Abstract
DNA cytosine methylation represents an intrinsic modification signal of the genome that plays important roles in heritable gene silencing, heterochromatin formation and certain transgenerational epigenetic inheritance. In contrast to the process of DNA methylation that is catalyzed by specific classes of methyltransferases, molecular players underlying active DNA demethylation have long been elusive. Emerging biochemical and functional evidence suggests that active DNA demethylation in vertebrates can be mediated through DNA excision repair enzymes, similar to the well-known repair-based DNA demethylation mechanism in Arabidopsis. As key regulators, non-enzymatic Gadd45 proteins function to recruit enzymatic machineries and promote coupling of deamination, base and nucleotide-excision repair in the process of DNA demethylation. In this article, we review recent findings and discuss functional and evolutionary implications of such mechanisms underlying active DNA demethylation.
Keywords: DNA demethylation, Gadd45, Gadd45a, Gadd45b, Gadd45g, 5-methylcytosine, deaminase, glycosylase, base excision repair, nucleotide excision repair
Introduction
Addition of a methyl moiety to the 5th position of DNA cytosine base (5-methycytosine) is an evolutionarily conserved feature of most vertebrate and plant genomes.1, 2 Such DNA methylation yields a fifth coding element for the genome, and plays diverse roles in modulation and expression of the associated genomic information. In mammals, DNA methylation occurs predominantly at CpG dinucleotides, and underlies a variety of transcriptional regulatory phenomena, including imprinting, X-chromosome inactivation, transgenerational epigenetic inheritance and stable silencing of gene activity or parasitic genetic elements (Figure 1).3–5 Long known to occupy silenced genomic regions, DNA methylation is also recently found to present in actively transcribed gene bodies, where it might play a role in suppressing cryptic transcriptional initiation from interior of genes.6–8 A conserved family of DNA methyltransferases (DNMT) has been identified to catalyze DNA methylation, which consequently prevents binding of transcription factors or recruits gene silencing complex to ensure local suppressive chromatin environment.5, 9 DNMTs are essential for normal development, and DNA methylation has also been implicated in numerous diseases including cancer.10, 11 Due to the symmetric nature of CpG and maintenance methyltransferases, specific patterns of DNA methylation can be inherited during DNA replication of cell cycle. The silencing effect of DNA methylation at specific genomic loci, therefore, is believed to provide an ideal substrate for long-term propagation of gene activity states.5, 12 Such an epigenetic role of DNA methylation has gained substantial experimental evidence, and indicates that DNA methylation is not readily reversible in most circumstances.4, 13–15
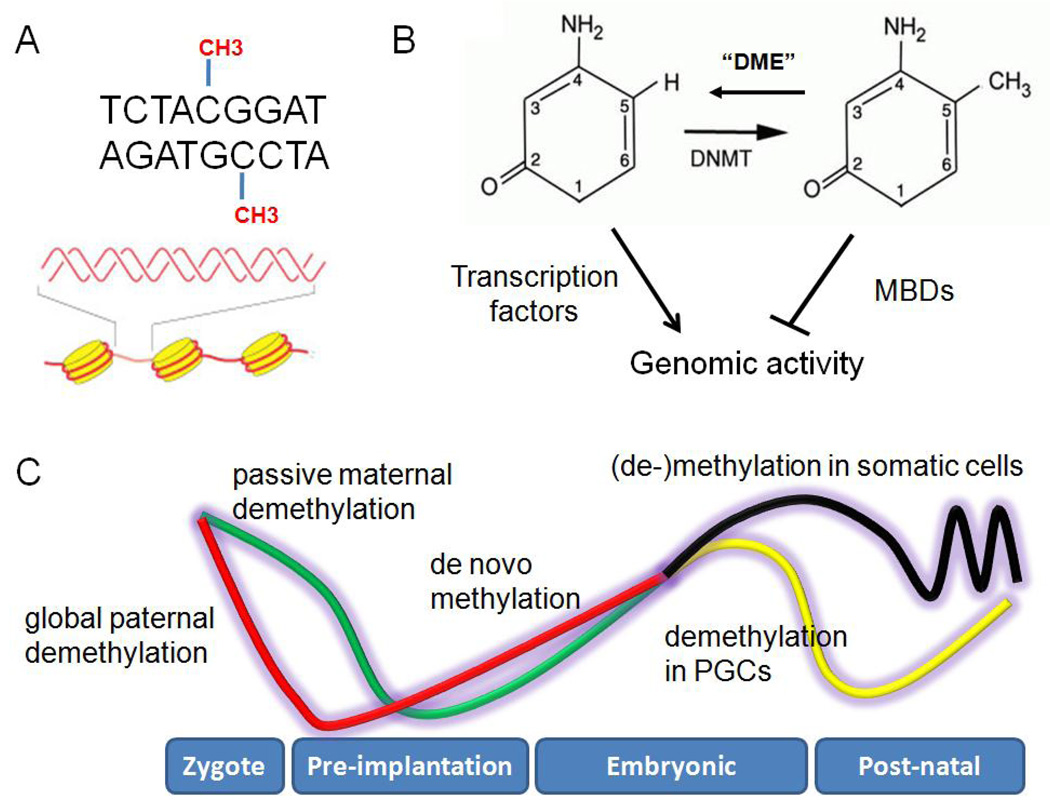
Figure 1. Overview of DNA methylation and demethylation processes.
DNA methylation occurs predominantly at cytosines of CpG dinucleotides in mammalian genomes (Figure 1A). Such methylation is catalyzed by DNA methyltransferases (DNMT) and can be reversed by DNA repair-related “demethylase” (DME) (Figure 1B). DNA methylation silences genomic activity through either preventing binding of transcription factors or recruiting methyl-CpG binding proteins (MBDs) and associated repressive complexes. During early embryonic development, the paternal genome is globally demethylated in the zygote (red line) and the maternal genome is passively demethylated during cleavage divisions (green line), followed by de novo DNA methylation in the diploid genome after implantation (Figure 1C). In primordial germ cells (PGCs), another round of global DNA demethylation takes place (yellow line) while somatic cells mostly maintain their methylation patterns during embryonic development. Certain adult somatic cells may undergo further dynamic changes of DNA methylation in response to physiological stimuli (black line).
During embryonic development and in adult somatic cells, however, DNA methylation undergoes dynamic patterns of erasure and re-establishment.16, 17 Indeed, DNA demethylation can occur through either passive mode during DNA replication, or even independently of cell cycle in many types of post-mitotic cells. Although not immediately predicted from its epigenetic function, the reversibility of DNA methylation would potentially provide long-lasting modulation of specific patterns of gene expression in response to developmental stage signals, physiological cues and environmental stimuli.
Active DNA demethylation in diverse biological systems
In mammals, specific methylation patterns of DNA in somatic cells and germ-line are set up during early development (Figure 1C). Soon after fertilization, the paternal genome of the zygote undergoes global DNA demethylation including CpG sites from many intergenic regions and single-copy genes, except most imprinted loci.18–20 The short time-window (6–8 hours after fertilization) and its DNA replication-independent nature strongly suggest an active mechanism in place. In contrast, the maternal genome undergoes gradual cell cycle-dependent demethylation during the cleavage stage. The genome-wide demethylation has been observed in most mammalian species including humans, and to a less extent, also occurs during epigenetic reprogramming of somatic cell nuclei.21–23
Soon after implantation of the blastocyst, DNA methyltransferases are deployed to re-establish cell type-specific patterns of methylation in both somatic and germ-line cells.5, 9, 16, 24 Subsequently around embryonic stage E12 in mice, primordial germ cells (PGCs) undergo genome-wide erasure of DNA methylation from many genomic CpG sites, including those from imprinted genes to set the stage for parent-of-origin specific formation of imprints and generation of totipotent zygotes.25, 26 Similar to the paternal genome demethylation in zygotes, such DNA demethylation occurring in PGCs has been shown to be an active process.25, 27 Analysis of associated changes in the chromatin demonstrates that DNA demethylation in PGCs is strongly correlated with, and likely precedes extensive histone modification and exchange that is facilitated by histone chaperone protein HIRA and NAP-1. It was proposed that such extensive chromatin modification may reflect a causal consequence of compromised DNA integrity during active DNA demethylation through DNA repair.27
Active DNA demethylation appears to occur in somatic cells as well. Once methylation patterns are established, maintenance DNA methyltransferase, mainly DNMT1, functions to preserve such patterns during somatic cell division.24 During somatic cell differentiation, most germline-specific genes are repressed by DNA methylation, while a further wave of DNA demethylation is invoked to permit expression of many tissue-specific genes, involving both passive and active mechanisms of DNA demethylation, through inhibition of DNMTs and DNA repair pathway, respectively.28–30 In fully differentiated somatic cells, active DNA demethylation has been shown to occur in T lymphocytes in response to cytokine or antibody treatment.31–33 In neurons, DNA methylation status of certain plasticity-related genes becomes dynamically regulated after animal behavioral experience or in response to electric activity stimuli.34–37 Distinct to global demethylation in early embryo and PGCs, demethylation in mature somatic cells appears to be highly region-specific.34, 35, 37
In search for mechanisms underlying active DNA demethylation
While accumulative evidence supports the existence of active DNA demethylation in mammals, searching for the DNA “demethylase” or any defined biochemical mechanisms seems to be a much more challenging task. Over the past three decades, some candidate “demethylases” have been proposed, yet few of them have yielded convincing evidence to fulfill a bona fide enzyme that can directly remove 5-methyl group off the cytosine base. Instead, the still viable mechanism to date appears to use a DNA repair-like process to remove the 5-methycytosine-containing base or a stretch of nucleotides rather than the methyl group directly (Figure 2A).16, 38 Intriguingly, this mechanism shares similarities to the well-characterized demethylation pathway in the experimental model plant Arabidopsis.39, 40
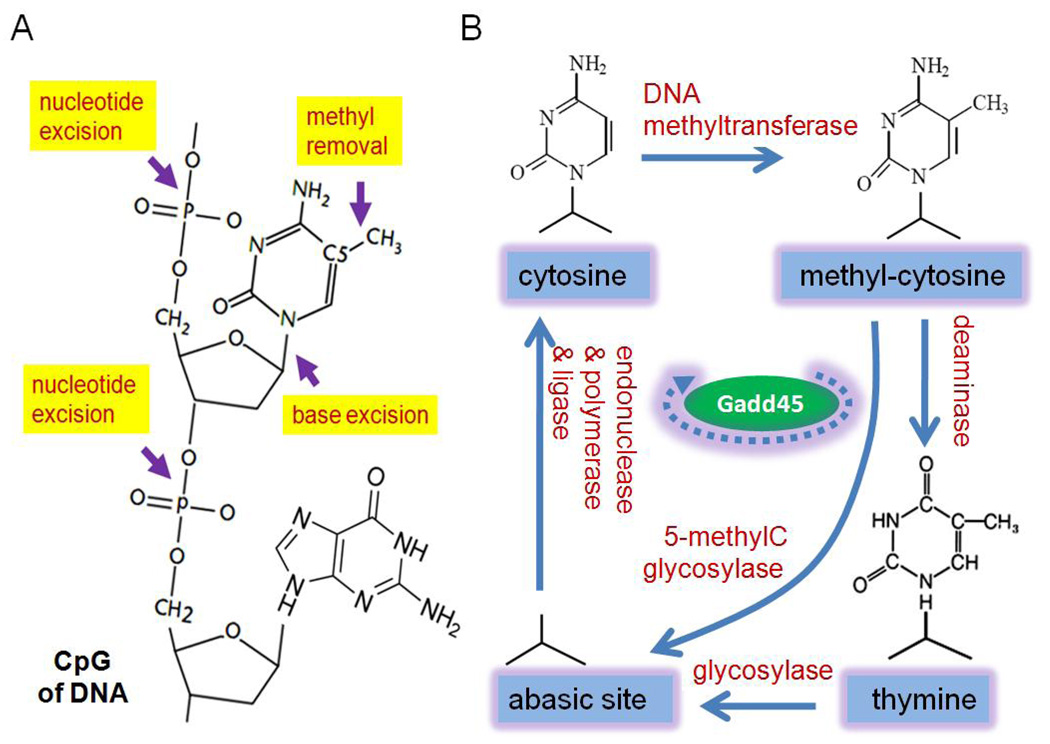
Figure 2. A mechanistic model for active DNA demethylation.
Active DNA demethylation can be potentially carried out through three mechanisms: direct removal of methyl group from the 5-methylcytosine base; base excision of 5-methylcytosine through DNA glycosylases; nucleotide excision of 5-methylcytosine through endonucleases (Figure 2A). Base and nucleotide excisions are followed by similar subsequent fill-in repair with unmethylated cytosine. A schematic illustration of DNA base structures indicates the DNA methylation cycle, each step of which is catalyzed by specific classes of enzymes in the proposed BER model (Figure 2B). 5-methylcytosine initially formed by DNA methyltransferases might be directly removed by DNA glycosylases or indirectly by thymine glycosylase after being converted by cytidine deaminases. Abasic sites are then filled-in through DNA polymerases and ligases. Gadd45 proteins promote the coupling of deamination, glycosylation and fill-in repair resulted from both base and nucleotide excisions. In addition, it may also play a more general role in gating or recruiting DNA demethylation machineries to specific genomic loci.
The earliest effort towards identifying a mammalian demethylase used nuclear extracts of murine erythroleukemia cells that evidently removed tritiated methyl signals from DNA.41 The active component in this nuclear extract was protease-sensitive, though its molecular identity was not characterized and might be explained by later studies as DNA glycosylases.42 From lysates of chick embryos, Jost and colleagues purified a 5-methylcytosine DNA glycosylase activity, and further identified thymine DNA glycosylase (TDG) as the active component.43 The activity of 5-methylcytosine glycosylase might be highly regulated in vivo, since the efficiency and specific activity of TDG on the methylated CpG substrate in vitro is insufficient to account for active DNA demethylation in vivo.44 The mammalian genome encodes a variety of DNA glycosylases, most of which are involved in DNA repair and might also function redundantly.44 In addition to TDG, methyl-CpG binding protein 4 (MBD4) has been examined to exhibit low levels of glycosylase activity on 5-methylcytosine.45 Furthermore, mechanisms have been proposed based on a ribozyme-like RNA-protein complex, or MBD2 that directly removed the methyl group from the pyrimidine ring of 5-methylcytosine.46, 47 Such findings, however, remain to be reproduced.14, 48
More recently, a series of studies have provided strong evidence supporting the DNA repair-like process for active DNA demethylation. In an expression cloning screen for genes that activate methylated reporter plasmids, Barreto and colleagues identified Gadd45a as a non-enzymatic factor that promotes active DNA demethylation, seemingly in a global fashion in cultured cells.49 The mechanism might be mediated by nucleotide excision repair (NER) since Gadd45a interacts with the NER endonuclease XPG, and the demethylation appears to require transcription-coupled NER. Though the study was challenged,50 the discrepancy could be explained by different experimental conditions (Ma et al., unpublished observation). In the proposed mechanism, 5-methylcytosine containing nucleotides are recognized and removed through Gadd45-XPG complex, and the resulting gap in DNA is filled by DNA polymerase δ/ε and ligase with resulting CpGs unmethylated.49 Since NER is preferably initiated by distortion of the DNA double-helical structure, or in the transcription-coupled repair by an arrested RNA polymerase, it remains to be determined how NER is initiated in the actively demethylated genomic regions. Notably, Gadd45 and XPG have also been implicated in base excision repair (BER), thus such demethylation process might be equally explained by BER-related mechanisms as elaborated below.51–53
Similar to NER during the nucleotide fill-in step, BER has been shown to excise methylated cytosine through distinct mechanisms. 5-methylcytosine glycosylase studied by Jost et al. would provide a direct means for removing the whole 5-methylcytosine base by breaking the β-N glycosidic bond to create an apurinic/apyrimidinic (AP) site and excision of AP mononucleotide by endonucleases, which is followed by fill-in of unmethylated nucleotides through DNA polymerase and ligases (Figure 2B).42 Alternatively, a cytidine deaminase would convert the 5-methylcytosine into a thymine, followed by glycosylase removal and replacement of unmethylated nucleotides. Metivier and colleagues provided evidence supporting such mechanism in cell lines and in vitro.54 Surprisingly, the enzyme responsible for DNA methylation itself, DNMT3, appears to possess deaminase activity in the absence of its methyltransferase substrate S-adenosyl methionine (SAM). While it remains to be shown whether SAM concentration would ever reach an optimal local concentration for deamination in vivo, such molecular Jekyll-and-Hyde aspects of DNMT3 would be appealing due to the economic molecular design, yet would require exceedingly exquisite regulation of its dual enzymatic activities under physiological conditions.
The emerging consensus of current biochemical and functional studies has pointed to DNA excision repair (NER or BER) as critical steps during active DNA demethylation (Figure 2A). Such mechanisms are apparently evolutionarily conserved since 5-methylcytosine glycosylases DME and ROS1 in Arabidopsis coupled with DNA repair protein complexes are characterized as bona fide “demethylases”.40 The exact molecular players involved in the mammalian pathway, however, seem to be more diverged. Both NER and BER may be involved, and depending on the context, the manifestation of DNA demethylation is likely dynamic and strand-specific during transcription.34, 54 Although TDG and MBD4 can exhibit certain activities on 5-methylcytosine, no mammalian 5-methylcytosine specific glycosylases have been unequivocally identified. The deaminase in the first step of the pathway is also unlikely exclusive to DNMTs. Rai and colleagues demonstrate that in zebrafish embryo, a cytidine deaminase AID is coupled with DNA glycosylase TDG in directing active DNA demethylation.55 Such coupling of the two enzymes is promoted by a non-enzymatic factor Gadd45, reminiscent of its role in facilitating DNA demethylation through NER (Figure 2B).
Gadd45 family proteins as regulators for active DNA demethylation
Gadd45 family proteins were originally identified as stress-inducible factors that play important roles in regulation of cell cycle arrest.56 There are three members of this family (Gadd45a, b, g) that share high amino acid identity. Gadd45 proteins are small (~17kD), highly acidic, and present in both cytoplasm and nucleus. A variety of proteins have been shown to interact with Gadd45, including p21, Cdc2, Histone 3/4, p38 MAPKs, Proliferating Cell Nuclear Antigen (PCNA), nuclear receptors, and a myriad of DNA repair proteins such as XPG and TDG.57 The functional roles of Gadd45 proteins have been shown to promote DNA excision repair, cell cycle arrest, maintain genome stability and perpetuate immune stimuli in lymphocytes.57–60 Molecular functions of Gadd45a, b and g are similar, yet their expression patterns are distinct in different contexts. In the nervous system, Gadd45b is a neuronal activity-inducible immediate early gene whereas Gadd45a is preferably activated by injury signals.34, 61 Recently, it has been shown that Gadd45b-induction was essential for region-specific, active DNA demethylation in mature neurons in response to electric activity stimuli.34 Such transient stimuli evoked up-regulation of certain neurotrophic factors and elicited long-lasting modulation of adult neurogenesis, providing genetic evidence in an animal model for a plausible biological function of active DNA demethylation in vivo.
How might Gadd45b promote active DNA demethylation? As discussed above, Gadd45 proteins strongly promote both BER and NER by direct coupling of deamination, glycosylation and fill-in repair machineries (Figure 2B). The effects of Gadd45 on excision repair pathway have been supported by both gain-of-function and loss-of-function evidence.49, 53 Gadd45 proteins can also oligomerize, thus may facilitate the coupling of multiple enzymatic steps required for excision repair-based DNA demethylation.62 In addition, its known interaction with acetylated histones at high affinity might function to sequester the nucleosomal histone away from its target genomic regions, allowing enhanced gene accessibility to demethylation complexes.63, 64 Interestingly, Gadd45 proteins share a stretch of 10-amino-acid identity with the histone chaperone nucleophosmin, suggesting a similar role of Gadd45 in chromatin decondensation.63
Recent studies have showed remarkable target specificity of Gadd45b-mediated demethylation in vivo.34 One likely mechanism is through binding of Gadd45 to sequence-specific transcription factors. It is known that Gadd45 proteins directly interact with many nuclear hormone receptors, including RXRα, RARα, ERα, PPARα, PPARβ, and PPARγ2.65 Another plausible mechanism is through its avid binding to nucleic acids. Gadd45 belongs to ribosomal protein L7Ae/L30e/S12e/Gadd45 superfamily and presumably binds to RNA or DNA directly. Interestingly, a recent study has shown that active ribosomal DNA demethylation is mediated by Gadd45a, raising the possibility that local transcribed RNA may recruit Gadd45 and associated complex for region-specific DNA demethylation.65
Given their inducible nature and their low expression levels in most basal conditions, Gadd45 proteins might play a key role in linking external stimuli to epigenetic DNA demethylation and subsequent adaptive gene expression. The abundance of Gadd45 proteins in vivo is generally low, and induced Gadd45 proteins may be ideally suited to serve as rate-limiting factors for triggering the activation of DNA demethylation pathway. Consistently, DNA methylation patterns in both Gadd45a and Gadd45b null mice are reported to be largely indistinguishable.34, 66 Only stimuli-dependent, region-specific demethylation is impaired in the Gadd45 null mice. The Gadd45 family proteins are multi-functional in different cellular processes, and appear to have also evolved a novel function in epigenetic control of gene expression in response to developmental, physiological and environmental stimuli.
Perspectives
Emerging evidence suggests that Gadd45-coupled DNA excision repair-like demethylation pathway is utilized to promote region-specific DNA demethylation. It remains unclear whether the DNA excision repair-like process is also involved in global DNA demethylation observed in early embryos, though such scenario would engender considerable compromise of DNA integrity. Zygotes and PGCs contain an unusually high DNA repair activity, indicating that a robust surveillance mechanism would be in place to ensure genomic integrity during repair-mediated demethylation.67 In addition, the AP endonuclease during the excision repair is inhibited by its product from the other strand to prevent DNA double strand break.68 Intriguingly, the link between DNA methylation and repair is evolutionarily ancient.69 A “demethylase” that directly removes the methyl group from 5-methylcytosine at a genome-wide global level might still exist. Potential candidates include certain types of oxygenase, decarboxylase and carbon-carbon hydroxylase, with fulfilling criteria that would be required to establish at both biochemical and genetic levels. Given the known interaction partners of Gadd45, it is likely that Gadd45 may also play a general role in gating or recruitment of demethylation machineries (Figure 2B).
The widespread presence of DNA methylation in almost all vertebrates and plants suggest its evolutionarily conserved roles in modulation of genomic activity. Many invertebrate eukaryotes, including budding yeasts and nematode worms, do not possess DNA methylation. Such facts may support the so-called Gilbert’s conjecture that DNA methylation is not one of the primary, top-level controls on gene expression or developmental program.69 Instead, vertebrate DNA methylation in a region-specific manner may have evolved to enable extra levels of fail-safe locking and more long-lasting control of genomic activity, secondary to primary transcriptional repressor and small RNA-mediated initiation of gene silencing. DNA demethylation would further provide the corresponding key to reverse such silencing in eliciting long-term gene activation in appropriate developmental or physiological conditions. The evolutionary significance of global DNA demethylation observed in early developing embryos of most mammals remains obscure, though it would possibly serve to further sculpt paternal-specific imprinting loci and to restore totipotency for finer execution of a mammalian-specific genetic program to generate the diversity of cell types.
Acknowledgement
The research in Drs. Ming and Song’s laboratories was supported by the National Institute of Health (AG024984, NS047344, MH084018, NS048271), March of Dimes, NARSAD, and MSCRF.
A list of abbreviations and acronyms
- (DNMT)
DNA methyltransferases
- (PGCs)
primordial germ cells
- (MBD)
methyl-CpG binding protein
- (NER)
nucleotide excision repair
- (BER)
base excision repair
- (AP)
apurinic/apyrimidinic
- (SAM)
S-adenosyl methionine
References
- 1.Holliday R, Pugh JE. DNA modification mechanisms and gene activity during development. Science. 1975;187:226–232. [PubMed] [Google Scholar]
- 2.Razin A, Cedar H. DNA methylation and gene expression. Microbiol Rev. 1991;55:451–458. doi: 10.1128/mr.55.3.451-458.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jones PA, Takai D. The role of DNA methylation in mammalian epigenetics. Science. 2001;293:1068–1070. doi: 10.1126/science.1063852. [DOI] [PubMed] [Google Scholar]
- 4.Youngson NA, Whitelaw E. Transgenerational epigenetic effects. Annu Rev Genomics Hum Genet. 2008;9:233–257. doi: 10.1146/annurev.genom.9.081307.164445. [DOI] [PubMed] [Google Scholar]
- 5.Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 6.Zilberman D, Gehring M, Tran RK, Ballinger T, Henikoff S. Genome-wide analysis of Arabidopsis thaliana DNA methylation uncovers an interdependence between methylation and transcription. Nat Genet. 2007;39:61–69. doi: 10.1038/ng1929. [DOI] [PubMed] [Google Scholar]
- 7.Hellman A, Chess A. Gene body-specific methylation on the active X chromosome. Science. 2007;315:1141–1143. doi: 10.1126/science.1136352. [DOI] [PubMed] [Google Scholar]
- 8.Zhang X, Yazaki J, Sundaresan A, Cokus S, Chan SW, Chen H, et al. Genome-wide high-resolution mapping and functional analysis of DNA methylation in arabidopsis. Cell. 2006;126:1189–1201. doi: 10.1016/j.cell.2006.08.003. [DOI] [PubMed] [Google Scholar]
- 9.Goll MG, Bestor TH. Eukaryotic cytosine methyltransferases. Annu Rev Biochem. 2005;74:481–514. doi: 10.1146/annurev.biochem.74.010904.153721. [DOI] [PubMed] [Google Scholar]
- 10.Li E, Bestor TH, Jaenisch R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell. 1992;69:915–926. doi: 10.1016/0092-8674(92)90611-f. [DOI] [PubMed] [Google Scholar]
- 11.Ohm JE, Baylin SB. Stem cell chromatin patterns: an instructive mechanism for DNA hypermethylation? Cell Cycle. 2007;6:1040–1043. doi: 10.4161/cc.6.9.4210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jaenisch R, Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat Genet. 2003;33 Suppl:245–254. doi: 10.1038/ng1089. [DOI] [PubMed] [Google Scholar]
- 13.Wigler M, Levy D, Perucho M. The somatic replication of DNA methylation. Cell. 1981;24:33–40. doi: 10.1016/0092-8674(81)90498-0. [DOI] [PubMed] [Google Scholar]
- 14.Ooi SK, Bestor TH. The colorful history of active DNA demethylation. Cell. 2008;133:1145–1148. doi: 10.1016/j.cell.2008.06.009. [DOI] [PubMed] [Google Scholar]
- 15.Probst AV, Dunleavy E, Almouzni G. Epigenetic inheritance during the cell cycle. Nat Rev Mol Cell Biol. 2009;10:192–206. doi: 10.1038/nrm2640. [DOI] [PubMed] [Google Scholar]
- 16.Morgan HD, Santos F, Green K, Dean W, Reik W. Epigenetic reprogramming in mammals. Hum Mol Genet. 2005;14:R47–R58. doi: 10.1093/hmg/ddi114. Spec No 1. [DOI] [PubMed] [Google Scholar]
- 17.Reik W, Dean W, Walter J. Epigenetic reprogramming in mammalian development. Science. 2001;293:1089–1093. doi: 10.1126/science.1063443. [DOI] [PubMed] [Google Scholar]
- 18.Mayer W, Niveleau A, Walter J, Fundele R, Haaf T. Demethylation of the zygotic paternal genome. Nature. 2000;403:501–502. doi: 10.1038/35000656. [DOI] [PubMed] [Google Scholar]
- 19.Oswald J, Engemann S, Lane N, Mayer W, Olek A, Fundele R, et al. Active demethylation of the paternal genome in the mouse zygote. Curr Biol. 2000;10:475–478. doi: 10.1016/s0960-9822(00)00448-6. [DOI] [PubMed] [Google Scholar]
- 20.Yao H, Li B, Wang S, Kou Z, Zhang Y, Gao S, et al. Genome-wide and gene specific paternal demethylation in androgenetic embryos. Front Biosci. 2009;14:3884–3891. doi: 10.2741/3497. [DOI] [PubMed] [Google Scholar]
- 21.Fulka H, Mrazek M, Tepla O, Fulka J., Jr DNA methylation pattern in human zygotes and developing embryos. Reproduction. 2004;128:703–708. doi: 10.1530/rep.1.00217. [DOI] [PubMed] [Google Scholar]
- 22.Simonsson S, Gurdon J. DNA demethylation is necessary for the epigenetic reprogramming of somatic cell nuclei. Nat Cell Biol. 2004;6:984–990. doi: 10.1038/ncb1176. [DOI] [PubMed] [Google Scholar]
- 23.Ma DK, Chiang CH, Ponnusamy K, Ming GL, Song H. G9a and Jhdm2a regulate embryonic stem cell fusion-induced reprogramming of adult neural stem cells. Stem Cells. 2008;26:2131–2141. doi: 10.1634/stemcells.2008-0388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen T, Li E. Establishment and maintenance of DNA methylation patterns in mammals. Curr Top Microbiol Immunol. 2006;301:179–201. doi: 10.1007/3-540-31390-7_6. [DOI] [PubMed] [Google Scholar]
- 25.Hajkova P, Erhardt S, Lane N, Haaf T, El-Maarri O, Reik W, et al. Epigenetic reprogramming in mouse primordial germ cells. Mech Dev. 2002;117:15–23. doi: 10.1016/s0925-4773(02)00181-8. [DOI] [PubMed] [Google Scholar]
- 26.Lee J, Inoue K, Ono R, Ogonuki N, Kohda T, Kaneko-Ishino T, et al. Erasing genomic imprinting memory in mouse clone embryos produced from day 11.5 primordial germ cells. Development. 2002;129:1807–1817. doi: 10.1242/dev.129.8.1807. [DOI] [PubMed] [Google Scholar]
- 27.Hajkova P, Ancelin K, Waldmann T, Lacoste N, Lange UC, Cesari F, et al. Chromatin dynamics during epigenetic reprogramming in the mouse germ line. Nature. 2008;452:877–881. doi: 10.1038/nature06714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Song F, Mahmood S, Ghosh S, Liang P, Smiraglia DJ, Nagase H, et al. Tissue specific differentially methylated regions (TDMR): Changes in DNA methylation during development. Genomics. 2009;93:130–139. doi: 10.1016/j.ygeno.2008.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jost JP, Oakeley EJ, Zhu B, Benjamin D, Thiry S, Siegmann M, et al. 5-Methylcytosine DNA glycosylase participates in the genome-wide loss of DNA methylation occurring during mouse myoblast differentiation. Nucleic Acids Res. 2001;29:4452–4461. doi: 10.1093/nar/29.21.4452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Frank D, Keshet I, Shani M, Levine A, Razin A, Cedar H. Demethylation of CpG islands in embryonic cells. Nature. 1991;351:239–241. doi: 10.1038/351239a0. [DOI] [PubMed] [Google Scholar]
- 31.Bruniquel D, Schwartz RH. Selective, stable demethylation of the interleukin-2 gene enhances transcription by an active process. Nat Immunol. 2003;4:235–240. doi: 10.1038/ni887. [DOI] [PubMed] [Google Scholar]
- 32.Kim ST, Fields PE, Flavell RA. Demethylation of a specific hypersensitive site in the Th2 locus control region. Proc Natl Acad Sci U S A. 2007;104:17052–17057. doi: 10.1073/pnas.0708293104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Murayama A, Sakura K, Nakama M, Yasuzawa-Tanaka K, Fujita E, Tateishi Y, et al. A specific CpG site demethylation in the human interleukin 2 gene promoter is an epigenetic memory. Embo J. 2006;25:1081–1092. doi: 10.1038/sj.emboj.7601012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ma DK, Jang MH, Guo JU, Kitabatake Y, Chang ML, Pow-Anpongkul N, et al. Neuronal activity-induced Gadd45b promotes epigenetic DNA demethylation and adult neurogenesis. Science. 2009;323:1074–1077. doi: 10.1126/science.1166859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Miller CA, Sweatt JD. Covalent modification of DNA regulates memory formation. Neuron. 2007;53:857–869. doi: 10.1016/j.neuron.2007.02.022. [DOI] [PubMed] [Google Scholar]
- 36.Lubin FD, Roth TL, Sweatt JD. Epigenetic regulation of BDNF gene transcription in the consolidation of fear memory. J Neuron. 2008;28:10576–10586. doi: 10.1523/JNEUROSCI.1786-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Weaver IC, Cervoni N, Champagne FA, D'Alessio AC, Sharma S, Seckl JR, et al. Epigenetic programming by maternal behavior. Nat Neurosci. 2004;7:847–854. doi: 10.1038/nn1276. [DOI] [PubMed] [Google Scholar]
- 38.Gehring M, Reik W, Henikoff S. DNA demethylation by DNA repair. Trends Genet. 2009;25:82–90. doi: 10.1016/j.tig.2008.12.001. [DOI] [PubMed] [Google Scholar]
- 39.Huh JH, Bauer MJ, Hsieh TF, Fischer RL. Cellular programming of plant gene imprinting. Cell. 2008;132:735–744. doi: 10.1016/j.cell.2008.02.018. [DOI] [PubMed] [Google Scholar]
- 40.Kapoor A, Agius F, Zhu JK. Preventing transcriptional gene silencing by active DNA demethylation. FEBS Lett. 2005;579:5889–5898. doi: 10.1016/j.febslet.2005.08.039. [DOI] [PubMed] [Google Scholar]
- 41.Gjerset RA, Martin DW., Jr Presence of a DNA demethylating activity in the nucleus of murine erythroleukemic cells. J Biol Chem. 1982;257:8581–8583. [PubMed] [Google Scholar]
- 42.Jost JP. Nuclear extracts of chicken embryos promote an active demethylation of DNA by excision repair of 5-methyldeoxycytidine. Proc Natl Acad Sci U S A. 1993;90:4684–4688. doi: 10.1073/pnas.90.10.4684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhu B, Zheng Y, Hess D, Angliker H, Schwarz S, Siegmann M, et al. 5-methylcytosine-DNA glycosylase activity is present in a cloned G/T mismatch DNA glycosylase associated with the chicken embryo DNA demethylation complex. Proc Natl Acad Sci U S A. 2000;97:5135–5139. doi: 10.1073/pnas.100107597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cortazar D, Kunz C, Saito Y, Steinacher R, Schar P. The enigmatic thymine DNA glycosylase. DNA Repair (Amst) 2007;6:489–504. doi: 10.1016/j.dnarep.2006.10.013. [DOI] [PubMed] [Google Scholar]
- 45.Zhu B, Zheng Y, Angliker H, Schwarz S, Thiry S, Siegmann M, et al. 5-Methylcytosine DNA glycosylase activity is also present in the human MBD4 (G/T mismatch glycosylase) and in a related avian sequence. Nucleic Acids Res. 2000;28:4157–4165. doi: 10.1093/nar/28.21.4157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Weiss A, Keshet I, Razin A, Cedar H. DNA demethylation in vitro: involvement of RNA. Cell. 1996;86:709–718. doi: 10.1016/s0092-8674(00)80146-4. [DOI] [PubMed] [Google Scholar]
- 47.Bhattacharya SK, Ramchandani S, Cervoni N, Szyf M. A mammalian protein with specific demethylase activity for mCpG DNA. Nature. 1999;397:579–583. doi: 10.1038/17533. [DOI] [PubMed] [Google Scholar]
- 48.Ng HH, Zhang Y, Hendrich B, Johnson CA, Turner BM, Erdjument-Bromage H, et al. MBD2 is a transcriptional repressor belonging to the MeCP1 histone deacetylase complex. Nat Genet. 1999;23:58–61. doi: 10.1038/12659. [DOI] [PubMed] [Google Scholar]
- 49.Barreto G, Schafer A, Marhold J, Stach D, Swaminathan SK, Handa V, et al. Gadd45a promotes epigenetic gene activation by repair-mediated DNA demethylation. Nature. 2007;445:671–675. doi: 10.1038/nature05515. [DOI] [PubMed] [Google Scholar]
- 50.Jin SG, Guo C, Pfeifer GP. GADD45A does not promote DNA demethylation. PLoS Genet. 2008;4 doi: 10.1371/journal.pgen.1000013. e1000013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bessho T. Nucleotide excision repair 3' endonuclease XPG stimulates the activity of base excision repairenzyme thymine glycol DNA glycosylase. Nucleic Acids Res. 1999;27:979–983. doi: 10.1093/nar/27.4.979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Klungland A, Hoss M, Gunz D, Constantinou A, Clarkson SG, Doetsch PW, et al. Base excision repair of oxidative DNA damage activated by XPG protein. Mol Cell. 1999;3:33–42. doi: 10.1016/s1097-2765(00)80172-0. [DOI] [PubMed] [Google Scholar]
- 53.Jung HJ, Kim EH, Mun JY, Park S, Smith ML, Han SS, et al. Base excision DNA repair defect in Gadd45a-deficient cells. Oncogene. 2007;26:7517–7525. doi: 10.1038/sj.onc.1210557. [DOI] [PubMed] [Google Scholar]
- 54.Metivier R, Gallais R, Tiffoche C, Le Peron C, Jurkowska RZ, Carmouche RP, et al. Cyclical DNA methylation of a transcriptionally active promoter. Nature. 2008;452:45–50. doi: 10.1038/nature06544. [DOI] [PubMed] [Google Scholar]
- 55.Rai K, Huggins IJ, James SR, Karpf AR, Jones DA, Cairns BR. DNA demethylation in zebrafish involves the coupling of a deaminase, a glycosylase, and gadd45. Cell. 2008;135:1201–1212. doi: 10.1016/j.cell.2008.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fornace AJ, Jr, Alamo I, Jr, Hollander MC. DNA damage-inducible transcripts in mammalian cells. Proc Natl Acad Sci U S A. 1988;85:8800–8804. doi: 10.1073/pnas.85.23.8800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hoffman B, Liebermann DA. Gadd45 modulation of intrinsic and extrinsic stress responses in myeloid cells. J Cell Physiol. 2009;218:26–31. doi: 10.1002/jcp.21582. [DOI] [PubMed] [Google Scholar]
- 58.Smith ML, Chen IT, Zhan Q, Bae I, Chen CY, Gilmer TM, et al. Interaction of the p53-regulated protein Gadd45 with proliferating cell nuclear antigen. Science. 1994;266:1376–1380. doi: 10.1126/science.7973727. [DOI] [PubMed] [Google Scholar]
- 59.Lu B, Ferrandino AF, Flavell RA. Gadd45beta is important for perpetuating cognate and inflammatory signals in T cells. Nat Immunol. 2004;5:38–44. doi: 10.1038/ni1020. [DOI] [PubMed] [Google Scholar]
- 60.Hollander MC, Sheikh MS, Bulavin DV, Lundgren K, Augeri-Henmueller L, Shehee R, et al. Genomic instability in Gadd45a-deficient mice. Nat Genet. 1999;23:176–184. doi: 10.1038/13802. [DOI] [PubMed] [Google Scholar]
- 61.Befort K, Karchewski L, Lanoue C, Woolf CJ. Selective up-regulation of the growth arrest DNA damage-inducible gene Gadd45 alpha in sensory and motor neurons after peripheral nerve injury. Eur J Neurosci. 2003;18:911–922. doi: 10.1046/j.1460-9568.2003.02827.x. [DOI] [PubMed] [Google Scholar]
- 62.Kovalsky O, Lung FD, Roller PP, Fornace AJ., Jr Oligomerization of human Gadd45a protein. J Biol Chem. 2001;276:39330–39339. doi: 10.1074/jbc.M105115200. [DOI] [PubMed] [Google Scholar]
- 63.Carrier F, Georgel PT, Pourquier P, Blake M, Kontny HU, Antinore MJ, et al. Gadd45, a p53-responsive stress protein, modifies DNA accessibility on damaged chromatin. Mol Cell Biol. 1999;19:1673–1685. doi: 10.1128/mcb.19.3.1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.D'Alessio AC, Weaver IC, Szyf M. Acetylation-induced transcription is required for active DNA demethylation in methylation-silenced genes. Mol Cell Biol. 2007;27:7462–7474. doi: 10.1128/MCB.01120-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yi YW, Kim D, Jung N, Hong SS, Lee HS, Bae I. Gadd45 family proteins are coactivators of nuclear hormone receptors. Biochem Biophys Res Commun. 2000;272:193–198. doi: 10.1006/bbrc.2000.2760. [DOI] [PubMed] [Google Scholar]
- 66.Engel N, Tront JS, Erinle T, Nguyen N, Latham KE, Sapienza C, et al. Conserved DNA methylation in Gadd45a(−/−) mice. Epigenetics. 2009:4. doi: 10.4161/epi.4.2.7858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zeng F, Baldwin DA, Schultz RM. Transcript profiling during preimplantation mouse development. Dev Biol. 2004;272:483–496. doi: 10.1016/j.ydbio.2004.05.018. [DOI] [PubMed] [Google Scholar]
- 68.David-Cordonnier MH, Cunniffe SM, Hickson ID, O'Neill P. Efficiency of incision of an AP site within clustered DNA damage by the major human AP endonuclease. Biochemistry. 2002;41:634–642. doi: 10.1021/bi011682l. [DOI] [PubMed] [Google Scholar]
- 69.Smith SS. Gilbert's conjecture: the search for DNA (cytosine-5) demethylases and the emergence of new functions for eukaryotic DNA (cytosine-5) methyltransferases. J Mol Biol. 2000;302:1–7. doi: 10.1006/jmbi.2000.4046. [DOI] [PubMed] [Google Scholar]