Abstract
Interruptions in the repeating (Gly-X1-X2)n amino acid sequence pattern are found in the triple-helix domains of all non-fibrillar collagens, and perturbations to the triple-helix at such sites are likely to play a role in collagen higher order structure and function. This report defines the sequence features and structural consequences of the most common interruption, where one residue is missing in the tripeptide pattern, Gly-X1-X2-Gly-AA1-Gly-X1-X2, designated as G1G interruptions. Residues found within G1G interruptions are predominantly hydrophobic (70%), followed by a significant amount of charged residues (16%), and the Gly-X1-X2 triplets flanking the interruption are atypical. Studies on peptide models indicate the degree of destabilization is much greater when a Pro is in the interruption, GP, than when hydrophobic residues (GF, GY) are present, and a rigid Gly-Pro-Hyp tripeptide adjacent to the interruption leads to greater destabilization than a flexible Gly-Ala-Ala sequence. Modeling based on NMR data indicates the Phe residue within a GF interruption is located on the outside of the triple-helix. The G1G interruptions resemble a previously studied collagen interruption GPOGAAVMGPO, designated as a G4G type, in that both are destabilizing, but allow continuation of rod-like triple-helices and maintenance of the 1-residue stagger throughout the imperfection, with a loss of axial register of the superhelix on both sides. Both kinds of interruptions result in a highly localized perturbation in hydrogen bonding and dihedral angles, but the hydrophobic residue of a G4G interruption packs near the central axis of the superhelix while the hydrophobic residue of a G1G interruption is located on the triple-helix surface. The different structural consequences of G1G and G4G interruptions in the repeating tripeptide sequence pattern suggest a physical basis for their differential susceptibility to matrix metalloproteinases in type X collagen.
Keywords: collagen, triple-helix, interruptions, structure, peptides
Introduction
Collagens are structural proteins in the extracellular matrix, and their defining feature is the presence of a triple-helix structure. Twenty-eight types of collagens have been described in vertebrates, and different types show characteristic tissue distributions, distinct higher order structures and specific biological roles 1,2,3. The most abundant collagens are found in characteristic periodic fibrils, while the others are designated as non-fibrillar collagens. The collagen triple-helix motif has three extended polyproline II-like chains which are staggered by one residue and supercoiled about a common axis. This conformation is characterized by a (Gly-X1-X2)n repeating sequence. Proline and its post-translationally modified hydroxyproline (Hyp) stabilize the polyproline II-like chain conformation and are frequently found in the X1 and X2 positions respectively, e.g. Gly-Pro-Hyp sequences. The requirement for the presence of Gly as every third residue has been a basic feature from the earliest models of the triple-helix. Only this small residue can fit near the central axis where the three chains pack tightly without distortion and allow the formation of direct backbone hydrogen bonds between the chains 4,5,6. However, such a strict regularity of Gly as every third residue is not found in the triple-helix domains of non-fibrillar collagens, and this report investigates the effects of an interruption missing one residue in the tripeptide pattern.
The abundant fibril-forming collagens, found in fibrils with a D=670Å axial period (types I, II, III, V, XI), all maintain a precise Gly-X1-X2 repeat and a standard triple-helix throughout their ∼1000 residue triple-helix domain 1. A pathological condition results when even one Gly is replaced by another residue as a result of a single base mutation. Such Gly substitutions in either of the type I collagen chains, α1(I) or α2(I), lead to osteogenesis imperfecta (OI) and mutations are located at many sites along the length of the chains in different OI cases 7,8. All types of collagen other than fibril forming collagens contain interruptions in the Gly-X1-X2 repeating tripeptide pattern. These non-fibrillar types include collagens found in networks, such as type IV and type X collagens; membrane proteins, such as type XVII and type XIII; FACIT collagens (types IX, XII, and XIV) found on the surface of periodic collagen fibrils; and type VII collagen in anchoring fibrils 1,2. Thus, the perfect (Gly-X1-X2)n pattern is a feature only of fibril forming collagens and is likely to relate to requirements for the higher order molecular association in D-periodic fibrils, while interruptions in this pattern are a normal feature of non-fibrillar collagens. Structural perturbations to the standard triple-helix must occur at such interruption sites.
Interruptions of varying lengths are found within the triple-helix of non-fibrillar collagens and they can be classified by the number of residues between normal repeating tripeptide sequences. In many cases, Gly residues are separated by 1 or 4 residues rather than the normal 2 residues, while in other instances, there are larger stretches of residues separating the Gly-X1-X2 repeats. Interruptions of the form Gly-X1-X2-Gly-AA1-Gly-X1-X2, where one residue appears to be missing are designated here as G1G, while an interruption with 4 non-Gly residues between two glycines Gly-X1-X2-Gly-AA1-AA2-AA3-AA4-Gly-X1-X2 is denoted as G4G.
Many non-fibrillar collagens are homotrimers with interruptions at the same location in all three chains. Examples of homotrimers include the anchoring fibril type VII collagen with 20 interruptions and type X collagen with 8 interruptions, which is found at the mineralizing front in calcifying cartilage. A small number of non-fibrillar collagens are heterotrimers. For instance, type IV collagen in basement membranes consists of a family of six chains, α1(IV), α2(IV), α3(IV), α4(IV), α5(IV), and α6(IV), and combinations of these chains can form three distinct types of heterotrimeric collagen molecules 9,10,11. Each type IV chain has more than 20 interruptions. The location of interruptions is often similar for the three chains within a heterotrimer while there may be different lengths of the interruptions at a given site in each of the three chains. For instance, at one site in the α3(IV) α4(IV) α5(IV) heterotrimer, there is a G6G in α3(IV), α G4G in α4(IV) and a G1G in α5(IV).
Despite the natural presence of interruptions in non-fibrillar collagens, the replacement of one Gly residue in a (Gly-X1-X2)n sequence of their triple-helix can lead to a clinical phenotype. More than 100 Gly substitution mutations leading to Alport Syndrome with progressive kidney failure have been defined within the X-linked α5(IV) chain 11,12 and a predisposition to hemorrhagic stroke has been associated with a mutation in the α1(IV) chain 13. Gly substitution mutations in type VII collagen lead to the dystrophic form of epidermolysis bullosa, with scarring and blistering skin 14,15,16.
Peptides have served as useful models for defining the structure of the normal collagen triple helix and its variations 17. Recently, the effect of a G4G imperfection in the α5(IV) chain on triple-helix stability, conformation and hydrogen bonding was studied in a peptide model 18 and a model structure was derived using NMR spectroscopy 19. A peptide with a G1G interruption in the most stable peptide (Pro-Hyp-Gly)10, (POG)3POGPGPO(POG)4 was observed to be highly destabilizing 20, and its high resolution structure obtained by x-ray crystallography was reported recently 21. F.N.1 [F.N.1: The amino acid sequence of this peptide could be defined as the loss of one Hyp residue in the (Pro-Hyp-Gly)10 peptide, and was previously denoted as Hyp-; here it is considered as an example of a G1G interruption, with Pro in the interruption, and denoted as the GP peptide.]
Although the collagen triple-helix is considered to be a well-characterized conformation and motif, it is not known what occurs at interruption sites within the triple-helix domains of non-fibrillar collagens where the canonical structure cannot be maintained. Here, sequence analysis and peptide studies are presented to further investigate the occurrence and structural consequences of G1G interruptions within the triple-helix. Analyses of non-fibrillar collagens show G1G to be the most common kind of interruptions and indicate they predominantly incorporate a hydrophobic residue. Peptides were studied with varied residues within the G1G as well as with distinctive sequences surrounding it. The degree of destabilization is found to depend on the residue within the interruption as well as the Gly-X1-X2 sequence environment. Modeling based on NMR data indicates the Phe residue within a GF interruption is located on the outside of the triple-helix, which contrasts with the incorporation of a hydrophobic residue near the core of the triple-helix within a G4G interruption.
Results
Analysis of G1G interruptions in collagens
Occurrence of G1G interruptions
The amino acid sequences of the triple-helix domains of the 42 chains which make up the 28 types of human collagens were taken from SwissProt and used as a database for analyzing the occurrence of G1G interruptions in the repeating (Gly-X1-X2)n pattern. The nine chains in the five fibril forming collagens (types I, II, III, V, and XI collagens) contain perfect (Gly-X1-X2)n sequences with no interruptions. The recently discovered types XXIV and XXVII collagens, which appear to be part of the fibril-forming family, each contain two interruptions, but none are of a G1G nature. The chains of all non-fibrillar collagens contain at least one interruption, with a total number of 354 interruptions in the triple-helix domains of the 33 chains of non-fibrillar collagens (Table 1). The G1G type is the most common kind of interruption in these non-fibrillar collagens, with a total of 91 G1G found in all chains (26% of all interruptions in human collagen chains). The next most common is G4G, which constitutes 19% of all interruptions. The interruptions in the α1 chain of type VIII are all G1G (8 G1Gs), while the interruptions found in the α2 chain of type VIII, the type X chain, and the type XXVIII chain are all G1G and G4G. Type XIII and type XX have G4G and other types of interruptions, but contain no G1G.
Table 1. The total number of interruptions in each non-fibrillar collagen, together with the numbers of G1G and G4G types.
| COLLAGEN | TOTAL # INTERRUPTIONS | # G1G | # G4G |
|---|---|---|---|
| Type IV a | 21,23,23,27,22,25 | 7,3,2,4,4,4 | 5,2,7,5,7,2 |
| Type VI b | 2,2,2 | 1,0,1 | 0,1,0 |
| Type VII | 20 | 5 | 5 |
| Type VIII c | 8,8 | 8,6 | 0,2 |
| Type IX d | 5,5,5 | 2,2,2 | 1,0,1 |
| Type X | 8 | 5 | 3 |
| Type XII | 5 | 1 | 2 |
| Type XIII | 6 | 0 | 4 |
| Type XIV | 4 | 1 | 2 |
| Type XV | 12 | 1 | 3 |
| Type XVI | 21 | 7 | 2 |
| Type XVII | 20 | 2 | 1 |
| Type XVIII | 20 | 3 | 3 |
| Type XIX | 14 | 2 | 2 |
| Type XX | 4 | 0 | 1 |
| Type XXI | 1 | 0 | 0 |
| Type XXII | 11 | 5 | 1 |
| Type XXIII | 4 | 0 | 0 |
| Type XXIV | 2 | 0 | 0 |
| Type XXV | 5 | 1 | 2 |
| Type XXVI | 1 | 0 | 0 |
| Type XXVII | 2 | 0 | 0 |
| Type XXVIII | 16 | 12 | 4 |
| TOTAL | 354 | 91 | 67 |
six values are given for α1(IV), α2(IV), α3(IV), α4(IV), α5(IV) and α6(IV) chains respectively;
three values are given for α1(VI), α2(VI) and α3(VI) chains respectively;
wo values are given for α1(VIII) and α2(VIIII) chains respectively;
three values are given for α1(IX), α2(IX) and α3(IX) chains respectively
Nature of a G1G interruption
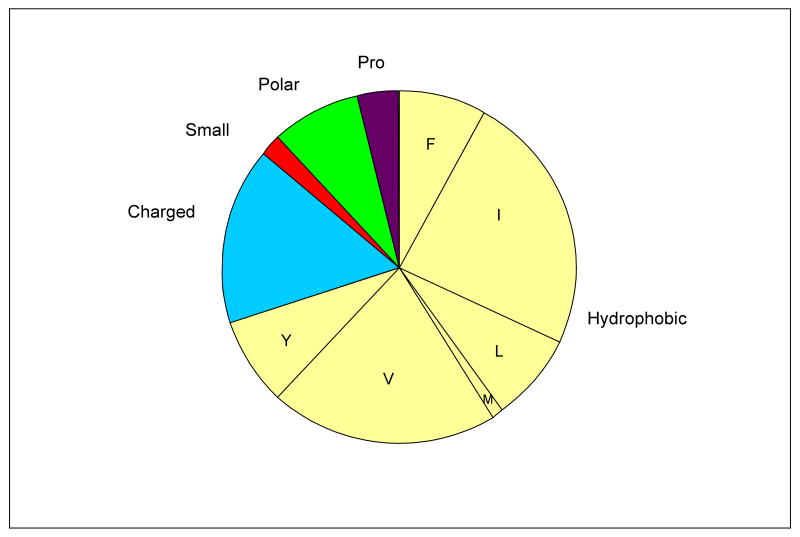
The identity of the residue within the G1G was examined, to see if this differs from the distribution of residues in the X1 and X2 positions of (Gly-X1-X2)n sequences. As seen in Figure 1, the residues within G1G are predominantly hydrophobic (70%), followed by a significant amount of charged residues (16%), and small amounts of Pro (4%), polar residues (8%) and small residues (2%). Among G1G interruptions, Gly-Ile is most common (22/91, 24%) and Gly-Val is the second most common residue (19/91, 21%). For comparison, residues in X1 and X2 positions of the (Gly-X1-X2)n sequences in all six chains of type IV collagen were analyzed. A chi-squared test indicates the distribution of residues observed within this interruption is significantly different from residues observed in the X1 and X2 positions in the (Gly-X1-X2)n repeats of type IV chains (p< 0.001), and the major contributions are due to the high frequency of hydrophobic residues and lower frequency of imino acid residues within G1G sequences. Examination of the type IV Gly-X1-X2 repeating sequences shows a higher proportion of hydrophobic residues (35%) and lower proportion of imino acids (21%) in the X1 position compared with the X2 position (10% hydrophobic, 48% imino acid). This information was used in designing control peptides (see below).
Figure 1.
(a) Diagram showing the nature of residues within all 91 G1G interruptions found in non-fibrillar collagens, where Hydrophobic ( ) = F,I,L,M,V,Y; Charged (
) = F,I,L,M,V,Y; Charged ( ) = D,E,H,K,R; Polar (
) = D,E,H,K,R; Polar ( ) = Q,N,T; Small (
) = Q,N,T; Small ( ) = A,C,G,S; Pro (
) = A,C,G,S; Pro ( ).
).
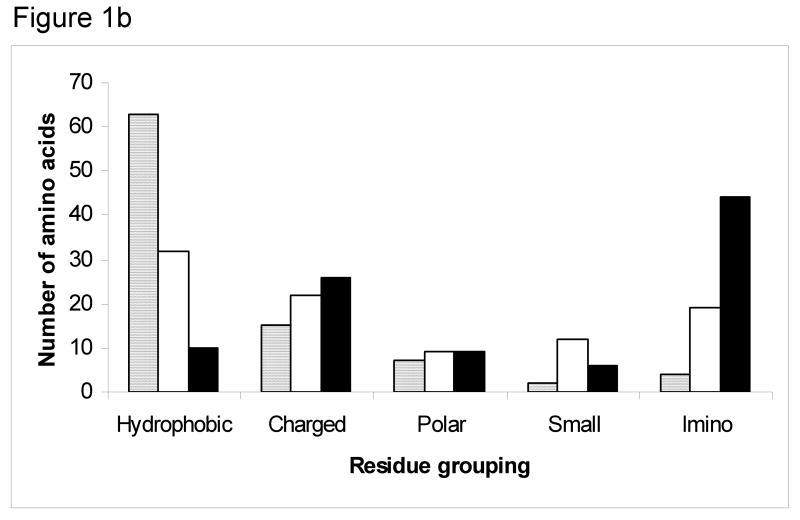
(b) Histogram showing the observed (gray) versus expected frequencies in X1 position (white) and X2 position (black) of groups of amino acids in G1G interruptions in all non-fibrillar collagens. The expected frequencies were calculated based on the identity of amino acids in the X1 and X2 positions of (Gly-X1-X2)n sequences of all 6 chains of type IV collagen. A chi square analysis indicates the observed distribution is very different from that expected for the X1 position (p<0.001) and the X2 position (p<0.001), and the major chi square terms come from the high number of hydrophobic residues and lower than expected number of imino acids.
Environment around a G1G interruption
To see if there were atypical features in the flanking Gly-X1-X2 sequences, three triplets on either side of all G1G interruptions were analyzed. Statistical comparison of these flanking tripeptide sequences with the residues found in the X1 and X2 positions of the 2515 Gly-X1-X2 tripeptides of the six human type IV chains indicates that there are significant differences. In the immediate triplet N-terminal to a G1G, there is a significantly higher percentage of imino acids (P) and small residues (A, C, G, S) in the X1 position (p<0.001) and a higher percentage of charged residues and imino acids in the X2 position (p<0.001). In the immediate triplet at the C-terminal side of the G1G, there is a higher hydrophobic/lower imino acid content in the X2 position (p<0.001), while the X1 position does not show any atypical residue features.
Conservation of G1G interruptions in α1(IV)
Multiple sequence alignment was used on the α1(IV) chain from 6 invertebrate and vertebrate species to investigate whether the G1G interruptions sites are conserved. The α1(IV) chain is the most suitable sequence for studies of conservation of interruptions, since basement membrane type IV collagen and this chain in particular, is found in all multicellular organisms. There a total of 21 interruptions, with 7 of the G1G type and 5 of the G4G type, in the α1 chain of human type IV collagen. Using multiple sequence alignment, the sites of G1G imperfections observed in human a1(IV) collagen were compared with two vertebrate species, chick (Gallus gallus) and mouse (Mus musculus) and with three invertebrate orthologues, Strongylocentrotus purpuratus (sea urchin), Drosophila melanogaster, and Ceanorhabditis elegans (Figure 2). The mouse and chick sequences show close similarity to the human chain, as do all vertebrates, with the same total number and location of interruptions in α1(IV). The 7 G1G interruptions are located at the same sites as found in the human α1(IV) chain, but the mouse sequence has an additional G1G which aligns with a human G4G. At 4 out of the 7 sites, there is a variation in the identity of the residue within the G1G for mouse and/or chick, which are conservative hydrophobic replacements in all cases except one (Site 1, chick).
Figure 2. Multiple sequence alignment of the seven G1G interruptions in the α1 chain of human type IV collagen, together with two vertebrates mouse and chick, and three invertebrates, C. elegans, Drosophila and sea urchin.
A small section of the Clustal W multiple alignment of different species is shown depicting the sequence that aligns with each of the seven G1G interruptions in human. The interruptions are underlined and in bold. The Swiss-Prot accession numbers are: Human (Q5VWF6), Mouse (P02463), Chick (Q9I9K3), C. elegans (P17139), D. melanogaster (P08120), and Sea urchin (Strongylocentrotus purpuratus – Q07265).
The three invertebrate chains studied have a smaller numbers of G1G sites than seen in vertebrates, and, in two cases an increased number of G4G sites: sea urchin α1(IV) chain has a total of 23 interruptions with 3 G1G, 8 G4G; C. elegans has a total of 18 interruptions with 1 G1G, 7 G4G; fruit-fly has a total of 18 interruptions, with 1 G1G and 5 G4G. Thus, more G4G sites than G1G sites are found in all of the invertebrate chains, compared with the larger number of G1G in vertebrates. The G1G sites observed in human α1(IV) are not conserved in the three invertebrate α1(IV) chains (Figure 2). The sites corresponding to human G1G sites are most often occupied by uninterrupted Gly-X1-X2 sequences, and in almost all cases, there is a suggestion of a larger deletion of one or two tripeptide units, in addition to a single residue. It appears that a number of uninterrupted sequences in invertebrates correspond to G1G sites in the vertebrate α1(IV) orthologues. Most of the other human G1G sites are G4G sequences in the invertebrate chains. Only in one case (Site 4, C. elegans) is there a G1G at a site homologous to the human G1G. There appears to be some relation between G1G and G4G sites, in both their large numbers and in the presence of G4G at sites in invertebrates corresponding to human G1G sites.
Peptide models of G1G interruptions: conformation, stability, and hydrogen bonding
Peptide design
Four peptides modeling G1G imperfections were studied. Three peptides contain hydrophobic residues within the G1G site (GF, GY, and GV), since hydrophobic residues are found most frequently in natural G1G sequences. It was desirable to include as many Gly-Pro-Hyp (GPO) triplets as possible in the peptide to ensure stability, and the sequences flanking natural G1G sites are often of this nature (Gly-X1-Hyp triplets comprise 67% of triplets N-terminal to and 57% of triplets C-terminal to G1G sites, while GPO triplets comprise 44% of triplets N-terminal to and 2% of triplets C-terminal to G1G sites). Peptides were designed to include G1G interruptions from human type IV collagen chains which naturally contain a GPO sequence on one or both sides. For instance, the sequence GPOGYGPQ found in the α3(IV) chain (residues 614-622) is incorporated into the peptide (GPO)3GPOGYGPQ(GPO)4, designated as the GY peptide. Similarly, the α5(IV) sequence GPOGFGPO (residues 621-628) is incorporated into the peptide (GPO)3-GPOGFGPO-(GPO)4GY designated as the GF peptide (Table 2). The previously described GP peptide 20,21 was also studied. Control peptides were synthesized for comparison with each of these peptides. Since the residue within G1G sequences most closely resembles residues in the X1 position, a Hyp (O), the most common residue in the X2 position, is introduced to create a control with a repeating tripeptide pattern for the peptides GY (GYO), GF (GFO), and GP (control is (POG)10) (Table 2). A GV sequence, one of the most common G1G sequences found in non-fibrillar collagens, is incorporated into a peptide GPOGAAGVGPO, designated as the GV peptide. This peptide was designed to complement previous studies on a model peptide with the GAAVM interruption (G4G) found in the type IV α5 chain18,19, where GAAGVM is the control peptide.
Table 2. Thermal stability, mean residual ellipticity at 225 nm and calorimetric enthalpy values for peptides containing a G1G interruption and homologous control peptides, in PBS (pH 7).
| Peptide | Peptide Sequence | Tm | MRE225nm | ΔHcal |
|---|---|---|---|---|
| Name | (°C) | (deg cm2 dmol-1) | (kJ/mol) | |
| GF a | (GPO)3GPOG*F*GPO(GPO)2G*POGPOGYb | 21 | 2950 | 100 |
| GFO a | (GPO)3GPOGFOGPO(GPO)4GY | 44 | 5500 | 221 |
| GY | Ac-(GPO)3GPOGYGPQ(GPO)4G-NH2 | 26.5 | 3966 | 310 |
| GYO | Ac-(GPO)3GPOGYOGPQ(GPO)4G-NH2 | 48.5 | 5120 | 357 |
| GP | (POG)3POGPGPO(GPO)4G | <5 (15c) | n.a | n.a |
| GPO | (POG)10 | 58 | 4012 | 390 |
| GV | Ac-(GPO)3GPOGAAGVGPO(GPO)3GY-NH2 | 27.5 | 3006 | 187 |
| GAAVM d | Ac-(GPO)3GPOGAAVMGPO(GPO)3GY-NH2 | 29.1 | 2360 | 188 |
| GAAGVM | Ac-(GPO)3GPOGAAGVMGPO(GPO)3GY-NH2 | 39.7 | 4950 | 354 |
Effect of G1G interruptions on triple-helix content, stability, and calorimetric enthalpy
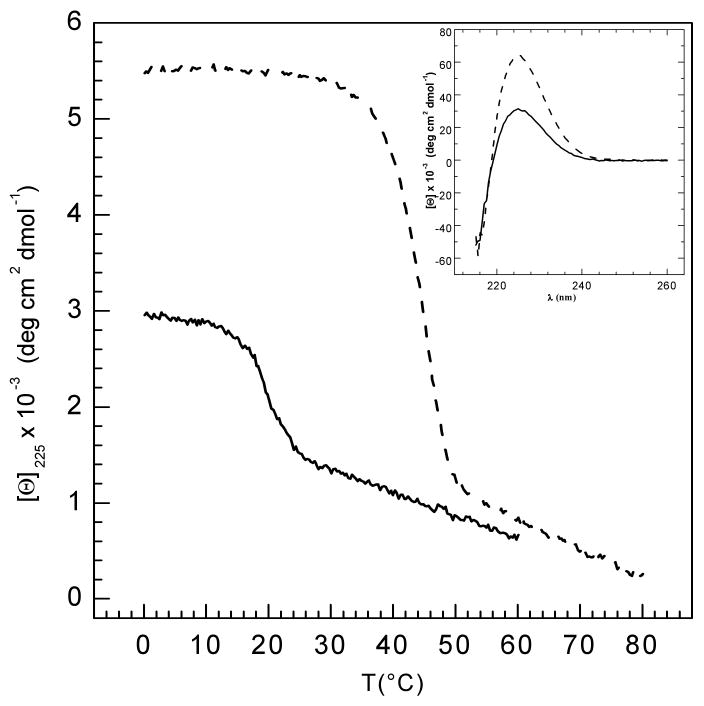
Circular dichroism (CD) spectroscopy was used to characterize the conformation and thermal stability of G1G sites. The control peptide GYO forms a stable triple helix at low temperature in PBS (pH 7), with a characteristic collagen triple-helix CD spectrum (MRE225 nm = 5120 deg cm2 dmol-1) and a sharp thermal transition with Tm = 48.5°C. The homologous peptide with GY in PBS shows a similar spectrum but with a substantial decrease in the 225 nm magnitude (MRE225 nm = 3966 deg cm2 dmol-1) and decreased Tm = 26.5°C. Peptides GF and GFO were insoluble in PBS and were studied in glycine buffer containing 0.6M GuHCl at pH 2.0. In this buffer, the control GFO peptide forms a stable triple-helical structure (MRE225 nm = 5500 deg cm2 dmol-1, Tm = 44°C) while the GF peptide shows a drop in ellipticity by almost half (MRE225 nm = 2950 deg cm2 dmol-1) and a reduced stability Tm = 21°C (Figure 3a). As a comparison, peptides GY and GYO were also studied in 0.6M GuHCl, pH 2.0, and both form stable triple-helices with a ∼5°C drop in Tm compared with PBS. This suggests that this low concentration of GuHCl acts largely as a solubilizing agent and not a denaturant, leading to some destabilization without influencing the native structure. The GP peptide was previously reported to form a triple-helix with a Tm ∼ 15°C in 0.1M acetic acid 20, but in PBS no triple-helix is formed (Tm <5°C). This inability to form a triple-helix contrasts with the high stability of its control peptide (POG)10, Tm ∼ 58°C.
Figure 3.
(a) Temperature-induced denaturation of GF (solid line) and GFO (dotted line) peptides monitored by circular dichroism spectroscopy at 225 nm. Both peptides (1mg/ml) were dissolved in buffer containing 20 mM glycine, 150 mM NaCl and 0.6M GuHCl, pH 2.0. Inset shows wavelength scans for both peptides recorded at 0°C, with the characteristic maximum at 225 nm. All samples were run under the same standard conditions (average heating rate = 0.1°C/min) 30.
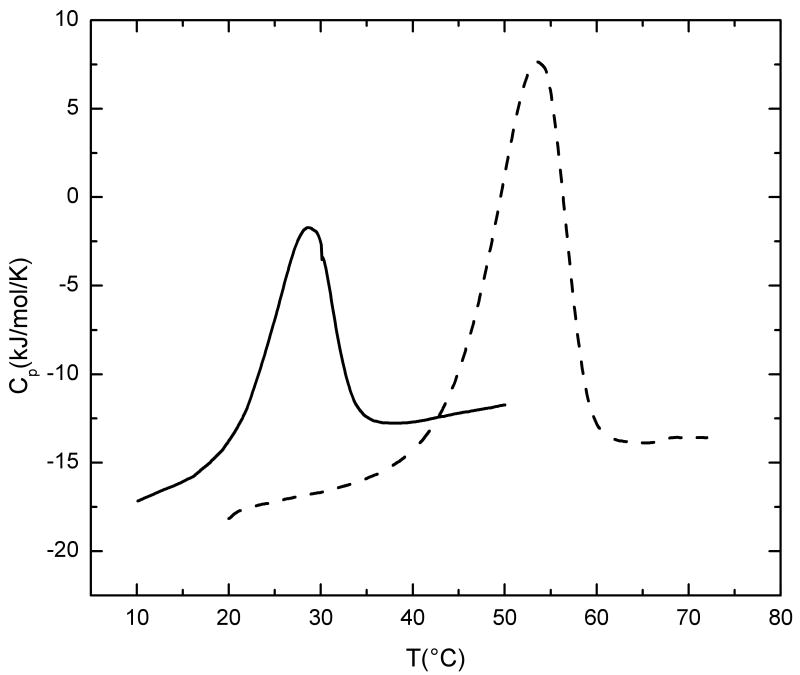
(b) Temperature dependence of the excess partial molar heat capacity (heating rate = 1°C/min) for GF (solid line) and GFO (dotted line) peptides monitored by differential scanning calorimetry. The calorimetric enthalpy values shown in table 2 represent the integration of the peaks. Both peptides (1mg/ml) were dissolved in buffer containing 20 mM glycine, 150 mM NaCl and 0.6M GuHCl, pH 2.0.
The GV peptide forms a stable triple-helix, with MRE225 nm = 3006 deg cm2 dmol-1 and Tm = 27.5°C. Comparison of the G1G peptide (GV) with the G4G (GAAVM) shows very similar effects on ellipticity and thermal stability compared with the GAAGVM control peptide (MRE225 nm = 4950 deg cm2 dmol-1; Tm = 39.7°C) (Table 2) 18. The degree of destabilization due to the introduction of a G1G in the GV peptide is less than seen for other G1G peptides, with aΔTm of 12.2°C.
Differential scanning calorimetry on these peptide sets shows that introduction of a G1G site leads to a loss of calorimetric enthalpy (Table 2; Figure 3b). The calorimetric enthalpy (ΔHcal) decreases from ΔHcal = 357 kJ/mol for the GYO peptide to ΔHcal = 310 kJ/mol for GY (PBS, pH 7), and from 221 kJ/mol for the GFO peptide (glycine buffer, 0.6 M GuHCl, pH 2.0) to 100 kJ/mol for GF. The ΔHcal of both GYO and GY peptides in PBS, pH 7.0 is similar to that in glycine buffer, 0.6M GuHCl, pH 2.0 with a comparable ΔΔHcal between the control and G1G peptide. A large decrease is observed for the GV peptide, going from ΔHcal=354 kJ/mol for the control peptide GAAGVM to a value of ΔHcal=187 kJ/mol for GV peptide, and this decrease is comparable to the ΔHcal=188 kJ/mol observed for the G4G model peptide GAAVM 18. Decreases in thermal stability measured by DSC are also observed for all G1G-containing peptides, but the Tm values are always higher in the DSC compared to the CD due to the faster scan rate under non-equilibrium conditions.
NMR studies on a peptide model with a G1G interruption
NMR chain assignment and conformational characterization
To represent one prototype of a G1G interruption, NMR studies were carried out on the GF peptide containing three 15N labeled residues for Gly-Phe at the imperfection site and one Gly in the C-terminal (Pro-Hyp-Gly)4 region. In the heteronuclear single quantum coherence (HSQC) spectrum of the GF peptide (Figure 4a), each labeled residue shows one or more trimer peaks, in addition to one or more monomer peaks, consistent with the formation of trimers at all positions 19. Residues G13 and F14 have three well separated trimer peaks, indicating the presence of a well-defined conformation with three non-equivalent chains at the G1G site. Residue G24 in the GPO repeating region at the C-terminus shows only a single trimer resonance with the typical triple-helix chemical shifts for Gly in the GPO repeating region 18. Trimer resonances for G13 and F14 are assigned to specific chains of the triple helix from the strong sequential NHi-Hαi-1 NOEs of the NOESY-HSQC experiment, and the chain stagger is derived from NOEs that define interchain interactions. For example, the observation of an NOE between 2G13NH of chain 2 and 3O12Hα of chain 3 indicates a 1-residue stagger between these chains (Figure 4b).
Figure 4.
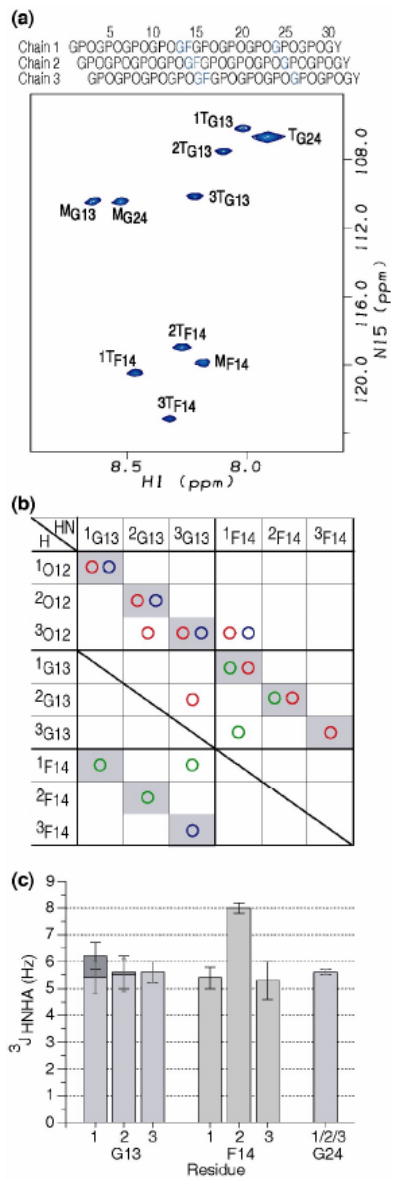
(a) 1H-15N HSQC spectrum of peptide GF at 10°C. Sequence diagram of the peptide above the HSQC shows the characteristic one residue stagger. The isotope-labeled residues are colored in blue. All labeled residues Gly13, Phe14 and Gly24 have trimer peaks as well as monomer peaks, showing the GF interruption is incorporated into a trimer. The peaks corresponding to the monomer and trimer state are denoted with a superscript M or T, respectively. Leading, middle, or trailing chain stagger assignment is indicated as chain 1, 2, or 3 by a number in front of the superscript T.
(b) Contact map generated from NH-H experimental NOEs from the NOESY-HSQC experiment for the GF peptide. Experimental NOEs for GF peptide are represented by circles (HN-HN( ), HN-Hα(
), HN-Hα( ), and HN-side chain protons(
), and HN-side chain protons( )). Intrachain NOEs are shaded in gray. Interchain contacts are consistent with one residue staggering of triple helix.
)). Intrachain NOEs are shaded in gray. Interchain contacts are consistent with one residue staggering of triple helix.
(c) Experimental 3JHNHa coupling constants of peptide GF. 3JHNHa coupling constants for Gly13, Phe14 and Gly24, with the two Hα Gly residues shown as dark gray and light gray bars.
An NOE contact map diagram (Figure 4b) was constructed using the experimental NOE data for GF in order to characterize interchain contacts and chain stagger 19. The experimental NOEs of GF are represented as circles. The boxes containing intrachain NOEs are shaded gray. Additional interchain backbone NOEs (2G13NH to 3O12Hα, 1F14NH to 3O12Hα, 3G13NH to 2G13Hα, 3G13NH to 1F14NH) are observed, supporting the one residue stagger of the triple-helix throughout the GF region even though a standard triple-helix is not possible due to the absence of one residue in the G-X1-X2 triplet. The limited number of NOEs is due to resonance overlap in the NOESY spectrum.
3JHNHa coupling constants which can be related to the dihedral angle φ are obtained for the trimers of the GF peptide from HNHA experiments (Figure 4c). All 3JHNHa coupling constants are uniform in the range of 5-6 Hz except 2F14, which has a large J coupling value of 8 Hz. The difference in the J coupling constants between the three Phe residues in different chains within a molecule indicates that non-equivalent distortions of backbone conformations are adopted by three chains at the G1G site. Multiple solutions of the angle φ are possible from the parameterized Karplus equation 22, and selection of the correct solution cannot be made from the 3JHNHa measurement alone. But the four allowed φ values put constraints on possible conformations within the GF region.
NMR Hydrogen exchange and NH temperature gradient studies
Hydrogen exchange experiments were performed on the three labeled residues within the GF peptide to explore the protection of labile amide protons from solvent (Figure 5a). The experimental monomer rate is too fast to be measured accurately, and therefore the theoretical monomer rate is used to calculate protection factors. The protection factors (P) were calculated by taking the ratio of the theoretical monomer rate to the experimental trimer rate for each residue. A very high degree of protection from exchange (P = 457) is seen for G24 in the stable C-terminal GPO region. The G13 residues in the three chains show an average protection factor P = 27, which is a 15-25 fold reduction relative to G24. The F14 residues show an even lower protection factor than G13 (average P = 11). Low protection factors could be related to decreased strength of hydrogen bonding, shielding from solvent, or local unfolding/breathing of the trimers.
Figure 5.
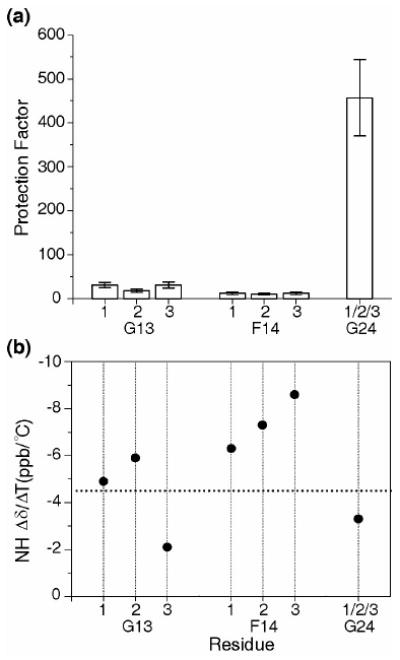
(a) Histogram of hydrogen/deuterium protection factors for the labeled residues in the GF peptide. Protection factors were calculated by taking the ratio of the theoretical monomer rate to the observed trimer rate for each residue. The low protection factors show that the G13 and F14 are more exposed, less hydrogen bonded, or more flexible than the control G24 in the GPO region.
(b) Amide NH Δδ/ΔT plot for GF peptide. The dashed horizontal line corresponds to Δδ/ΔT = -4.5 ppb/°C which provides the cutoff line for hydrogen bonding, with values less negative than this cutoff indicative of hydrogen bonding. G24 and 3G13 appear to have NH temperature gradients consistent with hydrogen bonding.
To complement hydrogen exchange studies, amide proton temperature gradients were performed on the GF peptide (Figure 5b). The NH temperature gradient for G24 is more positive than -4.5 ppd/°C indicating the existence of hydrogen bonds at the GPO rich C-terminal end 23. Similarly, 3G13 has a more positive temperature gradient than -4.5 ppd/°C indicating the formation of a hydrogen bond at the Gly13 of chain 3. The NH temperature gradients of the G13 residues in chains 1 and 2 are more negative than -4.5 ppd/°C supporting the non-equivalence in hydrogen bonding for three chains at G13. The more negative values of the NH temperature gradient for the three F14 residues suggest that they are not involved in hydrogen bonding.
Molecular modeling of the GF peptide
In order to define the conformation in solution for a GF interruption, molecular modeling was performed with the incorporation of NMR data using the strategy described previously 19. Models were generated based on an x-ray structure of a peptide where the central GP sequence of the crystal structure of the peptide (POG)3-PO-GPG-(POG)5 was replaced by GF 21. The structure was energy minimized incorporating φ angle restraints from 3JHNHa measurements and distance restraints from NOESY experiments. Back calculation of NMR parameters from the resulting structures was performed to eliminate structures that were not consistent with experimental data. Three structures were obtained that gave good agreement with NMR data.
All three models present a rod like structure without a kink or obvious bulge at the G1G site and with standard triple helical structures on both sides of the imperfection. The 1-residue stagger between the 3 chains is preserved throughout the GF sequence (Figure 6a). The G13 residues in all 3 chains are closely packed near the central axis although a standard triple helix cannot be formed because of the residue deletion. The three F14 residues are located on the outside of the triple-helix, similar to residues found at X1 positions, but with a certain deformation (Figure 6b). The Ramachandran plot of the GF models indicates a local disruption of the PPII dihedral angles (Figure 6c). G13 for chain 1 always falls within PPII, in contrast the G13 residues in chain 2 for all three models are below the PPII region while G13 residues in chain 3 for two models are in the β region. In all three models, the F14 residues of chain 1 fall within the α-helix region and the F14 residues of chain 2 are on the edge of the polyproline II region; F14 residues of chain 3 are in the PPII range. The G15 residue of chain 1 falls in the left-handed α-helix in two models, with G15 of chain 2 in the β region for one of the models (Figure 6d). In addition, there are dihedral angles of P11 and O12 for 1 or 2 chains which are non-PPII in some, but not all of the model structures. Although the structure was minimized for residues 6 - 19, the deformation at the GF site is highly localized, and only residues 11 to 15 have dihedral angles which deviate from the standard triple helical conformation. These models indicate a well-defined perturbation which is absorbed by a small number of residues. A significant variation among the three acceptable model structures can be observed, particularly around residue G15 in chain 1, as seen for the three red chains in Figure 6a and in the Ramachandran plot (Figure 6c). There is insufficient NMR data on the two labeled residues G13 and F14 to determine whether these variations arise from flexibility at that site.
Figure 6. Model structures of peptide GF.
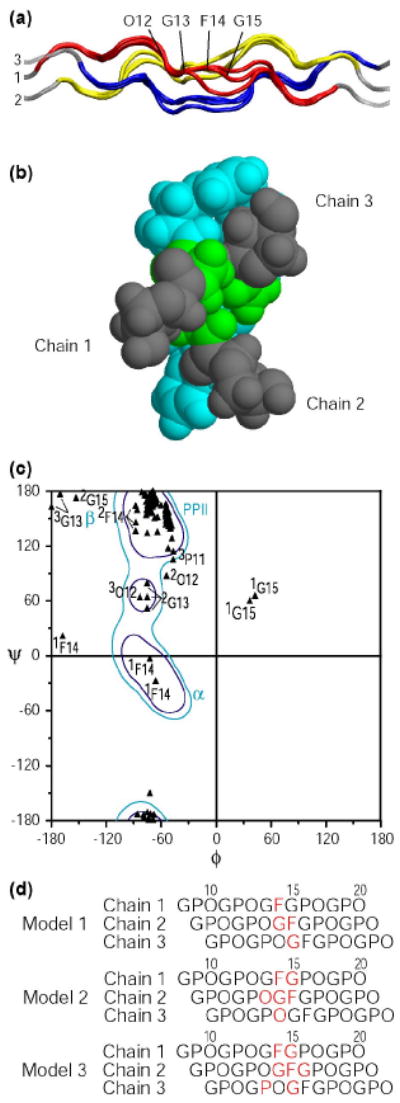
(a) Ribbon diagram of three GF models represented with the N-terminal at the left and chain numbers. The NMR parameters are used as restraints in the energy minimization, including the phi angle restraints from J coupling constants and distance restraints from NOEs. A set of three model structures that are consistent with all NMR experimental data were obtained. Leading, middle and trailing chains are represented as 1 (red), 2 (yellow) and 3 (blue), for the energy minimized segment of residues 6 to 19. The O12 to G15 residues in chain 1 of one structure are labeled for clarification.
(b) Space filling model of the cross section view from the N terminus to the C terminus of one model structure of GF. This view shows that at the GF region the G13 residues are closely packed at the center while the F14 residues are on the outside. The OGF segment is colored as O12( ), G13(
), G13( ), and F14(
), and F14( ).
).
(c) Ramachandran plot for three possible model structures of GF. To highlight the central region of the peptides, only residues 6 to 19 in all 3 chains of GF are plotted and shown in black triangles. Residues which fall outside the polyproline II region are labeled with the residue number and the chain (superscript). Values for the three models are given, so the presence of three values of 1F14 indicates that the Phe in this leading chain is outside the PPII region in all three models. The Ramachandran contour map for Pro residues is shown in the background and typical secondary structures are indicated, where α denotes α-helix; β denotes the β-sheet region; and PPII denotes the polyproline II and collagen region.
(d) Diagram of the 3 staggered chains near the GF sequence, showing the residues in red which fall outside the PPII dihedral angle region on the Ramachandran plot for the 3 acceptable models.
Discussion
In contrast with the structural homogeneity of fibrillar collagens, the non-fibrillar collagens show a wide diversity of architecture in their supramolecular forms. The presence of interruptions in the (Gly-X1-X2)n sequence pattern in all non-fibrillar collagens indicates that the canonical triple-helix structure must be discontinued or distorted in some way at these interruption sites, and these structural perturbations are likely to play a role in higher order structure, function, or degradation. For example, there are 8 interruption sites in human type X collagen, 5 G1G and 3 G4G, and it has been shown that two of the G4G sequences represent sites susceptible to specific matrix metalloproteinase degradation 24. In addition, interruptions are considered to play an important role in the network-like structure of type IV collagen in basement membranes, which is required for its filtration function, and in formation of the large fanned arrays of antiparallel type VII collagen molecules that make up anchoring fibrils 1,2,11. This report classifies and defines the structural consequences of the most frequent type of interruptions in the triple-helix found in human collagens, G1G sites, where the Gly-X1-X2 repeating pattern is interrupted by the absence of one residue.
All peptides with G1G interruptions form stable triple-helices in PBS except for the GP peptide, which formed some triple-helix in acetic acid (Tm of ∼15°C), but not in PBS. The G1G peptides that form triple helices show a substantial decrease in MRE225 nm compared with control Gly-X1-X2 repeating peptides, with their MRE values ranging from 55%-80% of the control. The introduction of a G1G also causes a drop in thermal stability and the degree of destabilization depends on the residue within the G1G and on the rigidity of the surrounding sequences. A decrease in Tm of ∼ 20°C is observed for GF and GY, compared with >50°C destabilization for GP within a GPO repeating environment. It is likely that the rigidity of the Pro residue within the interruption requires a greater degree of distortion than more flexible residues with the side chain attached to the β-carbon to be accommodated within a surrounding triple-helix framework. A decrease of only 12°C is seen in the GV peptide compared to its control. This is likely to reflect the more flexible GAA triplet N-terminal to the GV compared to GPO preceding the GF and GY sequences. It has been suggested that the Gly-X1-X2 residues surrounding interruptions have some characteristic features with respect to charges and imino acids 25, and the immediate tripeptide environment of G1G sites is found to be atypical. Such sequences might play a crucial role by acting to promote triple-helix propagation or renucleation when an interruption is encountered during the folding process.
The models obtained from NMR data on the GF peptide indicate a localized change in dihedral angles through the Gly-Phe region in all three chains. The residues outside the PPII region adopt allowed dihedral angles reported for Gly, Pro, and pre-Pro residues in crystal structures of large numbers of globular proteins (Fig. 6) 26. A diagram of the residues with dihedral angles outside the PPII region (Fig. 6d) shows the effect of the 1-residue staggering of the 3 chains on the conformational perturbation. In all three possible models, there are 2-3 residues distorted from PPII dihedral angles at the cross-sectional level when a Phe is first encountered (Phe14, chain 1; Gly13, chain 2; Hyp12, chain 3) and at the next level (Gly15, chain 1; Phe14, chain 2; Gly13, chain 3).
An examination of hydrogen bonding indicates that the G13 residues of chain 3 adopts a non-standard hydrogen bond with the carbonyl group of G13 in chain 2 in all three acceptable models. The amide protons of 1G13 and 2G13 form standard Rich-Crick hydrogen bonds to the carbonyl group of P11 in the neighboring chain in some but not all of the models. The non-equivalence in hydrogen bonds of three G13 residues is consistent with the amide proton temperature gradients of G13, with 3G13 showing a more positive temperature gradient than G13 residues in the other two chains. All three G13 residues have low protection factors compared with G24 in the C-terminal Gly-Pro-Hyp repeating region, indicating an increased flexibility that may be due to local breathing at the interruption site. The Phe chains are exposed on the outside of the helix, consistent with their very low protection factors and some flexibility. Both NMR and x-ray studies support multiple conformations for the structure within the G1G site 21. The increased number of conformational states near the interruption would lead to more favorable entropy, which may compensate in part for the decreased enthalpy due to less favorable hydrogen bonding. The NMR results on the GF peptide are in good agreement with the high resolution structure of the GP peptide solved by x-ray crystallography in showing a straight triple-helical molecule with localized changes in H-bonding and dihedral angles 21. Although GP and GF structures show no indication of a bend in the crystal structure or in solution, such a kink has been proposed to occur at a GQ sequence in the collagen triple-helix domain of the mannose binding lectin on the basis of electron microscopy 27.
Analysis of interruptions in human non-fibrillar collagen chains indicates the most common type are G1G, with G4G the next most frequent type. The preponderance of hydrophobic residues within G1G sequences supports functional significance and suggests G1G sites will introduce the same structural perturbation wherever found. Evolutionary analysis of a type IV collagen chain indicates that the location and nature of G1G sites are highly conserved among vertebrates. In contrast, sites correponding to human G1G sequences in the homologous chains of sea urchin, C. elegans, and Drosophila are frequently occupied by an uninterrupted Gly-X1-X2 pattern, suggesting G1G interruptions may have evolved in vertebrates to perform specific functions, such as binding, degradation or supramolecular structure. Some G1G sites in the vertebrate α1(IV) chains are occupied by G4G sequences in invertebrate orthologues. Consideration of a G4G interruption Gly-AA1-AA2-AA3-AA4-Gly shows that it can become a G1G when position AA2 or AA3 is occupied by Gly, or by the loss of three residues. The presence of G1G or G4G interruptions at corresponding sites within α1(IV) chains may suggest some evolutionary and/or functional relationship between these two types of small and common imperfections.
Although G1G and G4G breaks have some features in common, they produce distinct conformational consequences. Comparison of a G1G peptide with a related G4G peptide shows many similarities, including their thermal stability, enthalpy, formation of rod-like helices, maintenance of the 1-residue stagger throughout the imperfection site and a localized perturbation in hydrogen bonding and dihedral angles. Both lead to a loss of axial register of the superhelix on both sides. However, the G1G versus G4G amino acid sequence leads to a dramatic difference in the placement of hydrophobic residues with respect to the triple-helix structure. In the G4G case studied, the hydrophobic residue (Val) in the AA3 position packs near the central axis of the superhelix 19 while in the GF case reported here, the Phe is located on the outside of the helix. We propose that G1G and G4G interruptions introduce distinctive structural features within a triple-helix, which could serve as specific recognition sites. Such structural differences may help explain why two of the G4G sites, but none of the G1G sites within type X collagen are susceptible to matrix metalloproteinases 24,28. The results here all deal with homotrimer G1G sequences, as found in type VII or type X collagens, but the relationship between G1G and G4G may shed light on cases where these interruptions are found opposite each other in heterotrimeric molecules.
Materials and Methods
Sequence Analyses
Sequences of all non-fibrillar collagens were obtained from the protein database at www.expasy.ch. The χ2 test of statistical significance at a level of p< 0.001 was applied to test whether residues within the G1G interruption differs from the usual amino acids at X1 and X2 positions in (Gly-X1-X2)n regions found in all six chains of type IV collagen. The amino acids were grouped into hydrophobic (F, I, L, M, V, Y), charged (D, E, K, R, H), small (A, G, S, C), polar (T, Q, N) and imino acids (P). The χ2 test was also applied to test whether the types of residues (using the above groupings) in the tripeptide sequences immediately adjacent to G1G interruptions were different from average residues found in the X1 and X2 positions of the type IV (Gly-X1-X2)n sequences.
Evolutionary conservation of breaks in α1(IV) collagen chain was studied by multiple sequence alignments performed using the Clustal W program. The basic alignment of the α1(IV) chains in different organisms is largely determined by the highly conserved C-terminal NC1 globular domain 29. The alignment within the triple-helix domain is dominated by the presence of Gly as every third residue, and it is difficult to determine the precise position of gaps within that region. However, small shifts in the alignment do not affect the overall conclusions.
Peptides
All peptides used in the study were synthesized by Tufts University Core Facility (Boston, MA) and are shown in Table 2, using the single letter amino acid code, where hydroxyproline (Hyp) is denoted by O. For accurate concentration values, a Tyr was included at the C-terminus for all peptides except GYO and GY, and the concentration was determined using a molar extinction coefficient of 1400 (M-1cm-1) at 275 nm on a Beckman DU640 spectrophotometer. Peptides were purified on a Shimadzu reverse-phased HPLC system and the purity of the peptides confirmed by laser desorption mass spectrometry. All peptides were dissolved in phosphate-buffered saline (20 mM sodium phosphate, 150 mM NaCl, pH 7.0), except the GF and GFO peptides, which, because of solubility problems, were dissolved in glycine buffer (20 mM glycine, 150 mM NaCl, pH 2.0) containing 0.6M GuHCl.
Circular Dichroism (CD) Spectroscopy
CD spectra were recorded on an Aviv model 62DS spectropolarimeter. Cuvettes with 1- or 0.2-mm path length were used and the temperature was controlled using a Peltier temperature controller. Peptide solutions were made using either PBS, pH 7.0 or glycine buffer with 0.6M GuHCl, pH 2.0, and equilibrated at 0°C for at least 48h prior to measurements. Wavelength scans were collected in 0.5 nm steps with 4s averaging time and repeated three times. For temperature-induced denaturation, ellipticity was monitored at 225 nm. The peptides were equilibrated at each temperature for 2 min and the temperature was increased at an average rate of 0.1°C/min. The melting curves were obtained under standard conditions used in our laboratory for comparison, and, although not far from equilibrium, these conditions reflect a non-equilibrium state 30. The Tm was calculated as the temperature at which the fraction folded was equal to 0.5 when fit to a trimer-monomer transition.
Differential Scanning Calorimetry (DSC)
DSC transition curves were recorded on a NANO-DSC II model 6100 Calorimeter (Calorimetry Sciences Corp). Samples were equilibrated at 0°C for at least 48h prior to measurements. Samples were dialyzed against the respective buffers before each run. The rates of heating and cooling were maintained at 1°C/min. Since this heating rate is ten times faster than used for the CD melting curves, the denaturation temperature (Tm) observed is scan rate-dependent and higher than that seen from the CD scans. It has been shown earlier that the calorimetric enthalpy determined by DSC for similar collagenous peptides is independent of the scan rate 30. The enthalpy was calculated from the first scan (heating) since the scans were not reversible upon cooling.
NMR of peptide models of G1G interruptions
The GF peptide was synthesized with 15N amino acids at positions G13, F14, and G24. The sample was prepared in 0.4M GuHCl in 10% D2O/90% H2O at pH 2.0, c = 4.7 mM. NMR experiments were performed on a Varian VNMRS 600 MHz spectrometer equipped with a cold probe. 1H-15N heteronuclear single quantum coherence (HSQC) were obtained at 10°C. 3D 15N edited TOCSY-HSQC with a mixing time of 45ms and 3D 15N edited NOESY-HSQC with mixing times of 30-50ms were performed at 5°C and 10°C 31,32. Short mixing times (30ms) in NOESY-HSQC were used to eliminate spin diffusion and the data at various temperatures were used to help resolve overlapped resonances. 3D HNHA experiments 22 were performed to measure homonuclear 3JHNHa coupling constants at 10°C, with a H-H coupling period of 25ms. The correction factor for the 3JHNHa coupling constants was obtained as described 19. Hydrogen exchange experiments were carried out at 10°C, pDcorrect 2.5, as described 18. For measurements of amide proton temperature gradients, 1H-15N HSQC spectra were obtained at 0-20°C with an interval of 5°C. The sample was equilibrated at each temperature for at least 3 hours. Amide proton temperature gradients were obtained by linear regression analysis of the amide proton chemical shifts versus temperature.
All data were processed using the FELIX 2004 software package (MSI, San Diego, CA), and/or NMRPipe 33 and analyzed with FELIX 2004 or NMRView 34 as described previously19.
Generation of NOE contact map and molecular modeling
The NOE contact map for peptide GF was made from observed NH-H NOEs in the 3D 1H-15N NOESY-HSQC experiment and classified as NH-NH, NH-Hα, and NH-side chain (Hβ, Hγ, Hδ).
A computer model structure of GF was generated based on the x-ray crystal structure of GPG (PDB ID: 1EI8) 21 using the Molecular Operating Environment 2006.07 (Chemical Computing Group Inc., Montreal, Canada). The resulting model was energy minimized from residue 6 to residue 19 with dihedral angle φ and NOE distance restraints as described previously 19. The multiple solutions of dihedral angle φ derived from the 3JHNHa coupling constants using the Karplus equation are incorporated by using one solution at a time, resulting in a set of model structures, each obtained from a different combination of possible φ angles. Back calculation of 3JHNHa values and NOEs was used to eliminate models that were not consistent with the experimental data. Three representative structures consistent with experimental 3JHNHa values and all 1H-1H NOEs were selected for GF.
Acknowledgments
This work was supported in part by grants from the National Institutes of Health (GM060048 to BB and GM45302 to JB) and by a postdoctoral fellowship from the NIH Interdisciplinary Workforce (5 T90 DK070135) to GT. We would also like to thank Jordi Bella for helpful discussions on interruptions in collagen and Dr. Danny Chan for information on type X collagen.
Abbreviations Used
- Hyp
Hydroxyproline (three letter code)
- O
Hydroxyproline (single letter code)
- DSC
Differential Scanning Calorimetry
- HSQC
Heteronuclear single quantum coherence
Footnotes
Supported by a postdoctoral fellowship from the NIH Interdisciplinary Workforce, grant # 5 T90 DK070135
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Kielty CM, Grant ME. Connective tissue and its heritable disorders molecular, genetic, and medical aspect. In: Royce PM, S B, editors. The Collagen Family: Structure, Assembly and Organization in the Extracellular Matrix. Wiley-Liss, Inc; New York: 2002. [Google Scholar]
- 2.Myllyharju J, Kivirikko KI. Collagens, modifying enzymes and their mutations in humans, flies and worms. Trends Genet. 2004;20:33–43. doi: 10.1016/j.tig.2003.11.004. [DOI] [PubMed] [Google Scholar]
- 3.Veit G, Kobbe B, Keene DR, Paulsson M, Koch M, Wagener R. Collagen XXVIII, a novel von Willebrand factor A domain-containing protein with many imperfections in the collagenous domain. J Biol Chem. 2006;281:3494–504. doi: 10.1074/jbc.M509333200. [DOI] [PubMed] [Google Scholar]
- 4.Bella J, Eaton M, Brodsky B, Berman HM. Crystal and molecular structure of a collagen-like peptide at 1.9 A resolution. Science. 1994;266:75–81. doi: 10.1126/science.7695699. [DOI] [PubMed] [Google Scholar]
- 5.Rich A, Crick FH. The molecular structure of collagen. J Mol Biol. 1961;3:483–506. doi: 10.1016/s0022-2836(61)80016-8. [DOI] [PubMed] [Google Scholar]
- 6.Ramachandran GN. Treatise on collagen. New York: Academic Press; London: 1967. [Google Scholar]
- 7.Byers PH, Cole WG. Connective tissue and its heritable disorders molecular, genetic, and medical aspect. In: Royce PM, S B, editors. Osteogenesis imperfecta. Wiley-Liss, Inc; New York: 2002. [Google Scholar]
- 8.Marini JC, Forlino A, Cabral WA, Barnes AM, San Antonio JD, Milgrom S, Hyland JC, Korkko J, Prockop DJ, De Paepe A, Coucke P, Symoens S, Glorieux FH, Roughley PJ, Lund AM, Kuurila-Svahn K, Hartikka H, Cohn DH, Krakow D, Mottes M, Schwarze U, Chen D, Yang K, Kuslich C, Troendle J, Dalgleish R, Byers PH. Consortium for osteogenesis imperfecta mutations in the helical domain of type I collagen: regions rich in lethal mutations align with collagen binding sites for integrins and proteoglycans. Hum Mutat. 2007;28:209–21. doi: 10.1002/humu.20429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hostikka SL, Eddy RL, Byers MG, Hoyhtya M, Shows TB, Tryggvason K. Identification of a distinct type IV collagen alpha chain with restricted kidney distribution and assignment of its gene to the locus of X chromosome-linked Alport syndrome. Proc Natl Acad Sci U S A. 1990;87:1606–10. doi: 10.1073/pnas.87.4.1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boutaud A, Borza DB, Bondar O, Gunwar S, Netzer KO, Singh N, Ninomiya Y, Sado Y, Noelken ME, Hudson BG. Type IV collagen of the glomerular basement membrane. Evidence that the chain specificity of network assembly is encoded by the noncollagenous NC1 domains. J Biol Chem. 2000;275:30716–24. doi: 10.1074/jbc.M004569200. [DOI] [PubMed] [Google Scholar]
- 11.Hudson BG, Tryggvason K, Sundaramoorthy M, Neilson EG. Alport's syndrome, Goodpasture's syndrome, and type IV collagen. N Engl J Med. 2003;348:2543–56. doi: 10.1056/NEJMra022296. [DOI] [PubMed] [Google Scholar]
- 12.Jais JP, Knebelmann B, Giatras I, De Marchi M, Rizzoni G, Renieri A, Weber M, Gross O, Netzer KO, Flinter F, Pirson Y, Verellen C, Wieslander J, Persson U, Tryggvason K, Martin P, Hertz JM, Schroder C, Sanak M, Krejcova S, Carvalho MF, Saus J, Antignac C, Smeets H, Gubler MC. X-linked Alport syndrome: natural history in 195 families and genotype-phenotype correlations in males. J Am Soc Nephrol. 2000;11:649–57. doi: 10.1681/ASN.V114649. [DOI] [PubMed] [Google Scholar]
- 13.Gould DB, Phalan FC, van Mil SE, Sundberg JP, Vahedi K, Massin P, Bousser MG, Heutink P, Miner JH, Tournier-Lasserve E, John SW. Role of COL4A1 in small-vessel disease and hemorrhagic stroke. N Engl J Med. 2006;354:1489–96. doi: 10.1056/NEJMoa053727. [DOI] [PubMed] [Google Scholar]
- 14.Hammami-Hauasli N, Schumann H, Raghunath M, Kilgus O, Luthi U, Luger T, Bruckner-Tuderman L. Some, but not all, glycine substitution mutations in COL7A1 result in intracellular accumulation of collagen VII, loss of anchoring fibrils, and skin blistering. J Biol Chem. 1998;273:19228–34. doi: 10.1074/jbc.273.30.19228. [DOI] [PubMed] [Google Scholar]
- 15.Hovnanian A, Duquesnoy P, Blanchet-Bardon C, Knowlton RG, Amselem S, Lathrop M, Dubertret L, Uitto J, Goossens M. Genetic linkage of recessive dystrophic epidermolysis bullosa to the type VII collagen gene. J Clin Invest. 1992;90:1032–6. doi: 10.1172/JCI115916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Varki R, Sadowski S, Uitto J, Pfendner E. Epidermolysis bullosa. II. Type VII collagen mutations and phenotype-genotype correlations in the dystrophic subtypes. J Med Genet. 2007;44:181–92. doi: 10.1136/jmg.2006.045302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brodsky B, Persikov AV. Molecular structure of the collagen triple helix. Adv Protein Chem. 2005;70:301–39. doi: 10.1016/S0065-3233(05)70009-7. [DOI] [PubMed] [Google Scholar]
- 18.Mohs A, Popiel M, Li Y, Baum J, Brodsky B. Conformational features of a natural break in the type IV collagen Gly-X-Y repeat. J Biol Chem. 2006;281:17197–202. doi: 10.1074/jbc.M601763200. [DOI] [PubMed] [Google Scholar]
- 19.Li Y, Brodsky B, Baum J. NMR Shows Hydrophobic Interactions Replace Glycine Packing in the Triple Helix at a Natural Break in the (Gly-X-Y)n Repeat. J Biol Chem. 2007;282:22699–706. doi: 10.1074/jbc.M702910200. [DOI] [PubMed] [Google Scholar]
- 20.Long CG, Braswell E, Zhu D, Apigo J, Baum J, Brodsky B. Characterization of collagen-like peptides containing interruptions in the repeating Gly-X-Y sequence. Biochemistry. 1993;32:11688–95. doi: 10.1021/bi00094a027. [DOI] [PubMed] [Google Scholar]
- 21.Bella J, Liu J, Kramer R, Brodsky B, Berman HM. Conformational effects of Gly-X-Gly interruptions in the collagen triple helix. J Mol Biol. 2006;362:298–311. doi: 10.1016/j.jmb.2006.07.014. [DOI] [PubMed] [Google Scholar]
- 22.Vuister GW, Bax A. Quantitative J correlation: a new approach for measuring homonuclear three-bond J(HNHA) coupling constants in 15N-enriched proteins. J Am Chem Soc. 1993;115:7772–7777. [Google Scholar]
- 23.Baxter NJ, Williamson MP. Temperature dependence of 1H chemical shifts in proteins. J Biomol NMR. 1997;9:359–69. doi: 10.1023/a:1018334207887. [DOI] [PubMed] [Google Scholar]
- 24.Welgus HG, Fliszar CJ, Seltzer JL, Schmid TM, Jeffrey JJ. Differential susceptibility of type X collagen to cleavage by two mammalian interstitial collagenases and 72-kDa type IV collagenase. J Biol Chem. 1990;265:13521–7. [PubMed] [Google Scholar]
- 25.Long CG, Thomas M, Brodsky B. Atypical Gly-X-Y sequences surround interruptions in the repeating tripeptide pattern of basement membrane collagen. Biopolymers. 1995;35:621–8. doi: 10.1002/bip.360350608. [DOI] [PubMed] [Google Scholar]
- 26.Lovell SC, Davis IW, Arendall WB, 3rd, de Bakker PI, Word JM, Prisant MG, Richardson JS, Richardson DC. Structure validation by Calpha geometry: phi,psi and Cbeta deviation. Proteins. 2003;50:437–50. doi: 10.1002/prot.10286. [DOI] [PubMed] [Google Scholar]
- 27.Thiel S, Reid KB. Structures and functions associated with the group of mammalian lectins containing collagen-like sequences. FEBS Lett. 1989;250:78–84. doi: 10.1016/0014-5793(89)80689-1. [DOI] [PubMed] [Google Scholar]
- 28.Gadher SJ, Schmid TM, Heck LW, Woolley DE. Cleavage of collagen type X by human synovial collagenase and neutrophil elastase. Matrix. 1989;9:109–15. doi: 10.1016/s0934-8832(89)80028-9. [DOI] [PubMed] [Google Scholar]
- 29.Leinonen A, Mariyama M, Mochizuki T, Tryggvason K, Reeders ST. Complete primary structure of the human type IV collagen alpha 4(IV) chain. Comparison with structure and expression of the other alpha (IV) chains. J Biol Chem. 1994;269:26172–7. [PubMed] [Google Scholar]
- 30.Persikov AV, Xu Y, Brodsky B. Equilibrium thermal transitions of collagen model peptides. Protein Sci. 2004;13:893–902. doi: 10.1110/ps.03501704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fesik SW, Zuiderweg ER. Heteronuclear three-dimensional nmr spectroscopy. A strategy for the simplification of homonuclear two-dimensional NMR spectra. J Magn Reson. 1988;78:588–593. [Google Scholar]
- 32.Messerle BA, Wider G, Otting G, Weber C, Wüthrich K. Solvent suppression using a spin-lock in 2D and 3D NMR spectroscopy with H2O solutions. J Magn Reson. 1989;85:608–613. [Google Scholar]
- 33.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–93. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 34.Johnson BA, Blevins RA. NMR View - a Computer-Program for the Visualization and Analysis of NMR Data. J Biomol NMR. 1994;4:603–614. doi: 10.1007/BF00404272. [DOI] [PubMed] [Google Scholar]