Abstract
Recessive mutations in the Phenylalanine hydroxylase (PAH) gene predispose to phenylketonuria (PKU) in conjunction with dietary exposure to phenylalanine. Previous studies have suggested PAH variations could confer risk for schizophrenia, but comprehensive follow-up has not been reported. We analyzed 15 common PAH “tag” SNPs and 3 exonic variations that are rare in Caucasians but common in African-Americans among four independent samples (total n = 5,414). The samples included two US Caucasian cohorts (260 trios, 230 independent cases, 474 controls), Bulgarian families (659 trios), and an African-American sample (464 families, 401 controls). Analyses of both US Caucasian samples revealed associations with five SNPs; most notably the common allele (G) of rs1522305 from case-control analyses (z = 2.99, p = 0.006). This SNP was independently replicated in the Bulgarian cohort (z = 2.39, p = 0.015). A non-significant trend was also observed among African-American families (z = 1.39, p = 0.165), and combined analyses of all four samples were significant (rs1522305: χ2 = 23.28, 8 d.f., p = 0.003). These results for this SNP met our a priori criteria for statistical significance, namely an association that was robust to multiple testing correction in one sample, a replicated risk allele in multiple samples, and combined analyses that were nominally significant. Case-control results in African-Americans detected an association with L321L (p = 0.047, OR = 1.46). Our analyses suggest several associations at PAH, with consistent evidence for rs1522305. Further analyses, including additional variations and environmental influences such as phenylalanine exposure are warranted.
Introduction
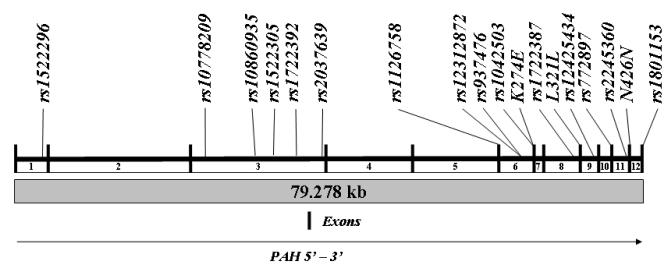
Phenylalanine hydroxylase (PAH) catalyses the conversion of phenylalanine (Phe) to tyrosine. This reaction is the rate limiting step in the synthesis of catecholamines and accounts for approximately 75% of the disposal of dietary Phe. The gene encoding PAH is localized to chromosome 12q23.2, contains 13 exons, and the genomic sequence spans approximately 79.3 kilobases. PAH is expressed in the liver and kidney.
Mutations in PAH can lead to phenylketonuria (PKU) in the presence of a diet that includes Phe. PKU manifests as mental retardation (MR), associated with peculiarities of gait and posture, eczema, epilepsy, light pigmentation, cataracts, brain calcification and a ‘mousy’ odor (Følling 1934). These manifestations have been attributed to hyperphenylalaninemia resulting from impaired PAH activity. Early postnatal and long term use of a low Phe diet enables near normal cognitive development (Donlon et al., 2004). PKU is inherited as an autosomal recessive disorder, with an average birth incidence of 1 / 10,000 in European populations. Despite the increased frequency of several rare mutations in African-Americans compared to Caucasians, the incidence of PKU in African Americans is about one-third that in Caucasians (National Institute of Child Health and Human Development). The aggregate mutant allele frequency in these groups is estimated at 0.01. There is considerable allelic heterogeneity, with over 500 catalogued mutations leading to a spectrum of disease ranging from benign hyperphenylalaninemia to classical PKU (www.pahdb.mcgill.ca) (Scriver et al., 2003). Genetic heterogeneity is also present, as PKU can occur due to mutations in tetrahydrobiopterin (BH4), an essential PAH co-factor (Thony and Blau 2006).
Penrose first suggested co-segregation of psychiatric illnesses and PKU, raising the possibility that PAH mutations may contribute to psychopathology other than MR (Penrose 1935). Studies to explore this hypothesis have been conducted among PKU probands and their relatives, as well as psychiatric patients and their relatives, particularly schizophrenia (SZ) patients. The severe MR observed among individuals with untreated PKU would preclude a diagnosis of SZ using current criteria, though some case reports with such co-morbidity have been published in the past (Fisch et al., 1979). More recent case-reports suggesting co-occurrence of PKU among individuals with psychoses have also been published (Shiwach and Sheikha 1998). A large scale survey among institutionalized psychotic individuals did not detect any individuals with PKU (Cares 1956). On the other hand, early studies of SZ patients found elevated fasting Phe levels, as well as abnormal responses to Phe tolerance tests (Poisner 1960), suggesting that some SZ patients could be carriers of mutant PAH alleles.
Recent genetic studies have investigated a connection between PAH polymorphisms and increased susceptibility to SZ. Sobell et al. first examined two point mutations (R408W and IVS12nt1) known to be associated with PKU in a case-control study design (190 SZ cases, 336 controls), but did not detect a significant association (Sobell et al., 1993). A linkage study of three quantitative traits in a sample of European and African-American schizophrenia affected siblings identified modest evidence for linkage with a marker at 109.5 cM overlapping PAH (LOD = 2.12). Linkage with negative symptoms bolstered linkage evidence somewhat for this sample (LOD score = 2.97 at 104 cM), as well as an association between this marker and schizophrenia (Wilcox et al., 2002). A series of studies previously conducted by Dr. Mary Richardson and colleagues have suggested associations between several PAH mutations and psychiatric illness among African-Americans but not Caucasians (Richardson et al., 1999a) (Richardson et al., 1999b) (Chao and Richardson 2002). Richardson and colleagues also reported on 9 exonic variants at PAH among 123 psychiatrically ill individuals and 34 controls (Richardson et al., 2003). One exonic variant (K274E) was noted among African-Americans and was over-represented among SZ patients (cases: 4/24; controls: 1/13). The K274E mutation was associated with altered Tyr levels following a Phe loading test. Recently, linkage was detected using short tandem repeat polymorphisms near PAH in an island population from Palau when mothers of schizophrenia patients were treated as the affected generation (Devlin et al., 2007). These results are intriguing, because they suggest maternal-fetal interaction in SZ genesis. If true, such a mechanism might account for variability in conventional association and linkage analyses.
The published studies suggest a link between common and / or rare PAH polymorphisms and SZ. To investigate this hypothesis, we evaluated 18 PAH variations in four independent samples. Our analyses included 15 common polymorphisms and three additional exonic variations reported on previously (Richardson et al., 2003).
Methods
Study design
We tested the hypothesis that common and / or rare PAH variations increase risk for schizophrenia (SZ). We analyzed 15 single nucleotide polymorphisms (SNPs) that tagged common variations in Caucasians (Figure 1, details below). These SNPs were evaluated in four independent samples of either European or African-American ancestry. We also selected three variations based on published analyses with psychosis that were monomorphic in Caucasians but polymorphic in African-Americans (K274E, N426N, and L321L, referred to as ‘rare variants’ herein for clarity) (Richardson et al., 1999a), (Richardson et al., 1999b), (Chao and Richardson 2002). Our primary study included only SNP based analyses, first in each sample individually then combined across samples. Associations with the ‘rare variants’ were conducted next, followed by exploratory analyses to evaluate covariates such as gender and maternal genotypes.
Figure 1. Genomic organization of PAH and variations evaluated.
The vertical bars represent exons. The numbers below the line represent the introns. The polymorphisms analyzed are listed above the line.
Samples
Caucasians
US
Unrelated patients were recruited at Western Psychiatric Institute and Clinic, Pittsburgh, Pennsylvania and surrounding regions (n = 490). Diagnoses were based on the Diagnostic Interview for Genetic Studies (Nurnberger et al., 1994), supplemented by medical records and informant interviews. Consensus DSM-IV diagnoses of schizophrenia or schizoaffective disorder were assigned by board-certified psychiatrists / psychologists following review of all these sources of information. Both parents of 260 patients were ascertained for family based analyses, but diagnostic evaluations were not conducted for the parents (260 trios). Control DNA samples were collected from the cord blood of 474 unscreened Caucasian neonates born at Magee-Women’s Hospital, Pittsburgh, PA. Only ancestry and gender was available for these samples.
Bulgaria
SZ patients and their parents were recruited in Bulgaria as described previously (Kirov et al., 2004). Diagnoses among probands were made according to DSM-IV criteria, following assessment by a psychiatrist using the Schedules for Clinical Assessment in Neuropsychiatry (Wing JK 1990 Jun) , which has been validated for use in the Bulgarian language, and inspection of hospital discharge summaries. All patients and their parents received written information on the project and signed an informed consent form. The Bulgarian sample included 659 trios (total n = 1,977). Probands were diagnosed with SZ (n = 576) or SZA (n = 83).
African Americans
African-American patients and their parents were ascertained as part of an ongoing collaborative study to investigate risks for schizophrenia in an African-American sample (Aliyu et al., 2006). Families were chosen for genotyping from the overall consortium, and analyses were carried out based on phenotype data as of January 19th, 2008. The sample was composed of 464 total families ascertained for both linkage and association studies, including 73 complete trios (proband + 2 parents), 181 “duo + sibs” (proband + 1 parent + unaffected siblings), 122 “case + sibs” (affected proband + unaffected siblings, no parents), 53 affected sibling pairs without parents, 27 affected sibling pairs with 1 parent, 5 affected sibling pairs + both parents, and 3 “duos” (affected proband + 1 parent, no siblings). From these family configurations, most but not all individuals were informative for family-based association tests. For the three ‘rare variants’ (see below), 551 African-American cases were contrasted with 401 adult controls. The cases included one patient with SZ or SZA randomly chosen from each of the 464 families and 87 singleton cases where no parents were available. The controls were screened for absence of psychoses and current substance abuse using the same procedures as the cases (Aliyu et al., 2006).
The University of Pittsburgh Institutional Review Board (IRB) approved the study. Approval from appropriate IRBs was also obtained at each collaborating US site. Ethics committee approval was obtained from ethics committees in all regions of Bulgaria where families were recruited. Written informed consent was obtained from all participants, except neonatal controls, in accordance with IRB guidelines.
Polymorphism Selection
We chose tag SNPs to represent all common variations among 60 unrelated Caucasians available in release 20 (phase II, January, 2006) of the International HapMap Project (HapMap 2003). To accomplish this, we selected all available SNPs within PAH and 5 kb of flanking sequence 5′ and 3′ to the gene. Genotypes were obtained from CEPH samples (US residents collected in 1980 by the Centre d’Etude du Polymorphisme Humain). These participants have ancestry from Northern and Western Europe. Tag SNPs were identified to represent common variation with a minor allele frequency (MAF) greater than 5% in Caucasians using Hclust software (Rinaldo et al., 2005). Hclust computes a similarity matrix from the square of Pearson’s correlation (r2) between allele counts at pairs of loci then uses hierarchical clustering to group correlated SNPs. We selected a SNP as a tag if the correlation between loci was below a threshold of r2 < 0.9. Thus, 21 SNPs were identified. When SNPs were initially rejected by Applied Biosystems in the assay design (8 SNPs), surrogates were sought. If no surrogates were available, we re-analyzed the dataset to identify another SNP with a lower LD threshold to use as a proxy (r2 > 0.8 between surrogate and failed marker). Using this procedure, only two tag SNPs were not represented at a minimum correlation threshold of r2 = 0.8 in our analyses (rs1281013 and rs1851381).
Previous research by Richardson et al. suggested associations between several exonic variations and psychosis among African-Americans (Richardson et al., 2003). Those analyses indicated that the SNPs had minor allele frequencies (MAF) greater than 1% among African-Americans, but had MAF < 0.01 in European Americans. We chose three such variants (K274E, L321L, N426N; referred to in this study as ‘rare variants’ for clarity) to be genotyped in all of our family samples. An additional set of case-control analyses were conducted for only these SNPs among the African-American probands, as well as cases not included in the family based analyses, and an adult African-American control sample typed exclusively for these polymorphisms.
Genotyping Assays
All 18 variants were included in assays for all four independent samples using the hybridization based SNPlex assay (ABI Biosystems, Inc), as described elsewhere (Tobler et al., 2005). The assay utilizes custom designed oligonucleotide pools of up to 48 SNPs, which can be genotyped in a single reaction. The three ‘rare variants’ were genotyped among the African-American controls using the ABI SNaPshot assay (Applied Biosystems, Inc). The assay involves a multiplexed PCR reaction followed by single base extension (Mansour et al., 2005). The genomic organization of PAH and the selected polymorphisms are shown in Figure 1. All molecular genetic analyses were conducted at the University of Pittsburgh.
Quality control
All genotype assays included duplicated samples and/or CEPH individuals genotyped by HapMap (HapMap 2003). Negative control samples (water) were also included in each assay plate. A random subset of 34 African-American samples were selected from all individuals found to carry at least one copy of the rare alleles of K274E, L321L, and N426N and individually sequenced to confirm the SNPlex and SNaPshot genotype calls. Tests for Mendelian inconsistencies were conducted in all family-based samples using PEDCHECK (O’Connell and Weeks 1998) and tests of Hardy-Weinberg Equilibrium (HWE) were carried out for probands, parents, and controls separately in each population using GENEPOP software (version 1.31) (Raymond and Rousset 1995).
Statistical Analysis
Transmission distortion was analyzed using FBAT software (Laird et al., 2000), which can appropriately handle families of mixed configuration such as those in the African-American sample analyzed here. Differences in genotype distributions between cases and controls were evaluated with the Armitage Trends test (SAS software) (Devlin and Roeder 1999) or Fisher’s exact test, as appropriate. Test statistics were converted to z scores for case-control analyses for ease of comparison regarding risk alleles (i.e., z positive or negative) across samples. We estimated the effective number of independent tests among these SNPs using the statistical package R based on published methods (Conneely and Boehnke 2007). We estimated the number of effective tests in the Caucasians and African-Americans separately due to the expected differences in LD patterns between these populations. Our analyses suggested 7.9 effective tests in the Caucasians and 12.6 effective tests in the African-Americans. We analyzed each SNP for association in each sample individually. To evaluate evidence against the null hypothesis across the four independent samples, we combined results based on Fisher’s combined probability test (Fisher, 1948).
Exploratory analyses
We conducted exploratory analyses to determine if risk conferred by individual polymorphisms was modified by gender. To carry out these analyses, we analyzed allele transmissions to male and female probands separately in family based analyses, and performed logistic regression among male cases / controls and female cases / controls separately.
Previous analyses in an island population detected linkage to the maternal generation of affected SZ cases at 12q23.2 (Devlin et al., 2007). We therefore hypothesized that SZ liability was conferred by maternal PAH genotypes. To test this hypothesis, we compared allele frequencies for all 15 common SNPs between mothers and fathers in all three samples (Armitage trends test).
Interpretation of statistical significance
We considered an association with SZ significant if (1) an individual SNP test exceeded an alpha threshold of 0.0063 in any Caucasian sample (0.05 / 7.9 tests) or 0.0040 (0.05 / 12.6 tests) in the African-American samples, (2) a nominally significant replication for an individual SNP (and allele) was detected (p ≤ 0.05 in two or more samples), or (3) combined analyses provided evidence of an association. Exploratory analyses were considered significant only if replication was detected (p ≤ 0.05).
Results
Quality Control
All 18 SNPs were genotyped in the Caucasians, but rs124125434 could not be assayed in the African-American samples. The mean genotype call rate was 95% or greater in all four samples for the SNPlex assays and 96.8% in the SNaPshot assays. Using duplicated samples and CEPH individuals to compare with HapMap, we estimated our genotyping accuracy to range between 99.95% - 99.88% in all 4 samples. These data are comparable to HapMap estimates and our previous analyses in these Caucasian samples (Talkowski et al., 2008). We sequenced 34 African-Americans for the three rare variations to confirm their genotype. We found 100% concordance between the sequencing genotypes and SNPlex / SNaPshot genotypes for these individuals.
Linkage Disequilibrium
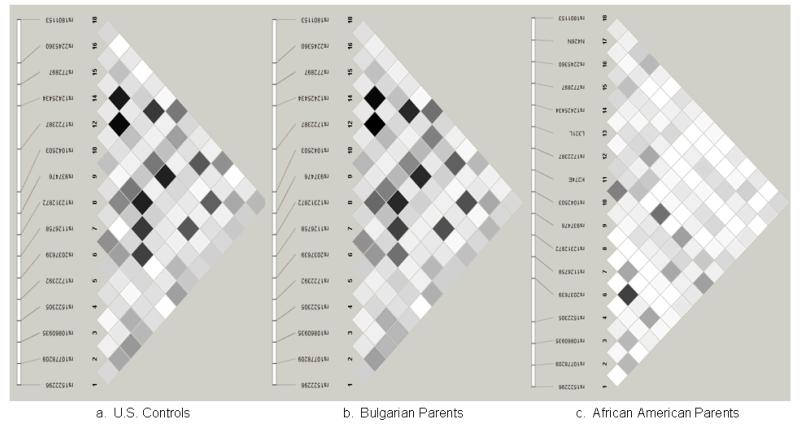
Linkage disequilibrium (LD) was estimated using Haploview software among unrelated Caucasian controls from the US (n = 474), unrelated parents from Bulgaria (n = 1318), and unrelated African-American parents (n = 367). As expected, pairwise LD (r2) was similar between Caucasian samples, but differed among African-Americans (Figure 2).
Figure 2. Linkage disequilibrium analysis.
Linkage disequilibrium (r2) was estimated between SNPs among (a) unrelated US Caucasian controls, (b) parents of affected Caucasian probands from Bulgaria, and (c) parents of affected African-American probands.
Primary association analyses
Caucasians
In the US case-control sample (230 cases independent of the trios, 474 controls), two SNPs were associated with SZ (rs1522305, z = 2.74, p = 0.006, OR = 1.64, 95% CI = 1.15 – 2.32; rs12312872, z = 1.98, p = 0.050, OR = 1.34, 95% CI = 1.84 – 0.99; all p-values uncorrected). In the US family sample (260 trios), transmission distortion was detected with three SNPs, including rs1042503 (z = -2.0, p = 0.05), rs12425434 (z = -2.2, p = 0.03), and rs10860935 (z = 2.3, p = 0.02).
In the Bulgarian families (659 trios), the most significant association in the US case-control analyses (common G allele of rs1522305) was replicated in this independent cohort (z = 2.4, p = 0.015). Three other SNPs were nominally significant (uncorrected p < 0.05; rs2245360, rs937476, rs152296). Transmission distortion that did not reach statistical significance was noted for two SNPs that was consistent with associations in the US sample, namely rs12312872 (Bulgarian p = 0.06, US case-control p = 0.05) and rs10860935 (Bulgarian p = 0.09, US family-based analyses p = 0.02) (see Table 1). The ‘rare variants’ were monomorphic among all Caucasian samples genotyped.
Table I.
Association analyses of PAH variations
| US Cases / Controls (n = 230 / 474) |
US Trios (n = 260) |
Bulgarian Trios (n = 659) |
African-American Families (n = 464) |
Combined Analysis (All samples) |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SNP | Position | N | Case Freq |
Control Freq |
Z1 | p1 | Z2 | p2 | Allele Freq |
Z3 | p3 | Allele Freq |
Z4 | p4 | X82 | Pall |
| rs1522296 | 101834917 | G | 0.707 | 0.686 | 0.77 | 0.450 | 1.61 | 0.11 | 0.68 | 2.23 | 0.025 | 0.49 | 0.01 | 1.00 | 13.4 | 0.099 |
| rs10778209 | 101808102 | G | 0.765 | 0.751 | 0.58 | 0.565 | 0.76 | 0.45 | 0.72 | -0.24 | 0.813 | 0.92 | -0.58 | 0.56 | 4.3 | 0.826 |
| rs10860935 | 101807061 | Ta | 0.850 | 0.868 | -0.94 | 0.371 | 2.27 | 0.02 | 0.84 | 1.67 | 0.094 | 0.66 | -0.63 | 0.53 | 15.5 | 0.051 |
| rs1522305 | 101804886 | G a | 0.898 | 0.843 | 2.99 | 0.006 | 0.46 | 0.65 | 0.88 | 2.39 | 0.015 | 0.82 | 1.39 | 0.17 | 23.3 | 0.003 |
| rs1722392 | 101802875 | C | 0.539 | 0.558 | -0.67 | 0.528 | -0.20 | 0.84 | 0.52 | -1.38 | 0.167 | 0.57 | 0.01 | 0.99 | 5.2 | 0.734 |
| rs2037639 | 101795480 | G | 0.733 | 0.738 | -0.23 | 0.818 | -1.40 | 0.16 | 0.77 | 0.60 | 0.550 | 0.87 | -0.27 | 0.79 | 5.7 | 0.680 |
| rs1126758 | 101773054 | A | 0.557 | 0.582 | -0.90 | 0.376 | -1.36 | 0.17 | 0.58 | 1.23 | 0.218 | 0.80 | -0.07 | 0.95 | 8.6 | 0.376 |
| rs12312872 | 101771783 | Aa | 0.858 | 0.818 | 1.98 | 0.050 | 0.63 | 0.53 | 0.86 | 1.85 | 0.064 | 0.63 | -0.77 | 0.44 | 14.4 | 0.072 |
| rs937476 | 101771694 | Ab | 0.586 | 0.575 | 0.38 | 0.722 | 0.57 | 0.57 | 0.56 | -2.07 | 0.039 | 0.55 | 1.39 | 0.17 | 11.9 | 0.157 |
| rs1042503 | 101770830 | G | 0.714 | 0.727 | -0.51 | 0.614 | -2.00 | 0.05 | 0.74 | 0.44 | 0.664 | 0.95 | -0.72 | 0.47 | 9.5 | 0.305 |
| K274E | 101770744 | A | 0.98 | 0.00 | 1.00 | |||||||||||
| rs1722387 | 101765200 | G | 0.843 | 0.855 | -0.59 | 0.572 | 0.91 | 0.37 | 0.84 | 1.38 | 0.167 | 0.85 | 0.32 | 0.75 | 7.3 | 0.506 |
| L321L | 101764809 | C | 0.94 | 0.62 | 0.54 | |||||||||||
| rs12425434 | 101764197 | C | 0.716 | 0.723 | -0.30 | 0.769 | -2.20 | 0.03 | 0.73 | -0.22 | 0.829 | N/A | ||||
| rs772897 | 101761598 | Gc | 0.843 | 0.848 | -0.23 | 0.822 | 1.08 | 0.28 | 0.83 | 1.11 | 0.269 | 0.82 | 1.25 | 0.21 | 8.7 | 0.368 |
| rs2245360 | 101758674 | G | 0.642 | 0.638 | 0.17 | 0.870 | 0.72 | 0.47 | 0.62 | -2.18 | 0.030 | 0.81 | -0.23 | 0.82 | 9.2 | 0.323 |
| N426N | 101757710 | T | 0.86 | 0.67 | 0.50 | |||||||||||
| rs1801153 | 101756896 | G | 0.811 | 0.787 | 1.06 | 0.312 | 0.18 | 0.86 | 0.74 | 0.84 | 0.399 | 0.39 | 0.53 | 0.60 | 5.5 | 0.703 |
“Results from association analyses of 18 PAH variations in four independent samples. SNPs are provided in the direction of PAH transcription 5′ to 3′. N = nucleotide for which frequency data are listed. Freq = frequency of allele for which nucleotide is provided (common allele). Z = test statistic for common allele (negative = risk conferred by minor allele). Combined analysis using Fisher’s method of combining probabilities from independent tests of significance (distributed as χ22N statistic). Reference allele nomenclature consistent with HapMap and reference sequence designations.
SNP genotyped on “+” strand, allele provided is “reference” allele.
SNP genotyped on “-“ strand, allele provided is the “reference” allele.
SNP genotyped on “-“ strand, allele provided is ““other”” allele. SNP positions based on dbSNP build 129.
African-Americans
In the African-American family sample, no SNPs were significantly associated with schizophrenia, but a trend for over-transmission of the G allele at rs1522305 was noted (z = 1.39, p = 0.167). The over-transmitted allele was consistent with the US and Bulgarian samples and its frequency was similar across samples (US cases 0.898, US cords 0.843, Bulgarian cases 0.875, African-American cases 0.819). All three ‘rare variants’ (K274E, L321L, N426N) were present at a frequency greater than 1% in the African-Americans. None were significantly over-transmitted to probands, however minor allele frequencies for K274E (0.014) did not enable meaningful analyses of transmission distortion given the size and configuration of the present sample. Case-control comparisons in the African-American samples were therefore conducted for only these three SNPs (551 cases, 402 controls). None of the rare alleles were found to be associated with SZ risk, however a nominally significant association was detected with the common allele (non-mutant allele) of L321L (p = 0.047, OR = 1.46, 95% CI = 1.03 - 2.14), replicating previous results by Richardson and colleagues (Table 2).
Table II.
Comparison of three PAH variations among African-Americans
| Case Genotype | Control Genotype | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| SNP | Nuc | 11 | 12 | 22 | 11 | 12 | 22 | Case Freq |
Control Freq |
Y | p-value |
| K274E | 1 = A 2 = G |
523 | 16 | 0 | 369 | 17 | 0 | 0.985 | 0.978 | 1.35 | 0.246 |
| L321L | 1 = C 2 = T |
483 | 52 | 2 | 326 | 57 | 0 | 0.948 | 0.926 | 3.96 | 0.047 |
| N426N | 1 = C 2 = T |
9 | 138 | 386 | 9 | 83 | 287 | 0.146 | 0.133 | 0.63 | 0.428 |
Case-control analyses of 3 exonic PAH variations (referred to in text as ‘rare variants’) among an African-American case-control sample. Nuc = nucleotide. Freq = allele frequency provided for allele 1. Y, p-value: results of trends test from distribution of genotypes.
Fisher’s exact test p-value: K274E, p = 0.207, L321L, p = 0.017.
Combined analyses
We combined the observed probabilities for each of the four independent samples at each of the 14 SNPs tested across all samples (rs12425434 and each of the three ‘rare variants’ were not informative for associations in all four samples). As expected from the initial findings in three of the four samples, combined analyses suggested a significant association with the common allele of rs1522305 (χ2 = 23.28, 8 d.f., p = 0.003). A nominally significant association was also detected with rs10860935 (χ2 = 15.47, 8 d.f., p = 0.05). Another SNP, rs12312872, was significant among European samples (χ2 = 12.76, 6 d.f., p = 0.047), but not when African-Americans were included in combined analyses (p = 0.072).
Exploratory analyses
Gender specific associations were detected in the Bulgarian trios with nine SNPs. None were associated in both males and females. Over-transmission to affected male patients was observed for six SNPs, the most significant being rs937476 (p = 0.004, OR = 1.4). Three SNPs were associated among females, most notably rs1522305 (G allele, p = 0.002, OR = 1.84) and rs152296 (G allele, p = 0.007, OR = 1.43) (Supplementary Table 1).
Replicate analyses in the US and African-American samples detected a significant association with the common allele of rs1522305 when US Caucasian female cases were compared with female controls (US case-control p = 0.05). However, an association was not detected among the US Caucasian trios or the African-American family sample. Consistent replication was also detected between Bulgarian male patients and US male patients (p ≤ 0.05 in both samples) with rs1042503, rs12425434, and rs2037639. None were replicated among US Caucasian male probands or African-American males. (Supplementary Table 1).
We compared the allele frequencies of the 18 common polymorphisms between the mothers and fathers in all three available family samples. No significant differences were found for any of these comparisons (data not shown).
Interpretation of statistical significance
Our analyses found the equivalent of 7.9 effective tests in each individual Caucasian sample and 12.6 effective tests in the African-American samples. There were thus 36.3 effective tests across all four samples for the primary analyses and 132.6 total tests across all primary and exploratory analyses. The associations at rs1522305 fulfilled all three pre-established criteria for significance. The initial analyses in the US case-control sample exceeded the individual experiment correction for multiple testing (uncorrected p = 0.006, corrected p = 0.047) (criterion #1 above). This SNP was significant in two independent samples (US case-control p = 0.006, Bulgarian p = 0.015) (criterion #2), and was associated following combined analyses from all four samples (p = 0.003) (criterion #3). No other SNP associations were robust to correction for multiple testing in individual samples, nor were any other SNPs replicated in more than one sample, although rs10860935 was significant in combined analysis of all samples (p = 0.05) (Table 1).
Discussion
We tested associations between PAH variants and schizophrenia by evaluating tag SNPs to represent all available common PAH SNPs among Caucasians, as well as three ‘rare variants’ previously suggested as risk factors for schizophrenia. We detected several associations of modest effect size in individual samples, with one replicated association in multiple cohorts. The magnitude of the effects detected here were similar to those reported with other genes in complex disorders (odds ratios 1.10 – 1.50). Simulation studies, as well as analyses of the association between apoE variants and Alzheimer disease suggest that variable patterns of association can be observed in independent samples of varying size, particularly if the primary risk variant is not investigated (Bacanu et al., 2002) (Yu et al., 2007). Thus, it is often difficult to replicate associations with genetically complex disorders consistently across samples, especially if the magnitude of the association is modest. To reduce the probability of rejecting associations prematurely, we conducted analyses in four individual samples, followed by combined analyses. Using this approach, a consistent association was detected at rs1522305. The association was nominally significant in two of the three Caucasian samples and combining the results across all four samples revealed a significant association. Similarly, exploratory analyses yielded replicable results related to gender between European samples at this locus. Our analytic strategy combined test statistics from multiple independent samples (even those with modest power) in an effort to identify meaningful SZ risk conferred by the same allele that may not reach nominal significance in individual samples.
Prior studies have suggested that PAH mutations or exonic polymorphisms may be risk factors for SZ among African-Americans (Richardson et al., 2003). We evaluated three such variants in all our samples. We detected one nominally significant association with L321L, a synonymous substitution among African-Americans. The associated allele is the common allele, consistent with the results of Richardson et al., however our results failed to support the findings of risk conferred by the rare allele of N426N. These variants appeared to be monomorphic in the Caucasian samples, although it is possible that rare alleles were present in individuals that failed the SNPlex assays for these SNPs. More comprehensive analyses of other known PAH mutations and / or deep sequencing of the region are indicated.
It is not known if allelic variation at the associated SNPs alters PAH activity, so the functional impact of the associations is uncertain. It is possible that the associated SNPs serve as surrogates for unidentified primary risk allele(s). There is modest LD between rs1522305 and two other SNPs, namely rs12312872 and rs1042503 (Figure 2). Analysis of available HapMap data also suggested LD with more remote SNPs, e.g., an intergenic region 100.2 kb 3′ to rs1522305 (rs1722400, D’ = 0.75, r2 = 0.52). If the associated SNPs have demonstrable effects on transcription, there are plausible mechanisms for the genetic associations. Hyperphenylalaninemia (HPA) following PAH deficiency can enhance competition between phenylalanine and tyrosine for transport across the blood brain barrier (BBB) (Pardridge and Choi 1986). Reduced transport of tyrosine across the BBB may decrease catecholamine synthesis (Fernstrom and Fernstrom 2007). The reduced synthesis may lead to altered dopamine function, a well known mechanism proposed for SZ genesis (Carlsson 1988) (Snyder 1973) (Seeman et al., 1976). HPA may also increase Phe catabolism through alternative pathways, such as increased synthesis of phenylethylamine (PEA), a putative psychotogenic compound (Jeste et al., 1981). This hypothesis has been investigated extensively previously, albeit with conflicting results (O’Reilly and Davis 1994).
Several other lines of investigations may prove helpful in order to further explore the present results. Since current DSM IV criteria preclude a diagnosis of SZ in the presence of MR, it would be of interest to estimate the prevalence of psychoses among PKU patients who have undergone rigid dietary control. Unfortunately, most published follow-up studies have involved children prior to the modal age at onset for SZ (Weglage et al., 2000) (Corcoran et al., 2005). Interestingly, several investigators have reported that frontal lobe dependent cognitive functions are impaired into young adulthood even among PKU patients who were treated early and aggressively (Welsh et al., 1990) (Diamond et al., 1994) (Corcoran et al., 2005). Similar cognitive impairment has been noted among patients with SZ and their relatives (Gur et al., 2007) (Greenwood et al., 2007). Evaluation of cognitive function among patients with the putative risk alleles may prove insightful in this regard. To follow up Penrose’s early analyses, re-examination of psychiatric disorders among obligate carriers of PAH mutations (e.g., parents of individuals with PKU) may also be informative (Penrose, 1935).
The clinical features of PKU are manifested only when individuals with PAH mutations consume a diet that includes Phe. The present study did not evaluate such dietary risk factors. Confirmation of a link between SZ and PAH mutations or polymorphisms opens the possibility of use of one of a growing number of therapeutic options for treating PKU (including supplementation with biopterin derivatives and large neutral amino acids) to examine their effect on the development of psychiatric disease. A prior linkage study suggested a role for maternal PAH variation in pathogenesis (Devlin et al., 2007). We did not find differences in allele frequencies between mothers of Caucasian probands and controls or fathers of the probands at the associated SNPs. This hypothesis needs to be explored further. The mechanism for the gender related associations noted here is unclear. It is possible that gender serves as a proxy for other variables.
Improvement on the current analyses could be made in future studies by considering a denser set of polymorphisms in African-American samples. The tag SNPs analyzed in the present study represented common variations in Caucasian samples only. Analysis of the Nigerian sample from HapMap suggests that up to 43 SNPs may have been required to comprehensively represent all available SNPs in African-Americans sample (HapMap 2003). Moreover, the power of our African-American samples was relatively low, owing to both a smaller number of samples and incomplete family configurations. Therefore, further analyses of African-American samples are required. Despite the decreased power in the African-American and US family samples, our combined analyses considered the p-values from each independent sample equally and could be conservative. It is noteworthy that analyses of the joint distribution of test statistics across groups weighted by sample size also suggested a significant deviation from the null hypothesis at rs1522305 (data not shown).
Our analyses of four independent samples of Caucasian and African-American ancestry identified replicable associations between schizophrenia and an intronic PAH polymorphism. The functional role for the associated polymorphisms is unknown. It remains possible that risk is conferred primarily by as yet unidentified polymorphism(s). Further analyses of rare exonic variations, population specific tag SNPs for African-Americans, and additional ethnic groups are warranted, preferably in conjunction with environmental risk factors.
Supplementary Material
Acknowledgements
This work was funded by grants from NIMH (MH56242, MH063420 and MH66263 to VLN, MH080582 to MET) and Jannsen Research Foundation, Beerse, Belgium to GK, MJO and MO. The PAARTNERS sample collection was supported by MH66181 to RGo, MH066050 to JM, MH66049 to NE, MH66004 to AS, MH66006 to DB, MH66005 to JK, and MH66121 to RG.
References
- Aliyu MH, Calkins ME, Swanson CL, Jr., Lyons PD, Savage RM, May R, Wiener H, Devlin B, Nimgaonkar VL, Ragland JD, Gur RE, Gur RC, Bradford LD, Edwards N, Kwentus J, McEvoy JP, Santos AB, McCleod-Bryant S, Tennison C, Go RC. Project among African-Americans to explore risks for schizophrenia (PAARTNERS): Recruitment and assessment methods. Schizophr Res. 2006;87(1-3):32–44. doi: 10.1016/j.schres.2006.06.027. [DOI] [PubMed] [Google Scholar]
- Bacanu SA, Devlin B, Chowdari KV, DeKosky ST, Nimgaonkar VL, Sweet RA. Linkage analysis of Alzheimer disease with psychosis. Neurology. 2002;59(1):118–20. doi: 10.1212/wnl.59.1.118. [DOI] [PubMed] [Google Scholar]
- Cares RM. Absence of phenylketonuria in adult psychotics; a survey of 4246 inmates of a state mental hospital. Am J Psychiatry. 1956;112(11):938–9. doi: 10.1176/ajp.112.11.938. [DOI] [PubMed] [Google Scholar]
- Carlsson A. The current status of the dopamine hypothesis of schizophrenia. Neuropsychopharmacology. 1988;1(3):179–86. doi: 10.1016/0893-133x(88)90012-7. [DOI] [PubMed] [Google Scholar]
- Chao HM, Richardson MA. Aromatic amino acid hydroxylase genes and schizophrenia. Am J Med Genet. 2002;114(6):626–30. doi: 10.1002/ajmg.10606. [DOI] [PubMed] [Google Scholar]
- Conneely KN, Boehnke M. So Many Correlated Tests, So Little Time! Rapid Adjustment of P Values for Multiple Correlated Tests. Am J Hum Genet. 2007;81(6) doi: 10.1086/522036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corcoran C, Whitaker A, Coleman E, Fried J, Feldman J, Goudsmit N, Malaspina D. Olfactory deficits, cognition and negative symptoms in early onset psychosis. Schizophr Res. 2005;80(2-3):283–93. doi: 10.1016/j.schres.2005.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devlin B, Klei L, Myles-Worsley M, Tiobech J, Otto C, Byerley W, Roeder K. Genetic liability to schizophrenia in Oceanic Palau: a search in the affected and maternal generation. Hum Genet. 2007;121(6):675–84. doi: 10.1007/s00439-007-0358-7. [DOI] [PubMed] [Google Scholar]
- Devlin B, Roeder K. Genomic control for association studies. Biometrics. 1999;55:997–1004. doi: 10.1111/j.0006-341x.1999.00997.x. [DOI] [PubMed] [Google Scholar]
- Diamond A, Ciaramitaro V, Donner E, Djali S, Robinson MB. An animal model of early-treated PKU. J Neurosci. 1994;14(5 Pt 2):3072–82. doi: 10.1523/JNEUROSCI.14-05-03072.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donlon J, Levy H, Scriver CR. Hyperphenylalaninemia: Phenylalanine Hydroxylase Deficiency. McGraw-Hill; New York: 2004. The Metabolic and Molecular Bases of Inherited Disease. [Google Scholar]
- Fernstrom JD, Fernstrom MH. Tyrosine, phenylalanine, and catecholamine synthesis and function in the brain. J Nutr. 2007;137(6 Suppl 1):1539S–1547S. doi: 10.1093/jn/137.6.1539S. discussion 1548S. [DOI] [PubMed] [Google Scholar]
- Fisch RO, Hosfield WB, Chang PN, Barranger J, Hastings DW. An adult phenylketonuric with schizophrenia. Clinical and biochemical similarities and possible genetic connection between the two diseases. Minn Med. 1979;62(4):243–6. [PubMed] [Google Scholar]
- Følling A. Ueber Ausscheidung von Phenylbrenztraubensaeure in den Harn als Stoffwechselanomalie in Verbindung mit Imbezillitaet. Hoppe-Seyler’s Zeitschrift Fuer Physiologische Chemie. 1934;227:169–176. [Google Scholar]
- Greenwood TA, Braff DL, Light GA, Cadenhead KS, Calkins ME, Dobie DJ, Freedman R, Green MF, Gur RE, Gur RC, Mintz J, Nuechterlein KH, Olincy A, Radant AD, Seidman LJ, Siever LJ, Silverman JM, Stone WS, Swerdlow NR, Tsuang DW, Tsuang MT, Turetsky BI, Schork NJ. Initial heritability analyses of endophenotypic measures for schizophrenia: the consortium on the genetics of schizophrenia. Arch Gen Psychiatry. 2007;64(11):1242–50. doi: 10.1001/archpsyc.64.11.1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gur RE, Nimgaonkar VL, Almasy L, Calkins ME, Ragland JD, Pogue-Geile MF, Kanes S, Blangero J, Gur RC. Neurocognitive Endophenotypes in a Multiplex Multigenerational Family Study of Schizophrenia. American Journal of Psychiatry. 2007;164:813–819. doi: 10.1176/ajp.2007.164.5.813. [DOI] [PubMed] [Google Scholar]
- HapMap The International HapMap Project. Nature. 2003;426(6968):789–96. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- Jeste DV, Doongaji DR, Panjwani D, Datta M, Potkin SG, Karoum F, Thatte S, Sheth AS, Apte JS, Wyatt RJ. Cross-cultural study of a biochemical abnormality in paranoid schizophrenia. Psychiatry Res. 1981;5(3):341–52. doi: 10.1016/0165-1781(81)90082-2. [DOI] [PubMed] [Google Scholar]
- Kirov G, Ivanov D, Williams NM, Preece A, Nikolov I, Milev R, Koleva S, Dimitrova A, Toncheva D, O’Donovan MC, Owen MJ. Strong evidence for association between the dystrobrevin binding protein 1 gene (DTNBP1) and schizophrenia in 488 parent-offspring trios from Bulgaria. Biol Psychiatry. 2004;55(10):971–5. doi: 10.1016/j.biopsych.2004.01.025. [DOI] [PubMed] [Google Scholar]
- Laird NM, Horvath S, Xu X. Implementing a unified approach to family-based tests of association. Genet Epidemiol. 2000;19(Suppl 1):S36–42. doi: 10.1002/1098-2272(2000)19:1+<::AID-GEPI6>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- Mansour HA, Talkowski ME, Wood J, Pless L, Bamne M, Chowdari KV, Allen M, Bowden CL, Calabrese J, El-Mallakh RS, Fagiolini A, Faraone SV, Fossey MD, Friedman ES, Gyulai L, Hauser P, Ketter TA, Loftis JM, Marangell LB, Miklowitz DJ, Nierenberg AA, Patel J, Sachs GS, Sklar P, Smoller JW, Thase ME, Frank E, Kupfer DJ, Nimgaonkar VL. Serotonin gene polymorphisms and bipolar I disorder: focus on the serotonin transporter. Ann Med. 2005;37(8):590–602. doi: 10.1080/07853890500357428. [DOI] [PubMed] [Google Scholar]
- Nurnberger JI, Jr., Blehar MC, Kaufmann CA, York-Cooler C, Simpson SG, Harkavy-Friedman J, Severe JB, Malaspina D, Reich T. Diagnostic interview for genetic studies. Rationale, unique features, and training. NIMH Genetics Initiative. Archives of General Psychiatry. 1994;51(11):849–59. doi: 10.1001/archpsyc.1994.03950110009002. discussion 863-4. [DOI] [PubMed] [Google Scholar]
- O’Connell JR, Weeks DE. PedCheck: a program for identification of genotype incompatibilties in linakge analysis. American Journal of Human Genetics. 1998;63(1):259–266. doi: 10.1086/301904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Reilly RL, Davis BA. Phenylethylamine and schizophrenia. Prog Neuropsychopharmacol Biol Psychiatry. 1994;18(1):63–75. doi: 10.1016/0278-5846(94)90024-8. [DOI] [PubMed] [Google Scholar]
- Pardridge WM, Choi TB. Neutral amino acid transport at the human blood-brain barrier. Fed Proc. 1986;45(7):2073–8. [PubMed] [Google Scholar]
- Penrose LS. The Detection of Autosomal Linkage in Data Which Consists of Pairs of Brothers and Sisters of Unspecified Parentage. Annals of Eugenics. 1935;6:133–138. [Google Scholar]
- Poisner AM. Serum phenylalanine in schizophrenia: biochemical genetic aspects. Journal of Nervous and Mental Disease. 1960;131:74–6. doi: 10.1097/00005053-196007000-00009. [DOI] [PubMed] [Google Scholar]
- Raymond M, Rousset F. GENEPOP (version 1.2): population genetics software for exact tests and ecumenicism. Journal of Heredity. 1995;86:248–249. [Google Scholar]
- Richardson MA, Guttler F, Guldberg P, Read L, Clelland J, Chao H, Reilly M, Suckow R. Functional and psychiatric associations of the phenylalanine hydroxylase gene. Molecular Psychiatry. 1999a;4:S41. [Google Scholar]
- Richardson MA, Guttler F, Guldberg P, Reilly M, Suckow R, Read L, Chao H, Clelland J. Phenylalanine hydroxylase gene mutation associated with schizophrenia and African-American ethnic status. Schizophrenia Research. 1999b:95. [Google Scholar]
- Richardson MA, Read LL, Clelland JD, Chao HM, Reilly MA, Romstad A, Suckow RF. Phenylalanine hydroxylase gene in psychiatric patients: screening and functional assay of mutations. Biol Psychiatry. 2003;53(6):543–53. doi: 10.1016/s0006-3223(02)01528-7. [DOI] [PubMed] [Google Scholar]
- Rinaldo A, Bacanu SA, Devlin B, Sonpar V, Wasserman L, Roeder K. Characterization of multilocus linkage disequilibrium. Genet Epidemiol. 2005;28:193–206. doi: 10.1002/gepi.20056. [DOI] [PubMed] [Google Scholar]
- Scriver CR, Hurtubise M, Konecki D, Phommarinh M, Prevost L, Erlandsen H, Stevens R, Waters PJ, Ryan S, McDonald D, Sarkissian C. PAHdb 2003: what a locus-specific knowledgebase can do. Hum Mutat. 2003;21(4):333–44. doi: 10.1002/humu.10200. [DOI] [PubMed] [Google Scholar]
- Seeman P, Lee T, Chau-Wong M, Wong K. Antipsychotic drug doses and neuroleptic/dopamine receptors. Nature. 1976;261(5562):717–9. doi: 10.1038/261717a0. [DOI] [PubMed] [Google Scholar]
- Shiwach RS, Sheikha S. Delusional disorder in a boy with phenylketonuria and amine metabolites in the cerebrospinal fluid after treatment with neuroleptics. J Adolesc Health. 1998;22(3):244–6. doi: 10.1016/S1054-139X(97)00158-4. [DOI] [PubMed] [Google Scholar]
- Snyder SH. Amphetamine psychosis: a “model” schizophrenia mediated by catecholamines. Am J Psychiatry. 1973;130(1):61–7. doi: 10.1176/ajp.130.1.61. [DOI] [PubMed] [Google Scholar]
- Sobell JL, Heston LL, Sommer SS. Novel association approach for determining the genetic predisposition to schizophrenia: case-control resource and testing of a candidate gene. American Journal of Medical Genetics. 1993;48(1):28–35. doi: 10.1002/ajmg.1320480108. [DOI] [PubMed] [Google Scholar]
- Talkowski ME, Kirov G, Bamne M, Georgieva L, Torres G, Mansour H, Chowdari KV, Milanova V, Wood J, McClain L, Prasad K, Shirts B, Zhang J, O’Donovan MC, Owen MJ, Devlin B, Nimgaonkar VL. A network of dopaminergic gene variations implicated as risk factors for schizophrenia. Hum Mol Genet. 2008;17(5):747–58. doi: 10.1093/hmg/ddm347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thony B, Blau N. Mutations in the BH4-metabolizing genes GTP cyclohydrolase I, 6-pyruvoyl-tetrahydropterin synthase, sepiapterin reductase, carbinolamine-4a-dehydratase, and dihydropteridine reductase. Hum Mutat. 2006;27(9):870–8. doi: 10.1002/humu.20366. [DOI] [PubMed] [Google Scholar]
- Tobler AR, Short S, Andersen MR, Paner TM, Briggs JC, Lambert SM, Wu PP, Wang Y, Spoonde AY, Koehler RT, Peyret N, Chen C, Broomer AJ, Ridzon DA, Zhou H, Hoo BS, Hayashibara KC, Leong LN, Ma CN, Rosenblum BB, Day JP, Ziegle JS, De La Vega FM, Rhodes MD, Hennessy KM, Wenz HM. The SNPlex genotyping system: a flexible and scalable platform for SNP genotyping. J Biomol Tech. 2005;16(4):398–406. [PMC free article] [PubMed] [Google Scholar]
- Weglage J, Grenzebach M, Pietsch M, Feldmann R, Linnenbank R, Denecke J, Koch HG. Behavioural and emotional problems in early-treated adolescents with phenylketonuria in comparison with diabetic patients and healthy controls. J Inherit Metab Dis. 2000;23(5):487–96. doi: 10.1023/a:1005664231017. [DOI] [PubMed] [Google Scholar]
- Welsh MC, Pennington BF, Ozonoff S, Rouse B, McCabe ER. Neuropsychology of early-treated phenylketonuria: specific executive function deficits. Child Dev. 1990;61(6):1697–713. [PubMed] [Google Scholar]
- Wilcox MA, Faraone SV, Su J, Van Eerdewegh P, Tsuang MT. Genome scan of three quantitative traits in schizophrenia pedigrees. Biol Psychiatry. 2002;52(9):847–54. doi: 10.1016/s0006-3223(02)01465-8. [DOI] [PubMed] [Google Scholar]
- Wing JKBT, Brugha T, Burke J, Cooper JE, Giel R, Jablenski A, Regier D, Sartorius N. SCAN. Schedules for Clinical Assessment in Neuropsychiatry. Arch Gen Psychiatry. 1990 Jun;47(6):589–93. doi: 10.1001/archpsyc.1990.01810180089012. [DOI] [PubMed] [Google Scholar]
- Yu CE, Seltman H, Peskind ER, Galloway N, Zhou PX, Rosenthal E, Wijsman EM, Tsuang DW, Devlin B, Schellenberg GD. Comprehensive analysis of APOE and selected proximate markers for late-onset Alzheimer’s disease: Patterns of linkage disequilibrium and disease/marker association. Genomics. 2007;89(6):655–665. doi: 10.1016/j.ygeno.2007.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.