymal stem cell gene expression. Ann Biomed Eng. 2009;37:492. doi: 10.1007/s10439-008-9629-2. [DOI] [PubMed] [Google Scholar]
- 45.Kisiday J., Frisbie D.D., McIlwraith W., Grodzinsky A. Dynamic compression stimulates proteoglycan synthesis by mesenchymal stem cells in the absence of chondrogenic cytokines. Tissue Eng A. 2009 doi: 10.1089/ten.tea.2008.0357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ward D.F., Jr. Salasznyk R.M. Klees R.F. Backiel J. Agius P. Bennett K. Boskey A. Plopper G.E. Mechanical strain enhances extracellular matrix-induced gene focusing and promotes osteogenic differentiation of human mesenchymal stem cells through an extracellular-related kinase-dependent pathway. Stem Cells Dev. 2007;16:467. doi: 10.1089/scd.2007.0034. [DOI] [PubMed] [Google Scholar]
- 47.Kook S.H. Son Y.O. Choi K.C. Lee H.J. Chung W.T. Hwang I.H. Lee J.C. Cyclic mechanical stress suppresses myogenic differentiation of adult bovine satellite cells through activation of extracellular signal-regulated kinase. Mol Cell Biochem. 2008;309:133. doi: 10.1007/s11010-007-9651-y. [DOI] [PubMed] [Google Scholar]
- 48.Shimizu N. Yamamoto K. Obi S. Kumagaya S. Masumura T. Shimano Y. Naruse K. Yamashita J.K. Igarashi T. Ando J. Cyclic strain induces mouse embryonic stem cell differentiation into vascular smooth muscle cells by activating PDGF receptor beta. J Appl Physiol. 2008;104:766. doi: 10.1152/japplphysiol.00870.2007. [DOI] [PubMed] [Google Scholar]
- 49.Simmons C.A. Matlis S. Thornton A.J. Chen S. Wang C.Y. Mooney D.J. Cyclic strain enhances matrix mineralization by adult human mesenchymal stem cells via the extracellular signal-regulated kinase (ERK1/2) signaling pathway. J Biomech. 2003;36:1087. doi: 10.1016/s0021-9290(03)00110-6. [DOI] [PubMed] [Google Scholar]
- 50.Liu J. Zhao Z. Li J. Zou L. Shuler C. Zou Y. Huang X. Li M. Wang J. Hydrostatic pressures promote initial osteodifferentiation with ERK1/2 not p38 MAPK signaling involved. J Cell Biochem. 2009;107:224. doi: 10.1002/jcb.22118. [DOI] [PubMed] [Google Scholar]
- 51.Griffith L.G. Swartz M.A. Capturing complex 3D tissue physiology in vitro. Nat Rev Mol Cell Biol. 2006;7:211. doi: 10.1038/nrm1858. [DOI] [PubMed] [Google Scholar]
- 52.Rhee S. Grinnell F. Fibroblast mechanics in 3D collagen matrices. Adv Drug Deliv Rev. 2007;59:1299. doi: 10.1016/j.addr.2007.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Emerman J.T. Pitelka D.R. Maintenance and induction of morphological differentiation in dissociated mammary epithelium on floating collagen membranes. In Vitro. 1977;13:316. doi: 10.1007/BF02616178. [DOI] [PubMed] [Google Scholar]
- 54.Lund A.W. Stegemann J.P. Plopper G.E. Inhibition of ERK promotes collagen gel compaction and fibrillogenesis to amplify the osteogenesis of human mesenchymal stem cells in three dimensional, collagen I culture. Stem Cells Dev. 2008;18:331. doi: 10.1089/scd.2008.0075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Alsberg E. Anderson K.W. Albeiruti A. Franceschi R.T. Mooney D.J. Cell-interactive alginate hydrogels for bone tissue engineering. J Dent Res. 2001;80:2025. doi: 10.1177/00220345010800111501. [DOI] [PubMed] [Google Scholar]
- 56.Martino M.M. Mochizuki M. Rothenfluh D.A. Rempel S.A. Hubbell J.A. Barker T.H. Controlling integrin specificity and stem cell differentiation in 2D and 3D environments through regulation of fibronectin domain stability. Biomaterials. 2009;30:1089. doi: 10.1016/j.biomaterials.2008.10.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Batorsky A. Liao J. Lund A.W. Plopper G.E. Stegemann J.P. Encapsulation of adult human mesenchymal stem cells within collagen-agarose microenvironments. Biotechnol Bioeng. 2005;92:492. doi: 10.1002/bit.20614. [DOI] [PubMed] [Google Scholar]
- 58.Rowe S.L. Stegemann J.P. Interpenetrating collagen-fibrin composite matrices with varying protein contents and ratios. Biomacromolecules. 2006;7:2942. doi: 10.1021/bm0602233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zisch A.H. Schenk U. Schense J.C. Sakiyama-Elbert S.E. Hubbell J.A. Covalently conjugated VEGF—fibrin matrices for endothelialization. J Control Release. 2001;72:101. doi: 10.1016/s0168-3659(01)00266-8. [DOI] [PubMed] [Google Scholar]
- 60.Moon J.J. Lee S.H. West J.L. Synthetic biomimetic hydrogels incorporated with ephrin-A1 for therapeutic angiogenesis. Biomacromolecules. 2007;8:42. doi: 10.1021/bm060452p. [DOI] [PubMed] [Google Scholar]
- 61.Silva E.A. Mooney D.J. Spatiotemporal control of vascular endothelial growth factor delivery from injectable hydrogels enhances angiogenesis. J Thromb Haemost. 2007;5:590. doi: 10.1111/j.1538-7836.2007.02386.x. [DOI] [PubMed] [Google Scholar]
- 62.Hadjipanayi E. Brown R.A. Mudera V. Interface integration of layered collagen scaffolds with defined matrix stiffness: implications for sheet-based tissue engineering. J Tissue Eng Regen Med. 2009;3:230. doi: 10.1002/term.157. [DOI] [PubMed] [Google Scholar]
- 63.Hadjipanayi E. Mudera V. Brown R.A. Guiding cell migration in 3D: a collagen matrix with graded directional stiffness. Cell Motil Cytoskeleton. 2009;66:121. doi: 10.1002/cm.20331. [DOI] [PubMed] [Google Scholar]
- 64.Albrecht D.R. Underhill G.H. Wassermann T.B. Sah R.L. Bhatia S.N. Probing the role of multicellular organization in three-dimensional microenvironments. Nat Methods. 2006;3:369. doi: 10.1038/nmeth873. [DOI] [PubMed] [Google Scholar]
- 65.Lee S.H. Moon J.J. West J.L. Three-dimensional micropatterning of bioactive hydrogels via two-photon laser scanning photolithography for guided 3D cell migration. Biomaterials. 2008;29:2962. doi: 10.1016/j.biomaterials.2008.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Papavasiliou G. Songprawat P. Perez-Luna V. Hammes E. Morris M. Chiu Y.C. Brey E. Three-dimensional patterning of poly(ethylene glycol) hydrogels through surface-initiated photopolymerization. Tissue Eng C Methods. 2008;14:124. doi: 10.1089/ten.tec.2007.0355. [DOI] [PubMed] [Google Scholar]
- 67.Albrecht D.R. Underhill G.H. Mendelson A. Bhatia S.N. Multiphase electropatterning of cells and biomaterials. Lab Chip. 2007;7:702. doi: 10.1039/b701306j. [DOI] [PubMed] [Google Scholar]
- 68.Albrecht D.R. Tsang V.L. Sah R.L. Bhatia S.N. Photo- and electropatterning of hydrogel-encapsulated living cell arrays. Lab Chip. 2005;5:111. doi: 10.1039/b406953f. [DOI] [PubMed] [Google Scholar]
- 69.Alsberg E. Kong H.J. Hirano Y. Smith M.K. Albeiruti A. Mooney D.J. Regulating bone formation via controlled scaffold degradation. J Dent Res. 2003;82:903. doi: 10.1177/154405910308201111. [DOI] [PubMed] [Google Scholar]
- 70.Heidemann W. Jeschkeit S. Ruffieux K. Fischer J.H. Wagner M. Kruger G. Wintermantel E. Gerlach K.L. Degradation of poly(D,L)lactide implants with or without addition of calciumphosphates in vivo. Biomaterials. 2001;22:2371. doi: 10.1016/s0142-9612(00)00424-5. [DOI] [PubMed] [Google Scholar]
- 71.Lee S.H. Miller J.S. Moon J.J. West J.L. Proteolytically degradable hydrogels with a fluorogenic substrate for studies of cellular proteolytic activity and migration. Biotechnol Prog. 2005;21:1736. doi: 10.1021/bp0502429. [DOI] [PubMed] [Google Scholar]
- 72.Seliktar D. Zisch A.H. Lutolf M.P. Wrana J.L. Hubbell J.A. MMP-2 sensitive, VEGF-bearing bioactive hydrogels for promotion of vascular healing. J Biomed Mater Res A. 2004;68:704. doi: 10.1002/jbm.a.20091. [DOI] [PubMed] [Google Scholar]
- 73.Nelson C.M. Geometric control of tissue morphogenesis. Biochim Biophys Acta. 2009;1793:903. doi: 10.1016/j.bbamcr.2008.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chung I.M. Enemchukwu N.O. Khaja S.D. Murthy N. Mantalaris A. Garcia A.J. Bioadhesive hydrogel microenvironments to modulate epithelial morphogenesis. Biomaterials. 2008;29:2637. doi: 10.1016/j.biomaterials.2008.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Rosines E. Schmidt H.J. Nigam S.K. The effect of hyaluronic acid size and concentration on branching morphogenesis and tubule differentiation in developing kidney culture systems: potential applications to engineering of renal tissues. Biomaterials. 2007;28:4806. doi: 10.1016/j.biomaterials.2007.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ehrbar M. Metters A. Zammaretti P. Hubbell J.A. Zisch A.H. Endothelial cell proliferation and progenitor maturation by fibrin-bound VEGF variants with differential susceptibilities to local cellular activity. J Control Release. 2005;101:93. doi: 10.1016/j.jconrel.2004.07.018. [DOI] [PubMed] [Google Scholar]
- 77.Lund A.W. Bush J.A. Plopper G.E. Stegemann J.P. Osteogenic differentiation of mesenchymal stem cells in defined protein beads. J Biomed Mater Res B Appl Biomater. 2008;87:213. doi: 10.1002/jbm.b.31098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bonzani I.C. George J.H. Stevens M.M. Novel materials for bone and cartilage regeneration. Curr Opin Chem Biol. 2006;10:568. doi: 10.1016/j.cbpa.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 79.Bonzani I.C. Adhikari R. Houshyar S. Mayadunne R. Gunatillake P. Stevens M.M. Synthesis of two-component injectable polyurethanes for bone tissue engineering. Biomaterials. 2007;28:423. doi: 10.1016/j.biomaterials.2006.08.026. [DOI] [PubMed] [Google Scholar]
- 80.Stevens B. Yang Y. Mohandas A. Stucker B. Nguyen K.T. A review of materials, fabrication methods, and strategies used to enhance bone regeneration in engineered bone tissues. J Biomed Mater Res B Appl Biomater. 2008;85:573. doi: 10.1002/jbm.b.30962. [DOI] [PubMed] [Google Scholar]
- 81.Stegemann J.P. Hong H. Nerem R.M. Mechanical, biochemical, and extracellular matrix effects on vascular smooth muscle cell phenotype. J Appl Physiol. 2005;98:2321. doi: 10.1152/japplphysiol.01114.2004. [DOI] [PubMed] [Google Scholar]
- 82.Niklason L.E. Langer R.S. Advances in tissue engineering of blood vessels and other tissues. Transpl Immunol. 1997;5:303. doi: 10.1016/s0966-3274(97)80013-5. [DOI] [PubMed] [Google Scholar]
- 83.Liu Y. Ramanath H.S. Wang D.A. Tendon tissue engineering using scaffold enhancing strategies. Trends Biotechnol. 2008;26:201. doi: 10.1016/j.tibtech.2008.01.003. [DOI] [PubMed] [Google Scholar]
- 84.Rotini R. Fini M. Giavaresi G. Marinelli A. Guerra E. Antonioli D. Castagna A. Giardino R. New perspectives in rotator cuff tendon regeneration: review of tissue engineered therapies. Chir Organi Mov. 2008;91:87. doi: 10.1007/s12306-007-0015-2. [DOI] [PubMed] [Google Scholar]
- 85.An Y. Tsang K.K. Zhang H. Potential of stem cell based therapy and tissue engineering in the regeneration of the central nervous system. Biomed Mater. 2006;1:R38. doi: 10.1088/1748-6041/1/2/R02. [DOI] [PubMed] [Google Scholar]
- 86.Schmidt C.E. Leach J.B. Neural tissue engineering: strategies for repair and regeneration. Annu Rev Biomed Eng. 2003;5:293. doi: 10.1146/annurev.bioeng.5.011303.120731. [DOI] [PubMed] [Google Scholar]
- 87.Stoddart M.J. Grad S. Eglin D. Alini M. Cells and biomaterials in cartilage tissue engineering. Regen Med. 2009;4:81. doi: 10.2217/17460751.4.1.81. [DOI] [PubMed] [Google Scholar]
- 88.Kerker J.T. Leo A.J. Sgaglione N.A. Cartilage repair: synthetics and scaffolds: basic science, surgical techniques, and clinical outcomes. Sports Med Arthrosc. 2008;16:208. doi: 10.1097/JSA.0b013e31818cdbaa. [DOI] [PubMed] [Google Scholar]
- 89.Stosich M.S. Moioli E.K. Wu J.K. Lee C.H. Rohde C. Yoursef A.M. Ascherman J. Diraddo R. Marion N.W. Mao J.J. Bioengineering strategies to generate vascularized soft tissue grafts with sustained shape. Methods. 2009;47:116. doi: 10.1016/j.ymeth.2008.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Chan B.P. Leong K.W. Scaffolding in tissue engineering: general approaches and tissue-specific considerations. Eur Spine J. 2008;17(Suppl 4):467. doi: 10.1007/s00586-008-0745-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Morgan S.M. Ainsworth B.J. Kanczler J.M. Babister J.C. Chaudhuri J.B. Oreffo R.O. Formation of a human-derived fat tissue layer in P(DL)LGA hollow fibre scaffolds for adipocyte tissue engineering. Biomaterials. 2009;30:1910. doi: 10.1016/j.biomaterials.2008.12.033. [DOI] [PubMed] [Google Scholar]
- 92.Lutolf M.P. Hubbell J.A. Synthetic biomaterials as instructive extracellular microenvironments for morphogenesis in tissue engineering. Nat Biotechnol. 2005;23:47. doi: 10.1038/nbt1055. [DOI] [PubMed] [Google Scholar]
- 93.Zisch A.H. Lutolf M.P. Ehrbar M. Raeber G.P. Rizzi S.C. Davies N. Schmokel H. Bezuidenhout D. Djonov V. Zilla P. Hubbell J.A. Cell-demanded release of VEGF from synthetic, biointeractive cell ingrowth matrices for vascularized tissue growth. FASEB J. 2003;17:2260. doi: 10.1096/fj.02-1041fje. [DOI] [PubMed] [Google Scholar]
- 94.Bissell M.J. Radisky D.C. Rizki A. Weaver V.M. Petersen O.W. The organizing principle: microenvironmental influences in the normal and malignant breast. Differentiation. 2002;70:537. doi: 10.1046/j.1432-0436.2002.700907.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Shields J.D. Fleury M.E. Yong C. Tomei A.A. Randolph G.J. Swartz M.A. Autologous chemotaxis as a mechanism of tumor cell homing to lymphatics via interstitial flow and autocrine CCR7 signaling. Cancer Cell. 2007;11:526. doi: 10.1016/j.ccr.2007.04.020. [DOI] [PubMed] [Google Scholar]
- 96.Grinnell F. Ho C.H. Tamariz E. Lee D.J. Skuta G. Dendritic fibroblasts in three-dimensional collagen matrices. Mol Biol Cell. 2003;14:384. doi: 10.1091/mbc.E02-08-0493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Cukierman E. Pankov R. Yamada K.M. Cell interactions with three-dimensional matrices. Curr Opin Cell Biol. 2002;14:633. doi: 10.1016/s0955-0674(02)00364-2. [DOI] [PubMed] [Google Scholar]
- 98.Erler J.T. Weaver V.M. Three-dimensional context regulation of metastasis. Clin Exp Metastasis. 2008 doi: 10.1007/s10585-008-9209-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Damianova R. Stefanova N. Cukierman E. Momchilova A. Pankov R. Three-dimensional matrix induces sustained activation of ERK1/2 via Src/Ras/Raf signaling pathway. Cell Biol Int. 2008;32:229. doi: 10.1016/j.cellbi.2007.08.029. [DOI] [PubMed] [Google Scholar]
- 100.Kang X. Xie Y. Powell H.M. James L.L. Belury M.A. Lannutti J.J. Kniss D.A. Adipogenesis of murine embryonic stem cells in a three-dimensional culture system using electrospun polymer scaffolds. Biomaterials. 2007;28:450. doi: 10.1016/j.biomaterials.2006.08.052. [DOI] [PubMed] [Google Scholar]
- 101.Nur-E-Kamal A.X. Kamal J. Schindler M. Meiners S. Three-dimensional nanofibrillar surfaces promote self-renewal in mouse embryonic stem cells. Stem Cells. 2006;24:426. doi: 10.1634/stemcells.2005-0170. [DOI] [PubMed] [Google Scholar]
- 102.Choe M.M. Tomei A.A. Swartz M.A. Physiological 3D tissue model of the airway wall and mucosa. Nat Protoc. 2006;1:357. doi: 10.1038/nprot.2006.54. [DOI] [PubMed] [Google Scholar]
- 103.Dixon J.B. Raghunathan S. Swartz M.A. A tissue-engineered model of the intestinal lacteal for evaluating lipid transport by lymphatics. Biotechnol Bioeng. 2009;103:1224. doi: 10.1002/bit.22337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Moreau J.E. Anderson K. Mauney J.R. Nguyen T. Kaplan D.L. Rosenblatt M. Tissue-engineered bone serves as a target for metastasis of human breast cancer in a mouse model. Cancer Res. 2007;67:10304. doi: 10.1158/0008-5472.CAN-07-2483. [DOI] [PubMed] [Google Scholar]
- 105.Fischbach C. Chen R. Matsumoto T. Schmelzle T. Brugge J.S. Polverini P.J. Mooney D.J. Engineering tumors with 3D scaffolds. Nat Methods. 2007;4:855. doi: 10.1038/nmeth1085. [DOI] [PubMed] [Google Scholar]
- 106.Blinov M.L. Ruebenacker O. Moraru I.I. Complexity and modularity of intracellular networks: a systematic approach for modelling and simulation. IET Syst Biol. 2008;2:363. doi: 10.1049/iet-syb:20080092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Bennett K.P. Bergeron C. Acar E. Klees R.F. Vandenberg S.L. Yener B. Plopper G.E. Proteomics reveals multiple routes to the osteogenic phenotype in mesenchymal stem cells. BMC Genomics. 2007;8:380. doi: 10.1186/1471-2164-8-380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Yener B. Acar E. Aguis P. Bennett K. Vandenberg S.L. Plopper G.E. Multiway modeling and analysis in stem cell systems biology. BMC Syst Biol. 2008;2:63. doi: 10.1186/1752-0509-2-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Rangamani P. Iyengar R. Modelling spatio-temporal interactions within the cell. J Biosci. 2007;32:157. doi: 10.1007/s12038-007-0014-3. [DOI] [PubMed] [Google Scholar]
- 110.Karlebach G. Shamir R. Modelling and analysis of gene regulatory networks. Nat Rev Mol Cell Biol. 2008;9:770. doi: 10.1038/nrm2503. [DOI] [PubMed] [Google Scholar]
- 111.Aldridge B.B. Burke J.M. Lauffenburger D.A. Sorger P.K. Physicochemical modelling of cell signalling pathways. Nat Cell Biol. 2006;8:1195. doi: 10.1038/ncb1497. [DOI] [PubMed] [Google Scholar]
- 112.Gao S.Y. Lees J.G. Wong J.C. Croll T.I. George P. Cooper-White J.J. Tuch B.E. Modeling the adhesion of human embryonic stem cells to poly(lactic-co-glycolic acid) surfaces in a 3D environment. J Biomed Mater Res A. 2009 doi: 10.1002/jbm.a.32401. [ePub head of print.] [DOI] [PubMed] [Google Scholar]
- 113.Giurumescu C.A. Sternberg P.W. Asthagiri A.R. Intercellular coupling amplifies fate segregation during Caenorhabditis elegans vulval development. Proc Natl Acad Sci USA. 2006;103:1331. doi: 10.1073/pnas.0506476103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Kodiha M. Brown C.M. Stochaj U. Analysis of signaling events by combining high-throughput screening technology with computer-based image analysis. Sci Signal. 2008;1:l2. doi: 10.1126/scisignal.137pl2. [DOI] [PubMed] [Google Scholar]
- 115.Albeck J.G. Burke J.M. Aldridge B.B. Zhang M. Lauffenburger D.A. Sorger P.K. Quantitative analysis of pathways controlling extrinsic apoptosis in single cells. Mol Cell. 2008;30:11. doi: 10.1016/j.molcel.2008.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Lazzara M.J. Lauffenburger D.A. Quantitative modeling perspectives on the ErbB system of cell regulatory processes. Exp Cell Res. 2009;315:717. doi: 10.1016/j.yexcr.2008.10.033. [DOI] [PubMed] [Google Scholar]
- 117.Checa S. Prendergast P.J. A mechanobiological model for tissue differentiation that includes angiogenesis: a lattice-based modeling approach. Ann Biomed Eng. 2009;37:129. doi: 10.1007/s10439-008-9594-9. [DOI] [PubMed] [Google Scholar]
- 118.Isaksson H. van Donkelaar C.C. Huiskes R. Ito K. A mechano-regulatory bone-healing model incorporating cell-phenotype specific activity. J Theor Biol. 2008;252:230. doi: 10.1016/j.jtbi.2008.01.030. [DOI] [PubMed] [Google Scholar]
- 119.Isaksson H. Wilson W. van Donkelaar C.C. Huiskes R. Ito K. Comparison of biophysical stimuli for mechano-regulation of tissue differentiation during fracture healing. J Biomech. 2006;39:1507. doi: 10.1016/j.jbiomech.2005.01.037. [DOI] [PubMed] [Google Scholar]
- 120.Anchang B. Sadeh M.J. Jacob J. Tresch A. Vlad M.O. Oefner P.J. Spang R. Modeling the temporal interplay of molecular signaling and gene expression by using dynamic nested effects models. Proc Natl Acad Sci USA. 2009;106:6447. doi: 10.1073/pnas.0809822106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Chen Y. Ladi E. Herzmark P. Robey E. Roysam B. Automated 5-D analysis of cell migration and interaction in the thymic cortex from time-lapse sequences of 3-D multi-channel multi-photon images. J Immunol Methods. 2009;340:65. doi: 10.1016/j.jim.2008.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Shen Q. Wang Y. Kokovay E. Lin G. Chuang S.M. Goderie S.K. Roysam B. Temple S. Adult SVZ stem cells lie in a vascular niche: a quantitative analysis of niche cell-cell interactions. Cell Stem Cell. 2008;3:289. doi: 10.1016/j.stem.2008.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Jungreuthmayer C. Donahue S.W. Jaasma M.J. Al-Munajjed A.A. Zanghellini J. Kelly D.J. O'Brien F.J. A comparative study of shear stresses in collagen-glycosaminoglycan and calcium phosphate scaffolds in bone tissue-engineering bioreactors. Tissue Eng A. 2009;15:1141. doi: 10.1089/ten.tea.2008.0204. [DOI] [PubMed] [Google Scholar]
- 124.Nelson C.M. Vanduijn M.M. Inman J.L. Fletcher D.A. Bissell M.J. Tissue geometry determines sites of mammary branching morphogenesis in organotypic cultures. Science. 2006;314:298. doi: 10.1126/science.1131000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Bilgin C. Demir C. Nagi C. Yener B. Cell-graph mining for breast tissue modeling and classification. Conf Proc IEEE Eng Med Biol Soc. 2007;2007:5311. doi: 10.1109/IEMBS.2007.4353540. [DOI] [PubMed] [Google Scholar]
- 126.Demir C. Gultekin S.H. Yener B. Learning the topological properties of brain tumors. IEEE/ACM Trans Comput Biol Bioinform. 2005;2:262. doi: 10.1109/TCBB.2005.42. [DOI] [PubMed] [Google Scholar]
- 127.Demir C. Gultekin S.H. Yener B. Augmented cell-graphs for automated cancer diagnosis. Bioinformatics. 2005;21(Suppl 2):ii7. doi: 10.1093/bioinformatics/bti1100. [DOI] [PubMed] [Google Scholar]
- 128.Gunduz C. Yener B. Gultekin S.H. The cell graphs of cancer. Bioinformatics. 2004;20(Suppl 1):i145. doi: 10.1093/bioinformatics/bth933. [DOI] [PubMed] [Google Scholar]