Abstract
Hepatitis B virus X protein (HBx) plays a crucial role in the development of hepatocellular carcinoma (HCC). However, the significance of circulating antibody to hepatitis B virus X antigen (anti-HBx) in sera remains unclear. In the present study, we examined the titers of anti-HBx (IgG) in the sera from 173 patients with chronic hepatitis B (CHB), 106 liver cirrhosis (LC), and 61 HCC by enzyme-linked immunosorbent assay (ELISA), respectively. Our data showed that the positive rates of anti-HBx were higher in sera of LC (40.6%) and HCC (34.4%) than those of CHB (10.4%), P < .05. In all 40 patients with anti-HBx+ out of 340 patients, 39 (97.5%) were HBsAg/HBeAg/anti-HBc+ and 1 (2.5%) was anti-HBs+ (P < .01), suggesting that anti-HBx in sera is a marker of HBV replication rather than a protective antibody. Thus, our findings reveal that circulating anti-HBx in sera is one of the markers of development of LC and HCC mediated by HBV.
1. Introduction
Infection of hepatitis B virus (HBV) always leads to liver cirrhosis (LC) and hepatocellular carcinoma (HCC) [1–3]. The proportion of people infected with HBV in China is higher than that in other countries. HBV is the prototype member of the hepadnaviridae family and consists of a circular partially double-stranded DNA molecule of 3.2 kb in length which contains four open reading frames (ORFs) that code for surface proteins (HBsAg), core proteins (HBcAg/HBeAg), the viral polymerase, and the transcriptional transactivator X protein [1]. Previously, we found that the positive rate of hepatitis B virus X antigen (HBxAg) was 76.5% in HCC tissues by immunohistochemistry [4]. The integration of HBV DNA into the host genome may be associated with the development of HCC [5, 6]. The hepatitis B virus X protein (HBx) is a 154 amino acid polypeptide, which has a molecular weight of 17 kDa. It has been reported that HBx plays an important role in the development of HCC. The HBx protein has been implicated in many functions associated with liver diseases such as chronic hepatitis B (CHB), LC, and HCC. The antibodies to HBxAg (anti-HBx) may serve as a preneoplastic marker for HCC [7]. However, the studies of the correlation of HBxAg and anti-HBx antibodies with the intensity of HBV replication or the clinical status of HBV-infected patients are conflicting in reports [8–10]. Hwang et al. reported that the positive rate of anti-HBx in sera of HCC patients was 70%, while 5% of sera from CHB patients contained antibodies with significant binding to the HBx protein [9]. The detection of HBxAg in patients' sera or in liver tissues also has been reported [4, 11–13]. Several researches have reported that HBx gene was detectable in HCC tissues [14, 15]. However, at present few data show the relationships between HBxAg/anti-HBx in sera and development of liver diseases with HBV infection, such as CHB, LC, and HCC.
In our present study, we examined HBxAg and anti-HBx (IgG) in a large amount of serum samples from patients suffering from CHB, LC, and HCC by enzyme-linked immunosorbent assay (ELISA). HBx gene was detected by PCR in the genome of HCC tissues as well. Our findings show that the anti-HBx in sera is a marker of HBV replication rather than a protective antibody, particularly it is one of markers of development of LC and HCC mediated by HBV.
2. Materials and Methods
2.1. Materials
Serum samples were taken from 173 patients with CHB (116 males and 57 females aged 14–69 years, with an average age of 38), 106 patients with LC (72 males and 34 females aged 23–81 years, with an average age of 53), and 61 patients with HCC (48 males and 13 females aged 23–76 years, with an average age of 57). All of the samples were obtained from Tianjin Third Central Hospital, Tianjin and Affiliated Hospital, Chengde Medical College, Chengde, China, respectively. Forty five cases of HCC tissues were taken from Tianjin First Central Hospital, Tianjin, China (totally, 42 males and 3 females aged 21–70 years, with an average age of 51.9). According to the hospital records, all patients underwent total or subtotal hepatectomy followed by pathologic diagnosis showed the examination of HBV markers, such as HBsAg, antibody to HBsAg (anti-HBs), HBeAg, antibody to HBeAg (anti-HBe), and antibody to HBcAg (anti-HBc). Normal sera of 213 individuals were taken from healthy examination (Tianjin, China). We obtained the ethics approve for using the materials of sera and HCC tissues from Hospital's Ethics Committees.
2.2. Methods
2.2.1. Plasmid Constructions and Protein Purification
Full-length of HBx gene PCR product was obtained by PCR using pCMV-X plasmid (kindly provided by Dr. A. Graessmann) as template. According to the known HBx sequence (GenBank accession number AB104894.1), the primer sequences were forward, 5′-CAGAATTCATGGCTGCTAGGCTGTGC-3′, including the restriction site of EcoRI and ATG; reverse, 5′-ATCTCGAGAGAAGTCGTCGTCGTCC-3′, including the restriction site of XhoI and TAA. The amplification condition consisted of 30 cycles at 94°C for 30 seconds, 60°C for 30 seconds, and 72°C for 45 seconds. The resulting PCR product was inserted into the pMD18-T vector (Takara, China) to yield pMD18-T-HBx. This construct was digested with EcoRI and XhoI, and the HBx gene fragment was ligated into the pET-30a vector containing His-Tag (Novagen, USA) to generate pET-30a-HBx, followed by confirmation using sequence. His-HBx fusion proteins were expressed in Escherichia coli BL21 (DE3) by induction with 0.5 mmol/L isopropyl-β-D-thiogalactopyranoside (IPTG) at 37°C for 4 hours. Cells were harvested and sonicated in Phosphate Buffered Saline (PBS; 50 mmol/L NaH2PO4, 30 mmol/L NaCl, 20 mmol/L Imidazole, pH 8.0). After centrifugation the sedimentation was collected. For purification, the extracts were denatured by buffer B (100 mmol/L NaH2PO4, 10 mmol/L Tris-HCl, 8 mol/L Urea, 20 mmol/L Imidazole, 75 mg/mL Phenylmethanesulfonyl fluoride, pH 8.0), and then incubated with Ni-NTA agarose (QIAGEN, USA) at room temperature for 1 hour. The beads were precipitated and washed according to the instruction. The purified His-HBx fusion protein was detected by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE). Thus, the eluted proteins were divided.
2.2.2. Generation of Rabbit Anti-HBx Antibodies
His-HBx fusion protein was used to raise rabbit anti-HBx antibodies. Briefly, approximately 1 mg of His-HBx fusion protein was mixed with complete Freund's adjuvant (Sigma, USA) and was subcutaneously injected into three rabbits, respectively. Booster injections consisting of 500 μg of proteins with incomplete Freund's adjuvant (Sigma) were given three times at 7-day intervals. Sera were taken by bleeding from the ear vein each time before injection. After 6 weeks, the rabbits were killed and sera were taken by bleeding from the carotid. The antibodies of rabbit against HBx were purified from the immune sera by affinity purification. The characteristics of rabbit anti-HBx antibodies were identified by Western blot analysis as the above protocol in a model of H7402-X cells [4], in which HBx was expressed.
2.2.3. Identification of Rabbit Anti-HBx Antibodies
To identify the generated rabbit anti-HBx antibodies, we examined HBx protein in H7402-X cells stably transfected with HBx gene [4] by Western blot analysis, using generated rabbit anti-HBx polyclonal antibodies and mouse anti-HBx monoclonal antibody, respectively. H7402-P cells stably transfected with empty pcDNA3 vector [4] and H7402 cells were used as controls. The cells were lysed in lysis buffer (62.5 mmol/L Tris-HCl, pH 6.8, 2% SDS, 5% β-mercaptoethanol, 10% glycerol). Equal amounts of protein (30 μg) were separated by 15% SDS-PAGE and transferred onto a nitrocellulose membrane for 1 hour. The membrane was blocked in blocking buffer (PBS, 5% skim milk, 0.1% Tween 20) at room temperature for 2 hours, and then incubated with the appropriate primary antibody (diluted in blocking buffer). The primary antibodies were mouse anti-HBx (1: 5000 dilution, Chemicon, USA), our generated rabbit anti-HBx (1: 1000 dilution) and β-actin (1: 5000 dilution, Sigma, USA). The membranes were then washed three times in PBS with 0.1% Tween 20 and incubated for 1 hour in the secondary antibody (peroxidase-conjugated antirabbit or antimouse IgG). The membranes were then washed three times, and the bands were visualized by ECL (Amersham, USA).
2.2.4. Examination of HBxAg and Circulating Anti-HBx (IgG) in Sera
HBxAg and circulating anti-HBx (IgG) in all sera from 340 patients with CHB, LC and HCC as well as 213 healthy individuals were detected by enzyme-linked immunosorbent assay (ELISA). We examined the circlating anti-HBx (IgG) in sera by an indirect ELISA method [12]. The following examination was used as positive control. Purified 100 μL of His-HBx fusion protein was immobilized on the bottoms of wells (10 μg/well) of a 96-well plate in triplicate overnight at 4°C. After treatment with 3% bovine serum albumin (BSA) at 37°C for 1 hour and washed, then reacted with 50 μL purified antibodies of rabbit anti-HBx at 37°C for 1 hour and washed, finally the antigen-antibody complex was detected by a peroxidase-conjugated goat antihuman IgG (1: 2000 dilution) and the oxidation of ortho-phenyldiamine. The degree of oxidation was measured at 490 nm. Meanwhile, we examined the circlating anti-HBx (IgG) in sera from patients, such as CHB, LC and HCC as above protocol.
Then, we examined the HBxAg in sera from patients, such as CHB, LC, and HCC by an indirect ELISA method [12]. Mouse anti-HBx monoclonal antibody (Chemicon, USA) was immobilized on the bottoms of wells of a 96-well plate in triplicate (10 μg/well) at 4°C overnight. After treatment with 3% BSA, washed with PBS, and reacted with the patients' sera at 37°C for 1 hour, washed with PBS, then reacted with purified rabbit anti-HBx polyclonal antibody (1: 250 dilution) at 37°C for 1 hour, washed with PBS, finally the antigen-antibody complex was detected by a peroxidase-conjugated goat antirabbit IgG (1: 2,000 dilution) and the oxidation of ortho-phenyldiamine. The degree of oxidation was measured at 490 nm.
2.2.5. Examination of HBx Gene in the Genome of HCC Tissues
The DNA from HCC tissues was extracted using a standard proteinase K digestion and phenol/chloroform method [16]. The resultant DNA was PCR-amplified in a final volume of 50 μL containing 1 μL DNA as template and deoxyribonucleoside triphosphates (10 mmol/L each), 2.5 U of Ex Taq (Takara, China), and 0.2 μmol/L of each primer. The HBx gene specific primers were designed according to the sequence of HBx gene (Genbank: AB 104894.1, National Institutes of Health, Bethesda, Md). The primer sequences were sence, 5′-GCGCACCTCTCTTTACGC-3′, and antisence, 5′-CGTTGGCCGATTCATTAATG-3′. The PCR program is 94°C for 20 minutes, followed by 10 cycles at 94°C for 30 seconds, 65–55°C for 45 seconds, 72°C for 1 minute, and drop the annealing temperature for 1°C in each cycle, followed by 20 cycles at 94°C for 30 seconds, 55°C for 45 seconds, 72°C for 2 minutes and elongate at 72°C for 10 minutes. The PCR products were separated by electrophoresis on a 1.5% agarose gel and visualized with the use of ethidium bromide staining. The prospected PCR product is 350 bp. The template was not added in the negative control in the PCR system. The plasmid of pCMV-X cloned HBx gene [4] was used as positive control in the PCR system.
2.2.6. Statistical Analysis
Multiple comparisons for statistically significant differences in HBxAg or anti-HBx titers, measured in optical density (OD), were performed in the sera among patients with CHB, LC, HCC, and healthy individuals by a Tukey test [12]. The significant difference between the positive rates of HBsAg/HBeAg/anti-HBc and anti-HBs in the group of HBxAg-positive patients was performed by X2 test. The result are considered as significant when the statistical test is ≤ 0.05.
3. Results
3.1. Identification of Rabbit Anti-HBx Antibodies
The HBx gene was successfully constructed into the pET-30a vector, termed pET-30a-HBx, in which the sequence was confirmed by sequencing. To examine HBxAg and circulating anti-HBx in sera, we generated rabbit anti-HBx antibodies specific to HBx. The characteristics of the anti-HBx antibodies were confirmed by Western blot analysis. The data showed that the HBx could be recognized either by monoclonal anti-HBx antibody or generated polyclonal rabbit anti-HBx antibodies in H7402-X cells stably transfected with HBx gene (Figure 1), suggesting that the specificity of the polyclonal rabbit anti-HBx antibodies is well.
Figure 1.
Identification of rabbit anti-HBx polyclonal antibody. HBx in H7402-X, H7402-P, and H7402 cells was examined by Western blot analysis, using generated rabbit anti-HBx polyclonal antibody and mouse anti-HBx monoclonal antibody, respectively. The results showed that the generated rabbit anti-HBx polyclonal antibody was effectively able to recognize HBx in H7402-X cells as mouse anti-HBx monoclonal antibody did.
3.2. Titers of Circulating Anti-HBx Are Higher in Sera of LC and/or HCC Than CHB
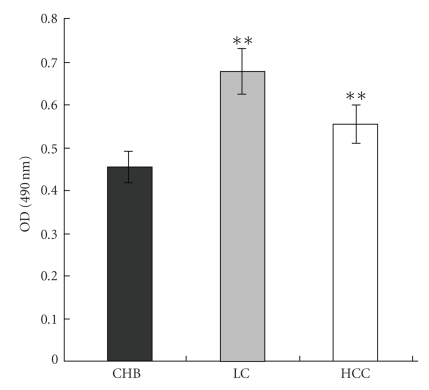
After the generation of rabbit anti-HBx, the quantities of HBxAg and anti-HBx (IgG) in sera were demonstrated by ELISA (Figure 2). The results showed that the positive rates of HBxAg/anti-HBx (IgG) were 8.7%/10.4% in 173 CHB sera, 17.9%/40.6% in 106 LC sera (P < .01, versus CHB anti-HBx+, Tukey test), and 9.8%/34.4% in 61 HCC sera (P < .01, versus CHB anti-HBx+, Tukey test), respectively, (Table 1) suggesting that the titers of anti-HBx (IgG) in sera of LC and/or HCC patients were higher than those of CHB patients. Table 2 showed that 21 out of 23 HBxAg-positive patients were HBsAg/HBeAg/anti-HBc-positive (91.3%), but only 2 out of 23 ones were anti-HBs positive (8.7%) (P < .01, X2 test). In all 40 patients with anti-HBx (IgG) positive, 39 were HBsAg/HBeAg/anti-HBc positive (97.5%) and 1 was anti-HBs positive (2.5%) (P < .01, X2 test), suggesting that the circulating anti-HBx (IgG) in sera is a marker of HBV replication rather than a protective antibody, and the circulating anti-HBx in sera is one of the markers of development of LC and HCC mediated by HBV. In control, both HBxAg and anti-HBx (IgG) were totally negative in all 213 sera of healthy individuals. Figure 2 showed that the titers of anti-HBx (IgG) were higher in sera of 106 LC patients, 61 HCC patients than those of 173 CHB patients by ELISA (P < .01, Tukey test). In sex, 65.0% (26/40) HBxAg-positive patients were males (8/15 CHB, 13/19 LC, 5/6 HCC), while 67.0% (55/82) anti-HBx-positive (IgG) patients were males (14/18 CHB, 23/43 LC, 18/21 HCC).
Figure 2.
ELISA showed the quantities of anti-HBx (IgG) in sera from clinical CHB patients, LC patients, HCC patients. The value in each group was mean OD (**P < .01, versus CHB, Tukey test).
Table 1.
Examination of HBV serum markers in CHB, LC, and HCC patients.
| Diseases | No. | HBsAg (%) | Anti-HBs (%) | HBeAg (%) | Anti-HBe (%) | Anti-HBc (%) | HBxAg (%) | Anti-HBx (IgG) (%) |
|---|---|---|---|---|---|---|---|---|
| CHB | 173 | 172 (99.4) | 5 (2.9) | 71 (41.0) | 120 (69.3) | 173 (100) | 15 (8.7) | 18 (10.4 ) |
| LC | 106 | 86 (81.1) | 9 (8.5) | 48 (45.3) | 82 (77.4) | 105 (99.1) | 19 (17.9) | 43 (40.6)* |
| HCC | 61 | 46 (75.4) | 5 (8.2) | 17 (27.9) | 45 (73.8) | 56 (91.8) | 6 (9.8) | 21 (34.4)* |
CHB chronic hepatitis B; LC liver cirrhosis; HCC hepatocellular carcinoma. X2 test, *P < .01 (versus CHB).
Table 2.
Clinical significance of HBxAg and anti-HBx in sera.
| Clinical significance | HBxAg+ (%) | anti-HBx+ (IgG) (%) |
|---|---|---|
| HBsAg/HBeAg/anti-HBc+ | 21 (91.3)* | 39 (97.5)* |
| Anti-HBs+ | 2 (8.7) | 1 (2.5) |
| Total | 23 (100) | 40 (100) |
X2 test *P < .01 (versus anti-HBs+).
3.3. Persistence of HBx Gene in HCC Tissues
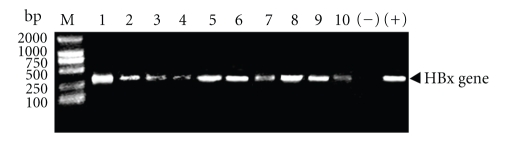
We examined the HBx gene in the 45 HCC tissues by PCR. HBx gene was detectable in 35 cases (77.8%). The PCR products were shown in Figure 3, suggesting that HBx gene was persistent in HCC tissues, which may integrate into the genome of liver cells or work as dissociative virus in the cells.
Figure 3.
Examination of HBx gene in HCC tissues by PCR. The results revealed that the HBx gene-positive rate was 77.8% (35/45) in HCC tissues. The PCR products of no. 1–10 HCC patients were shown. (−) means negative control without adding template in the PCR system. (+) means positive control using the plasmid of pCMV-X cloned HBx gene as template in the PCR system.
4. Discussion
During the past few decades, the HBV markers, such as HBsAg, anti-HBs, HBeAg, anti-HBe, and anti-HBc, in sera of patients infected with HBV were routinely examined for general diagnosis of HBV infection [17, 18]. Up to now, the clinical significance of HBxAg and anti-HBx in sera has not been thoroughly clarified. In our present study, we used the full-length HBx protein as antigen to generate rabbit anti-HBx antibodies. Thus, it is necessary to use purified full-length HBx protein as an antigen to test the accurate titers of anti-HBx in sera from various sources [9, 19]. In our experiment, the purified recombinant His-HBx protein was used as an antigen to immune rabbits, and got a good immune response. Using mouse anti-HBx monoclonal antibody as positive control, we found that the rabbit anti-HBx antibodies have good specificity and sensitivity by Western blot analysis. Based on the established ELISA method, we examined HBxAg and titers of anti-HBx (IgG) in sera of many patients infected with HBV. All of the carriers were carefully selected from different relevant stages in the natural history of chronic HBV infection with examination of HBV serum markers. However, in different reports the positive rates of HBxAg and anti-HBx in the sera of the patients were varied with the used HBx protein or HBx oligopeptides [5].
Currently, the clinical serum markers of HBV infection include HBsAg/anti-HBs, HBeAg/anti-HBe, anti-HBc [20, 21], HBV DNA, and so forth [22, 23]. Usually, the detection of above five serum markers and HBc-IgM was performed by ELISA method. However, at present the clinical serological detection of HBxAg and anti-HBx is not launched for hepatitis, cirrhosis, and liver cancer. Therefore, the clinical significance of serum HBxAg and anti-HBx remains unclear. In our present study, for HBxAg detection we found that the HBxAg-positive rate was only 9.8% (6/61) in sera of HCC patients. While for anti-HBx detection, one report demonstrated that the anti-HBx-positive rate was 70% in sera of 20 HCC patients [9]. In contrast, our data showed that the anti-HBx (IgG)-positive rates were 40.6% in sera of LC patients and 34.4% in sera of HCC patients (Table 1), respectively. Moreover, the quantities of anti-HBx (IgG) in the sera of LC or HCC patients were higher than those in sera of CHB patients as well (Figure 2), suggesting that the titers of anti-HBx in sera were closely related to the development of LC or/and HCC.
As we know, the anti-HBs in sera is a protective antibody after HBV infection or vaccination for hepatitis B vaccine. The HBsAg/HBeAg/anti-HBc-positive in sera means the replication of the viruses, and the patients are infectious. However, our data (Table 2) suggest that the anti-HBx in sera is not a protective antibody. The anti-HBx-positive patients are infectious. In this study, we found that 65.0% HBxAg-positive patients and 67.0% anti-HBx-positive (IgG) patients were males, suggesting that the male patients may be the risk population for the development of HCC. As we know, the integration of HBV DNA in the host genome was a frequent event in HCC tissues [24, 25]. It has been reported that the detection rate of HBV DNA integration was increased in parallel with the progress of liver histology towards the neoplastic transformation [26]. However, it has been reported that the HBV DNA integration-positive rate is low in chronic hepatitis tissues, but high in liver tumor tissues (66.7%) [27]. Another study which measured the intrahepatic HBV DNA and covalently closed circular DNA (cccDNA) levels in tumor and nontumor tissues in HCC patients has reported that the levels of HBV replication in the tumor tissues appeared to be lower compared to the non-tumor tissues. But the tumor tissues had significantly higher proportion of intrahepatic HBV DNA in the form of cccDNA than the non-tumor tissues [28]. In our present study, we found that the HBx gene-positive rate was 77.8% in HCC tissues by PCR, suggesting that HBx gene was persistent in HCC tissues, which may integrate into the genome of liver cells or as dissociative virus in the cells.
Taken together, our findings show that anti-HBx in sera is a marker of HBV replication rather than a protective antibody. The circulating anti-HBx in sera is one of the markers of development of LC and HCC mediated by HBV.
Acknowledgments
The project was supported by Grants of the National Basic Research Program of China (973 Program, no. 2007CB914804, no. 2007CB914802, no. 2009CB521702), the National Natural Scientific Foundation (no. 30670959), and the Chinese State Key Projects for High-Tech Program (no. 2006AA02A247).
References
- 1.Zhang X, Zhang H, Ye L. Effects of hepatitis B virus X protein on the development of liver cancer. Journal of Laboratory and Clinical Medicine. 2006;147(2):58–66. doi: 10.1016/j.lab.2005.10.003. [DOI] [PubMed] [Google Scholar]
- 2.Zhang X, Zhang W, Ye L. Pathogenesis of hepatitis B virus infection. Future Virology. 2006;1(5):637–647. [Google Scholar]
- 3.McMahon BJ. Epidemiology and natural history of hepatitis B. Seminars in Liver Disease. 2005;25(supplement 1):3–8. doi: 10.1055/s-2005-915644. [DOI] [PubMed] [Google Scholar]
- 4.Zhang X, Dong N, Yin L, et al. Hepatitis B virus X protein upregulates survivin expression in hepatoma tissues. Journal of Medical Virology. 2005;77(3):374–381. doi: 10.1002/jmv.20466. [DOI] [PubMed] [Google Scholar]
- 5.Hann H-WL, Lee J, Bussard A, et al. Preneoplastic markers of hepatitis B virus-associated hepatocellular carcinoma. Cancer Research. 2004;64(20):7329–7335. doi: 10.1158/0008-5472.CAN-04-1095. [DOI] [PubMed] [Google Scholar]
- 6.Levrero M, Stemler M, Pasquinelli C, et al. Significance of anti-HBx antibodies in hepatitis B virus infection. Hepatology. 1991;13(1):143–149. [PubMed] [Google Scholar]
- 7.Vitvitski-Trepo L, Kay A, Pichoud C, et al. Early and frequent detection of HBxAg and/or anti-HBx in hepatitis B virus infection. Hepatology. 1990;12(6):1278–1283. doi: 10.1002/hep.1840120605. [DOI] [PubMed] [Google Scholar]
- 8.Pál J, Nyárády Z, Marczinovits I, et al. Comprehensive regression analysis of hepatitis B virus X antigen level and anti-HBx antibody titer in the sera of patients with HBV infection. Pathology and Oncology Research. 2006;12(1):34–40. doi: 10.1007/BF02893429. [DOI] [PubMed] [Google Scholar]
- 9.Hwang G-Y, Lin C-Y, Huang L-M, et al. Detection of the hepatitis B virus X protein (HBx) antigen and anti-HBx antibodies in cases of human hepatocellular carcinoma. Journal of Clinical Microbiology. 2003;41(12):5598–5603. doi: 10.1128/JCM.41.12.5598-5603.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pál J, Somogyi C, Szmolenszky A, et al. Immunohistochemical assessment and prognostic value of hepatitis B virus X protein in chronic hepatitis and primary hepatocellular carcinomas using anti-HBxAg monoclonal antibody. Pathology and Oncology Research. 2001;7(3):178–184. doi: 10.1007/BF03032346. [DOI] [PubMed] [Google Scholar]
- 11.Pál J, Czömpöly T, Nyárády Z, et al. Determination of the fine epitope specificity of an anti-hepatitis B virus X protein monoclonal antibody using microanalytical and molecular biological methods. Molecular Immunology. 2003;40(5):241–246. doi: 10.1016/s0161-5890(03)00146-9. [DOI] [PubMed] [Google Scholar]
- 12.Pál J, Pálinkás L, Nyárády Z, et al. Sandwich type ELISA and a fluorescent cytometric microbead assay for quantitative determination of hepatitis B virus X antigen level in human sera. Journal of Immunological Methods. 2005;306(1-2):183–192. doi: 10.1016/j.jim.2005.08.011. [DOI] [PubMed] [Google Scholar]
- 13.Yeh C-T, Shen C-H, Tai D-I, Chu C-M, Liaw Y-F. Identification and characterization of a prevalent hepatitis B virus X protein mutant in Taiwanese patients with hepatocellular carcinoma. Oncogene. 2000;19(46):5213–5220. doi: 10.1038/sj.onc.1203903. [DOI] [PubMed] [Google Scholar]
- 14.Chen GG, Li MY, Ho RLK, Chak ECW, Lau WY, Lai PBS. Identification of hepatitis B virus X gene mutation in Hong Kong patients with hepatocellular carcinoma. Journal of Clinical Virology. 2005;34(1):7–12. doi: 10.1016/j.jcv.2005.01.006. [DOI] [PubMed] [Google Scholar]
- 15.Zhu P, Tan D, Peng Z, Liu F, Song L. Polymorphism analyses of hepatitis B virus X gene in hepatocellular carcinoma patients from Southern China. Acta Biochimica et Biophysica Sinica. 2007;39(4):265–272. doi: 10.1111/j.1745-7270.2007.00279.x. [DOI] [PubMed] [Google Scholar]
- 16.Lu W, Takahashi H, Furusato B, et al. Allelotyping analysis at chromosome arm 8p of high-grade prostatic intraepithelial neoplasia and incidental, latent, and clinical prostate cancers. Genes Chromosomes and Cancer. 2006;45(5):509–515. doi: 10.1002/gcc.20314. [DOI] [PubMed] [Google Scholar]
- 17.Ribeiro RM, Lo A, Perelson AS. Dynamics of hepatitis B virus infection. Microbes and Infection. 2002;4(8):829–835. doi: 10.1016/s1286-4579(02)01603-9. [DOI] [PubMed] [Google Scholar]
- 18.Tedder RS, Ijaz S, Gilbert N, et al. Evidence for a dynamic host-parasite relationship in e-negative hepatitis B carriers. Journal of Medical Virology. 2002;68(4):505–512. doi: 10.1002/jmv.10241. [DOI] [PubMed] [Google Scholar]
- 19.Kumar V, Jayasuryan N, Reddi H, Sahal D, Panda SK. A monoclonal antibody against the X protein of hepatitis B virus: fine mapping of its epitope and application in a quantitative ELISA of the X protein in sera of hepatitis B patients. Hybridoma. 1998;17(2):157–164. doi: 10.1089/hyb.1998.17.157. [DOI] [PubMed] [Google Scholar]
- 20.Keeffe EB, Dieterich DT, Han SB, et al. A treatment algorithm for the management of chronic hepatitis B virus infection in the United States: an update. Clinical Gastroenterology and Hepatology. 2006;4(8):936–962. doi: 10.1016/j.cgh.2006.05.016. [DOI] [PubMed] [Google Scholar]
- 21.Ali HY. Trial of lamivudine in hepatitis B surface antigen carriers with persistent hepatitis B core IgM antibody. Saudi Medical Journal. 2003;24(9):996–999. [PubMed] [Google Scholar]
- 22.Chen T, Luk JM, Cheung S-T, Yu W-C, Fan S-T. Evaluation of quantitative PCR and branched-chain DNA assay for detection of hepatitis B virus DNA in sera from hepatocellular carcinoma and liver transplant patients. Journal of Clinical Microbiology. 2000;38(5):1977–1980. doi: 10.1128/jcm.38.5.1977-1980.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gordillo RM, Gutierrez J, Casal M. Evaluation of the COBAS TaqMan 48 real-time PCR system for quantitation of hepatitis B virus DNA. Journal of Clinical Microbiology. 2005;43(7):3504–3507. doi: 10.1128/JCM.43.7.3504-3507.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lupberger J, Hildt E. Hepatitis B virus-induced oncogenesis. World Journal of Gastroenterology. 2007;13(1):74–81. doi: 10.3748/wjg.v13.i1.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang J-M, Huang T-H, Qiu H-Y, Fang X-W, Zhuang T-G, Qiu J-W. Studies on the integration of hepatitis B virus DNA sequence in human sperm chromosomes. Asian Journal of Andrology. 2002;4(3):209–212. [PubMed] [Google Scholar]
- 26.Tamori A, Nishiguchi S, Kubo S, et al. HBV DNA integration and HBV-transcript expression in non-B, non-C hepatocellular carcinoma in Japan. Journal of Medical Virology. 2003;71(4):492–498. doi: 10.1002/jmv.10514. [DOI] [PubMed] [Google Scholar]
- 27.Huang H-P, Tsuei D-J, Wang K-J, et al. Differential integration rates of hepatitis B virus DNA in the liver of children with chronic hepatitis B virus infection and hepatocellular carcinoma. Journal of Gastroenterology and Hepatology. 2005;20(8):1206–1214. doi: 10.1111/j.1440-1746.2005.03789.x. [DOI] [PubMed] [Google Scholar]
- 28.Wong DK-H, Yuen M-F, Poon RT-P, Yuen JC-H, Fung J, Lai C-L. Quantification of hepatitis B virus covalently closed circular DNA in patients with hepatocellular carcinoma. Journal of Hepatology. 2006;45(4):553–559. doi: 10.1016/j.jhep.2006.05.014. [DOI] [PubMed] [Google Scholar]