Abstract
We assessed a set of biological (HDL, LDL, SGOT, SGPT, GGT, HTc, Hb and T levels) and psychometric variables (investigated through HAM-D, HAM-A, GAS, Liebowitz Social Anxiety Scale, Mark & Mathews Scale, Leyton scale, and Pilowski scale) in a sample of 64 alcohol dependent patients, at baseline and after a detoxification treatment. Moreover, we recruited 47 non-consanguineous relatives who did not suffer alcohol related disorders and underwent the same tests. In both groups we genotyped 11 genetic variations (rs1800587; rs3087258; rs1799724; 5-HTTLPR; rs1386493; rs1386494; rs1487275; rs1843809; rs4570625; rs2129575; rs6313) located in genes whose impact on alcohol related behaviors and disorders has been hypothesized (IL1A, IL1B, TNF, 5-HTTLPR, TPH2 and HTR2A). We analyzed the epistasis of these genetic variations upon the biological and psychological dimensions in the cases and their relatives. Further on, we analyzed the effects of the combined genetic variations on the short – term detoxification treatment efficacy. Finally, being the only not yet investigated variation within this sample, we analyzed the impact of the rs6313 alone on baseline assessment and treatment efficacy. We detected the following results: the couple rs6313 + rs2129575 affected the Leyton -Trait at admission (p = 0.01) (obsessive-compulsive trait), whilst rs1800587 + 5-HTTLPR impacted the Pilowski test at admission (p = 0.01) (hypochondriac symptoms). These results did not survive Bonferroni correction (p ≤ 0.004). This lack of association may depend on the incomplete gene coverage or on the small sample size which limited the power of the study. On the other hand, it may reflect a substantial absence of relevance of the genotype variants toward the alcohol related investigated dimensions. Nonetheless, the marginal significance we detected could witness an informative correlation worth investigating in larger samples.
Keywords: gene, alcohol dependence, IL1A, IL1B, TNF, 5HTPR, HTR2A, TPH2, association
1. Introduction
Genes coding for: interleukin 1, alpha (IL1A), interleukin 1, beta (IL1B), tumor necrosis factor (TNF), serotonin receptor 2A (HTR2A), the promoter of the serotonin transporter gene (5-HTTLPR) and tryptophan hydroxylase 2 (TPH2) are candidate genes whose deregulation is thought to be central to psychiatric disorders [1]. In particular, the relevance of the serotonin system has been widely investigated and supported in psychopathology [2,3], and related to the molecular disruptions associated with alcohol related disorders [4,5]. Consistently, the genetic variations located in the promoter of the serotonin transporter has been convincingly found to regulate mood disorders [6] and alcohol related disorders [7–9]. Conflicting results [10,11] have further stimulated research in this direction. HTR2A plays as a key serotonin receptor: it is responsible for the post-synaptic activation upon serotonin transmission, a role that may modulate the psychiatric-related neuronal deregulations [12]. Recent evidence supports its connection with alcohol addiction [13–15]. In particular, HTR2A A-1438G variation, which is in completely LD with the variation we investigated here (reviewed in [12]), was found to be associated with alcohol dependence [15] and tobacco smoking combined or not with alcohol dependence [13]. Furthermore, HTR2A rs6313, which regulates the expression rate of HTR2A (reviewed in [12]), was found to be associated with alcohol abuse in male patients [14]. TPH2 is the brain specific rate limiting enzyme in the synthesis of serotonin [16], a role that candidates it as a modulator of the serotonin system orchestration. Recent reports discouraged the hypothesis of its participation to alcohol related behaviors [17–19]. IL1A and IL1B are mediators of the acute phase response, some genetic variations located in these genes have been associated with depressive symptomatology, antidepressant outcome and alcoholism [20,21]. TNF is a white cell produced pro-inflammatory cytokine likely involved in depressive disorder [16] which has been poorly investigated as a mediator of alcohol related psychiatric disorders. We yet investigated a set of variations located in the IL1A (rs1800587), IL1B (rs3087258), TNF (rs1799724), 5-HTTLPR (5-HTTLPR) and TPH2 (rs1386493; rs1386494; rs1487275; rs1843809; rs4570625; rs2129575) as modulators of alcohol related disruptions, both enzymatic and psychometric in previous published papers [22,23]: despite the relevance of these receptors and enzymes in the modulation of alcohol related phenotypes, we reported a lack of association. The reasons of this may be attributable to: lack of power, incomplete coverage of genetic mutations, incomplete analysis of epigenetic events, incomplete analysis of socio-demographic events and so on. In particular, epistasis between gene may have been played a fundamental role which remained undiscovered during the analysis of single mutations in previous papers we published on this sample. The aim of this paper is to challenge this particular limit of the previous publications on this sample [6,22] through the analysis of the epistasis between the set of variations yet singularly analyzed, also including a not yet investigated mutation (HTR2A rs6313) in the model. Further on, given that HTR2A rs6313 was not investigated as a single mutation in this sample of patients, we investigated the HTR2A rs6313 alone impact as a regulator of the biological and psychometric alcohol related variables.
2. Methods
2.1. Inclusion and Exclusion Criteria
To be included in the study patients had to fulfill the DSM-IV (Diagnostic and Statistical Manual of mental disorders) diagnostic criteria for alcohol abuse/dependence – “primary alcoholism” – they had to have no recent alcohol intake (average period of abstinence prior to the admission to the clinic = 24.0 ± 12.2 h), and to give full voluntary consent to the study. Other inclusion criteria were: absence of serious physical illness (as assessed through physical examination and routine laboratory screening), absence of other drug abuse, age between 20 and 75 years old, absence of DSM-IV axis I co-morbidity. The presence of affective symptoms was not considered to be an exclusion criterion. Alcohol abusers who fulfilled the DSM-IV diagnosis of depressive disorder were excluded from the study in the case that a full diagnosed depressive episode was precedent to the onset of the alcohol dependence. Relatives (non-consanguineous) were included in the study if they did not meet criteria for past or current alcohol abuse/dependence and major medical disturbances. Written informed consent was obtained from each participant after approval from the local ethical committee. Patients and their relatives were enrolled at the Drug and Alcohol Addiction Clinic of the Athens University Psychiatric Clinic at the Eginition Hospital in Athens, Greece. For patients, alcohol detoxification program included vitamin replacement (B, C, E) and oral administration of diazepam (10–40 mg daily in divided doses), with gradual taper off over a week. The overall period of detoxification was completed in 4–5 weeks. Patients then followed an inpatient standard treatment program with a short-term psychotherapy of cognitive-behavioral orientation.
2.2. Assessment
The biochemical profile of cholesterol, high density lipoprotein (HDL), low density lipoprotein (LDL), glutamic oxaloacetic transaminase (SGOT or AST), serum glutamic pyruvic transaminase (SGPT or ALT) and gamma-glutamyltransferase (GGT), were assessed once in relatives and twice in patients. Hematocrit, hemoglobin and thyroid hormonal levels (Triiodothyronine = T3, Thyroxine = T4, Thyrotropin = TSH) were measured once in both relatives and alcohol dependents. Patients were evaluated by a set of psychological tests: at admission they were evaluated by the Schedules for Clinical Assessment in Neuropsychiatry (SCAN [24]) and the Composite International Diagnostic Interview (CIDI - section on alcohol consumption [25]). Sociodemographic data were collected as well. Hamilton Depression Rating Scale (HAM-D) the Hamilton Anxiety Rating Scale (HAM-A) the Global Assessment Scale (GAS), the Liebowitz Social Anxiety Scale (social fear and avoidance) [26], the Mark & Mathews Scale (general fear) [27], the Leyton Obsessional Inventory [28], and the Pilowski scale (hypochondriac symptoms) [29] were administered at admission and discharge. Genotyping was performed as previously described [22].
2.3. Statistics
Principal analysis
In the principal analysis the independent variable was the epistasis between the investigated variations (rs1800587; rs3087258; rs1799724; 5-HTTLPR; rs1386493; rs1386494; rs1487275; rs1843809; rs4570625; rs2129575; rs6313). Diagnosis, biological and psychometric tests’ scores were treated as dependent variables of the principal analysis. Correlation analysis was employed in order to assess whether socio-demographic variables were associated with the dependent variables. In the event of significant association, socio-demographic variables were treated as covariates in the principal analysis. We employed GMDR Software Beta version 0.7 to investigate the gene to gene interactions [30], possible confounders were included in the analysis accordingly to GMDR manual [30]. Gender and age were included as possible confounders when analyzing cases versus controls. Alpha value was conservatively set to 0.004 (Bonferroni correction: 0.05/11 polymorphisms). Power was sufficient in our sample to detect an effect size of d = 0.88 or similar as previously detailed [22]. The multifactor dimensionality reduction (MDR) method [30] was employed by the GMDR Software Beta version 0.7 to investigate the gene–gene interactions. The MDR model computes the data reducing the n-dimensional space formed by a given set to a single dimension to analyze n-way interactions. Each class is then classified as either high risk or low risk according to the relative proportion of cases to controls in that class. This is performed for all possible combinations of SNPs, and the combination with the best predictive capacity is identified. Accordingly to the settings of the software, we tested all the possible two- to six-loci interactions using 10-fold cross-validation in an exhaustive search, thus considering all possible SNP combinations. As outcome parameters we considered the testing balanced accuracy, the significance test, along with the test sensitivity and specificity. The testing balanced accuracy measures the degree to which the interaction accurately predicts case–control status (label 1 indicating good prediction of the model, label 0.50 suggesting that the model is no better than chance in selecting cases from controls). The significance test gives the p value of the calculation.
Secondary analysis
In the secondary analysis the independent variable was: rs6313 genotypes. Diagnosis, biological and psychometric tests’ scores were treated as dependent variables. Chi-square test, ANOVA and ANCOVA analyses were employed when needed in order to infer the impact of rs6313 genotypes on the dependent variables (diagnosis, biological and psychometric tests’ scores).
3. Results
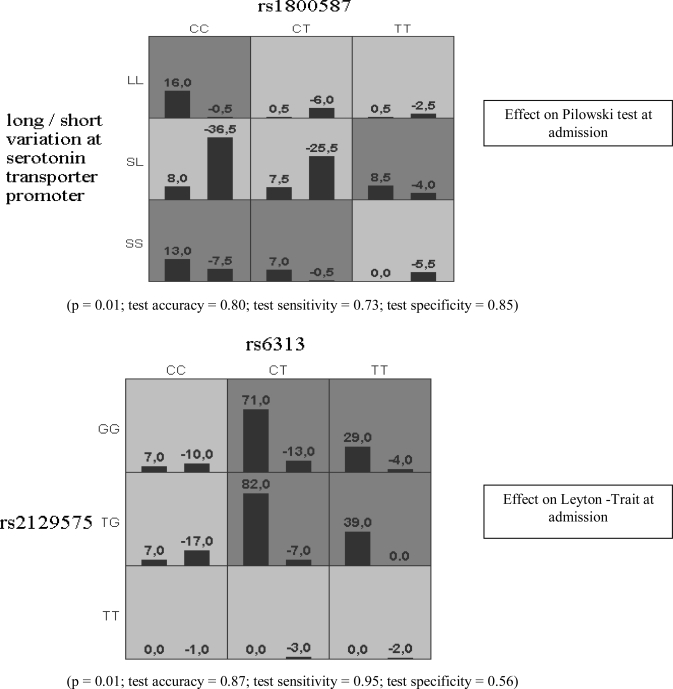
Sixty four alcohol dependent patients and 47 relatives (non-consanguineous) were included in the study. Patients’ and their relatives’ characteristics are reported in Table 1. Table 2 reports the psychological assessment at admission and discharge. Biochemical reports are detailed in [22]. As a result of the principal analysis, that is, the interaction between epistasis (independent variable) and diagnosis and psychometric tests’ scores (dependent variable), we found a marginal impact of HTR2A rs6313 + TPH2 rs2129575 on Leyton -Trait at admission (p = 0.01; test accuracy = 0.87; test sensitivity = 0.95; test specificity = 0.56) and a similar marginal impact of ILA rs1800587 + 5-HTTLPR on Pilowski test at admission (p = 0.01; test accuracy = 0.80; test sensitivity = 0.73; test specificity = 0.85).
Table 1.
Demographic data of patients.
| Variable | Total | Females | Males |
|---|---|---|---|
| Sex | Cases = 64 (100%) | Cases = 15 (21%) | Cases = 49 (79%) |
| Controls = 47 (100%) | Controls = 39 (52%) | Controls = 8 (48%) | |
| Age (years) | Cases = 45.60 ± 9.89 | Cases = 45.42 ± 9.96 | Cases = 45.64 ± 9.94 |
| Controls = 47 ± 11.52 | Controls = 48.79 ± 10.98 | Controls = 43.87 ± 13.96 | |
| Weight (kg) | 74.42 ± 11.45 | 62.94 ± 0.96 | 77.11 ± 9.83 |
| Alcohol (gr/day) | 265.27 ± 138.32 | 152.32 ± 55.66 | 291.77 ± 138.7 |
| Age of onset | 26.91 ± 10.14 | 29 ± 8.75 | 26.42 ± 10.45 |
| Smoking (cigs/day) | 11.40 ± 15.84 | 9.74 ± 10.47 | 11.79 ± 16.89 |
Table 2.
Psychopathological assessment: admission and discharge.
| Variable | Admission | 2nd week | 3rd week | 4th week | P (adm vs 4th week) |
|---|---|---|---|---|---|
| HDRS | 39.45 ± 6.86 | 31.35 ± 7.11 | 14.5 ± 6.81 | 6.65 ± 5.20 | < 0.001 |
| HARS | 34.44 ± 9.80 | 27.54 ± 8.47 | 14.35 ± 7.11 | 6.71 ± 5.29 | < 0.001 |
| GAS | 46.30 ± 5.06 | 56.7 ± 5.51 | 74.3 ± 8.68 | 83.90 ± 8.27 | < 0.001 |
| Liebowitz Social Anxiety Scale (social fear) | 51 ± 9.45 | – | – | 30.16 ± 6.04 | < 0.001 |
| Liebowitz Social Anxiety Scale (social avoidance) | 54.91 ± 11.04 | – | – | 31.89 ± 7.63 | < 0.001 |
| Mark & Mathews Scale | 49.30 ± 11.59 | – | – | 18.86 ± 5.91 | < 0.001 |
| Leyton Obsessional Inventory | 14.5 ± 3.34 | – | – | 10.04 ± 3.17 | < 0.001 |
| Pilowski scale | 9.14 ± 2.72 | – | – | 4.92 ± 2.16 | < 0.001 |
The results are shown in Figure 1: the frequencies of combined genotypes which have been found to be significantly associated with the scores of the dependent variables are in grey boxes; positive scores upon the right column in every box represent the frequencies of combined genotypes that have been associated with higher scores at the investigated test, negative scores upon each left column in each box represent the frequencies of the combined genotypes which have been associated with lower scores at the investigated test. Higher and lower scores are defined accordingly to the normal distribution obtained from the analysis of input data, “0” corresponding to the mean value of dependent variable, positive scores corresponding to scores upon the mean and negative scores corresponding to scores below the mean in the sample. Light boxes represent not significant associations. These results did not survive the Bonferroni correction. In the secondary analysis of the study, rs6313 did not impact the psychopathologic dimension and did not separate patients from their relatives (data not shown). In the pictures the grey shadowed boxes designate the combinations of genotypes associated with a statistically significant different distribution of the scores of the test under investigation. The light shadowed boxes designate the combinations of genotypes associated with a non statistically significant different distribution of the scores of the test under investigation.
Figure 1.
Marginal significant epistasis results.
Dark gray cells = the combinations for which the combined frequencies of genotypes can significantly predict the distribution of the dependent variable.
Light gray cells = the combinations that cannot predict the distributions of the dependent variable.
Bars in each box = the frequency of the combined genotypes are separated on the basis of the dependent variable into two groups, being the mean score of the dependent variable the threshold between them.
Numbers upon each column = given that the dependent variable was implemented as a normalized distribution with “0” as mean and threshold value, columns on the left in each box are labeled as positive, and columns on the right of each box are labeled as negative. Absolute numbers upon each column represent the frequency of each combined genotype.
To be implemented in the GMDR software the scores were normalized with “0” as the mean and threshold value. The GMDR software computes the frequencies of subjects who score higher (positive values on the left column on each box) or lower (negative values on the right column on each box) than “0” which is representative of the mean of the scores at the test under investigation. The scores are differentiated in positive and negative: the mean score given as a threshold, the positive scores stand for subjects who ranked higher than the mean, the negative stand for subjects who ranked lower than the mean. The difference in the frequencies of positive and negative scoring subjects is labeled as significant in the grey shadowed boxes. Light shadowed boxes stand for not significant difference in distribution of positive and negative scores. For example, in the first picture it is shown that IL1A rs1800587 and 5-HTTLPR combined variations are associated with a significant different score at Pilowski test at admission only in some specific combination of genotypes: the genotype CC at rs1800587 is associated with higher frequencies of subjects who ranked higher than the mean of the Pilowski test, only when it is found to be in combination with the LL or SS genotypes at the 5-HTTLPR. On the other hand, the combination with heterozygosis at the 5-HTTLPR with the same genotype at rs1800587 resulted in a not significant different distribution of subjects who ranked higher or lower than the mean of the Pilowski test. In the second picture, the combination of the CT genotype at rs6313 and the GG genotype at rs2129575 results into statistically significant higher scores at the Leyton – Trait test at admission, along with the other combinations of genotypes which are grey shadowed.
4. Discussion
We found a marginal impact of the epistatic effect between the investigated variations and the obsessive-compulsive traits and hypochondriac symptoms. The low level of significance which did not stand the Bonferroni correction, is likely mainly related to the small sample size we investigated here: as formerly reported, the power was sufficient to detect a large effect size. Moreover, the genetic analysis we performed here did not cover the genes that are under investigation, which results in a possible genetic bias: some variations remain hidden in the present investigation that could impact the functionality of the investigated mutations. This exposes our study to a biological stratification bias which is quite common in literature [31]. Finally, the most part of the investigated variations belong to the TPH2 gene which has been quite consistently reported to be not associated with alcohol dependence. In all facts, even though TPH2 is the rate limiting enzyme for the synthesis of serotonin in central nervous system [32], a molecular role that candidates it as a central regulator of psychiatric disorders such as major depressive disorder [33–35] and bipolar disorder [35–37], recent reports rejected the hypothesis that variations located in the TPH2 gene could modulate alcohol dependence [17–19]. This is in contrast with the inhibitory role played by the serotonin system toward the mesolimbic dopaminergic system [38,39] which is thought to be primarily involved in the disruptions that lead to alcohol dependence [5]. This is the biological evidence that steered our attention on the possible epistasis impact of some relevant and well replicated variants (5-HTTLPR and HTR2A rs6313) over a putative good candidate as the TPH2 is. Some recent reports on the significant epistasis interaction between genetic variables that did not cast informative results when singularly investigated [40,41], convinced us to re-analyze our previously published results. Unfortunately, we could only find not conclusive data in this specific case. The question arises as whether the marginal significant associations we report are false positive findings, which are a common concern in genetic association studies [42], or the first partial evidence of a significant association.
Acknowledgments
We thank the support of Fondazione del Monte di Bologna e Ravenna Prot. 915bis/2008.
Abbreviations:
- 5-HTTLPR
promoter of the gene of the serotonin transporter;
- CIDI
Composite International Diagnostic Interview;
- GAS
Global Assessment Scale;
- GGT
gamma-glutamyltransferase;
- GMDR
general multifactor dimensionality reduction;
- HAM-A
Hamilton Anxiety Rating Scale;
- HAM-D
Hamilton Depression Rating Scale;
- Hb
hemoglobin;
- HDL
high density lipoprotein;
- HTc
hematocrit;
- HTR2A
serotonin receptor 2A;
- IL1A
interleukin 1, alpha;
- IL1B
interleukin 1, beta;
- LDL
low density lipoprotein;
- MDR
multifactor dimensionality reduction;
- SCAN
Schedules for Clinical Assessment in Neuropsychiatry;
- SGOT (or AST)
glutamic oxaloacetic transaminase;
- SGPT (or ALT)
glutamic pyruvic transaminase;
- T levels
thyroid hormonal levels;
- T3
Triiodothyronine;
- T4
Thyroxine;
- TNF
tumor necrosis factor;
- TPH2
tryptophan hydroxylase 2;
- TSH
Thyrotropin.
References
- 1.Serretti A, Artioli P. The pharmacogenomics of selective serotonin reuptake inhibitors. Pharmacogenomics J. 2004;4:233–244. doi: 10.1038/sj.tpj.6500250. [DOI] [PubMed] [Google Scholar]
- 2.Fisher PM, Munoz KE, Hariri AR. Identification of neurogenetic pathways of risk for psychopathology. Am. J. Med. Genet. C Semin. Med. Genet. 2008;2:147–153. doi: 10.1002/ajmg.c.30173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.D’Souza UM, Craig IW. Functional polymorphisms in dopamine and serotonin pathway genes. Hum. Mutat. 2006;1:1–13. doi: 10.1002/humu.20278. [DOI] [PubMed] [Google Scholar]
- 4.Lesch KP. Alcohol dependence and gene x environment interaction in emotion regulation: Is serotonin the link? Eur. J. Pharmacol. 2005;1:113–124. doi: 10.1016/j.ejphar.2005.09.027. [DOI] [PubMed] [Google Scholar]
- 5.Vengeliene V, Bilbao A, Molander A, Spanagel R. Neuropharmacology of alcohol addiction. Br. J. Pharmacol. 2008;2:299–315. doi: 10.1038/bjp.2008.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Serretti A, Calati R, Mandelli L, de Ronchi D. Serotonin transporter gene variants and behavior: a comprehensive review. Curr. Drug Target. 2006;12:1659–1669. doi: 10.2174/138945006779025419. [DOI] [PubMed] [Google Scholar]
- 7.Pinto E, Reggers J, Gorwood P, Boni C, Scantamburlo G, Pitchot W, Ansseau M. The short allele of the serotonin transporter promoter polymorphism influences relapse in alcohol dependence. Alcohol Alcoholism. 2008;4:398–400. doi: 10.1093/alcalc/agn015. [DOI] [PubMed] [Google Scholar]
- 8.Dick DM, Plunkett J, Hamlin D, Nurnberger J, Jr, Kuperman S, Schuckit M, Hesselbrock V, Edenberg H, Bierut L. Association analyses of the serotonin transporter gene with lifetime depression and alcohol dependence in the Collaborative Study on the Genetics of Alcoholism (COGA) sample. Psychiatr. Genet. 2007;1:35–38. doi: 10.1097/YPG.0b013e328011188b. [DOI] [PubMed] [Google Scholar]
- 9.Marques FZ, Hutz MH, Bau CH. Influence of the serotonin transporter gene on comorbid disorders among alcohol-dependent individuals. Psychiatr. Genet. 2006;3:125–131. doi: 10.1097/01.ypg.0000199449.07786.7d. [DOI] [PubMed] [Google Scholar]
- 10.Kohnke MD, Kolb W, Lutz U, Maurer S, Batra A. The serotonin transporter promotor polymorphism 5-HTTLPR is not associated with alcoholism or severe forms of alcohol withdrawal in a German sample. Psychiatr. Genet. 2006;6:227–228. doi: 10.1097/01.ypg.0000218629.11375.b0. [DOI] [PubMed] [Google Scholar]
- 11.Rasmussen H, Bagger Y, Tanko LB, Christiansen C, Werge T. Lack of association of the serotonin transporter gene promoter region polymorphism, 5-HTTLPR, including rs25531 with cigarette smoking and alcohol consumption. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2009;150:575–580. doi: 10.1002/ajmg.b.30880. [DOI] [PubMed] [Google Scholar]
- 12.Serretti A, Drago A, De Ronchi D. HTR2A gene variants and psychiatric disorders: a review of current literature and selection of SNPs for future studies. Curr. Med. Chem. 2007;19:2053–2069. doi: 10.2174/092986707781368450. [DOI] [PubMed] [Google Scholar]
- 13.Polina ER, Contini V, Hutz MH, Bau CH. The serotonin 2A receptor gene in alcohol dependence and tobacco smoking. Drug Alcohol Depend. 2009;1:128–131. doi: 10.1016/j.drugalcdep.2008.11.001. [DOI] [PubMed] [Google Scholar]
- 14.Hwu HG, Chen CH. Association of 5HT2A receptor gene polymorphism and alcohol abuse with behavior problems. Am. J. Med. Genet. 2000;6:797–800. doi: 10.1002/1096-8628(20001204)96:6<797::aid-ajmg20>3.0.co;2-k. [DOI] [PubMed] [Google Scholar]
- 15.Nakamura T, Matsushita S, Nishiguchi N, Kimura M, Yoshino A, Higuchi S. Association of a polymorphism of the 5HT2A receptor gene promoter region with alcohol dependence. Mol. Psychiat. 1999;1:85–88. doi: 10.1038/sj.mp.4000474. [DOI] [PubMed] [Google Scholar]
- 16.Himmerich H, Fulda S, Linseisen J, Seiler H, Wolfram G, Himmerich S, Gedrich K, Kloiber S, Lucae S, Ising M, Uhr M, Holsboer F, Pollmacher T. Depression, comorbidities and the TNF-alpha system. Eur. Psychiat. 2008;6:421–429. doi: 10.1016/j.eurpsy.2008.03.013. [DOI] [PubMed] [Google Scholar]
- 17.Zill P, Preuss UW, Koller G, Bondy B, Soyka M. SNP- and haplotype analysis of the tryptophan hydroxylase 2 gene in alcohol-dependent patients and alcohol-related suicide. Neuropsychopharmacology. 2007;8:1687–1694. doi: 10.1038/sj.npp.1301318. [DOI] [PubMed] [Google Scholar]
- 18.Mokrovic G, Matosic A, Hranilovic D, Stefulj J, Novokmet M, Oreskovic D, Balija M, Marusic S, Cicin-Sain L. Alcohol dependence and polymorphisms of serotonin-related genes: association studies. Coll. Antropol. 2008;32:127–131. [PubMed] [Google Scholar]
- 19.Gacek P, Conner TS, Tennen H, Kranzler HR, Covault J. Tryptophan hydroxylase 2 gene and alcohol use among college students. Addict. Biol. 2008;3:440–448. doi: 10.1111/j.1369-1600.2008.00118.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yu YW, Chen TJ, Hong CJ, Chen HM, Tsai SJ. Association study of the interleukin-1 beta (C-511T) genetic polymorphism with major depressive disorder, associated symptomatology, and antidepressant response. Neuropsychopharmacology. 2003;6:1182–1185. doi: 10.1038/sj.npp.1300172. [DOI] [PubMed] [Google Scholar]
- 21.Pastor IJ, Laso FJ, Romero A, Gonzalez-Sarmiento R. Interleukin-1 gene cluster polymorphisms and alcoholism in Spanish men. Alcohol Alcoholism. 2005;3:181–186. doi: 10.1093/alcalc/agh153. [DOI] [PubMed] [Google Scholar]
- 22.Serretti A, Liappas I, Mandelli L, Albani D, Forloni G, Malitas P, Piperi C, Zisaki A, Tzavellas EO, Papadopoulou-Daifoti Z, Prato F, Batelli S, Pesaresi M, Kalofoutis A. Interleukin-1 alpha and beta, TNF-alpha and HTTLPR gene variants study on alcohol toxicity and detoxification outcome. Neurosci. Lett. 2006;1:107–112. doi: 10.1016/j.neulet.2006.07.003. [DOI] [PubMed] [Google Scholar]
- 23.Serretti A, Liappas I, Mandelli L, Albani D, Forloni G, Malitas P, Piperi C, Politis A, Tzavellas EO, Papadopoulou-Daifoti Z, Zisaki A, Prato F, Batelli S, Polito L, De Ronchi D, Kalofoutis A. TPH2 gene variants and anxiety during alcohol detoxification outcome. Psychiat. Res. 2009;1:106–114. doi: 10.1016/j.psychres.2007.12.006. [DOI] [PubMed] [Google Scholar]
- 24.Wing JK, Babor T, Brugha T, Burke J, Cooper JE, Giel R, Jablenski A, Regier D, Sartorius N. SCAN, schedules for clinical assessment in neuropsychiatry. Arch. Gen. Psychiat. 1990;6:589–593. doi: 10.1001/archpsyc.1990.01810180089012. [DOI] [PubMed] [Google Scholar]
- 25.Ye J, Wang L, Zhang X, Tantishaiyakul V, Rojanasakul Y. Inhibition of TNF-alpha gene expression and bioactivity by site-specific transcription factor-binding oligonucleotides. Am. J. Physiol. Lung Cell Mol. Physiol. 2003;2:L386–L394. doi: 10.1152/ajplung.00134.2002. [DOI] [PubMed] [Google Scholar]
- 26.Liebowitz MR. Social phobia. Mod. Probl. Pharm. 1987;22:141–173. doi: 10.1159/000414022. [DOI] [PubMed] [Google Scholar]
- 27.Marks IM, Mathews AM. Brief standard self-rating for phobic patients. Behav. Res. Ther. 1979;3:263–267. doi: 10.1016/0005-7967(79)90041-x. [DOI] [PubMed] [Google Scholar]
- 28.Cooper JE. The Leyton Obsessional Inventory. Psychol. Med. 1970;1:48–64. doi: 10.1017/s0033291700040010. [DOI] [PubMed] [Google Scholar]
- 29.Pilowsky I. Dimensions of hypochondriasis. Br. J. Psychiat. 1967;494:89–93. doi: 10.1192/bjp.113.494.89. [DOI] [PubMed] [Google Scholar]
- 30.Lou XY, Chen GB, Yan L, Ma JZ, Zhu J, Elston RC, Li MD. A generalized combinatorial approach for detecting gene-by-gene and gene-by-environment interactions with application to nicotine dependence. Am. J. Hum. Genet. 2007;6:1125–1137. doi: 10.1086/518312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Drago A, de Ronchi D, Serretti A. Incomplete coverage of candidate genes: a poorly considered bias. Curr. Genomics. 2007;7:476–483. doi: 10.2174/138920207783591681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Walther DJ, Peter JU, Bashammakh S, Hortnagl H, Voits M, Fink H, Bader M. Synthesis of serotonin by a second tryptophan hydroxylase isoform. Science. 2003;299:76. doi: 10.1126/science.1078197. [DOI] [PubMed] [Google Scholar]
- 33.Zill P, Baghai TC, Zwanzger P, Schule C, Eser D, Rupprecht R, Moller HJ, Bondy B, Ackenheil M. SNP and haplotype analysis of a novel tryptophan hydroxylase isoform (TPH2) gene provide evidence for association with major depression. Mol. Psychiat. 2004;11:1030–1036. doi: 10.1038/sj.mp.4001525. [DOI] [PubMed] [Google Scholar]
- 34.Zhou Z, Peters EJ, Hamilton SP, McMahon F, Thomas C, McGrath PJ, Rush J, Trivedi MH, Charney DS, Roy A, Wisniewski S, Lipsky R, Goldman D.Response to Zhang et al. 2005: loss-of-function mutation in tryptophan hydroxylase-2 identified in unipolar major depressionNeuron 45, 11–16. Neuron 200548704–705.; author reply 705–706. [DOI] [PubMed] [Google Scholar]
- 35.Zhou Z, Roy A, Lipsky R, Kuchipudi K, Zhu G, Taubman J, Enoch MA, Virkkunen M, Goldman D. Haplotype-based linkage of tryptophan hydroxylase 2 to suicide attempt, major depression, and cerebrospinal fluid 5-hydroxyindoleacetic acid in 4 populations. Arch. Gen. Psychiat. 2005;10:1109–1118. doi: 10.1001/archpsyc.62.10.1109. [DOI] [PubMed] [Google Scholar]
- 36.Harvey M, Shink E, Tremblay M, Gagne B, Raymond C, Labbe M, Walther DJ, Bader M, Barden N. Support for the involvement of TPH2 gene in affective disorders. Mol. Psychiat. 2004;11:980–981. doi: 10.1038/sj.mp.4001557. [DOI] [PubMed] [Google Scholar]
- 37.de Luca V, Likhodi O, van Tol HH, Kennedy JL, Wong AH. Tryptophan hydroxylase 2 gene expression and promoter polymorphisms in bipolar disorder and schizophrenia. Psychopharmacology (Berl) 2005;3:378–382. doi: 10.1007/s00213-005-0191-4. [DOI] [PubMed] [Google Scholar]
- 38.Hoebel BG, Hernandez L, McClelland RC, Schwartz D. Dexfenfluramine and feeding reward. Clin. Neuropharmacol. 1988;11:S72–S85. [PubMed] [Google Scholar]
- 39.Jadhav SA, Gaikwad RV, Gaonkar RK, Thorat VM, Gursale SC, Balsara JJ. Dose-dependent response of central dopaminergic systems to buspirone in mice. Indian J. Exp. Biol. 2008;10:704–714. [PubMed] [Google Scholar]
- 40.Urwin RE, Nunn KP. Epistatic interaction between the monoamine oxidase A and serotonin transporter genes in anorexia nervosa. Eur. J. Hum. Genet. 2005;3:370–375. doi: 10.1038/sj.ejhg.5201328. [DOI] [PubMed] [Google Scholar]
- 41.van Gestel S, Forsgren T, Claes S, Del-Favero J, van Duijn CM, Sluijs S, Nilsson LG, Adolfsson R, van Broeckhoven C. Epistatic effect of genes from the dopamine and serotonin systems on the temperament traits of novelty seeking and harm avoidance. Mol. Psychiat. 2002;5:448–450. doi: 10.1038/sj.mp.4001005. [DOI] [PubMed] [Google Scholar]
- 42.Sullivan PF. Spurious genetic associations. Biol. Psychiat. 2007;10:1121–1126. doi: 10.1016/j.biopsych.2006.11.010. [DOI] [PubMed] [Google Scholar]