Abstract
AIM: To investigate the effects of lentivirus vector mediated short hairpin RNA interference targeting methionine adenosyltransferase 2β gene (LV-shMAT2B) on hepatocellular carcinoma (HCC) cells.
METHODS: We constructed four plasmids of RNA interference targeting the MAT2B gene. After LV-shMAT2B was transfected with L-02 cells and two kinds of HCC cells, cell viability and proliferation were measured with MTT and [3H]thymidine assays respectively. Flow cytometry was used to assess cell apoptosis. The level of S-adenosyl methionine (SAMe) in HepG2 cells was evaluated. The expressions of cyclin D1, cyclin D2, bcl-xL and bcl-xS were detected with western blot.
RESULTS: We constructed LV-shMAT2B successfully. LV-shMAT2B was safe for human normal liver cells. LV-shMAT2B caused dramatic reduction in proliferation compared with controls in HCC cells Bel-7402 (P = 0.054) and HepG2 (P = 0.031). Flow cytometry analysis showed that cell apoptosis caused by LV-shMAT2B was greater in HCC cells Bel-7402 and HepG2 than in control induced by scrambled siRNA (P = 0.047), but apoptosis rates in L-02 induced by LV-shMAT2B and scrambled siRNA respectively had no significant difference. Moreover, LV-shMAT2B significantly suppressed expression of MAT2B leading to growth-inhibition effect on HCC cells by down-regulating cyclin D1. Apoptosis induced by LV-shMAT2B was involved in down-regulating bcl-xL and up- regulating bcl-xS.
CONCLUSION: LV-shMAT2B can induce cell apoptosis and growth-inhibition in HCC cells. MAT2B may be a therapy target in HCC in the future.
Keywords: Lentivirus, Methionine adenosyltransferase 2β gene, Growth inhibition, Apoptosis, Hepatocellular carcinoma
INTRODUCTION
In mammals, methionine adenosyltransferase 1A (MAT1A) and MAT2A encode two homologous MAT catalytic subunits, MATα1 and MATα2 respectively. MAT1A is expressed mostly in liver and encodes MATα1 found in two native MAT isozymes, which are either a dimer (MATIII) or tetramer (MATI) of this single subunit. MAT2A encodes MATα2 found in a native MAT isozyme (MATII) which is associated with a catalytically inactive regulatory subunit (β) in lymphocytes encoded by the MAT2B gene. MAT2A predominates in the fetal liver and is progressively replaced by MAT1A during liver development[1,2]. It has been demonstrated that a switch in MAT expression in liver cancer (from MAT1A to MAT2A) played an important pathogenetic role in facilitating the tumorigenesis of liver cancer[3]. The switch in MAT expression of liver cancer is important, because it offers cancerous cells a growth advantage as well as a decrease in intracellular S-adenosyl methionine (SAMe) content. The function of the β subunit is to regulate MATII activity by lowering its Km for L-methionine and by increasing the sensitivity of the enzyme to feedback inhibition by SAMe[4]. Therefore, regulation of the expression of the β subunit may be a mechanism to regulate the intracellular content of SAMe. The importance of MAT expression on SAMe level and liver phenotype has been confirmed in the MAT1A knockout mouse model. In this research, the replacement of MAT1A with MAT2A has resulted in chronic hepatic SAMe depletion and eventual development of hepatocellular carcinoma (HCC)[5,6]. SAMe has also been found to be antiapoptotic in cultured rat hepatocytes but proapoptotic in human hepatoma cells[7]. β subunit has been proved to be associated with cirrhosis and cancer by providing a proliferative advantage in hepatoma cells through its interaction with MATIIα2 and down-regulation of SAMe levels[8]. Since the decreased content of intracellular SAMe has been proved to be related with the increased expression of the MAT2B gene, we wanted to know whether cancerous cell growth advantage or SAMe depletion could be prevented by knockout of MAT2B. So we constructed lentivirus vector mediating RNA interference targeting MAT2B (LV-shMAT2B). The efficacy of siMAT2B plasmids in interference with MAT2B was confirmed by two different ways. We found that LV-shMAT2B promoted growth-inhibition and apoptosis in human hepatocellular cancer cells by markedly increasing the intracellular content of SAMe and the expression of bcl-xS and decreasing the expressions of cyclin D1 and bcl-xL. But it had no effect on normal liver cells.
MATERIALS AND METHODS
Construction and production of Lentivirus vectors
The construction and production of Lentivirus vectors were previously described[9]. We designed and cloned a short hairpin siRNA template into a lentivirus vector. A third generation of self-inactivating lentivirus vector containing a CMV driven GFP reporter and a U6 promoter upstream of cloning restriction sites (AgeI and EcoRII) to allow the introduction of oligonucleotides encoding shRNAs (Figure 1A) was as described previously[10]. We constructed the scrambled siRNA (as a control) and four siMAT2B lentivirus vectors, namely LV-shiMAT2B-1, LV-shMAT2B-2, LV-shMAT2B-3 and LV-shMAT2B-4, targeting human MAT2B from corresponding siMAT2B plasmids (pGCL-GFP-siMAT2B). Each hairpin consisted of a 20-21 nt sense sequence, a short spacer (CTCGAG), the antisense sequence, 5Ts (a stop signal for RNA polymerase III), and an AgeI site[10,11]. The oligos were annealed and inserted between the AgeI and EcoRII sites of the plasmid. Some mutations were introduced in the sense sequence of the hairpin structure to facilitate sequence and avoid destruction by bacteria during amplification in bacterial host[12]. Correct insertions of shRNA cassettes were confirmed by restriction mapping and direct DNA sequencing. Recombinant lentivirus vectors were produced by co-transfecting 293T cells with the lentivirus expression plasmid and packaging plasmids (pHelper 1.0 including gag/pol and pHelper 2.0 including VSVG) using the calcium phosphate method[13–15]. Infectious lentivirus vectors were harvested at 48 and 72 h post-transfection, centrifuged to get rid of cell debris, and then filtered through 0.22 cellulose acetate filters[16]. The infectious titer was determined by fluorescence-activated cell sorting analysis of GFP positive in 293 cells. The virus titers were at the range of 109 transducing units/mL medium.
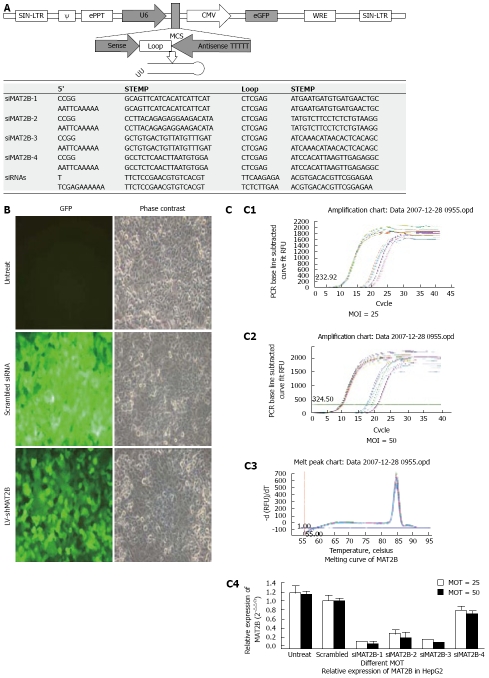
Figure 1.
A: Each consist of a hairpin as was demonstrated; B: HepG2 cells were without treatment (untreated), and infected with control lentivirus (scrambled siRNA) or LV-shMAT2B. The cells were infected (MOI = 25), GFP expression and the phase contrast images of the same areas were taken after 72 h. C: msRNA level of MAT2B after HepG2 cell was treated with LV-shMAT2B in different MOI (C1 and C2) detected by real-time PCR (C4), melting curve of MAT2 B (C3).
Cell culture and transfection
Human hepatocellular cancer HepG2 and Bel-7402 cell lines and normal liver cell L-02 were purchased from Classic Specimen Culture and Storage Centre at Wuhan University. Cells were grown in 5% CO2 saturated humidity, at 37°C and cultured as monolayers in RPMI 1640 supplemented with penicillin/streptomycin, 2 mmol/L glutamine and 10% FBS. Cells were subcultured at 1 × 105 cells per well into six-well tissue culture plates. After 72 h of culture, the cells were transfected with LV-shMAT2Bs (0.5 μL per well) formulated into liposomes according to the manufacturer’s instructions (TransMessengerTM, Qiagen, Valencia, CA; Figure 1B). The final volume of culture medium was 2 mL per well.
Knockdown of MAT2B in HepG2 cells by real time-PCR analysis
Total RNA was subjected to reverse transcription (RT) with Moloney murine leukemia virus reverse transcriptase (Invitrogen, Carlsbad, CA). Cells were then respectively prepared with scrambled siRNA, and lentivirus vectors such as LV-shMAT2B-1, LV-shMAT2B-2, LV-shMAT2B-3 and LV-shMAT2B-4. Three days after virus infection at the multiplicity of infection (MOI) 25 and 50. the RT products were subjected to quantitative real-time PCR analysis. The primers and TaqMan probes for human MAT2B and universal PCR master mix were purchased from Promega Company. GAPDH was used as a control housekeeping gene. The thermal profile consisted of 1 cycle at 95°C for 15 min followed by 45 cycles at 95°C for15 s and at 60°C for 30 s. The expression of MAT2B was checked by normalization of the cycle threshold (Ct) of these genes to that of the control housekeeping gene (GAPDH). The delta Ct (△Ct) = (Ct of MAT2B) - (Ct of GAPDH in each group). △△Ct obtained was used to find the relative expression of MAT2B gene according to the following formula: Relative expression = 2-△△Ct; △△Ct = (mean △Ct of MAT2B genes in siRNAs groups) - (△Ct of MAT2B genes in different treatment groups. Melting curve of MAT2B was drafted.
Western-blot analysis
pEGFP-C1-MAT2B was eukaryotic expression plasmid of the MAT2B gene (Figure 2C). Four siMAT2B plasmids (pGCL-GFP-siMAT2B) were co-transfected with pEGFP-C1-MAT2B in 293T cells, respectively. Forty- eight hours after transfection, cells were collected, rinsed in ice-cold PBS 15 min, and lysed in lysis buffer containing 50 mmol/L HEPES (pH 7.9), 0.4 mol/L NaCl, 1 mmol/L EDTA, 2 Ag/mL leupeptin, 2 Ag/mL aprotinin, 5 Ag/mL benzamidine, 0.5 mmol/L phenylmethylsulfonyl fluoride and 1% NP40. The lysates were centrifuged at 14 000 r/min to remove any cellular debris. Protein concentrations of the lysates were determined by the Bio-Rad Dc protein assay system which was 2 μg/μL. An equal amount of protein was separated by 12% SDS-PAGE, transferred to PVDF membrane, and blocked with 5% nonfat dry milk in TBS/Tween 20 (0.05% v/v) for 1 h at room temperature The membrane was incubated with primary antibody overnight. Anti-cyclin D1, anti-cyclin D2 and anti-GAPDH antibodies were obtained from Santa Cruz Biotech Anti-bcl-xL and Anti-bcl-xS antibodies were obtained from Dako Biotech. Anti-MAT2B was purchased from Abnova Company (Taiwan). After washing, the membrane was incubated with secondary antibody (1:4000, Amersham Pharmacia Biotech Arlington Heights, IL) for 1 h. After several washes, the blots were developed by enhanced chemiluminescence (Amersham Arlington Heights, IL). Each experiment was repeated at least twice with similar results.
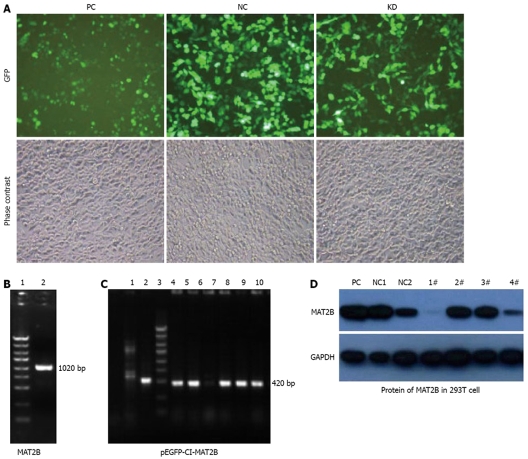
Figure 2.
MAT2B gene was cloned from the cDNA library with production of 1020 bp by PCR (B), pEGFP-C1-MAT2B was constructed after restricted enzyme cutting and connection with production of 420 bp by PCR as showed in Figure2C. Plasmids containing siMAT2B (pGCL-GFP-siMAT2B) were co-transfected with pEGFP-C1-MAT2B in 293T cell, phase contrast and GFP expression under a fluorescent microscope was taken after 72 h (A). Protein level of MAT2B in 293T cells was detected by western blot (D). pGCL-GFP-siMAT2B-1 and -4 can significantly knock down expression of MAT2B at the protein level (D). PC, pcDNA3.1; NC, non-silence control vector. Lanes 1-4: pGCL-GFP-siMAT2B-1, -2, -3 and -4, respectively.
Viability assay
Equal numbers of cells per well were seeded into 24-well plates and incubated until 60% confluent and treated with LV-shMAT2B (0.5 μL) or DMSO for 72 h. 3-(4, 5-dimethylthiazole-2-yl)-2, 5-diphenyltetrazolium bromide (MTT, Sigma, St. Louis, MO), 10% weight/volume in phosphate buffered saline was added to each well. After appropriate incubation, cells were washed, lysed with isopropanol and the solubilized product spectrophotometrically quantified at 490 nm.
Proliferation assay
Proliferation assay with [3H]thymidine. Equal numbers of cells per well were plated onto 24-well plates and cultured until 60% confluent. Cells were treated with LV-shMAT2B (0.5 μL) or DMSO as described above. [3H]thymidine was added to the medium for 72 h at 2.5 Ci/mL. Cells were fixed with 0.5μg LV-shMAT2B, rinsed, dried and 1 mL of 0.33 mol/L sodium hydroxide added. [3H]thymidine incorporation was measured in a liquid scintillation counting system (Beckman Coulter, Fullerton, CA).
Flow cytometry
The inducing apoptosis effects of LV-shMAT2B on human hepatocellular cancer cells and normal liver cell (L-02) were analyzed by Flow cytometry. Briefly, 1 × 105 cells were trypsinized, then washed with PBS, centrifuged at 800 r/min, fixed with 70% precooling alcohol to the second day, alcohol was extracted by trypsinization at 1000 r/min, the cells were incubated in the solution mixture with PI (100 μg), Triton-X100 (0.5%)and RNase (2000 μg/mL) 15 min then analyzed by FCM.
Assay of MATIIenzyme activity
Enzyme activity was measured as described previously[17]. Protein extracts were obtained from transfected cells by sonicating and then centrifuged at 13 000 g for 15 min. Two hundred and fifty μg of protein as determined by the method of Bradford[18] was added to the reaction mixture containing 80 mmol/L Tris-HCl (pH 7.4), 50 mmol/L KCl, 40 mmol/L MgCl2, 5 mmol/L ATP, 10 mmol/L dithiothreitol, 0.5mmol/L EDTA, 50 μmol/L methionine and 0.3 μCi L-[Methyl-3H] methionine. The mixture was applied to a phosphocellulose paper disc (HA 0.45 μm, Millipore) and placed on a filtering system for washing. The disc was added to 10 mL of ScintiVerse E for scintillation counting using a Beckman model LS6000TA Liquid Scintillation Counter (Beckman Instruments, Fullerton, CA, USA). MATII activity was reported as nmol SAMe formed per mg protein per 40 min.
Assay of hepatic SAMe levels
Hepatic SAMe levels were measured using a method described previously[19] with slight modifications. Liver specimens were homogenized in phosphate- buffered saline, and an aliquot was saved for protein assay. The rest was treated with 100 μL of 1 mol/L perchloric acid (PCA) on ice for 5 min and centrifuged at 1000 g for 15 min at 4°C. The aqueous layer was quantitatively removed, neutralized with 3 mol/L KOH and centrifuged at 3000 g for 10 min at 4°C. SAMe levels were determined in the neutralized PCA extracts by HPLC (LC-10ATVP pump, SCL-10AVPsystem control) with a SPD-10AVP UV detector and a SIL-10ADVP autosampler (Shimadzu) using a Partisil SCX 10 μm column (25 cm × 0.44 cm i.d.; Whatman Chem.Sep. Maidstone, Cleveland). SAMe was eluted isocratically at 1 mL/min with 0.19 mol/L NH4H2PO4 adjusted to pH 2.6 with 2mol/L H3PO4. SAMe levels were calculated using a standard curve of SAMe prepared at the same time as the samples and are reported as nmol/mg protein.
Statistical analysis
Data are expressed as mean ± SD, and statistical analyses of the data were performed using ANOVA Post Hoc Dunnett t test. Values of P < 0.05 were considered to be statistically significant.
RESULTS
Expression of MAT2B suppressed by LV-shMAT2B in HCC HepG2 cells in vitro
In this research, GFP expression was observed under a fluorescent microscope in HepG2 cells 72 h after infection with LV-shMAT2B at MOI 25 (Figure 1B). Then we performed real time-PCR[8] to determine the mRNA level of MAT2B in HCC HepG2 cells in vitro. We observed significant suppression of MAT2B by LV-shMAT2B-1 and LV-shMAT2B-3 at two different MOI 25 and 50 (Figure 1C).
Phase contrast and GFP expression was observed (Figure 1B), 72 h after 4 plasmids containing siMAT2B (pGCL-GFP-siMAT2B) co-transfected with pEGFP-C1-MAT2B in 293T cell respectively. Results of a quantitative RT-PCR assay showed that the expression of MAT2B mRNA was reduced by 90% (Figure 2C) when siMAT2B-1 or siMAT2B-3 cotransfected with pEGFP-C1-MAT2B in 293T cells. Results of western-blot assay showed that pGCL-GFP-siMAT2B-1 and pGCL-GFP-siMAT2B-4 could significantly suppress the expression of MAT2B at the protein level in 293T cells (Figure 2D). According to the results of the quantitative RT-PCR and western-blot assays, siMAT2B-1 was the most ideal siMAT2B of the four siMAT2B. Thus LV-shMAT2B-1 corresponding to siMAT2B-1 was used as LV-shMAT2B in the following research.
Cell viability and proliferation significantly inhibited by LV-shMAT2B in HCC cells
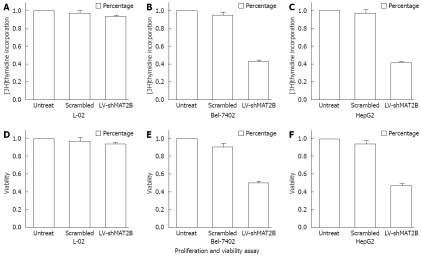
To assess the potential effects of Lentivirus RNAi-mediated MAT2B silencing, we investigated cell proliferation and viability 72 h after L-02 and HCC cells were transfected with LV-shMAT2B. We then examined thymidine incorporation as a measure of DNA synthesis and cell proliferation. MTT assay was used to evaluate cell viability. Value of absorption at 490 nm was obtained. Percentage of cell viability treated with LV-shMAT2B compared with controls was counted, As a result we found that scrambled siRNA had no effect on cell proliferation and viability in all cells (Figure 3), LV-shMAT2B caused dramatic reduction in proliferation (Figure 3B, C) compared with controls in HCC cells Bel-7402 (P = 0.054) and HepG2 (P = 0.031), but it had no effect on normal liver cells L-02 (Figure 3D). In all HCC cells, significant decrease in viability was observed by MTT assay (Figure 3E, F). We discovered that the growth-inhibition caused by LV-shMAT2B was involved in down-regulation of cyclin D1 by western-blot assay.
Figure 3.
Proliferation of L-02 cell and HCC cells Bel-7402, HepG2 were shown in Figure 3A-C. The relative value of [3H]thymidine incorporation was measured after cells were treated with LV-shMAT2B. Percentage cell viability of L-02 cells and HCC cells Bel-7402. HepG2 were counted with MTT after cells were treated with LV-shMAT2B Figure 2D-F. It was seen that LV-shMAT2B can inhibit cell viability and proliferation of HCC cells but had no effect on normal liver cells.
Apoptosis induced by LV-shMAT2B in HCC cells
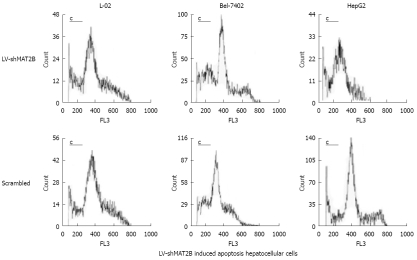
To investigate the effects of LV-shMAT2B on cell death, apoptosis rate was evaluated with flow cytometry analysis. Seventy-two hours after hepatoma cells were transfected with LV-shMAT2B, the sub-G1 population of apoptotic cells was determined. Results from analysis of L-02, Bel-7402 and HepG2 cells showed that apoptosis rates were 10.1% ± 1.9%, 26.3% ± 2.1% and 32.6% ± 3.7%, in the presence of LV-shMAT2B, respectively. Flow cytometry analysis also showed cell apoptosis caused by LV-shMAT2B was greater in HCC cells Bel-7402 and HepG2 than in control cells induced by scrambled siRNA (P = 0.047), but apoptosis rates in L-02 induced by LV-shMAT2B and scrambled siRNA, respectively, had no significant difference (Figure 4). Results of western-blot indicated that LV-shMAT2B caused the apoptosis in HCC cells by decreasing the expression of bcl-xL and increasing the expression of bcl-xS.
Figure 4.
Cell apoptosis caused by LV-shMAT2B was increased in human hepatocellular cancer cells Bel-7402 and HepG2 compared to controls induced by siRNAs, but they all had no impact on the normal hepatic cell (L-02) which does not express MAT2B.
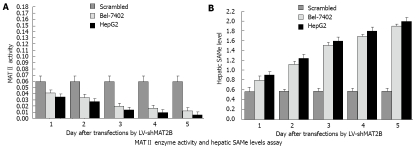
Decreased MAT II enzyme activity and increased intracellular SAMe content caused by LV-shMAT2B in HCC cells
To determine the effect of LV-shMAT2B on enzymatic activity of MATII, HepG2 and Bel-7402 cells were transfected with LV-shMAT2B. At different time points after transfection, protein extracts were prepared, enzyme activity assays were performed. Results from Liquid Scintillation Counter analysis indicated that MATII enzyme activity was gradually decreased from 1 d to 5 d in a time-dependent manner, but remained relatively unchanged in cells treated with scrambled siRNA (Figure 5A). These data demonstrated that LV-shMAT2B significantly inhibited MATII enzyme activity in Bel-7402 and HepG2 cells. The changes in MAT gene’s expression were likely to cause changes in the production of SAMe. To analyze the effect of LV-shMAT2B on SAMe production, we measured the contents of SAMe in HCC cells. Results from reverse-phase high performance liquid chromatography analysis showed that the intracellular content of SAMe in cells transfected with scrambled siRNAs was low at 0.58 ± 0.05 (nmol/mg protein), and increased to 1.89 ± 0.13 in HepG2 and 1.81 ± 0.17 in Bel-7402 after they were treated with LV-shMAT2B in a time-dependent manner (P = 0.0071, Figure 5B).
Figure 5.
MAT II enzyme activity was gradually decreased from 1 d to 5 d in a time-dependent manner after transfected with LV-shMAT2B, but remained relatively unchanged in cells treated with siRNAs. In human cancer cells Bel-7402 and HepG2 (A), Hepatic SAM level in the two HCC cells were increased by LV-shMAT2B in a time-dependent manner, but remained relatively unchanged in cells treated with siRNAs (B).
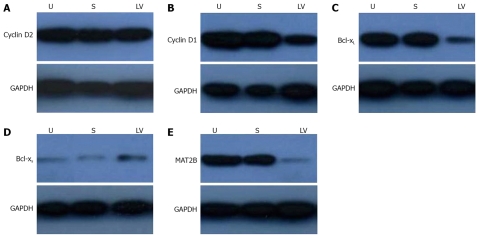
Down-regulation of cyclin D1 and bcl-xl by LV-shMAT2B in HCC cells
It has been indicated that the β subunit provided a proliferative advantage in HCC cells and this effect could be mediated through its interaction with MATIIα2 and down-regulating SAMe levels. SAMe inhibited HGF-dependent induction of cyclin D1 and cyclin D2 expression without affecting the activation of extracellular signal-regulated protein kinase (ERK)[20] and SAMe is proapoptotic in HCC cells in vitro by promoting the expressions of bcl-xL and bcl-xS[21]. Our results showed that LV-shMAT2B could down-regulate the protein levels of cyclin D1 (Figure 6B) and bcl-xL (Figure 6C) in human liver cancer cells. LV-shMAT2B increased the expression of bcl-xS (Figure 6D) but had no effect on cyclin D2 (Figure 6A).
Figure 6.
After HepG2 cells were transfected with LV-shMAT2B for 72 h, western blot was used to detect the protein levels of cyclin D1 and cyclin D2, bcl-xL and bcl-xs, results show the expressions of cyclin D1 (B)and Bcl-xL (C) were decreased compared to controls and untreated cells. The expression of bcl-xs (D) was increased but that of cyclin D2 did not change (A). The expression of MAT2B was suppressed by LV-shMAT2B significantly (E). U, untreated HepG2 cell; S, HepG2 cell treated with scrambled siRNA; LV, HepG2 cells treated with LV-shMAT2B. The protein level of MAT2B was measured by western blot.
Expression of MAT2B suppressed by LV-shMAT2B in protein level in HepG2 cells
To investigate the effect of LV-shMAT2B on HCC cells, we also detected the protein level of MAT2B in HepG2 cells. There was no expression of MAT2B after it was treated with LV-shMAT2B (Figure 6). This demonstrated that expression of MAT2B was suppressed by LV-shMAT2B.
DISCUSSION
It was demonstrated that a switch in MAT expression in liver cancer was accompanied with increasing expression of MAT2B which encoded the β subunit. The β subunit is associated with MATII and MAT2B is not expressed in normal liver. The expression of MAT2B is increased in liver cirrhosis and hepatocellular carcinoma, and chronic hepatic SAMe deficiency leads to malignant degeneration[22]. Recently there was also a report that hepatocyte growth factor (HGF) could promote proliferation of hepatoma cells by up-regulating the expression of MAT2B[23]. Leptin used to be taken as an adipokine shown to be mitogenic in human liver cancer cell lines HepG2 and Huh7, and its mitogenic effect was also related with the expression of MAT2B.
In order to find the role played by MAT2B in HCC, we suppressed MAT2B in HCC cells using the RNAi method. The inhibitory potency and the method of transferring siRNA into HCC cells were two critical factors for successful application of RNAi method. shRNAs were proved to provide long-lasting silencing and maximal inhibition of gene expression at lower concentration[24,25]. Inhibitory potency of shRNA was related to specificity of the targeting sequence, so we used two different methods to confirm the efficacy of siMAT2B in HepG2 cells and in 293T cells. Then we selected siMAT2B-1 which could knock out the expression of MAT2B in both methods. Thus we decided to use LV-siMAT2B-1 as LV-shMAT2B in following research. In this research we found that LV-shMAT2B could suppress the expression of MAT2B at the level of both mRNA and protein in HCC. LV-shMAT2B could dramatically decrease cell proliferation and viability as well as increase cell apoptosis in Bel-7402 and HepG2 cells, but it had no effect on normal liver cell L-02 which had no expression of MAT2B. We concluded that LV-shMAT2B had a high specificity for MAT2B.
In this research we chose lentivirus vector as our shRNAs delivery vehicle because lentivirus could transfect both dividing and nondividing cells at a high efficiency and sustain long-term gene expression by integrating into the host genome. It was shown that the suppression of MAT2B caused by LV-siMAT2B was in a MOI-dependent manner. Significant inhibitory effects were found at MOI of 25 and 50, where the expression of MAT2B was all reduced by 90%. Lentivirus vector is also safe for humans. Lentivirus vector encoding antisense targeting HIV envelope sequence has been used for HIV treatment in clinical trials with no obvious side effects[26,27]. Most recently, lentivirus vector containing beta-globin gene has been approved in phaseI/II clinical trials for human beta-thalassemia and sickle cell anemia gene therapy[28]. The MAT2B’s expression is very important because it not only can provide a proliferative advantage in hepatoma cells but can also decrease intracellular SAMe content. It was discovered that transfection with β subunit reduced the cellular content of SAMe in HuH7 cells, on the contrary we suppressed the expression of MAT2B by LV-shMAT2B, which resulted in decreased activity of MATII and increased content of SAMe. We found that their changes were all time-dependent. SAMe was antiapoptotic in cultured rat hepatocytes but proapoptotic in human hepatoma cells[7]. LV-shMAT2B could induce apoptosis of HCC cells but had no impact on normal liver cells, the reason might be that there was no expression of the MAT2B gene in normal liver cells. It was demonstrated that induction mRNA of bcl-xS by SAMe in HepG2 cells resulted in apoptosis, but SAMe had no effect on expression of bcl-xL[29]. We discovered that the protein level of bcl-xS was increased and the expression of bcl-xL was down-regulated by LV-shMAT2B. We supposed that it might be related to cellular stress brought on by LV-shMAT2B, bcl-xL was able to induce apoptosis in response to cellular stress[30]. We found that LV-shMAT2B prevented proliferation in HepG2 cells, Leptin could induce proliferation in human hepatocarcinoma cells by up-regulating cyclin D1[31] but Komal[30] found that Leptin could increase SAMe levels in HepG2 cells. On the contrary, SAMe was believed to inhibit the expressions of cyclin D1 and cyclin D2[20]. It was indicated in our research that the expression of cyclin D1 was decreased, so we considered that the down-regulation of cyclin D1 may be caused by knocking down of MAT2B, but not by increasing SAMe. We also found that the decreased activity of MATII was caused by LV-shMAT2B. The β subunit associated with cirrhosis and cancer could provide a proliferative advantage in hepatoma cells through its interaction with MATIIα2 and down-regulate SAMe level[8]. When the MAT2B gene was knocked out, the interaction between β and α2 subunit was interrupted. As a result, the activity of MATII decreased and SAMe level increased time-dependently, the proliferative advantage induced by MAT2B was prevented by LV-shMAT2B.
On the whole, we found that the suppression of MAT2B with LV-shMAT2B could lead to growth-inhibition and apoptosis in HCC cells. We concluded that the MAT2B gene could be used as a therapy target for hepatoma in the future.
COMMENTS
Background
It was demonstrated that a switch in methionine adenosyltransferase (MAT) expression in liver cancer played an important pathogenetic role by facilitating liver cancer growth. MAT2A encodes a catalytic subunit (α2) found in a native MAT isozyme (MATII) which is associated with a catalytically inactive regulatory subunit (β) encoded by MAT2B. It has been proved that β subunit is associated with cirrhosis and cancer providing a proliferative advantage in hepatoma cells through its interaction with MATIIα2 and down-regulation of S-adenosyl methionine (SAMe) levels. We supposed that suppression of MAT2B may result in growth-inhibition and increasing of SAMe which could induce hepatoma cells into apoptosis and we wanted to find the mechanism.
Research frontiers
We found that expression of MAT2B in hepatic cancer HepG2 cells were decreased by 90%. Suppression of MAT2B in hepatocellular carcinoma (HCC) cells significantly induced their growth-inhibition by down-regulation of cyclin D1. Apoptosis was induced by LV-shMAT2B involving down regulation of bcl-xL and up-regulation of bcl-xS.
Innovations and breakthroughs
LV-shMAT2B could induce HCC cells into apoptosis but had no impact on normal liver cells, which may occur because there is no expression of the MAT2B gene in the normal liver cell. It has been demonstrated that induction mRNA of bcl-xS by SAMe in HepG2 cells resulted in apoptosis but SAMe had no effects on expression of bcl-xL. We discovered that protein level of bcl-xS was increased and the expression of bcl-xL was down-regulated by LV-shMAT2B. We considered that the down- regulation of cyclin D1 may be caused by knocking down MAT2B but not because of increasing of SAMe.
Applications
Lentivirus vector is also safe for humans. Lentivirus vector encoding the antisense targeting HIV envelope sequence has been used for HIV treatment in clinical trials with no obvious side effects. Most recently, lentivirus vector containing beta-globin gene has been approved in phaseI/II clinical trials for human beta-thalassemia and sickle cell anemia gene therapy. What is, more LV-shMAT2B could induce HCC cells into apoptosis but had no impact on normal liver cells.
Peer review
The manuscript written by Wang et al reports that knockdown of MAT2B by transfection of lentivirus vector carrying siRNAs inhibits the growth of human hepatocellular cell lines but not that of human normal liver cells. The data are interesting and provide potentially important new clues for the future treatment of HCC. However, there are some points which need to be addressed.
Peer reviewer: Dr. Yukihiro Shimizu, Kyoto Katsura Hospital, 17 Yamada-Hirao, Nishikyo, Kyoto 615-8256, Japan
S- Editor Zhong XY L- Editor Alpini GD E- Editor Zhang WB
References
- 1.Horikawa S, Ozasa H, Ota K, Tsukada K. Immuno-histochemical analysis of rat S-adenosylmethionine synthetase isozymes in developmental liver. FEBS Lett. 1993;330:307–311. doi: 10.1016/0014-5793(93)80894-z. [DOI] [PubMed] [Google Scholar]
- 2.Gil B, Casado M, Pajares MA, Bosca L, Mato JM, Martin-Sanz P, Alvarez L. Differential expression pattern of S-adenosylmethionine synthetase isoenzymes during rat liver development. Hepatology. 1996;24:876–881. doi: 10.1002/hep.510240420. [DOI] [PubMed] [Google Scholar]
- 3.Halim AB, LeGros L, Geller A, Kotb M. Expression and functional interaction of the catalytic and regulatory subunits of human methionine adenosyltransferase in mammalian cells. J Biol Chem. 1999;274:29720–29725. doi: 10.1074/jbc.274.42.29720. [DOI] [PubMed] [Google Scholar]
- 4.Lu SC, Alvarez L, Huang ZZ, Chen L, An W, Corrales FJ, Avila MA, Kanel G, Mato JM. Methionine adenosyltransferase 1A knockout mice are predisposed to liver injury and exhibit increased expression of genes involved in proliferation. Proc Natl Acad Sci USA. 2001;98:5560–5565. doi: 10.1073/pnas.091016398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Martinez-Chantar ML, Corrales FJ, Martinez-Cruz LA, Garcia-Trevijano ER, Huang ZZ, Chen L, Kanel G, Avila MA, Mato JM, Lu SC. Spontaneous oxidative stress and liver tumors in mice lacking methionine adenosyltransferase 1A. FASEB J. 2002;16:1292–1294. doi: 10.1096/fj.02-0078fje. [DOI] [PubMed] [Google Scholar]
- 6.Mato JM, Corrales FJ, Lu SC, Avila MA. S-Adenosy-lmethionine: a control switch that regulates liver function. FASEB J. 2002;16:15–26. doi: 10.1096/fj.01-0401rev. [DOI] [PubMed] [Google Scholar]
- 7.Ansorena E, Garcia-Trevijano ER, Martinez-Chantar ML, Huang ZZ, Chen L, Mato JM, Iraburu M, Lu SC, Avila MA. S-adenosylmethionine and methylthioadenosine are antiapoptotic in cultured rat hepatocytes but proapoptotic in human hepatoma cells. Hepatology. 2002;35:274–280. doi: 10.1053/jhep.2002.30419. [DOI] [PubMed] [Google Scholar]
- 8.Martinez-Chantar ML, Garcia-Trevijano ER, Latasa MU, Martin-Duce A, Fortes P, Caballeria J, Avila MA, Mato JM. Methionine adenosyltransferase II beta subunit gene expression provides a proliferative advantage in human hepatoma. Gastroenterology. 2003;124:940–948. doi: 10.1053/gast.2003.50151. [DOI] [PubMed] [Google Scholar]
- 9.Liao W, Ning G. Knockdown of apolipoprotein B, an atherogenic apolipoprotein, in HepG2 cells by lentivirus-mediated siRNA. Biochem Biophys Res Commun. 2006;344:478–483. doi: 10.1016/j.bbrc.2006.03.164. [DOI] [PubMed] [Google Scholar]
- 10.Rubinson DA, Dillon CP, Kwiatkowski AV, Sievers C, Yang L, Kopinja J, Rooney DL, Zhang M, Ihrig MM, McManus MT, et al. A lentivirus-based system to functionally silence genes in primary mammalian cells, stem cells and transgenic mice by RNA interference. Nat Genet. 2003;33:401–406. doi: 10.1038/ng1117. [DOI] [PubMed] [Google Scholar]
- 11.Brummelkamp TR, Bernards R, Agami R. A system for stable expression of short interfering RNAs in mammalian cells. Science. 2002;296:550–553. doi: 10.1126/science.1068999. [DOI] [PubMed] [Google Scholar]
- 12.Miyagishi M, Sumimoto H, Miyoshi H, Kawakami Y, Taira K. Optimization of an siRNA-expression system with an improved hairpin and its significant suppressive effects in mammalian cells. J Gene Med. 2004;6:715–723. doi: 10.1002/jgm.556. [DOI] [PubMed] [Google Scholar]
- 13.Naldini L, Blomer U, Gallay P, Ory D, Mulligan R, Gage FH, Verma IM, Trono D. In vivo gene delivery and stable transduction of nondividing cells by a lentiviral vector. Science. 1996;272:263–267. doi: 10.1126/science.272.5259.263. [DOI] [PubMed] [Google Scholar]
- 14.Dull T, Zufferey R, Kelly M, Mandel RJ, Nguyen M, Trono D, Naldini L. A third-generation lentivirus vector with a conditional packaging system. J Virol. 1998;72:8463–8471. doi: 10.1128/jvi.72.11.8463-8471.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pfeifer A, Kessler T, Silletti S, Cheresh DA, Verma IM. Suppression of angiogenesis by lentiviral delivery of PEX, a noncatalytic fragment of matrix metalloproteinase 2. Proc Natl Acad Sci USA. 2000;97:12227–12232. doi: 10.1073/pnas.220399597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Naldini L, Blomer U, Gage FH, Trono D, Verma IM. Efficient transfer, integration, and sustained long-term expression of the transgene in adult rat brains injected with a lentiviral vector. Proc Natl Acad Sci USA. 1996;93:11382–11388. doi: 10.1073/pnas.93.21.11382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.LeGros HL Jr, Halim AB, Geller AM, Kotb M. Cloning, expression, and functional characterization of the beta regulatory subunit of human methionine adenosyltransferase (MAT II) J Biol Chem. 2000;275:2359–2366. doi: 10.1074/jbc.275.4.2359. [DOI] [PubMed] [Google Scholar]
- 18.Liu Q, Wu K, Zhu Y, He Y, Wu J, Liu Z. Silencing MAT2A gene by RNA interference inhibited cell growth and induced apoptosis in human hepatoma cells. Hepatol Res. 2007;37:376–388. doi: 10.1111/j.1872-034X.2007.00041.x. [DOI] [PubMed] [Google Scholar]
- 19.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 20.Chen L, Zeng Y, Yang H, Lee TD, French SW, Corrales FJ, Garcia-Trevijano ER, Avila MA, Mato JM, Lu SC. Impaired liver regeneration in mice lacking methionine adenosyltransferase 1A. FASEB J. 2004;18:914–916. doi: 10.1096/fj.03-1204fje. [DOI] [PubMed] [Google Scholar]
- 21.Garcia-Trevijano ER, Martinez-Chantar ML, Latasa MU, Mato JM, Avila MA. NO sensitizes rat hepatocytes to proliferation by modifying S-adenosylmethionine levels. Gastroenterology. 2002;122:1355–1363. doi: 10.1053/gast.2002.33020. [DOI] [PubMed] [Google Scholar]
- 22.Yang H, Magilnick N, Noureddin M, Mato JM, Lu SC. Effect of hepatocyte growth factor on methionine adenosyltransferase genes and growth is cell density-dependent in HepG2 cells. J Cell Physiol. 2007;210:766–773. doi: 10.1002/jcp.20891. [DOI] [PubMed] [Google Scholar]
- 23.Siolas D, Lerner C, Burchard J, Ge W, Linsley PS, Paddison PJ, Hannon GJ, Cleary MA. Synthetic shRNAs as potent RNAi triggers. Nat Biotechnol. 2005;23:227–231. doi: 10.1038/nbt1052. [DOI] [PubMed] [Google Scholar]
- 24.Kim DH, Behlke MA, Rose SD, Chang MS, Choi S, Rossi JJ. Synthetic dsRNA Dicer substrates enhance RNAi potency and efficacy. Nat Biotechnol. 2005;23:222–226. doi: 10.1038/nbt1051. [DOI] [PubMed] [Google Scholar]
- 25.Morris KV. VRX-496(VIRxSYS) Curr Opin Investig Drugs. 2005;6:209–215. [PubMed] [Google Scholar]
- 26.Manilla P, Rebello T, Afable C, Lu X, Slepushkin V, Humeau LM, Schonely K, Ni Y, Binder GK, Levine BL, et al. Regulatory considerations for novel gene therapy products: a review of the process leading to the first clinical lentiviral vector. Hum Gene Ther. 2005;16:17–25. doi: 10.1089/hum.2005.16.17. [DOI] [PubMed] [Google Scholar]
- 27.Bank A, Dorazio R, Leboulch P. A phase I/II clinical trial of beta-globin gene therapy for beta-thalassemia. Ann N Y Acad Sci. 2005;1054:308–316. doi: 10.1196/annals.1345.007. [DOI] [PubMed] [Google Scholar]
- 28.Yang H, Sadda MR, Li M, Zeng Y, Chen L, Bae W, Ou X, Runnegar MT, Mato JM, Lu SC. S-adenosylmethionine and its metabolite induce apoptosis in HepG2 cells: Role of protein phosphatase 1 and Bcl-x(S) Hepatology. 2004;40:221–231. doi: 10.1002/hep.20274. [DOI] [PubMed] [Google Scholar]
- 29.Chen C, Chang YC, Liu CL, Liu TP, Chang KJ, Guo IC. Leptin induces proliferation and anti-apoptosis in human hepatocarcinoma cells by up-regulating cyclin D1 and down-regulating Bax via a Janus kinase 2-linked pathway. Endocr Relat Cancer. 2007;14:513–529. doi: 10.1677/ERC-06-0027. [DOI] [PubMed] [Google Scholar]
- 30.Ramani K, Yang H, Xia M, Ara AI, Mato JM, Lu SC. Leptin's mitogenic effect in human liver cancer cells requires induction of both methionine adenosyltransferase 2A and 2beta. Hepatology. 2008;47:521–531. doi: 10.1002/hep.22064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Takehara T, Liu X, Fujimoto J, Friedman SL, Takahashi H. Expression and role of Bcl-xL in human hepatocellular carcinomas. Hepatology. 2001;34:55–61. doi: 10.1053/jhep.2001.25387. [DOI] [PubMed] [Google Scholar]