Abstract
AIM: To investigate the inhibitory effects of hepatitis B virus (HBV) replication and expression by transfecting artificial microRNA (amiRNA) into HepG2.2.15 cells.
METHODS: Three amiRNA-HBV plasmids were constructed and transfected into HepG2.2.15 cells. HBV antigen secretion was detected in the cells with transient and stable transfection by time-resolved fluoroimmunoassays (TRFIA). HBV DNA replication was examined by fluorescence quantitative PCR, and the level of HBV S mRNA was measured by semi-quantitative RT-PCR.
RESULTS: The efficiency of transient transfection of the vectors into 2.2.15 cells was 55%-60%. All the vectors had significant inhibition effects on HBsAg and HBeAg at 72 h and 96 h after transfection (P < 0.01 for all). The secretion of HBsAg and HBeAg into the supernatant was inhibited by 49.8% ± 4.7% and 39.9% ± 6.7%, respectively, at 72 h in amiRNA-HBV-S608 plasmid transfection group. The copy of HBV DNA within culture supernatant was also significantly decreased at 72 h and 96 h after transfection (P < 0.01 for all). In the cells with stable transfection, the secretion of HBsAg and HBeAg into the supernatant was significantly inhibited in all three transfection groups (P < 0.01 for all, vs negative control). The copies of HBV DNA were inhibited by 33.4% ± 3.0%, 60.8% ± 2.3% and 70.1% ± 3.3%, respectively.
CONCLUSION: In HepG2.2.15 cells, HBV replication and expression could be inhibited by artificial microRNA targeting the HBV S coding region. Vector-based artificial microRNA could be a promising therapeutic approach for chronic HBV infection.
Keywords: Hepatitis B virus, RNA interference, Artificial microRNA, HepG2.2.15 cell
INTRODUCTION
RNA interference (RNAi) has become a new promising approach to develop effective antiviral drugs in recent years, including hepatitis C virus (HCV), human immunodeficiency virus (HIV), poliovirus and hepatitis B virus (HBV)[1–3]. For HBV, RNAi was shown to have impressive inhibitory effects against viral gene transcription and expression[4–7]. One recent study further demonstrated the viral clearance from the liver of transgenic mice by RNAi targeted HBV[8], which sheds light on the use of RNAi in HBV gene therapy.
HBV replication and gene expression can be strongly inhibited with virus specific siRNA treatment. However, the high sequence specificity of siRNAs, combined with prolonged treatment, promoted the emergence of siRNA-resistant virus variants. Selection of RNAi escape mutants has been reported in vitro for HIV, HCV and HBV[9–11]. These findings indicate that the antiviral properties of specific siRNAs targeted virus are not as effective as expected.
RNAi can be triggered by small RNA molecules such as siRNA and microRNAs (miRNAs). MiRNAs endogenously expressed small ssRNA sequences of about 22 nucleotides in length, which naturally direct gene silencing through components shared with the RNAi pathway[12]. The mature miRNAs regulate gene expression by mRNA cleavage or translational repression[13,14]. Compared with siRNA, miRNA still can play a translational repression role when miRNAs partially complement with the target gene. Because of the flexibility of miRNA in binding with partially complementary mRNA targets, miRNA can serve as an anti-virus drug or vaccine to achieve a breakthrough in the treatment of virus mutation. Moreover, since miRNAs are single-stranded molecules insensitive to interferon systems, the utilization of this Pol-II-mediated miRNA generation can be safe both in vitro and in vivo without the cytotoxic effects of dsRNAs and siRNAs[15]. These findings indicate that miRNA mediated RNAi can be used as a tool for gene-specific therapeutics against viral infections.
In this study, we applied the recently developed artificial miRNA (amiRNA) expression vector based on the murine miR-155 sequence[16], to observe whether amiRNA could efficiently suppress the expression and replication of HBV in vitro.
MATERIALS AND METHODS
Design of amiRNA and plasmid construction
The pcDNA™6.2-GW/± EmGFP-miR plasmid (Invitrogen, Carlsbad, CA, USA), driving the expression of amiRNA with polymerase II and containing a spectinomycin resistance gene, was used in this study. The engineered pre-miRNA sequence structure is based on the murine miR-155. According to the sequences of conserved region in HBV genome, we designed three target sequences against the S region of the HBV genome using Invitrogen’s RNAi design algorithm at https://rnaidesigner.invitrogen.com/rnaiexpress/. The BLAST algorithm was also used to ensure the designed sequences would not target other gene transcripts to avoid off-target effects. These oligonucleotides (Table 1) were annealed and ligated into pcDNA6.2-GW/EmGFP-miR vector. The pcDNA6.2-GW/EmGFP-miR-negative control plasmid contains an insert that can form a hairpin structure which is processed into mature miRNA, but is predicted not to target any known vertebrate gene. Thus, this plasmid serves as a suitable negative control. Control cells were mock transfected with Lipofectamine 2000 alone.
Table 1.
Three pairs of two single-stranded DNA oligonucleotides
| Plasmid | Top oligo strand | Bottom oligo strand |
| amiRNA-HBV-S89 (89-109 nt) | 5’-TGCTGTCCACCACGAGTCTAGACTCTGTTTTGGCCACTGACTGACAGAGTCTACTCGTGGTGGA -3’ | 5’-CCTGTCCACCACGAGTAGACTCTGTCAGTCAGTGGCCAAAACAGAGTCTAGACTCGTGGTGGAC-3’ |
| amiRNA-HBV-S367 (367-387nt) | 5’-TGCTGTTGAGCAGTAGTCATGCAGGTGTTTTGGCCACTGACTGACACCTGCATCTACTGCTCAA -3’ | 5’-CCTGTTGAGCAGTAGATGCAGGTGTCAGTCAGTGGCCAAAACACCTGCATGACTACTGCTCAAC -3’ |
| amiRNA-HBV-S608 (608-628nt) | 5’-TGCTGTCAAGATGCTGTACAGACTTGGTTTTGGCCACTGACTGACCAAGTCTGCAGCATCTTGA -3’ | 5’-CCTGTCAAGATGCTGCAGACTTGGTCAGTCAGTGGCCAAAACCAAGTCTGTACAGCATCTTGAC -3’ |
Cell culture and transfection
The HepG2.2.15 cells were cultured in Dulbecco’s modified Eagle’s medium (Invitrogen) supplemented with 10% fetal bovine serum (FBS, Gibco), 380 mg/L antibiotic G-418 sulfate (Promega, USA), and 100 IU/mL penicillin and streptomycin, and 1% L-glutamine, at 37°C in the atmosphere of 5% CO2. Transfections were performed using Lipofectamine 2000 reagent (Invitrogen) according to the manufacturer’s instructions. Briefly, cells were trypsinized and plated in 12-well plates at a density of 5.0 × 104 cells/mL (1 mL/well) for 24 h before transfection. Four μL of lipofectamine was diluted dropwise into Opti-MEMI(Gibco, USA) for a final volume of 100 μL and incubated at room temperature for 5 min. Then 1.6 μg of amiRNA expression vector was added to the diluted lipofectamine and incubated for another 20 min. The normal HepG 2.2.15 cells and the cell transfection with negative control plasmid containing scrambled miRNA were used as the negative control. Each plasmid was repeated in three wells. After transfection, the medium was partly removed for analysis every 24 h and the cells were replenished with fresh medium. The cell culture supernatant was collected for detection of HBsAg, HBeAg and HBV-DNA.
Stable transfection was carried out at 24 h after transfection, the cells were passaged at a 1:10 dilution into fresh growth medium and selection was performed with 10 mg/L Blasticidin 30 h later. The Blasticidin resistant colonies were picked with 200 μL pipet tip and cultured with medium containing 10 mg/L Blasticidin to establish individual clone lines. Successfully transfected cell clones were obtained by a long-term culture in a selected medium containing 6 mg/L Blasticidin.
Detection of HBV-DNA in cell culture medium by real-time PCR
After transfection, 50 μL of the supernatant were mixed with an equal volume of the DNA extractant. Samples were incubated at 94°C for 10 min and then centrifuged at 10 000 × g for 5 min. The supernatant was used as template for real-time PCR. The forward primer of HBV-DNA is 5’-GGAGTATGGATTCGCACTCCTC-3’, the reverse primer is 5’-TTGTTGTTGTAGGGGACCTGCCT-3’, the fluorescent probe 5’-ACTTCCGGAAACTACTGTTAGACGA-3’, and the quenching probe 5’-GTAGTTTCCGGAAGT-3’. PCR amplification and analysis were performed using the ABI 7500 real-time PCR detector (ABI). Assays were repeated in triplicate and average threshold cycle values were used to determine the concentration of HBV-DNA. The inhibitory rate was calculated using the follow formula: inhibitory rate (%) = (C control-C tester)/C control × 100%. Control represents HBV-DNA copies in HepG2.2.15 cells transfected with Lipofectamine alone, C tester represents HBV-DNA copies in cells transfected with amiRNA.
HBsAg and HBeAg assay
To assess the effect of amiRNAs on HBV at the protein level, the viral proteins of hepatitis B surface antigen (HBsAg) and e antigen (HBeAg) of the culture supernatant from transfected cells at various times were measured with time-resolved fluoroimmunoassays kit (TRFIA) according to the supplier’s instructions. The inhibitory rates were calculated according to the following formula: inhibitory rate (%) = (C control-C tester )/C control × 100%. Assays were performed in triplicate, and the average inhibitory rate was expressed as a mean ± SD.
Semiquantitative RT-PCR analysis of HBS mRNA
Total RNA was isolated by Trizol Reagent (Invitrogen) following the manufacturer’s instructions. The quantity of total RNA was first determined by A260 measurement, and the quality of total RNA was estimated by 1.5% agarose gel electrophoresis. cDNA was synthesized from total RNA using the Reverse Transcription System (Promega) according to the manufacturer’s protocol. The first-strand cDNA product was used for semi-quantitative PCR. PCR reaction was performed in a single reaction of 25 μL volume. The forward (fp) and reverse (rp) primers used were: HBVS-FP: 5’-TAGACTCGTGGTGGACTTC-3’, HBVS-RP: 5’-ATTGGTAACAGCGGTAAA-3’, GAPDH-FP: 5’-ACCACAGTCCATGCCATCAC-3’, GAPDH-RP: 5’-TCCACCACCCTGTTGCTGTA-3’. The schedule consisted of incubation for 5 min at 94°C followed by 30 cycles of 94°C for 30 s, 55°C for 30 s and 72°C for 30 s, with a final cycle for 5 min at 72°C. PCR products were run on a 1% agarose gel and visualized by ethidium bromide staining, and the intensities were then measured by scanning the gel with JIEDA 801 (JIEDA, Nanjing, China). The products were quantified by densitometry and normalized with respect to GAPDH as internal control.
Statistical analysis
The data were expressed as mean ± SD. Statistical analysis was performed using Student’s t test, with P value less than 0.05 being considered statistically significant. All statistical analyses were performed using SPSS software.
RESULTS
Identification of recombinant plasmid amiRNA-HBV
RNAi was performed using the BLOCK-iTTM Pol II RNAi expression vector kit as recommended by the manufacturer (Invitrogen). Three plasmids containing pre-miRNA sequences were constructed and designated as amiRNA-HBV-S89, amiRNA-HBV-S367, and amiRNA-HBV-S608, respectively. In order to identify successful construction of recombinant plasmids, the miRNA forward sequencing primer 5’-GGCATGGACGAGCTGTACAA-3’ and miRNA reverse sequencing primer 5’-CTCTAGATCAACCACTTTGT-3’, (Invitrogen) were used to perform PCR. And the PCR products, which contained the miRNA insert fragments, were verified by DNA sequencing. The mutant in these inserted oligonucleotides was excluded from this experiment.
Inhibition of HBS mRNA expression by amiRNA
Once the recombinant plasmids were transfected into HepG2.2.15 cells, the expression of Emerald green fluoresecent protein (EmGFP) could be detected directly under fluorescence microscopy. By this way, we found that the transfection efficiency of these vectors into HepG2.2.15 cells was 55%-60% in the current study. At 72 h after transfection, compared with mock and negative control vector, amiRNA-mediated RNAi resulted in a significantly reduced level of HBS mRNA in three plasmids transfected cells (P = 0.008, 0.0015 and 0.00074, respectively). Among them, amiR-HBV-S367 and amiRNA-HBV-S608 were more efficient with 38.7% and 47.4% average inhibitory rate in viral mRNA (Figure not shown).
For cell clones with stable transfection, amiRNA-mediated RNAi resulted in a higher decrease degree of HBS mRNA in three plasmid transfected groups than the cells with transient transfection. Among them, compared with negative control vector, the inhibitory rate of HBV S mRNA was 40.6% ± 3.8% in amiRNA-HBV-S89 group (P = 0.00094). amiR-HBV-S367 and amiRNA-HBV -S608 was much more efficient with an inhibitory rate of 63.4% ± 2.5% and 83.0% ± 4.3% in HBV S mRNA (P = 0.000047 and 0.000011, respectively; Figure 1).
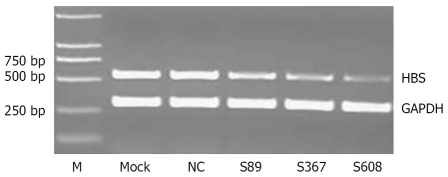
Figure 1.
Effect of amiRNA-HBV with stable transfection on HBV S mRNA levels, compared with negative control, all three plasmids had significant inhibitory effect on HBV S mRNA (P = 0.00094, 0.000047 and 0.000011, respectively).
Inhibition of HBeAg and HBsAg secretion by amiRNA
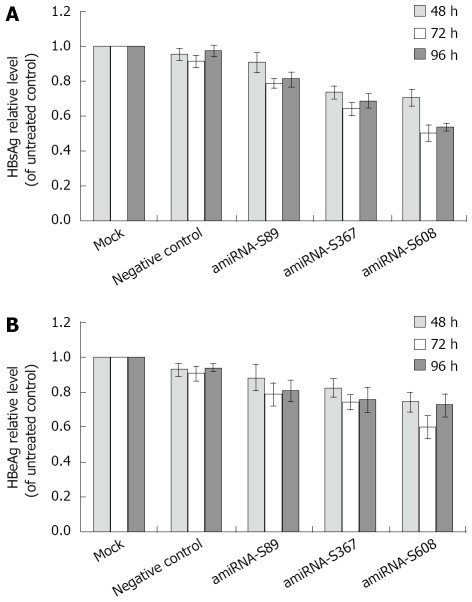
The concentrations of HBsAg and HBeAg in the culture supernatant were measured at 48 h, 72 h and 96 h, which were stable after transfection using TRFIA. As shown in Figure 2, our results demonstrated that three amiRNAs had distinct inhibition effects on HBsAg and HBeAg at 72 h and 96 h after transfection (HBsAg: P = 0.00039 and 0.0012 for S89 group, P = 0.000011 and 0.00021 for S367 group, P = 0.000016 and 0.000056 for S608 group; HBeAg: P = 0.02 and 0.035 for S89 group, P = 0.0093 and 0.0032 for S367 group, P = 0.007 and 0.0025 for S608 group). No significant reduction was measured when transfected with negative control vector, compared with mock control (P > 0.05). Transfection with amiRNA-HBV-S608 vector had the greatest reduction of HBsAg and HBeAg compared with the negative vector control. At 48 h, 72 h and 96 h after amiRNA-HBV-S608 transfection compared with negative control, the inhibitory rates of HBsAg secretion were 29.5% ± 5.0%, 49.8% ± 4.7% and 39.9% ± 6.7%, and the inhibitory rates of HBeAg secretion were 25.5% ± 5.6%, 39.9% ± 6.7% and 37.4% ± 6.7%, respectively.
Figure 2.
A: Effect of amiRNA-HBV on HBsAg levels; B: Effect of amiRNA-HBV on HBeAg levels. HBsAg and HBeAg levels are expressed as mean ± SD. Compared with negative control, all three plasmids had significant inhibitory effect on HBsAg and HBeAg at 72 h and 96 h after transfection (HBsAg: P = 0.00039 and 0.0012 for S89 group, P = 0.000011 and 0.00021 for S367 group, P = 0.000016 and 0.000056 for S608 group; HBeAg: P = 0.02 and 0.035 for S89 group, P = 0.0093 and 0.0032 for S367 group, P = 0.007 and 0.0025 for S608 group).
HBsAg and HBeAg in the culture medium of stably transfected cells were also assayed. The HBsAg and HBeAg levels of all the three cell lines integrated with the pcDNA6.2-HBV-amiRNA vector were significantly reduced compared with negative control of transfected HepG2.2.15 cells, (HBsAg: P = 0.00019, 0.000035 and 0.000012, respectively; HBeAg: P = 0.003, 0.0002 and 0.000024, respectively). The greatest reduction of HBsAg in amiRNA-S608 was 81.5% ± 2.2% (P = 0.000012, Figure 3A), while the greatest reduction of HBeAg in amiRNA-S608 was 58.1% ± 5.2% (P = 0.000024, Figure 3B). No significant reduction was measured on the cells stably transfected with negative control plasmid compared with HepG2.2.15 cells (P > 0.05).
Figure 3.
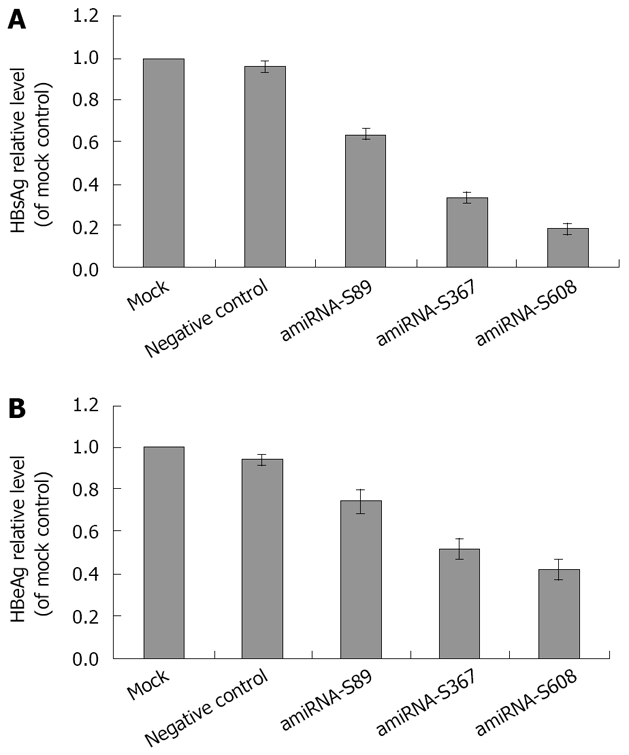
A: Effect of amiRNA-HBV with stable transfection on HBsAg levels; B: Effect of amiRNA-HBV with stable transfection on HBeAg levels. HBsAg and HBeAg levels are expressed as mean ± SD. Compared with negative control, all three plasmids had significant inhibitory effect on HBsAg and HBeAg (HBsAg: P = 0.00019, 0.000035 and 0.000012, respectively; HBeAg: P = 0.003, 0.0002 and 0.000024, respectively).
Inhibition of HBV DNA replication by amiRNA
Real-time fluorescence quantitative PCR was performed to determine whether transfection with amiRNA-HBV-S vector would result in reduction of HBV DNA level. Quantitative assay revealed that HBV DNA levels of all three plasmid transfection groups decreased at 48 h, 72 h and 96 h after transfection, compared with negative control (P = 0.049, 0.000021 and 0.0011 for S89 group; P = 0.0002, 0.000016 and 0.0012 for S367 group; P = 0.0003, 0.00006 and 0.00016 for S608 group). The greatest reduction was found in the amiRNA-HBV-S608 transfected group. The copies of HBV DNA in cells treated with amiRNA-HBV-S608 were reduced by 39.0% ± 4.5% at 72 h (P = 0.00006, vs negative control; Figure 4A). In the cell clones with stable transfection, compared with negative control vector, amiRNA-mediated RNAi resulted in a higher reduction level of HBV DNA in three plasmids transfected cells than those with transient transfection (Figure 4B). Among them, the HBV DNA level of amiRNA-HBV-S89 group decreased by 33.4% ± 3.0% (P = 0.00009); amiR-HBV-S367 and amiRNA-HBV-S608 was much more efficient with inhibitory rates of 60.8% ± 2.3% and 70.1% ± 3.3%, respectively (P = 0.000007 and 0.000006, respectively).
Figure 4.
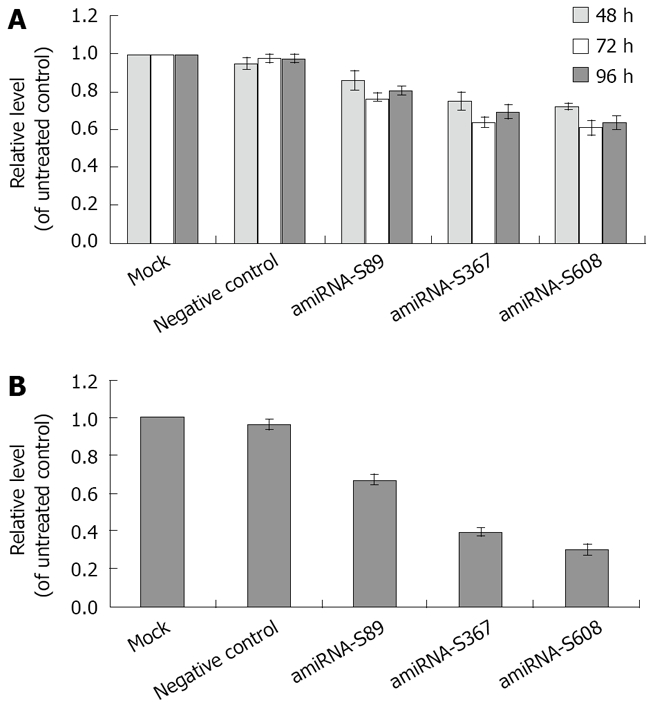
A: Effect of amiRNA-HBV with transient transfection on HBV DNA levels; B: Effect of amiRNA-HBV with stable transfection on HBV DNA levels, The amounts of HBV DNA are expressed as mean ± SD. Compared with negative control, all three plasmids had significant inhibitory effect on HBsAg and HBeAg at 48 h, 72 h and 96 h after transfection (P = 0.049, 0.000021 and 0.0011 for S89 group; P = 0.0002, 0.000016 and 0.0012 for S367 group; P = 0.0003, 0.00006 and 0.00016 for S608 group). All three plasmids had significant inhibitory effect on HBV DNA in stably transfected groups, (P = 0.00009, 0.000007 and 0.000006, respectively).
DISCUSSION
MicroRNA-induced RNA silencing has become a commonly used tool for the analysis of gene function[17,18]. The artificial microRNAs (amiRNAs) technology exploits endogenous miRNA precursors to generate sRNAs that direct gene silencing in either plants or animals[15,18–20]. AmiRNAs were first generated and used in human cell lines[15] and later in Arabidopsis[18], where they were shown to effectively interfere with reporter gene expression. Subsequently, it was demonstrated that not only reporter genes but also endogenous genes can be targeted with amiRNAs (also called synthetic miRNAs). A few tumor genes have been successfully targeted with artificial synthetic miRNAs[21–23].
In this paper, microRNA-induced RNA silencing was utilized to suppress HBV replication and expression. Three different miR-155 based vectors targeting HBV S gene were constructed. The three different plasmids all proved effective, but were not to the same extent at all three times. Among the three amiRNAs, amiRNA-HBV-S608 is the most potent in inhibiting HBV replication and expression in the transient transfection group. We found that HBsAg and HBeAg in the supernatant were inhibited by 49.8% ± 4.7% and 39.9% ± 6.7% at 72 h after tranfection with amiRNA-HBVS608 plasmid. The HBV DNA levels were also found to be decreased similarly. In view of transfection efficiency of 55%-60%, we concluded that actual antigen inhibition rate of amiRNA-HBV-S608 vector was above 80%. To further confirm the effect of amiRNA-HBV, we performed stable transfection of amiRNA-HBV plasmid to exclude the effect of transfection efficiency. In the stably transfected cells with amiRNA-S608 plasmid, amiRNA-mediated RNAi resulted in a higher reduction level of both HBV antigen and DNA than with transiently transfected cells. The greatest reduction of HBsAg and HBeAg in stably transfected cells with amiRNA-HBV-S608 was 81.5% ± 2.2% and 58.1% ± 5.2%, respectively in stably transfected cells. The copies of HBV DNA in cells with stable transfection of amiRNA-HBV-S608 were reduced by 70.1% ± 3.3% compared to negative controls. These data provided a strong indication that continuous expression of amiRNA could provoke stable and sequence-specific silencing of target genes of HBV.
The HepG2.2.15 cells can produce replicative viral DNA intermediates, mature Dane particles and high level of viral antigens constitutively, which may be similar to the behavior in HBV-infected human being. But the inhibitory effect of amiRNA-mediated RNAi in HepG2.2.15 cells is lower than that in other hepatocytes and mice using co-transfection method[24,25]. We believed that all the HepG2.2.15 cells contained HBV genome and could secrete HBV proteins and DNA, but only approximately 55%-60% of the cells received the amiRNA-HBV vector. So the low transfection efficiency on HepG2.2.15 cells resulted in the reduction of inhibition effect. When stable transfection was performed in our experiment with the same vectors, all of these vectors acted with a similar effect as against other cotransfection methods.
Our results showed that artificial miRNA-mediated RNAi could inhibit HBV protein expression and HBV DNA replication in HepG2.2.15 cells in vitro. Similar to the siRNA mediated RNAi, microRNA mediated RNAi was sufficient to disrupt the viral life cycle and inhibited HBV DNA replication. The inhibition was sequence-specific, because the transfection with the negative control had no such effects. This is the first time to our knowledge that a marked reduction of HBV replication induced by artificial miRNA has been noted.
In summary, we constructed three artificially expressed miRNA plasmids and used them as a tool to inhibit HBV replication. We systematically evaluated the effects of amiRNA-based RNAi on HBV expression and replication in HepG2.2.15 cells. The data from our experiments suggested that amiRNA mediated RNAi might represent an alternative approach for the treatment of chronic HBV infection, which can enhance the anti-HBV efficacy and overcome the drawbacks of current therapies.
COMMENTS
Background
MicroRNAs (miRNAs) endogenously expressed small ssRNA sequences with about 22 nucleotide, which naturally direct gene silencing through components shared with the RNAi pathway.
Research frontiers
Recently, it has been described how artificial miRNAs (amiRNAs) designed to target one or several genes of interest could provide a new and highly specific approach for effective post-transcriptional gene silencing.
Innovations and breakthroughs
The results of our study suggest that amiRNA-expressing vectors can be used as RNAi-based anti-HBV therapeutics. amiRNA mediated RNAi is more advantageous for treating chronic HBV infection, which is easy to mutate in vivo.
Applications
Vector-based amiRNA could be a promising approach for the treatment of chronic HBV infection.
Peer review
AmiRNA is a hot topic and the papers dealing with it is scarce. The importance of this strategy as a therapeutic tool of HBV infection could be outstanding.
Acknowledgments
The authors thank Yu Li for his technologic supports.
Supported by The National Natural Science Foundation of China, No. 30700698
Peer reviewers: Dr. Juan Ramón Larrubia, Gastroenterology Unit and Liver Research Unit, Guadalajara University Hospital, University of Alcalá, Donante de Sangre s/n, Guadalajara 19002, Spain; Dr. Carla Brady, Duke University Medical Center, DUMC Box 3913, Durham 27705, United States
S- Editor Zhong XY L- Editor Ma JY E- Editor Ma WH
References
- 1.Radhakrishnan SK, Layden TJ, Gartel AL. RNA interference as a new strategy against viral hepatitis. Virology. 2004;323:173–181. doi: 10.1016/j.virol.2004.02.021. [DOI] [PubMed] [Google Scholar]
- 2.Novina CD, Murray MF, Dykxhoorn DM, Beresford PJ, Riess J, Lee SK, Collman RG, Lieberman J, Shankar P, Sharp PA. siRNA-directed inhibition of HIV-1 infection. Nat Med. 2002;8:681–686. doi: 10.1038/nm725. [DOI] [PubMed] [Google Scholar]
- 3.Wilson JA, Jayasena S, Khvorova A, Sabatinos S, Rodrigue-Gervais IG, Arya S, Sarangi F, Harris-Brandts M, Beaulieu S, Richardson CD. RNA interference blocks gene expression and RNA synthesis from hepatitis C replicons propagated in human liver cells. Proc Natl Acad Sci USA. 2003;100:2783–2788. doi: 10.1073/pnas.252758799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Haasnoot PC, Cupac D, Berkhout B. Inhibition of virus replication by RNA interference. J Biomed Sci. 2003;10:607–616. doi: 10.1159/000073526. [DOI] [PubMed] [Google Scholar]
- 5.Arbuthnot P, Carmona S, Ely A. Exploiting the RNA interference pathway to counter hepatitis B virus replication. Liver Int. 2005;25:9–15. doi: 10.1111/j.1478-3231.2004.0966.x. [DOI] [PubMed] [Google Scholar]
- 6.Ying RS, Zhu C, Fan XG, Li N, Tian XF, Liu HB, Zhang BX. Hepatitis B virus is inhibited by RNA interference in cell culture and in mice. Antiviral Res. 2007;73:24–30. doi: 10.1016/j.antiviral.2006.05.022. [DOI] [PubMed] [Google Scholar]
- 7.Li GQ, Gu HX, Li D, Xu WZ. Inhibition of Hepatitis B virus cccDNA replication by siRNA. Biochem Biophys Res Commun. 2007;355:404–408. doi: 10.1016/j.bbrc.2007.01.163. [DOI] [PubMed] [Google Scholar]
- 8.Uprichard SL, Boyd B, Althage A, Chisari FV. Clearance of hepatitis B virus from the liver of transgenic mice by short hairpin RNAs. Proc Natl Acad Sci USA. 2005;102:773–778. doi: 10.1073/pnas.0409028102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Das AT, Brummelkamp TR, Westerhout EM, Vink M, Madiredjo M, Bernards R, Berkhout B. Human immuno-deficiency virus type 1 escapes from RNA interference-mediated inhibition. J Virol. 2004;78:2601–2605. doi: 10.1128/JVI.78.5.2601-2605.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Konishi M, Wu CH, Kaito M, Hayashi K, Watanabe S, Adachi Y, Wu GY. siRNA-resistance in treated HCV replicon cells is correlated with the development of specific HCV mutations. J Viral Hepat. 2006;13:756–761. doi: 10.1111/j.1365-2893.2006.00752.x. [DOI] [PubMed] [Google Scholar]
- 11.Wu HL, Huang LR, Huang CC, Lai HL, Liu CJ, Huang YT, Hsu YW, Lu CY, Chen DS, Chen PJ. RNA interference-mediated control of hepatitis B virus and emergence of resistant mutant. Gastroenterology. 2005;128:708–716. doi: 10.1053/j.gastro.2004.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 13.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 14.Yekta S, Shih IH, Bartel DP. MicroRNA-directed cleavage of HOXB8 mRNA. Science. 2004;304:594–596. doi: 10.1126/science.1097434. [DOI] [PubMed] [Google Scholar]
- 15.Ying SY, Lin SL. Current perspectives in intronic micro RNAs (miRNAs) J Biomed Sci. 2006;13:5–15. doi: 10.1007/s11373-005-9036-8. [DOI] [PubMed] [Google Scholar]
- 16.Zeng Y, Wagner EJ, Cullen BR. Both natural and designed micro RNAs can inhibit the expression of cognate mRNAs when expressed in human cells. Mol Cell. 2002;9:1327–1333. doi: 10.1016/s1097-2765(02)00541-5. [DOI] [PubMed] [Google Scholar]
- 17.Schwab R, Ossowski S, Riester M, Warthmann N, Weigel D. Highly specific gene silencing by artificial microRNAs in Arabidopsis. Plant Cell. 2006;18:1121–1133. doi: 10.1105/tpc.105.039834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chung KH, Hart CC, Al-Bassam S, Avery A, Taylor J, Patel PD, Vojtek AB, Turner DL. Polycistronic RNA polymerase II expression vectors for RNA interference based on BIC/miR-155. Nucleic Acids Res. 2006;34:e53. doi: 10.1093/nar/gkl143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Murchison EP, Hannon GJ. miRNAs on the move: miRNA biogenesis and the RNAi machinery. Curr Opin Cell Biol. 2004;16:223–229. doi: 10.1016/j.ceb.2004.04.003. [DOI] [PubMed] [Google Scholar]
- 20.Lee Y, Kim M, Han J, Yeom KH, Lee S, Baek SH, Kim VN. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004;23:4051–4060. doi: 10.1038/sj.emboj.7600385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tsuda N, Kawano K, Efferson CL, Ioannides CG. Synthetic microRNA and double-stranded RNA targeting the 3’-untranslated region of HER-2/neu mRNA inhibit HER-2 protein expression in ovarian cancer cells. Int J Oncol. 2005;27:1299–1306. [PubMed] [Google Scholar]
- 22.Liang Z, Wu H, Reddy S, Zhu A, Wang S, Blevins D, Yoon Y, Zhang Y, Shim H. Blockade of invasion and metastasis of breast cancer cells via targeting CXCR4 with an artificial microRNA. Biochem Biophys Res Commun. 2007;363:542–546. doi: 10.1016/j.bbrc.2007.09.007. [DOI] [PubMed] [Google Scholar]
- 23.Wu X, Fan J, Wang X, Zhou J, Qiu S, Yu Y, Liu Y, Tang Z. Downregulation of CCR1 inhibits human hepatocellular carcinoma cell invasion. Biochem Biophys Res Commun. 2007;355:866–871. doi: 10.1016/j.bbrc.2007.01.199. [DOI] [PubMed] [Google Scholar]
- 24.Shlomai A, Shaul Y. Inhibition of hepatitis B virus expression and replication by RNA interference. Hepatology. 2003;37:764–770. doi: 10.1053/jhep.2003.50146. [DOI] [PubMed] [Google Scholar]
- 25.McCaffrey AP, Nakai H, Pandey K, Huang Z, Salazar FH, Xu H, Wieland SF, Marion PL, Kay MA. Inhibition of hepatitis B virus in mice by RNA interference. Nat Biotechnol. 2003;21:639–644. doi: 10.1038/nbt824. [DOI] [PubMed] [Google Scholar]