Abstract
Fundamental reaction mechanism of cocaine esterase (CocE)-catalyzed hydrolysis of (−)-cocaine and the corresponding free energy profile have been studied by performing pseudobond first-principle quantum mechanical/molecular mechanical (QM/MM)-free energy (FE) calculations. Based on the QM/MM-FE results, the entire hydrolysis reaction consists of four reaction steps, including the nucleophilic attack on carbonyl carbon of (−)-cocaine benzoyl ester by hydroxyl group of Ser117, dissociation of (−)-cocaine benzoyl ester, nucleophilic attack on carbonyl carbon of (−)-cocaine benzoyl ester by water, and finally the dissociation between (−)-cocaine benzoyl group and Ser117 of CocE. The third reaction step involving the nucleophilic attack of a water molecule was found to be rate-determining, which is remarkably different from (−)-cocaine hydrolysis catalyzed by wild-type butyrylcholinesterase (where the formation of prereactive BChE-(−)-cocaine complex is rate-determining) or its mutants containing Tyr332Gly or Tyr332Gly mutation (where the first chemical reaction step is rate-determining). Besides, the role of Asp259 in the catalytic triad of CocE does not follow the general concept of the “charge-relay system” for all serine esterases. The free energy barrier calculated for the rate-determining step of CocE-catalyzed hydrolysis of (−)-cocaine is 17.9 kcal/mol, which is in good agreement with the experimentally derived activation free energy of 16.2 kcal/mol. In present study, where many sodium ions are present, the effects of counter ions are found to be significant in determining the free energy barrier. The finding of the significant effects of counter ions on the free energy barrier may also be valuable in guiding future mechanistic studies on other charged enzymes.
Keywords: Hydrolysis reaction, catalytic mechanism, enzyme, cocaine esterase, QM/MM, free energy perturbation
Introduction
Cocaine is a powerfully addictive stimulant that directly affects the brain and produces a number of toxic effects at high dose. Despite intensive efforts toward education, cocaine abuse continues to be a serious public health problem.1 Recent surveys in the United States show that cocaine was the first on the list of causes of illicit drug related emergency department visits2 and cocaine-related emergency department visits increased 212% between 1995 and 2006, rising from 58 to 181 visits per 100,000 people. The primary toxic effects of cocaine abuse include liver disease, myocardial ischemia, and acute myocardial infarction.3,4 The sequelae of cocaine overdose include generalized clonic-tonic seizures and status epilepticus capable of producing long-term neurological impairment and death.5,6 Unfortunately, there is no effective medication for cocaine abuse and toxicity, and the search for effective and safe treatments continues.7-11
In chemistry, cocaine can have two enantiomers. One is synthetic and biologically inactive (+)-cocaine, and the other is toxic and naturally occurring (−)-cocaine. Although the classic central nervous system receptor-antagonist approach has failed to yield an anticocaine therapeutic against (−)-cocaine, we have developed a proof of principle for a peripheral blocker to accelerate (−)-cocaine metabolism in the circulation12,13 producing biologically inactive metabolites via hydrolysis of (−)-cocaine benzoyl ester.9,11,14-18 The bacterial cocaine esterase (CocE),19 which was originally identified in the bacterium Rhodococcus sp. strain MB1, is particularly promising. This enzyme is one of the two known enzymes that have evolved under direct selection pressure for (−)-cocaine hydrolysis. It metabolizes (−)-cocaine through hydrolysis of (−)-cocaine benzoyl ester and is the most efficient native cocaine hydrolase yet identified.20 In rodent models, CocE can both prevent and reverse extreme cocaine toxicity,21,22 and even robustly protects rodents from the lethal effects of cocaine.23
A more recent mutagenesis study24 shows that although native CocE is unstable at physiological temperature, a designed CocE mutant by a novel computational approach aimed at increasing enzyme thermostability yielded ∼30-fold increase in plasma half-life both in vitro and in vivo, increasing the probability of clinical application of this enzyme for therapeutic use. The efficiency for CocE as an enzyme therapy for cocaine abuse may also be improved by rational design of CocE mutants aiming at accelerating the rate-determining step of the entire catalytic reaction process while the other steps are not impeded by the mutation. To achieve this end, one must first understand the fundamental mechanisms for CocE-catalyzed hydrolysis of (−)-cocaine. However, the detailed mechanistic issues including the rate-determining step as well as the nature of the transition state have not been addressed.
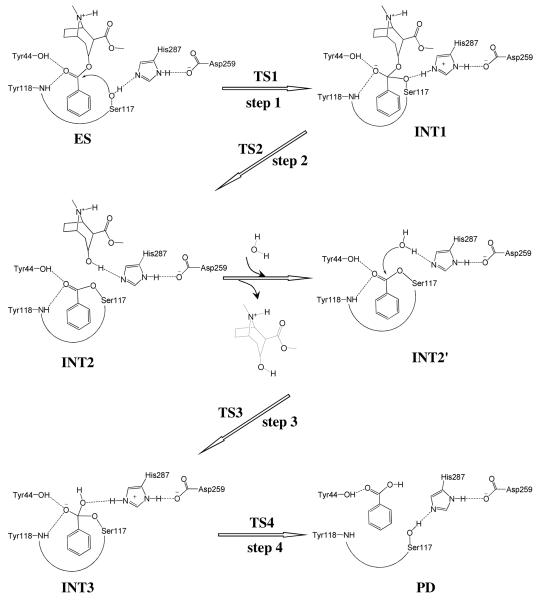
Recently reported X-ray crystallographic20 and mutagenesis25 studies have revealed CocE is a serine carboxylesterase with a catalytic triad formed by Ser117, His287, and Asp259, and with an oxyanion hole formed by the backbone amide of Tyr118 and the hydroxyl group of Tyr44. Mutations on residues in catalytic triad or oxyanion hole essentially remove catalytic activity against (−)-cocaine. The structural properties of the catalytic triad (Ser117, His287, and Asp259) are very similar to that in human butyrylcholinesterase (BChE) which consists of Ser198, His438, and Glu325, although the oxyanion hole (Tyr44 and Tyr118) is significantly different from that of BChE which consists of Gly116, Gly117, and Ala199. It is reasonable to assume (−)-cocaine hydrolysis in CocE undergoes a similar pathway as in BChE,15 where the (−)-cocaine hydrolysis consists of two major stages. The first stage is acylation, leading to covalent bond formation between (−)-cocaine and the enzyme and the departure of ecgonine methyl ester of (−)-cocaine. The second stage is deacylation, resulting in the dissociation of the (−)-cocaine benzoyl ester and enzyme in which a water molecule acts as the nucleophile and the free form of enzyme is restored. Based on our initial computational exploration of a possible CocE-catalyzed (−)-cocaine hydrolysis reaction pathway, we propose a more hypothesis for the detailed reaction mechanism of (−)-cocaine hydrolysis by CocE (Scheme 1).
Scheme 1.
Proposed catalytic mechanism for CocE-catalyzed hydrolysis of (−)-cocaine where Ser117 acts as nucleophile in the first reaction step.
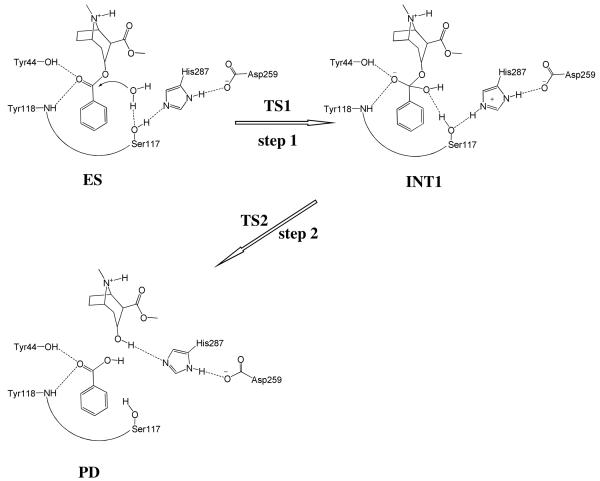
Although it is reasonable to assume that (−)-cocaine hydrolysis in CocE undergoes a similar pathway as in BChE, the proposed mechanism (Scheme 1) may not necessarily be the only choice. The role of the catalytic serine (Ser117) of CocE may not exactly be the same as that (Ser198) of BChE. Alternatively, the Ser117 may act as a general base to activate a water molecule and the activated water functions as the nucleophile to attack the carbonyl carbon of (−)-cocaine benzoyl ester. According to this alternative mechanistic hypothesis, the entire catalytic hydrolysis reaction consists of only two reaction steps, as depicted in Scheme 2. During the first reaction step, while the water oxygen gradually approaches the carbonyl carbon of (−)-cocaine benzoyl ester, a proton of the water molecule gradually transfers to the hydroxyl oxygen of Ser117 side chain and the hydroxyl proton of Ser117 side chain gradually transfers to a nitrogen atom of His287 side chain. During the second reaction step, the proton which the nitrogen atom of His287 side chain accepted from the Ser117 side chain gradually transfers to the ester oxygen of the (−)-cocaine benzoyl ester group while the benzoyl ester bond (C–O) is gradually broken.
Scheme 2.
Possible catalytic mechanism for CocE-catalyzed hydrolysis of (−)-cocaine where Ser117 acts as a general base to activate a water molecule and the activated water molecule acts as a nucleophile in the first reaction step.
In the present study, we have examined the feasibility of the two mechanistic hypotheses and, for the feasible mechanistic hypothesis, carried out first-principles quantum mechanical/molecular mechanical (QM/MM)-free energy (QM/MM-FE) calculations26-29 to uncover the detailed reaction pathway for the CocE-catalyzed (−)-cocaine hydrolysis reaction. In these QM/MM-FE calculations, first-principle QM/MM reaction coordinate calculations were followed by free energy perturbation (FEP) calculations on the protein environment in order to account for the dynamic effects of the protein environment on the free energy barriers for an enzymatic reaction. Our QM/MM simulations are based on the pseudobond first-principle QM/MM approach,26,27,29,30 which has been demonstrated to be a powerful tool in simulating a variety of enzymes,11,28,31-41 and some theoretical predictions31,33 were subsequently confirmed by experimental studies.42-44
In our current study, the computational results clearly reveal the detailed reaction pathway and the corresponding free energy profile for the CocE-catalyzed (−)-cocaine hydrolysis reaction process. The rate-determining reaction step is thereby identified and the role of essential residues including catalytic triad and oxyanion hole are discussed based on the QM/MM-optimized geometries of key configurations in the hydrolysis reaction pathway.
Methods
Structure Preparation
The X-ray crystal structure of CocE (PDB ID: 1JU3)20 and the structure of (−)-cocaine were used in the molecular docking to understand the detailed binding mode of CocE binding with (−)-cocaine. To dock (−)-cocaine molecule into the active site of CocE protein, we used a rigid docking method followed by molecular dynamics (MD) simulation. Molecular geometry of (−)-cocaine was optimized by performing ab initio quantum mechanical calculations using the Gaussian03 program45 at the HF/6-31G* level. The optimized geometry was used to calculate the electrostatic potentials on the molecular surfaces at the same HF/6-31G* level. The calculated electrostatic potentials were used to determine the partial atomic charges by using the standard restrained electrostatic potential (RESP) fitting procedure.46,47 The determined RESP charges were used for the calculation of electrostatic energies in the docking and MD simulation processes. The missing force field parameters for (−)-cocaine were taken from the General Amber Force Field (GAFF) implemented in AMBER8 program.48 The molecular docking for each ligand binding was carried out in the same way as we recently did for studying other protein-ligand binding systems.49,50 We first generated and energy-minimized various molecular orientations and conformations of (−)-cocaine by using Omega51 (Open Eye Scientific Software) and AMBER8 programs. Omega sampling is capable of selecting a ligand conformation similar to that of the targeted X-ray crystal structure by using an appropriate option (the default) including a low-energy cutoff to discard high-energy conformations and a low RMSD value below which two conformations are considered to be similar. FRED52 (OpenEye Scientific Software) docking calculations were carried out using protein structure with all hydrogen atoms and with the binding site definition provided by FRED Receptor program (Open Eye Scientific Software). FRED docking roughly consisted of two steps, i.e. shape fitting and optimization. During the shape fitting, the ligand was placed into a 0.5 Å-resolution grid box encompassing all active-site atoms (including hydrogen atoms) using a smooth Gaussian potential. A series of two optimization filters were then processed, consisting of rigid-body optimization and optimization of the ligand pose in the dihedral angle space. As found in the X-ray crystal structure20 of CocE, the phenyl group of the ligand should first fit between the side chain of Trp166 and Phe261 residues of the binding site. In separate docking runs, the conformational poses of (−)-cocaine that passed the shape-fitting, optimization filters, and interacting with the Trp166 and Phe261 residues were submitted to the MD simulation using AMBER8 program. During the simulation in vacuum, the non-bonded cutoff and the dielectric constant were set to group-based (20 Å cutoff distance) and distance-dependent (ε=4r),53,54 respectively, to mimic the solvent environment.
Finally, the pose with the lowest interaction energy (sum of the electrostatic and van der Waals interaction energy terms) was selected as the best binding mode and then refined by conducting ∼3 ns MD simulation in a fully solvated system. It was solvated in a rectangular box of TIP3P water molecules,55 with a minimum solute wall distance of 10 Å. Thirty-three sodium ions were added to neutralize the charge. As a detailed analysis of the MD-simulated CocE-(−)-cocaine (ES) complex demonstrated that Scheme 2 is not feasible (see below), our QM/MM reaction coordinate calculations were carried out to examine Scheme 1 only. As seen in Scheme 1, there are two reaction stages in the overall CocE-catalyzed hydrolysis of (−)-cocaine. The ecgonine group of (−)-cocaine leaves the system after acylation, resulting in different substrates in the acylation and deacylation stages, i.e. (−)-cocaine molecule in the acylation stage and benzoyl group in the deacylation stage. Consequently, we constructed the structure of INT2′ by removing the ecgonine group of (−)-cocaine out of the QM/MM-optimized INT2 structure. The constructed INT2′ was then relaxed by performing ∼3 ns MD simulation in which the system was also solvated in a rectangular box of TIP3P water molecules55 with a minimum solute wall distance of 10 Å and was neutralized by adding thirty-four sodium ions.
For both acylation and deacylation stages, the last snapshots of the MD simulations were used to prepare pseudobond first-principles QM/MM calculations, as the structures in the last snapshots were close to the average structures simulated. Since we are interested in the reaction center, the water molecules beyond 50 Å of the carbonyl carbon atom (Cζ, Figure 1) of (−)-cocaine benzoyl and all counter ions were removed, leaving the QM/MM system with 3,469 water molecules and a total of 18,967 atoms for acylation and 3,401 water molecules and a total of 18,777 atoms for deacylation. The QM/MM interface has been described by a pseudobond approach.26,27,30 The boundary of QM/MM systems for the two stages are defined in Figure 1. Prior to the QM/MM geometry optimizations, each initial reaction system was energy-minimized with MM method by using the revised AMBER8 program,48 where the convergence criterion is the root-mean-square (RMS) deviation of the energy gradient less than 0.1 kcal·mol−1·Å−1.
Figure 1.
Division of the QM/MM systems for simulating the CocE-catalyzed (−)-cocaine hydrolysis. Atoms in blue are treated by QM method. Three boundary carbon atoms (Cα, colored in red) are treated with the improved pseudobond parameters (ref. 26). All other atoms belong to the MM subsystem.
Minimum Energy Path of the Enzymatic Reaction
With a reaction coordinate driving method and an iterative energy minimization procedure,29 the enzyme reaction path was determined by the pseudobond QM/MM calculations at B3LYP/6-31G*:AMBER level, in which the QM calculations were performed at the B3LYP/6-31G* level of theory by using a modified version of Gaussian0345 and the MM calculations were performed by using a modified version of AMBER program.48 Normal mode analyses were performed to characterize the reactant, intermediates, transition states, and final product. In addition, single-point energy calculations were carried out at the QM/MM(B3LYP/6-31+G*:AMBER) level on the QM/MM-optimized geometries to evaluate the energy barriers. Throughout the QM/MM calculations, the boundary carbon atoms were treated with improved pseudobond parameters.26 No cutoff for nonbonded interactions was used in the QM/MM calculations. For QM subsystem, the convergence criterion for geometry optimizations follows the original Gaussian03 defaults. For MM subsystem, the geometry optimization convergence criterion is when the RMS energy gradient is less than 0.1 kcal·mol−1·Å−1. Prior to QM/MM calculations, the MM subsystem was relaxed by performing ∼500 steps of energy minimization with AMBER8 program. Then, atoms within 20 Å of the Cζ atom of (−)-cocaine benzoyl were allowed to move while all the other atoms outside this range were frozen in all QM/MM calculations, resulting in 2,981 movable atoms in QM/MM calculations for acylation and 3,089 movable atoms in QM/MM calculations for deacylation.
Free Energy Perturbation
After the minimum energy path was determined by the QM/MM calculations, the free energy changes associated with the QM-MM interactions were determined by using free energy perturbation (FEP) method.29 In FEP calculations, sampling of the MM subsystem was carried out with the QM subsystem frozen at different states along the reaction path. The point charges on the frozen QM atoms used in the FEP calculation are those determined by fitting the electrostatic potential (ESP) in the QM part of QM/MM calculation. The total free energy difference between the transition state and the reactant was obtained from the following formula:
| (1) |
where ΔFqm/mm (R → TS) is the free energy change associated with the QM-MM interaction. ΔEqm (R → TS) refers to the QM energy difference between the two QM subsystems, and is the change in contribution from QM subsystem fluctuation to the free energy difference.56 The FEP calculations enabled us to more reasonably determine relative free energy changes due to the QM-MM interaction. Technically, the final (relative) free energy determined by the QM/MM-FE calculations is the QM part of the QM/MM energy (excluding the Columbic interaction energy between the point charges of the MM atoms and the ESP charges of the QM atoms) plus the relative free energy change determined by the FEP calculations. In FEP calculations, the time step used was 2 fs, and bond lengths involving hydrogen atoms were constrained. In sampling of MM subsystem by MD simulations, the temperature was maintained at 300 K. Each of FEP calculation consisted of 50 ps of equilibration and 300 ps of sampling.
The MD simulations and QM/MM-FE calculations were performed on a supercomputer (e.g. IBM X-series Cluster with 340 nodes or 1,360 processors) at University of Kentucky Center for Computational Sciences. The other modeling and computations were carried out on SGI Fuel workstations and a 34-processor IBM x335 Linux cluster in our own lab.
Results and Discussion
Mechanistic insights from MD simulations
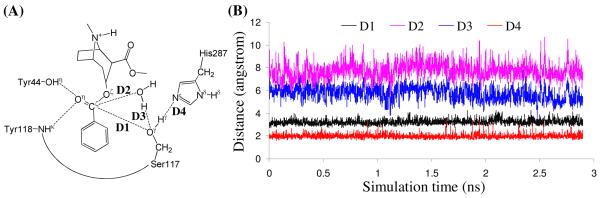
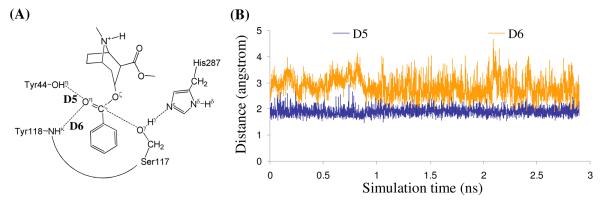
As shown clearly in Schemes 1 and 2, the key difference between the two mechanistic hypotheses exists in the nucleophile, i.e. Ser117 or the water molecule activated by Ser117. Therefore, it is important for examining the feasibility of the two possible catalytic mechanisms to track four most crucial internuclear distances, i.e. D1 to D4 depicted in Figure 2, associate with the two possible nucleophiles. Trace D1 is the distance between the carbonyl carbon (Cζ) of (−)-cocaine benzoyl ester and the hydroxyl oxygen (Oγ) of Ser117 side chain. Trace D2 is the distance between the Cζ atom of (−)-cocaine benzoyl ester and the oxygen of the water molecule closest to the Cζ atom for a given snapshot of the MD-simulated structure. We noted that the water molecule closest to the Cζ atom in one snapshot was not necessarily the same as that in another snapshot. Trace D3 in Figure 2 is the distance between the Oγ atom of Ser117 side chain and the water hydrogen atom closest to the Oγ atom. Trace D4 is the distance between the hydroxyl hydrogen (Hγ) of Ser117 side chain and the Nε atom of His287 side chain.
Figure 2.
Plots of the key internuclear distances (D1 to D4) vs the simulation time in the MD-simulated ES complex.
As seen in Figure 2B, in the MD-simulated most favorable binding structure of CocE-(−)-cocaine (ES) complex, the average value of D2 is ∼7.7±0.7 Å, showing that the water molecules are too far away from the Cζ atom to be the nucleophile. Furthermore, the average value of D3 is ∼5.7±0.6 Å, demonstrating that the interaction between the Ser117 hydroxyl group and the best available water molecule is too weak for Ser117 to activate the water molecule. On the contrary, trace D1 is more stable than D2 and the average value of trace D1 is ∼3.2±0.2 Å, indicating an appropriate distance for the Ser117 hydroxyl performing nucleophilic attack on the Cζ atom. Trace D4 is also more stable than D3 and the average value of D4 is ∼2.0±0.2 Å, showing a strong hydrogen bond between the Ser117 hydroxyl group and the Nε atom of His287 side chain. The MD-simulated CocE-(−)-cocaine (ES) complex revealed the feasibility of His287 being the general base to activate Ser117 in the nucleophilic attack process. Therefore, it is the hydroxyl group of Ser117 side chain, rather than a water molecule, that can act as the nucleophile to initiate the hydrolysis reaction.
We also tried to perform the MD simulations using different, less-favorable initial ES binding structures obtained from molecular docking. The MD simulations using different initial ES structures also eventually led to the similar ES structure (with similar ranges of the D1, D2, D3, and D4 values) suitable for the mechanism associated with Scheme 1. The MD simulations suggest that the (−)-cocaine hydrolysis in CocE may proceed in a similar reaction pathway (Scheme 1) as that in BChE. The possibility of reaction pathway associated with Scheme 2 may be excluded in light of the MD simulations.
Fundamental reaction pathway
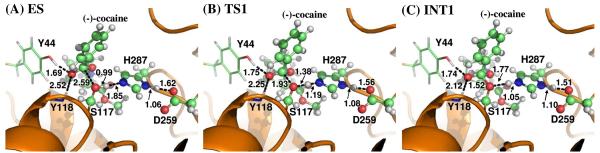
Molecular docking and MD simulation (see Figures 2 and 3) led to a dynamically stable CocE-(−)-cocaine (ES) complex. Our QM/MM reaction coordinate calculations at the B3LYP/6-31G*:AMBER level starting from the MD-simulated CocE-(−)-cocaine (ES) complex revealed that the CocE-catalyzed (−)-cocaine hydrolysis reaction consists of four reaction steps. The first reaction step is the nucleophilic attack on the carbonyl carbon (Cζ) of (−)-cocaine benzoyl ester by Oγ atom in Ser117. The second reaction step is the dissociation between (−)-cocaine benzoyl ester and (−)-cocaine ecgonine methyl ester. The third reaction step is the nucleophilic attack on the carbonyl carbon (Cζ) of (−)-cocaine benzoyl ester by a water molecule. The fourth reaction step is the dissociation between (−)-cocaine benzoyl group and Ser117 of CocE. The optimized geometries of the reactant, intermediates, transition states, and final product are shown in Figures 4 to 7. Below we discuss each of these reaction steps in detail.
Figure 3.
Plots of the key internuclear distances (D5 and D6) vs the simulation time in MD-simulated ES complex.
Figure 4.
Key configurations for step 1, the nucleophilic attack by Oγ atom of Ser117. The geometries were optimized at QM/MM(B3LYP/6-31G*:AMBER) level. The key distances in the figure are in angstroms. Carbon, oxygen, nitrogen, and hydrogen atoms are colored in green, red, blue, and white, respectively. The backbone of the protein is rendered as a cartoon and colored in orange. The QM atoms are represented as ball and stick, and the surrounding residues rendered as stick. The figures below are represented using the same method.
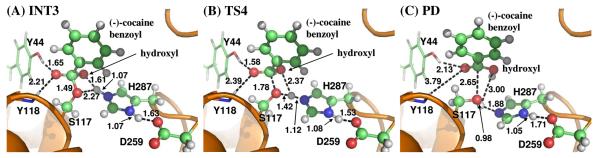
Figure 7.
Key configurations for step 4, the dissociation of (−)-cocaine benzoyl group and Ser117 of CocE. The geometries were optimized at QM/MM(B3LYP/6-31G*:AMBER) level.
Step 1: Nucleophilic attack on the Cζ atom by Oγ atom of Ser117
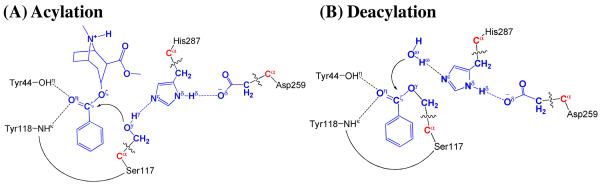
As shown in Figures 2B and 3B, the binding structure (ES) of (−)-cocaine molecule in CocE active site is quite stable. As discussed above, the stable trace D1 suggests (−)-cocaine is at an appropriate distance for nucleophilic attack by Ser117, the nucleophile in CocE catalytic triad. The strong hydrogen bond between hydroxyl hydrogen (Hγ) of Ser117 and Nε atom of His287 side chain indicated by D4 shows that His287, the general base in CocE catalytic triad, has been positioned and is ready for facilitating the nucleophilic attack process through accepting a proton from the nucleophile. The (−)-cocaine molecule is also stabilized by the oxyanion hole consisting of the backbone amide of Tyr118 and the hydroxyl group of Tyr44. The average value of D5 and D6 shown in Figure 3B are ∼1.9±0.2 Å and ∼2.8±0.4 Å, respectively, suggesting that two hydrogen bonds are formed between the carbonyl oxygen (Oη) of (−)-cocaine and the oxyanion hole. One is a strong hydrogen bond O–Hη⋯Oη between Tyr44 hydroxyl and Oη atom of (−)-cocaine, the other is a weak hydrogen bond N-Hκ⋯Oη between Tyr118 NH group and Oη atom of (−)-cocaine. Obviously, Tyr44 plays a more important role in stabilizing the substrate than Tyr118, the other residue in oxyanion hole, by providing stronger hydrogen bonding.
The nucleophilic attack process then proceeds as the serine hydroxyl oxygen, Oγ atom of Ser117, gradually approaches the Cζ atom of (−)-cocaine molecule. Meanwhile, the serine hydroxyl hydrogen, Hγ atom of Ser117, gradually moves towards the nitrogen (Nε) atom of His287 side chain. Since this reaction step involves the breaking of Oγ–Hγ bond and the forming of both Cζ–Oγ and Nε–Hγ bonds as shown in Scheme 1, the distance between Oγ and Hγ (ROγ–Hγ), the distance between Cζ and Oγ (RCζ–Oγ), and the distance between Nε and Hγ (RNε–Hγ) reflect the nature of chemical reaction step 1. Therefore the reaction coordinate for the current reaction step was set as ROγ–Hγ − RCζ–Oγ − RNε–Hγ. As shown in the QM/MM-optimized geometries (Figure 4), that as the Oγ atom of Ser117 gradually approaches the Cζ atom, the geometry of reactant (ES) in which the Cζ atom is sp2 hybridized and is in a planar geometry with its three attached groups gradually changes into a tetrahedral geometry centering on the sp3 hybridized Cζ atom in an intermediate (INT1) through a transition state (TS1).
Step 2: Dissociation of (−)-cocaine benzoyl ester
In this reaction step, the ecgonine group of (−)-cocaine gradually departs from the (−)-cocaine benzoyl ester group in which the benzoyl ester bond Cζ–Oζ is broken. Meanwhile, the proton (Hγ) attached to Nε atom of His287 side chain transfers to the benzoyl ester oxygen atom (Oζ) of (−)-cocaine. The changes of the distances of RCζ–Oζ, ROζ–Hγ and RNε–Hγ reflect the nature of a dissociation process. Thus the reaction coordinate for current reaction step can be expressed as RCζ–Oζ + RNε–Hγ − ROζ–Hγ.
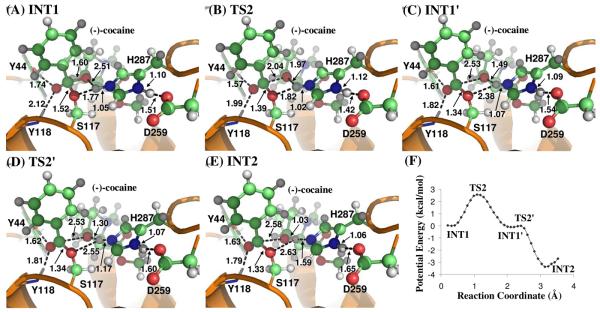
Contrary to what we purposed in Scheme 1 where only one transition state is hypothesized for reaction step 2, the potential energy surface (Figure 5F) which is determined by QM/MM reaction coordinate calculations at B3LYP/6-31G*:AMBER level shows clearly two transition states in current reaction process. The two transition states are labeled as TS2 and TS2′, respectively. The intermediate between the two transition states is labeled as INT1′. The intermediates and transition states of chemical reaction step 2 were verified by full geometry optimizations followed by harmonic normal mode calculations. The QM/MM-optimized geometries of the intermediates and transition states of current reaction process are given in Figure 5.
Figure 5.
(A) to (E) Key configurations for step 2, the dissociation of (−)-cocaine benzoyl ester. The geometries were optimized at QM/MM(B3LYP/6-31G*:AMBER) level. (F) Potential energy of step 2 is obtained at QM/MM(B3LYP/6-31G*:AMBER) level.
In the geometry of INT1 where the serine hydroxyl proton (Hγ) has been transferred to Nε atom of His287 in reaction step 1, the distance (ROγ–Hγ) between Oγ atom of Ser117 and Hγ atom of His287 side chain is 1.77 Å, indicating a very strong hydrogen bond of Nε–Hγ⋯Oγ between Ser117 terminal oxygen and His287 side chain. However, the distance (ROζ–Hγ) between Hγ and the leaving ester oxygen (Oζ) to which Hγ is about to be transferred is 2.51 Å, indicating a weak hydrogen bond between Hγ atom and Oζ atom and an unsuitable condition for proton transfer from Nε atom of His287 to the leaving ester oxygen Oζ atom. In changing from INT1 to INT1′, there are two major structural changes. One is the gradual breaking of the covalent bond Cζ–Oζ (RCζ–Oζ is 1.60 Å in INT1, 2.04 Å in TS2 and 2.53 Å in INT1′). Another is the formation of a hydrogen bond Nε–Hγ⋯Oζ indicated by the shorter and shorter distance ROζ–Hγ in going from INT1 to INT1′ (2.51 Å in INT1, 1.97 Å in TS2 and 1.49 Å in INT1′). In the mean time, the hydrogen bond Nε–Hγ ⋯Oγ formed between the transferring proton (Hγ) and the Oγ atom of Ser117 becomes progressively weaker (ROγ–Hγ is 1.77 Å in INT1, 1.82 Å in TS2 and 2.38 Å in INT1′), which is reasonable as the transferring proton (Hγ) is about to be transferred to the leaving ester oxygen (Oγ) in current reaction step.
The major difference between INT1′ and TS2′ is the position of transferring proton (Hγ) while the distance RCζ–Oζ remains unchanged, indicating that the reaction process associated with TS2′ is primarily the proton (Hγ) transfer from Nε atom of His287 side chain to the leaving ester oxygen (Oζ) atom (ROζ–Hγ is 1.49 Å in INT1′, 1.30 Å in TS2′, and 1.03 Å in INT2; RNε–Hγ is 1.07 Å in INT1′, 1.17 Å in TS2′, and 1.59 Å in INT2). Normally, the phenyl group of (−)-cocaine benzoyl ester is in the same plane of sp2 hybridized carbonyl planar geometry as there is more overlap between the π orbitals which makes the system more stable. In the geometry of INT2 where (−)-cocaine has been broken down into ecgonine methyl ester and benzoyl, the benzoyl carbonyl carbon (Cζ) atom becomes sp2 hybridization again and forms a co-planar shape with the three atoms attached to it. However, the phenyl group in INT2 does not lie in the same plane as the sp2 hybridized carbonyl. This is probably because the hydrophobic pocket where the (−)-cocaine benzoyl sits is too compact for (−)-cocaine benzoyl to rotate.
According to the two transition states given by the QM/MM potential energy surface, i.e., TS2 and TS2′, the proton transfer proceeds not simultaneously with but only after the breaking of C–O covalent bond. Technically, there should be one more reaction step in describing the entire hydrolysis reaction, namely five reaction steps instead of four reaction steps proposed in Scheme 1. However, the calculated energy barrier at B3LYP/6-31G*:AMBER level for the reaction process associated with TS2′ is only ∼0.1 kcal/mol. Such a small energy barrier is eliminated after the fluctuation of MM part is accounted for (see below). Therefore, in the present study we still use the notation used in Scheme 1 to describe the hydrolysis reaction pathway. The reaction step 2 involves both Cζ–Oζ bond breaking and the proton transfer from Nε atom to Oζ atom, and reaction step 3 is still the nucleophilic attack on Cζ atom by water.
Step 3: Nucleophilic attack on Cζ atom by a water molecule
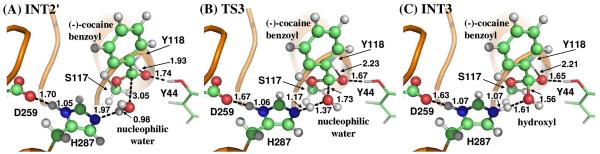
The ecgonine methyl ester was removed from the above-discussed QM/MM-optimized geometry of INT2 to construct the structure of INT2′, which was then relaxed by performing MD simulation. A water molecule close to carbonyl carbon (Cζ) atom was selected as the nucleophile and was treated by QM method. The QM/MM-optimized geometry of INT2′ (Figure 6A) at B3LYP/6-31G*:AMBER level shows that the phenyl group of (−)-cocaine is not completely on the same plane as the sp2 hybridized carbonyl. The dihedral angle between the plane of phenyl group and the plane of sp2 hybridized carbonyl is 23°. This is probably because of the same reason for the not-quite-planar shape of the benzoyl moiety in INT2 that the hydrophobic pocket is too compact for the benzoyl to rotate. Similarly as in the acylation stage, the benzoyl group is also stabilized by the oxyanion hole. As shown in Figure 6A, two strong hydrogen bonds are formed between carbonyl oxygen (Oη) atom of (−)-cocaine and the oxyanion hole in INT2′.
Figure 6.
Key configurations for step 3, the nucleophilic attack on the benzoyl carbonyl carbon atom by a water molecule. The geometries were optimized at QM/MM(B3LYP/6-31G*:AMBER) level.
The current nucleophilic process proceeds in a similar way as in the reaction step 1, which involves the breaking of Oω–Hω bond and the forming of both Cζ–Oω and Nε–Hω bonds. Thus the distances ROω–Hω, RCζ–Oω, and RNε–Hω were chosen to establish the reaction coordinate as ROω–Hω − RCζ–Oω − RNε–Hω for the current reaction step. In proceeding from INT2′ to INT3 through the transition state TS3 (Figure 6), the co-planar geometry changes into tetrahedral centering on the sp3 hybridized carbonyl carbon (Cζ) atom as the nucleophilic water gradually approaches Cζ atom with a spontaneous proton (Hω) transfer from Oω atom of the nucleophilic water to Nε atom of His287 side chain. The QM/MM-optimized geometry of INT3 shows that the nucleophilic attack process is completed with water dissociating into hydroxyl attaching to Cζ atom and a proton (Hω) attaching to Nε atom.
Step 4: Dissociation between (−)-cocaine benzoyl group and Ser117 of CocE
The proton transfer between Nε atom of His287 side chain and Oγ atom of Ser117 side chain and the breaking of covalent bond Cζ–Oγ are involved in the dissociation of benzoyl-enzyme. The changes of the distances RCζ–Oγ, ROγ–Hω, and RNε–Hω reflect the nature of reaction step 4. Thus the reaction coordinate for current reaction step was expressed as RCζ–Oγ + RNε–Hω − ROγ–Hω. Reaction step 4 is similar to reaction step 2, the dissociation of benzoyl ester. In both reaction steps, one type of C–O covalent bond is broken and one proton is transferred from Nε atom of His287 to the oxygen atom of broken C–O covalent bond. The difference between the two reaction steps is that the proton transfer in current reaction step proceeds spontaneously as C–O bond is gradually broken while that in reaction step 2 does not proceed spontaneously. As shown in Figure 7, in the same time of Cζ–Oγ covalent bond breaking, the distance between Oγ atom and Hω atom becomes closer and closer, illustrating a spontaneous proton transfer from Nε atom of His287 side chain to Oγ atom of Ser117.
Energetics
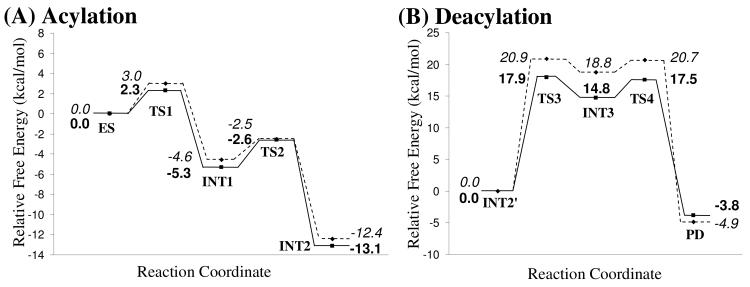
Using the QM/MM-optimized geometries at the QM/MM(B3LYP/6-31G*:AMBER) level, we carried out QM/MM single-point energy calculations at the QM/MM(MP2/6-31+G*:AMBER) level for each geometry along the minimum-energy path for both acylation and deacylation stages of CocE-catalyzed hydrolysis of (−)-cocaine. For each geometry along the minimum-energy path, the ESP charges determined in the QM part of the QM/MM single-point energy calculation were used in subsequent FEP simulations for estimating the free energy changes along the reaction path. Depicted in Figure 8 is the energy profile determined by the QM/MM-FE calculations excluding the zero-point and thermal corrections for the QM subsystem. The values given in the parentheses are the corresponding relative free energies with the zero-point and thermal corrections for the QM subsystem.
Figure 8.
Free energy profile determined by the MP2/6-31+G*:AMBER QM/MM-FE calculations excluding the zero-point and thermal corrections for the QM subsystem. The values in parenthesis are relative free energies including zero-point and thermal corrections for the QM subsystem.
Rate-determining step
As shown in Figure 8, the transition state TS2′ of reaction step 2 is eliminated after the free energy changes of MM part along the reaction path has been applied. The free energy barriers with zero-point and thermal corrections for the QM subsystem of the four reaction steps are 3.0, 2.1, 20.9, and 1.9 kcal/mol, respectively. Obviously, the reaction step 3 with free energy barrier of 20.9 kcal/mol is the rate-determining step of the entire CocE-catalyzed hydrolysis of (−)-cocaine.
It is remarkable to note that the rate-determining step for CocE-catalyzed hydrolysis of (−)-cocaine is different from that for (−)-cocaine hydrolysis catalyzed by BChE. Wild-type BChE has a low catalytic efficiency (kcat = 4.1 min−1 and KM = 4.5 μM)57 against (−)-cocaine because residue Tyr332 hinders the formation of prereactive BChE-(−)-cocaine complex.58 The combined computational and experimental data and analysis revealed that the rate-determining step of (−)-cocaine hydrolysis catalyzed by BChE mutants containing the Tyr332Gly or Tyr332Ala mutation is the first step of the chemical reaction process.9 For this reason, the computational design of high-activity mutants of BChE against (−)-cocaine has focused on the stabilization of the transition state for the first reaction step.9-11,17,58-64 Now that the rate-determining step for CocE-catalyzed hydrolysis of (−)-cocaine is the third reaction step, future computational design of high-activity mutants of CocE against (−)-cocaine should focus on the stabilization of the transition state for the third reaction step.
Effects of counter ions
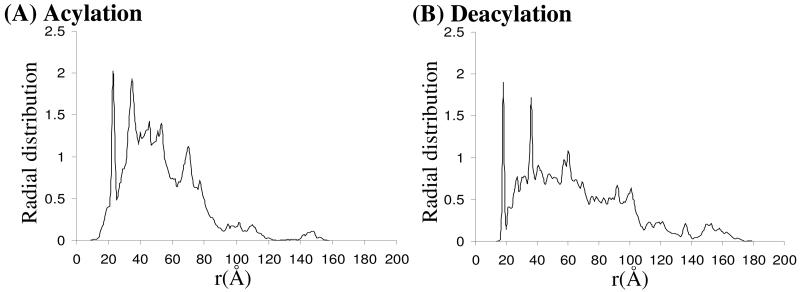
The activation free energy derived from the experimental rate constant (kcat =7.8 s−1)20 by using the conventional transition state theory (CTST)65,66 is 16.2 kcal/mol. By including zero-point and thermal corrections for QM subsystem, the free energy barrier of rate-determining step is ∼20.9 kcal/mol. There is ∼4.7 kcal/mol difference between the experimental activation free energy and the calculated free energy barrier. This is interesting as the free energy barrier determined by pesudobond first-principle QM/MM-FE method is usually very close to the experimentally derived activation free energy. The large number of the counter ions which were not presented in QM/MM calculations draws our attention. There are 33 sodium ions for the acylation stage and 34 sodium ions for the deacylation stage. This large number of counter ions should have effects on QM part, especially in determining energy barriers. A question arises whether the effects caused by counter ions are significant enough to change the hydrolysis pathway revealed by the counter-ions-excluded QM/MM calculations that is discussed above. In order to answer this question, a radial distribution of counter ions is defined as follows
| (2) |
where r is the radius with coordinates of Cζ atom as the origin. Index i runs over all the snapshots of the equilibrated MD trajectory. Index j runs from 0 to unlimited number and is increased by small step size of δ. Nj is the number of counter ions within rj Å of Cζ atom. nsnapshots is the number of snapshots. The radial distribution of counter ions for both acylation and deacylation stages are depicted in Figure 9. As shown clearly in Figure 9, the counter ions are mainly distributed in the range of ∼20 – ∼100 Å to Cζ atom that is far from CocE active site. For both reaction stages, counter ions are not found within ∼15 Å of Cζ atom and, therefore, we conclude that counter ions are not directly involved in the reaction pathway. Therefore, the hydrolysis mechanism described by counter-ions-excluded QM/MM calculations should not be altered in anyway by the effects of counter ions.
Figure 9.
Radial distribution of counter ions (Na+) centered around Cζ atom.
Since the counter ions are so far away from the active site, the van der Waals contributions to energy barrier of hydrolysis reaction is negligible. Only the electrostatic contributions from counter ions would be important to the energy barrier. Thus the electrostatic interactions between counter ions and QM subsystem for each key configuration were calculated with QM method. Such electrostatic interactions of each key configuration could be considered as the approximated correction of counter ions effects to the relative energies. In order to also take account of the fluctuation of counter ions, the coordinates of counter ions were taken out every 10 ps from MD trajectory where the initial structures for QM/MM calculations were taken. A total of 100 snapshots were used for estimating counter ions effects. The estimated shift caused by counter ions effects on relative energies for each key configuration are −0.7±0.4, −0.7±0.6, 0.0±0.4 and −0.7±0.8 kcal/mol in acylation stage from TS1 to INT2′, respectively, and −3.0±0.9, −4.0±1.1, −3.2±0.3 and 1.0±1.2 kcal/mol for each key configuration in deacylation stage from TS3 to PD, respectively. The free energy profile with electrostatic corrections from counter ions is re-plotted in Figure 10. The rate-determining step is still reaction step 3. By including the zero-point and thermal corrections for QM subsystem, the free energy barrier of rate-determining step is 17.9 kcal/mol, which is in good agreement with the experimentally derived activation energy of 16.2 kcal/mol.
Figure 10.
Free energy barriers with (solid line) or without (dashed line) electrostatics corrections from counter ions. The energies without electrostatic corrections from counter ions are in italics while those with corrections are in bold. The zero-point and thermal corrections for the QM subsystem are included.
The effects of counter ions could be important in studying chemical reaction by QM/MM method. Although counter ions may not be directly involved in the reaction mechanism, the interaction between counter ions and QM part is significant in determining free energy barrier of the reaction when many counter ions present in the system.
The Role of catalytic triad and oxyanion hole
According to the mechanism described by our pseudobond first-principle QM/MM-FE calculations, the catalytic triad and oxyanion hole are the most essential factors in the CocE-catalyzed hydrolysis of (−)-cocaine. The first residue in catalytic triad, Ser117, acts as a nucleophile in reaction step 1 and then forms a covalent bond with carbonyl carbon (Cζ) atom of (−)-cocaine benzoyl ester until the end of hydrolysis reaction. The second residue in catalytic triad, His287, serves as a general base accepting the proton from nucleophile to facilitate the two nucleophilic attack processes, i.e., reaction step 1 and step 3. The proton is then donated to the leaving group by His287 in the two dissociation processes, i.e., reaction step 2 and step 4. His287 facilitates either the forming or breaking of the C-O covalent bond in each reaction step. The observations for the two catalytic residues, Ser117 and His287, are basically the same as those of the catalytic triad in other widely studied enzymes. There has been a general concept regarding the role of the third residue in the catalytic triad during the hydrolysis reaction, i.e. Asp259 in CocE. Indeed, carboxylate of the third residue in catalytic triad has been suggested as a so called “charge-relay system”67,68 to serve as the general acid-base catalyst, which is involved in the proton transfer between the catalytic aspartic/glutamic acid and the catalytic histidine. It is of particular interest to know whether Asp259 in CocE is involved in a proton transfer between the catalytic histidine and aspartic acid during the CocE-catalyzed hydrolysis of (−)-cocaine concerned in the present study. The answer to this essential mechanistic question can be gleaned from examining two crucial internuclear distances within His287 and Asp259: one (RNδ–Hδ) is the distance between the Nδ atom and Hδ atom of His287 side chain and the other (RHδ–Oδ) is that between the Hδ atom of His287 side chain and the nearby oxygen Oδ of Asp259 side chain. As seen in Figures 4 to 7, RNδ–Hδ and RHδ–Oδ are no more than 1.12 Å and no less than 1.42 Å, respectively, among the key configurations along the reaction path, revealing a strong hydrogen bond between His287 and Asp259 but not a proton transfer. As suggested by the relatively shorter distance of hydrogen bond Nδ–Hδ⋯Oδ between His287 and Asp259 in transition states of each reaction step, Asp259 stabilizes the transition states through increasing the strength of its hydrogen bond with His287 during the whole hydrolysis reaction. Such catalytic role of Asp259 does not follow the general concept of the “charge-relay system”, but is similar to that of Glu334 in acetylcholinesterase-catalyzed hydrolysis of acetylcholine.32
It is also interesting to know the catalytic role of the oxyanion hole consisting of the backbone amide of Tyr118 and the hydroxyl group of Tyr44. Based on the QM/MM reaction coordinate calculations, throughout the hydrolysis reaction process, the carbonyl oxygen (Oη) of (−)-cocaine forms two hydrogen bonds with oxyanion hole. One is the hydrogen bond of O-Hη⋯Oη with hydroxyl hydrogen (Hη) atom of Tyr44 side chain and the other is the hydrogeon bond of N–Hκ⋯Oη with the backbone NH group (Hκ atom) of Tyr118. As one can see from Figures 4 to 7, the hydrogen bond O–Hη⋯Oη between Oη atom and Tyr44 hydroxyl is very strong throughout the hydrolysis reaction with the distance of ∼1.7 Å. The other hydrogen bond N–Hκ⋯Oη between Oη atom and Tyr118 backbone NH group is relatively weaker than the one with Tyr44 hydroxyl during the hydrolysis reaction. It is weak in ES with the distance of ∼2.5 Å and then becomes strong with the distance of ∼2.0 Å in the subsequent states of hydrolysis reaction. Therefore both hydrogen bonds stabilize the negative charge of carbonyl oxygen (Oη) developing during the hydrolysis reaction, where the primary contribution to the stabilization comes from Tyr44. This is also consistent with the mutagenesis study25 where Tyr44 was mutated to Phe causing the loss of the hydrogen bond with carbonyl oxygen (Oη) of (−)-cocaine and resulting in the complete loss of CocE catalytic activity.
Conclusion
The CocE-catalyzed hydrolysis of (−)-cocaine has been studied by using first-principle QM/MM-FE approach. The detailed reaction pathway has been elucidated. First, (−)-cocaine molecule binds in the CocE active site with the benzoyl group residing in the hydrophobic pocket. A nucleophilic attack on carbonyl carbon (Cζ) of (−)-cocaine benzoyl ester is then carried out by hydroxyl oxygen (Oγ) of Ser117. This process is facilitated by His287 through proton (Hγ) transfer from Ser117 hydroxyl to Nε atom of His287 side chain, which increases the nucleophilicity of the Ser117 hydroxyl. His287 is in turn stabilized by another hydrogen bond formation to Asp259. The Ser117 nucleophile attacks the electron-deficient Cζ atom forming a tetrahedral intermediate, in which the carbonyl oxygen (Oη) of (−)-cocaine with developing negative charge is stabilized by two tyrosine residues in the oxyanion hole. Then His287 donates the proton (Hγ) to the ester oxygen (Oζ) of leaving ecgonine group, completing the acylation stage. Then a water molecule which is activated by His287 initiates the deacylation stage. Similar reaction processes to the acylation stage are repeated in the deacylation stage, and the benzoic acid is released with CocE being restored to its free form.
There are four reaction steps in the hydrolysis reaction. The reaction step 3 where a water molecule attacks Cζ atom is found to be the rate-determining. Although we did not include effects of counter ions in QM/MM calculations, the counter ions are too far away to the active site and thus do not affect the reaction mechanism as described by our counter-ions-excluded QM/MM calculations. The calculated free energy barrier with both estimated correction of counter ions effects and zero-point and thermal corrections for QM subsystem is 17.9 kcal/mol, which is in good agreement with the experimentally derived activation free energy of 16.2 kcal/mol. The effects of counter ions in the present study, where many sodium ions are present, were found to be significant in determining the free energy barrier. The significant effects of counter ions on the free energy barrier found in the present study suggest that one should not ignore the effects of counter ions in computational prediction of energy barriers of an enzymatic reaction associated with a charged enzyme.
The role of catalytic triad and oxyanion hole has also been discussed. The QM/MM-optimized geometries of key configurations of hydrolysis reaction clearly show no proton transfer between His287 and Asp259, which does not follow the general concept of the “charge-relay system”. The QM/MM-optimized geometries indicate that oxyanion hole stabilizes the negative charge of carbonyl oxygen developing during hydrolysis reaction by providing two hydrogen bonds from Tyr44 and Tyr118, respectively. The hydrogen bond with Tyr44 is particularly strong and is the primary factor in stabilizing the carbonyl oxygen (Oη) of (−)-cocaine. This is also consistent with the mutagenesis study where Tyr44 was mutated to Phe causing the loss of the hydrogen bond with carbonyl oxygen (Oη) of (−)-cocaine and resulting in the complete loss of CocE catalytic activity.
The elucidated reaction mechanism of CocE-catalyzed hydrolysis of (−)-cocaine provides details of the chemical reaction, especially the rate-determining step and the nature of transition states. The rate-determining step for CocE-catalyzed hydrolysis of (−)-cocaine is remarkably different from that for (−)-cocaine hydrolysis catalyzed by wild-type BChE (where the formation of prereactive BChE-(−)-cocaine complex is rate determining) or its mutants containing Tyr332Gly or Tyr332Gly mutation (where the first chemical reaction step is rate-determining). Thus, unlike the computational design of high-activity mutants of BChE against (−)-cocaine, efforts aiming at increasing catalytic activity of CocE against (−)-cocaine should focus on the reaction step 3 where nucleophilic attack on Cζ atom is carried out by a water molecule.
Supplementary Material
Acknowledgments
This work was supported in part by NIH (grants R01DA013930 and R01DA025100 to C.-G.Z). J. Liu worked in C.-G. Zhan's laboratory at the University of Kentucky as an exchange graduate student from Central China Normal University. J. Liu thanks A. Goren of Transylvania University (who did sabbatical research in C.-G. Zhan's laboratory) for valuable discussion during the manuscript preparation. The authors also acknowledge the Center for Computational Sciences (CCS) at University of Kentucky for supercomputing time on IBM X-series Cluster with 340 nodes or 1,360 processors.
Footnotes
Supporting Information Available. Absolute QM/MM energies (in Hartree) calculated at the QM/MM(MP2/6-31+G*:AMBER) level, coordinates of QM part of the geometries optimized at the QM/MM(B3LYP/6-31G*:AMBER) level, and complete citations of references 45 and 48. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.National Institute on Drug Abuse Cocaine Abuse and Addiction. (NIH Publication No. 99-4342).NIDA Research Report Series. 2004 (Rev. Nov. 2004) [Google Scholar]
- 2.Substance Abuse and Mental Health Services Administration . Office of Applied Studies. Rockville, MD: 2006. Drug Abuse Warning Network, 2004: National Estimates of Drug-Related Emergency Department Visits. (DAWN Series D-28, DHHS Publication No. SMA 06-4143). [Google Scholar]
- 3.Boelsterli UA, Goldlin C. Arch. Toxicol. 1991;65(5):351–60. doi: 10.1007/BF02284256. [DOI] [PubMed] [Google Scholar]
- 4.Hahn IH, Hoffman RS. Emerg. Med. Clin. N. Am. 2001;19(2):493–510. doi: 10.1016/s0733-8627(05)70197-8. [DOI] [PubMed] [Google Scholar]
- 5.Crumb WJ, Clarkson CW. Biophys. J. 1990;57(3):589–599. doi: 10.1016/S0006-3495(90)82574-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Benowitz NL. Pharmacol. Toxicol. 1993;72(1):3–12. doi: 10.1111/j.1600-0773.1993.tb01331.x. [DOI] [PubMed] [Google Scholar]
- 7.Dickerson TJ, Janda KD. Recent Advances for the Treatment of Cocaine Abuse: Central Nervous System Immunopharmacotherapy. Drug Addiction. 2008:217–229. doi: 10.1208/aapsj070359. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 8.Vocci FJ, Acri J, Elkashef A. Am. J. Psychiat. 2005;162(8):1432–1440. doi: 10.1176/appi.ajp.162.8.1432. [DOI] [PubMed] [Google Scholar]
- 9.Pan Y, Gao D, Yang W, Cho H, Yang G, Tai H-H, Zhan C-G. Proc. Nat. Acad. Sci. U.S.A. 2005;102(46):16656–16661. doi: 10.1073/pnas.0507332102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gao D, Cho H, Yang W, Pan Y, Yang G, Tai H-H, Zhan C-G. Angew. Chem. Int. Ed. 2006;45(4):653–657. doi: 10.1002/anie.200503025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zheng F, Yang W, Ko M-C, Liu J, Cho H, Gao D, Tong M, Tai H-H, Woods JH, Zhan C-G. J. Am. Chem. Soc. 2008;130(36):12148–12155. doi: 10.1021/ja803646t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Landry DW, Zhao K, Yang GX, Glickman M, Georgiadis TM. Science. 1993;259(5103):1899–1901. doi: 10.1126/science.8456315. [DOI] [PubMed] [Google Scholar]
- 13.Mets B, Winger G, Cabrera C, Seo S, Jamdar S, Yang G, Zhao K, Briscoe RJ, Almonte R, Woods JH, Landry DW. Proc. Nat. Acad. Sci. U.S.A. 1998;95(17):10176–10181. doi: 10.1073/pnas.95.17.10176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gorelick DA. Drug Alcohol Depen. 1997;48(3):159–165. doi: 10.1016/s0376-8716(97)00119-1. [DOI] [PubMed] [Google Scholar]
- 15.Zhan C-G, Zheng F, Landry DW. J. Am. Chem. Soc. 2003;125(9):2462–2474. doi: 10.1021/ja020850+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Meijler MM, Kaufmann GF, Qi L, Mee JM, Coyle AR, Moss JA, Wirsching P, Matsushita M, Janda KD. J. Am. Chem. Soc. 2005;127(8):2477–2484. doi: 10.1021/ja043935e. [DOI] [PubMed] [Google Scholar]
- 17.Pan YM, Gao DQ, Yang WC, Cho H, Zhan C-G. J. Am. Chem. Soc. 2007;129(44):13537–13543. doi: 10.1021/ja073724k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rogers CJ, Mee JM, Kaufmann GF, Dickerson TJ, Janda KD. J. Am. Chem. Soc. 2005;127(28):10016–10017. doi: 10.1021/ja053086a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bresler MM, Rosser SJ, Basran A, Bruce NC. Appl. Environ. Microbiol. 2000;66(3):904–908. doi: 10.1128/aem.66.3.904-908.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Larsen NA, Turner JM, Stevens J, Rosser SJ, Basran A, Lerner RA, Bruce NC, Wilson IA. Nat. Struct. Mol. Biol. 2002;9(1):17–21. doi: 10.1038/nsb742. [DOI] [PubMed] [Google Scholar]
- 21.Cooper ZD, Narasimhan D, Sunahara RK, Mierzejewski P, Jutkiewicz EM, Larsen NA, Wilson IA, Landry DW, Woods JH. Mol. Pharmacol. 2006;70(6):1885–1891. doi: 10.1124/mol.106.025999. [DOI] [PubMed] [Google Scholar]
- 22.Ko M-C, Bowen LD, Narasimhan D, Berlin AA, Lukacs NW, Sunahara RK, Cooper ZD, Woods JH. J. Pharmacol. Exp. Ther. 2007;320(2):926–933. doi: 10.1124/jpet.106.114223. [DOI] [PubMed] [Google Scholar]
- 23.Emily MJ, Michelle GB, Ziva DC, Diwahar N, Roger KS, James HW. Ann. Emerg. Med. 2008 [Google Scholar]
- 24.Gao D, Narasimhan DL, Macdonald J, Brim R, Ko M-C, Landry DW, Woods JH, Sunahara RK, Zhan C-G. Mol. Pharmacol. 2009;75(2):318–323. doi: 10.1124/mol.108.049486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Turner JM, Larsen NA, Basran A, Barbas CF, Bruce NC, Wilson IA, Lerner RA. Biochemistry. 2002;41(41):12297–12307. doi: 10.1021/bi026131p. [DOI] [PubMed] [Google Scholar]
- 26.Zhang Y. J. Chem. Phys. 2005;122(2):024114. doi: 10.1063/1.1834899. [DOI] [PubMed] [Google Scholar]
- 27.Zhang Y, Lee T-S, Yang W. J. Chem. Phys. 1999;110(1):46–54. [Google Scholar]
- 28.Hu P, Zhang YK. J. Am. Chem. Soc. 2006;128(4):1272–1278. doi: 10.1021/ja056153+. [DOI] [PubMed] [Google Scholar]
- 29.Zhang Y, Liu H, Yang W. J. Chem. Phys. 2000;112(8):3483–3492. [Google Scholar]
- 30.Zhang Y. Theor. Chem. Acc. 2006;116(1):43–50. [Google Scholar]
- 31.Liu H, Zhang Y, Yang W. J. Am. Chem. Soc. 2000;122(28):6560–6570. [Google Scholar]
- 32.Zhang Y, Kua J, McCammon JA. J. Am. Chem. Soc. 2002;124(35):10572–10577. doi: 10.1021/ja020243m. [DOI] [PubMed] [Google Scholar]
- 33.Cisneros GA, Liu H, Zhang Y, Yang W. J. Am. Chem. Soc. 2003;125(34):10384–10393. doi: 10.1021/ja029672a. [DOI] [PubMed] [Google Scholar]
- 34.Zhang Y, Kua J, McCammon JA. J. Phys. Chem. B. 2003;107(18):4459–4463. [Google Scholar]
- 35.Cheng Y, Zhang Y, McCammon JA. J. Am. Chem. Soc. 2005;127(5):1553–1562. doi: 10.1021/ja0464084. [DOI] [PubMed] [Google Scholar]
- 36.Corminboeuf C, Hu P, Tuckerman ME, Zhang Y. J. Am. Chem. Soc. 2006;128(14):4530–4531. doi: 10.1021/ja0600882. [DOI] [PubMed] [Google Scholar]
- 37.Wang L, Yu X, Hu P, Broyde S, Zhang Y. J. Am. Chem. Soc. 2007;129(15):4731–4737. doi: 10.1021/ja068821c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang S, Hu P, Zhang Y. J. Phys. Chem. B. 2007;111(14):3758–3764. doi: 10.1021/jp067147i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xiao C, Zhang Y. J. Phys. Chem. B. 2007;111(22):6229–6235. doi: 10.1021/jp068657f. [DOI] [PubMed] [Google Scholar]
- 40.Hu P, Wang S, Zhang Y. J. Am. Chem. Soc. 2008;130(12):3806–3813. doi: 10.1021/ja075896n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cheng Y, Zhang Y, McCammon JA. Protein Sci. 2006;15(4):672–683. doi: 10.1110/ps.051852306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Poyner RR, Larsen TM, Wong SW, Reed GH. Arch. Biochem. Biophys. 2002;401(2):155–163. doi: 10.1016/S0003-9861(02)00024-3. [DOI] [PubMed] [Google Scholar]
- 43.Cisneros GA, Wang M, Silinski P, Fitzgerald MC, Yang W. Biochemistry. 2004;43(22):6885–6892. doi: 10.1021/bi049943p. [DOI] [PubMed] [Google Scholar]
- 44.Metanis N, Brik A, Dawson PE, Keinan E. J. Am. Chem. Soc. 2004;126(40):12726–12727. doi: 10.1021/ja0463841. [DOI] [PubMed] [Google Scholar]
- 45.Frisch MJ, et al. Gaussian 03. Gaussian, Inc.; Wallingford CT: 2004. Revision C.02. [Google Scholar]
- 46.Cieplak P, Cornell WD, Bayly C, Kollman PA. J. Comput. Chem. 1995;16(11):1357–1377. [Google Scholar]
- 47.Bayly CI, Cieplak P, Cornell WD, Kollman PA. J. Phys. Chem. 1993;97(40):10269–10280. [Google Scholar]
- 48.Case DA, et al. AMBER 8. University of California; San Francisco: 2004. [Google Scholar]
- 49.Bargagna-Mohan P, Hamza A, Kim YE, Ho YKA, Mor-Valknin N, Wendschlag N, Li JJ, Evans RM, Markovitz DM, Zhan C-G, Kim KB, Mohan R. Chem. Biol. 2007;14(6):623–634. doi: 10.1016/j.chembiol.2007.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hamza A, Zhan C-G. J. Phys. Chem. B. 2009;113(9):2896–2908. doi: 10.1021/jp8082612. [DOI] [PubMed] [Google Scholar]
- 51.Bostrom J, Greenwood JR, Gottfries J. J. Mol. Graphics Modell. 2003;21(5):449–462. doi: 10.1016/s1093-3263(02)00204-8. [DOI] [PubMed] [Google Scholar]
- 52.McGann MR, Almond HR, Nicholls A, Grant JA, Brown FK. Biopolymers. 2003;68(1):76–90. doi: 10.1002/bip.10207. [DOI] [PubMed] [Google Scholar]
- 53.Harvey SC. Proteins. 1989;5(1):78–92. doi: 10.1002/prot.340050109. [DOI] [PubMed] [Google Scholar]
- 54.Guenot J, Kollman PA. Protein Sci. 1992;1(9):1185–205. doi: 10.1002/pro.5560010912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. J. Chem. Phys. 1983;79(2):926–935. [Google Scholar]
- 56.Zhang Y, Liu H, Yang W. In: Methods for Macromolecular Modeling. Schlick T, Gan HH, editors. Springer-Verlag; New York: 2002. pp. 332–354. [Google Scholar]
- 57.Sun H, Pang Y-P, Lockridge O, Brimijoin S. Mol. Pharmacol. 2002;62(2):220–224. doi: 10.1124/mol.62.2.220. [DOI] [PubMed] [Google Scholar]
- 58.Hamza A, Cho H, Tai H-H, Zhan C-G. J. Phys. Chem. B. 2005;109(10):4776–4782. doi: 10.1021/jp0447136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gao DQ, Zhan C-G. J. Phys. Chem. B. 2005;109(48):23070–23076. doi: 10.1021/jp053736x. [DOI] [PubMed] [Google Scholar]
- 60.Zhan C-G, Gao DQ. Biophys. J. 2005;89(6):3863–3872. doi: 10.1529/biophysj.105.070276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gao DQ, Zhan C-G. Proteins. 2006;62(1):99–110. doi: 10.1002/prot.20713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zheng F, Zhan C-G. J. Comput. Aid. Mol. Des. 2008;22(9):661–671. doi: 10.1007/s10822-007-9144-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zheng F, Zhan C-G. Org. Biomol. Chem. 2008;6(5):836–843. doi: 10.1039/b716268e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhan C-G. Expert Rev. Clin. Pharmacol. 2009;2(1):1–4. doi: 10.1586/17512433.2.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pan YM, Gao DQ, Zhan C-G. J. Am. Chem. Soc. 2008;130(15):5140–5149. doi: 10.1021/ja077972s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Alvarez-Idaboy JR, Galano A, Bravo-Perez G, Ruiz ME. J. Am. Chem. Soc. 2001;123(34):8387–8395. doi: 10.1021/ja010693z. [DOI] [PubMed] [Google Scholar]
- 67.Blow DM, Birktoft JJ, Hartley BS. Nature. 1969;221(5178):337–40. doi: 10.1038/221337a0. [DOI] [PubMed] [Google Scholar]
- 68.Bachovchin WW, Roberts JD. J. Am. Chem. Soc. 1978;100(26):8041–8047. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.