Abstract
Complex gene regulatory networks are composed of genes, noncoding RNAs, proteins, metabolites, and signaling components. The availability of genome-wide mutagenesis libraries; large-scale transcriptome, proteome, and metabalome data sets; and new high-throughput methods that uncover protein interactions underscores the need for mathematical modeling techniques that better enable scientists to synthesize these large amounts of information and to understand the properties of these biological systems. Systems biology approaches can allow researchers to move beyond a reductionist approach and to both integrate and comprehend the interactions of multiple components within these systems. Descriptive and mathematical models for gene regulatory networks can reveal emergent properties of these plant systems. This review highlights methods that researchers are using to obtain large-scale data sets, and examples of gene regulatory networks modeled with these data. Emergent properties revealed by the use of these network models and perspectives on the future of systems biology are discussed.
Keywords: systems biology, Arabidopsis, transcription factors, emergent properties, modeling, genomics
INTRODUCTION: SYSTEMS THEORY AND GENE REGULATORY NETWORKS
A plant’s final form and ability to respond dynamically to the external environment are the outcome of many components interacting within complex gene regulatory networks. These networks exist at different scales: within a single cell, across multiple cells and cell types, and temporally in each of these contexts. Furthermore, it is these networks that are acted upon by evolutionary forces and whose modification gives rise to the vast diversity in morphological form among plant species. The emergence of high-throughput genomic technologies has resulted in an explosion of data, enabling the cataloging of genes, gene products, and their interactions. Plant biologists are now moving from the analysis of the role of a small number of genes to the monitoring of the global dynamics of entire biological circuits.
Ideker et al. (2001) proposed a systems biology framework to integrate genome-wide measurements in an effort to understand the properties of biological systems. Systems theory studies the organization of variables and is based on the belief that only through understanding the interaction of these components can one identify emergent properties of a system. Emergent properties are those that cannot be identified by examining interactions between individual components alone. First, the components and their interactions that act within the network must be identified and defined (Ideker et al. 2001). To facilitate a systems approach, however, these components are most appropriately characterized in a global manner. The next step is to perturb the system and monitor its response. Ideally, the effects of perturbations are monitored with genome-scale measurements. Finally, these data are integrated, and the system is modeled. The observed experimental values are compared with the predictions of the model. Novel interactions between components can be inferred, and alternative hypotheses are devised to address discrepancies (Ideker et al. 2001). New experiments can then be undertaken to test these new hypotheses. Ideally, systems approaches can provide insights into complex, regulatory networks and provide a framework for biologists to understand the organization of life.
The plant community has embraced the use of systems approaches (Gutierrez et al. 2005) and produced vast amounts of genomic-scale data and a variety of databases and tools. The primary aim of this review is to describe examples of gene regulatory networks and, in particular, plant transcriptional regulatory networks, whose properties have been elucidated by systems approaches. We begin with a description of the various components that act within gene regulatory networks and of the high-throughput techniques that are being used to elucidate these components. With a list of components and their interactions in hand, gene regulatory networks can be inferred by the use of several different methods. We describe some of these methods, including examples of theoretical modeling and integration of genome-scale data sets. Each of these methods has advantages and disadvantages, but all the methods share the ability to provide a framework with predictive power. We also provide examples of how comparisons of simulations with observed experimental data have revealed gaps in our understanding of molecular interactions. These discrepancies have provided the rationale for predicting novel components or interactions that can reconcile these inconsistencies. Finally, as mentioned above, identifying emergent network properties is a primary goal of the systems approach. We describe some emergent properties identified in network descriptions generated from high-throughput data as well as from smaller network descriptions generated with various modeling approaches. Once we understand and can correctly simulate the dynamics of these gene regulatory networks, we will be able to understand how these complex regulatory networks give rise to a plant’s form and function.
GENOME-SCALE DATA SETS: IDENTIFICATION OF COMPONENTS
Genes: Identification and Functional Characterization
A variety of methods have been developed in plants to identify components of plant regulatory networks in a high-throughput manner. Many of these methods have been described in other extensive reviews (Brady et al. 2006, Busch & Lohmann 2007, Jones-Rhoades et al. 2006, Willmann & Poethig 2007, Yazaki et al. 2007). Therefore, we highlight only a set that we feel will be essential for modeling the various molecular species that act to regulate gene expression.
Genome sequencing
Systems theory has been defined as the study of the nature of complex systems (Bertalanffy 1973). The use of systems approaches in the field of biological sciences has emerged only recently as a method to study, comprehend, and integrate large data sets generated during the genomics era. Although scientists have sequenced the genomes of a host of microbes and animals, to date only three plant genomes have been sequenced: Arabidopsis (The Arabidopsis Initiative 2000, Lin et al. 1999, Mayer et al. 1999, Theologis et al. 2000), rice (Int. Rice Genome Seq. Proj. 2005), and poplar (Tuskan et al. 2006). However, more than a dozen other plant species, including maize, potato, soybean, cassava, and grape, are in the sequencing pipeline (Pennisi 2007).
Mutant resources
Once the genome is sequenced and genes are predicted, the next goal is to determine the biological function of each gene. Systems biology is greatly facilitated by collections of sequence-indexed, genome-wide mutants (Alonso & Ecker 2006). This allows one to probe the function of genes, using reverse genetics approaches. These resources are integral to testing predictions made about gene regulatory networks. Although a range of methods can be used to generate collections of mutants, the preferred methods for generating sequence-indexed collections in plants include T-DNA and transposon insertional mutagenesis, TILLING (targeting-induced local lesions in genomes) of chemically induced mutants, ectopic expression, and gene silencing. The most extensive collections exist in Arabidopsis. Such collections include the Syngenta Arabidopsis Insertion Library (SAIL) lines, with insertions in the promoters and transcribed regions of ~15,000 to ~18,000 genes (Sessions et al. 2002); the SALK insertion collection, with insertions in more than 21,000 genes; and the GABI-KAT resource, which contains mutations in ~14,000 genes (Alonso et al. 2003, Rosso et al. 2003). Several groups have carried out, in rice and poplar, high-throughput T-DNA insertional mutagenesis and analysis of loci disrupted by these insertions (Chen et al. 2003, Groover et al. 2004, Hsing et al. 2007, Jeong et al. 2006, Sallaud et al. 2004, J. Zhang et al. 2006). Some of these lines allow functional characterization of target genes, easy detection of insertions using GUS fusions, and the possibility of obtaining ectopic expression of space-and time-specific genes (Groover et al. 2004, Hsing et al. 2007, J. Zhang et al. 2006).
However, insertional mutagenesis does have several disadvantages. Insertions can sometimes be biased toward intergenic regions (Hsing et al. 2007) and can cause dominant lethal mutations, which lead to incomplete saturation of the entire genome. To that end, scientists have begun TILLING plant populations after chemical mutagenesis (Till et al. 2004). This strategy begins with chemical mutagenesis of a population, followed by polymerase chain reaction (PCR) of target genes. Mutations are detected when the PCR products are denatured and reannealed, forming genetic lesions, which, when digested by a mismatch-specific endonuclease, are detectible by capillary gel electrophoresis. Although TILLING is labor intensive and therefore considerably more expensive than other methods, the advantages of TILLING are that it can be used in a high-throughput manner and that it produces heritable mutations in a genome independently of the genome size and of the plants’ transformability, reproductive system, and generation time (Gilchrist & Haughn 2005). In addition, TILLING can generate allelic series that allow the study of essential genes, for which complete loss of function can be lethal. Suzuki et al. (2007) used this method in single zygotic rice cells to generate a population with mutations every 135 kb, which will soon be available to the research community. The Maize TILLING Project (MTP), funded by the National Science Foundation, is a national effort to identify point mutations in specific genes in mutant populations for two inbred lines, B73 and W22 (http://genome.purdue.edu/maizetilling/). TILLING efforts are also taking place in Arabidopsis, Lotus, rice, and Brassica oleracea, as well as in other plants (Gilchrist & Haughn 2005, Till et al. 2007).
Targeted mutagenesis can also be employed in a high-throughput manner. RNA interference (RNAi), a defense mechanism that evolved to protect against viruses and transposons in plants, has been exploited as a strategy to disrupt target gene expression. In this method, specific small interfering RNA (siRNA) molecules, which degrade target RNA species, are generated. Efforts are currently under way to generate 150 to 500 base-pair gene-specific targets (GSTs) for at least 21,500 genes in Arabidopsis. These GSTs have been used for microarray transcript profiling and cloned into bacterial plasmid vectors for targeted RNAi (Hilson et al. 2004).
Large-scale gene expression analysis
Microarray technology has revolutionized our ability to monitor the output of transcriptional regulation at the level of the whole genome. These expression analyses have been further refined by our ability to monitor expression, at high resolution, within individual cell types, tissues, and organs; at different developmental time points; in different mutant backgrounds; and in response to hormones and various stress conditions (Birnbaum et al. 2003, 2005; Brady et al. 2007; Dembinsky et al. 2007; Kilian et al. 2007; Nawy et al. 2005; Nelson et al. 2006). These large-scale experiments have demonstrated that to obtain accurate representations of gene expression, one must sample at sufficient resolution to differentiate the full complement of transcriptional programs occurring within a plant. To accomplish this, investigators have developed two technologies to identify gene expression in individual cell types or tissues within the plant. The first is the use of fluorescence-activated cell sorting to isolate cell-type-specific green fluorescent protein (GFP)-marked root populations, and the subsequent isolation of RNA from these populations for use in microarray analysis (Birnbaum et al. 2003, 2005). This technique has been used to obtain the most comprehensive microarray expression map of an organ and its component cell types (Brady et al. 2007). The second technique is the use of laser capture microdissection to isolate cell-type populations in maize and Arabidopsis (Dembinsky et al. 2007, Nakazono et al. 2003, Spencer et al. 2007, Woll et al. 2005). Cells isolated by this technology have also been used to characterize the cell-type-specific proteome (Dembinsky et al. 2007).
Phylogenetic conservation
Recent efforts in the field of comparative genomics have also contributed to our understanding of gene regulatory networks. Researchers have identified potential cis-regulatory sequences in many organisms by searching for evolutionary conservation (Harbison et al. 2004, Kheradpour et al. 2007, Pritsker et al. 2004). Furthermore, natural variation can be exploited to make correlations concerning sequence diversity and its functional consequences for phenotype. Genome-wide detection of polymorphisms among many wild strains (accessions) of Arabidopsis also provides a useful resource for detecting evolutionary similarities or differences that regulate transcriptional networks (Borevitz et al. 2007, Clark et al. 2007).
Epigenetic modification
It is now well documented that gene expression can be regulated in a nonheritable fashion, through epigenetic modifications. Methylation of cytosine has been mapped throughout the Arabidopsis genome, and most genes that are methylated within their transcribed regions are highly expressed and constitutively active (X. Zhang et al. 2006, Zilberman et al. 2007). Histone methylation, particularly trimethylation of lysine 27 of histone H3 (H3K27me3), plays an important role in regulating animal development. Tiling arrays used to map H3K27me3 methylation sites in the Arabidopsis genome revealed a surprising number of genes that were silenced in a manner largely independent of other epigenetic pathways (Zhang et al. 2007). The evidence suggests that a complex network of epigenetic mechanisms acts to regulate gene expression in Arabidopsis and, presumably, in other plant species.
Posttranscriptional processing
Small RNAs, particularly microRNAs (miRNAs), are another component being investigated to understand the posttranscriptional control of gene regulatory networks (Schwab et al. 2005, Willmann & Poethig 2007, B. Zhang et al. 2006). Plant miRNAs are a large class of 20 to 24 base-pair noncoding RNAs. They bind to complementary sequences on target RNA and cause posttranscriptional gene silencing (PTGS) by cleaving or inhibiting translation of target mRNAs. miRNAs are a significant component of gene regulatory networks because they often modify the expression of transcription factors (Bartel 2004, Schwab et al. 2005). To date, more than 870 miRNAs have been identified in more than 70 plant species through genetic screens, cloning after isolation of small RNAs, and computational analyses (B. Zhang et al. 2006). Researchers are turning to several miRNA databases to identify and classify miRNAs. miRBase is a comprehensive repository for miRNAs and their predicted targets from 58 species (Griffiths-Jones et al. 2006). TarBase (Sethupathy et al. 2006) (http://www.diana.pcbi.upenn.edu/tarbase.html) is a collection of experimentally verified miRNA targets from human, mouse, fruit fly, worm, zebrafish, and plants. This database indicates whether an entry is a miRNA target protein–coding sequence or if it has tested negative for and is therefore unlikely to be a miRNA target. Related plant-specific databases include the Arabidopsis Small RNA Project (ASRP) (http://asrp.cgrb.oregonstate.edu/), which maintains a comprehensive database that seeks to functionally characterize all endogenous small RNAs, including miRNAs and siRNAs, in plants. Likewise, using high-throughput pyrosequencing, curators of the Cereal Small RNA Database (CSRDB) present a large data set of rice and maize small RNAs, along with their genomic location and predicted targets (Johnson et al. 2007).
Protein-DNA interactions
Proteins frequently interact with a large number of other gene products, and owing to the organization of the resulting complexes, genetic alterations in single genetic loci often do not yield phenotypes. Therefore, it is essential to understand how proteins interact to build gene regulatory networks. In a transcriptional regulatory network, proteins interact with cis-regulatory elements in target genes to control the expression of downstream targets. In plants, yeast-one-hybrid assays have identified interactions of several basic leucine zipper (bZIP) transcription factors with abscisic acid (ABA)-responsive elements (Kim et al. 1997, Uno et al. 2000). A Gateway™-compatible yeast-one-hybrid system that utilizes entire promoters has been developed in Caenorhabditis elegans (Deplancke et al. 2004), and this method should greatly facilitate the identification of protein-DNA interactions in plants.
Chromatin immunoprecipitation (ChIP) coupled with quantitative real-time PCR is another technique that plant researchers are increasingly employing. This technique allows one to isolate proteins bound to target chromatin (Orlando 2000). ChIP has been used to validate putative downstream targets of DELLA proteins, transcriptional regulators that control many aspects of signaling by the plant hormone gibberellin (Zentella et al. 2007). The technique has also been used to validate a physical interaction between TRICHOMELESS1, a MYB protein, and cis elements in the promoter of another MYB protein, GLABRA1 (GL1), which controls tri-chome formation (Wang et al. 2007), and to validate downstream targets of SHORTROOT, a protein essential for proper root development (Cui et al. 2007, Levesque et al. 2006). In a more systematic approach, researchers are also using ChIP in conjunction with high-density microarray platforms (ChIP-on-chip, or ChIP-chip, analyses). This allows global characterization of DNA associated with a target protein. For example, Thibaud-Nissen et al. (2006) used a whole-genome array as well as a high-density array with 2-kb promoter sequences for 27,166 putative Arabidopsis genes to determine the binding site for TGA2, a member of the TGA subclass of bZIP transcription factors, in salicylic acid–treated plants (Thibaud-Nissen et al. 2006). Characterization of LONG HYPOCOTYL5 (HY5) interactions also used this strategy, revealing more than 3800 putative targets (Lee et al. 2007).
Protein-protein interactions
In addition to binding to chromatin, proteins involved in gene regulatory networks form complexes with other proteins to regulate target gene expression. Various proteomics options are available for the global study of plant protein content, form, and activity (Thelen & Peck 2007). However, the most commonly used methods for protein interaction are yeast-two-hybrid assays and affinity purification of tagged protein complexes followed by mass spectrometry (AP-MS). Tandem affinity purification (TAP) of tagged proteins coupled to mass spectrometry has been used to study the cell cycle interactome in Arabidopsis cell culture and protein kinase interactions in rice (Rohila et al. 2006, Van Leene et al. 2007). Protein microarrays can also identify protein-protein interactions in a high-throughput manner. Popescu et al. (2007) used a high-density protein array to detect interactions between Arabidopsis calmodulins and other plant proteins (Popescu et al. 2007). In addition, new techniques such as fluorescence resonance energy transfer (FRET) and bioluminescence resonance energy transfer (BRET) are being used in plants to visualize protein-protein interactions in vivo (Xu et al. 2007, Zelazny et al. 2007).
Metabolite profiling
Obtaining data from the transcriptome, its epigenetic modifications, and the proteome provides the pieces that can then be assembled into a model of transcriptional regulatory networks. However, researchers are assembling one other large data source that, in part, can represent the terminal output of transcriptional regulation. The identification of small-molecule metabolites can provide a direct link to a cell’s physiology and, in some cases, signaling networks. In plants, a vast variety of metabolites are produced; their biosynthesis is the result of complex metabolic pathways. These metabolites are important in the differentiation of tissues and organs and in the responses of plants to the environment. In some cases, metabolites act in signaling cascades that influence transcriptional networks. Mass spectrometry–based and nuclear magnetic resonance–based methods have been applied to profile known metabolites within plants (Last et al. 2007, Ward et al. 2007). Moving from these metabolic fingerprints to high spatial and temporal resolution measurements of the dynamics of these metabolic pathways is a crucial next step and requires the measurement of metabolic flux, mathematical modeling, and the generation of dynamic biosensors (Lalonde et al. 2005, Morgenthal et al. 2006, Ratcliffe & Shachar-Hill 2006, Rios-Estepa & Lange 2007).
MODELING APPROACHES USED TO DESCRIBE GENE REGULATORY NETWORKS
To model regulatory networks, methods that describe nodes (genes) and their regulatory interactions (edges) are needed. Ideally, an edge will have support from multiple data sources. A number of methods commonly used to model transcriptional regulatory networks have been previously described (Brady & Benfey 2006) and are only briefly mentioned here. Many geneticists are familiar with one type of network model, diagram models, in which genetic interactions are indicated with a line between two nodes and the direction of the interaction is indicated by an arrowhead or a perpendicular line. These simple diagrams are further refined in circuit or wiring diagrams; a particularly elegant example is the sea urchin endomesoderm developmental regulatory network (Davidson et al. 2002). Relevance networks can use microarray expression data to infer gene interactions and to predict positive or negative interactions, using correlation and partial correlation methods (Ma et al. 2007). Boolean network models, one popular deterministic modeling approach, employ binary values (on/off states) and Boolean rules (e.g., AND, OR, NOR, etc.) to determine a target’s Boolean state (Price & Shmulevich 2007). The dynamics of the system can then be modeled by updating the Boolean functions either synchronously or asynchronously. In these cases a state is a binary representation of all variables’ activity within the system at a particular time point. The system can then transition from state to state over time (Price & Shmulevich 2007). However, gene expression is often stochastic, and measurements of gene expression on a global scale often contain noise. This uncertainty can be further incorporated into probabilistic Boolean networks, which have been used in a number of systems (Price & Shmulevich 2007).
Bayesian networks offer another opportunity to incorporate statistical uncertainty. These networks provide a directed, acyclic, graphical representation of the joint probability distribution between random variables (Pe’er 2005). Dynamic Bayesian networks (DBNs) can be used to model networks that include the dimension of time. Standard Bayesian networks infer directed relationships and therefore cannot incorporate feedback control mechanisms (Zou & Conzen 2005). Kinetic models are particularly useful in the modeling of dynamic regulatory networks. In these models, the kinetics of a series of transcriptional regulatory mechanisms are described by the use of differential equations (Locke et al. 2005a). However, this requires knowledge of the rates of regulatory interactions, as well as parameters describing the activity or association and disassociation constants of transcription factors with their partners.
PLANT TRANSCRIPTIONAL REGULATORY NETWORKS: RECENT ADVANCES
Modeling Regulatory Networks: Theoretical Approaches
Theoretical approaches, often mathematical in nature, have long been used as a framework to conceptually understand regulatory interactions that give rise to organismal form and function. At the organismal level, Turing mathematically modeled the set of possible phyllotactic arrangements in vegetative plant growth, given available mechanical restraints (Saunders 1992). In vertebrates, Cooke and Zeeman presented the clock-wavefront hypothesis to explain the regular production of somites. Indeed, when the genes responsible for somitogenesis were elucidated, and their interaction characterized, they appeared to interact according to Cooke and Zeeman’s hypothesis (Dequeant et al. 2006).
Recent studies have used these conceptual frameworks to try to understand the regulatory events underlying plant architecture. Mundermann et al. (2005) developed a model to capture quantitative aspects of Arabidopsis development. This model was intended to serve as a framework to identify the key attributes in plant form needed to specify observed growth and as a path for incremental development of mechanistic models of the whole plant (Mundermann et al. 2005). This study utilized imaging to capture data, including the size, shape, and growth rates in shoot organs from the seedling to the maturation stage of development. The input components included four types of modules—apices, internodes, leaves, and flowers. Each flower was further decomposed into modules representing individual floral organs and supporting tissue—the pedicel, carpel, sepals, petals, stamens, and carpel. Components were assembled into a three-dimensional structure, which, when simulated, described a growing plant and remarkably recapitulated Arabidopsis growth (Mundermann et al. 2005). An example of an emergent property revealed by modeling plant growth was the inverse correlation between the divergence angles of leaves and of the lateral inflorescences subtended by them. This inverse correlation suggested that the timing and positioning of primordia are interdependent, such that primordia that are initiated close together in time are positioned far apart in space.
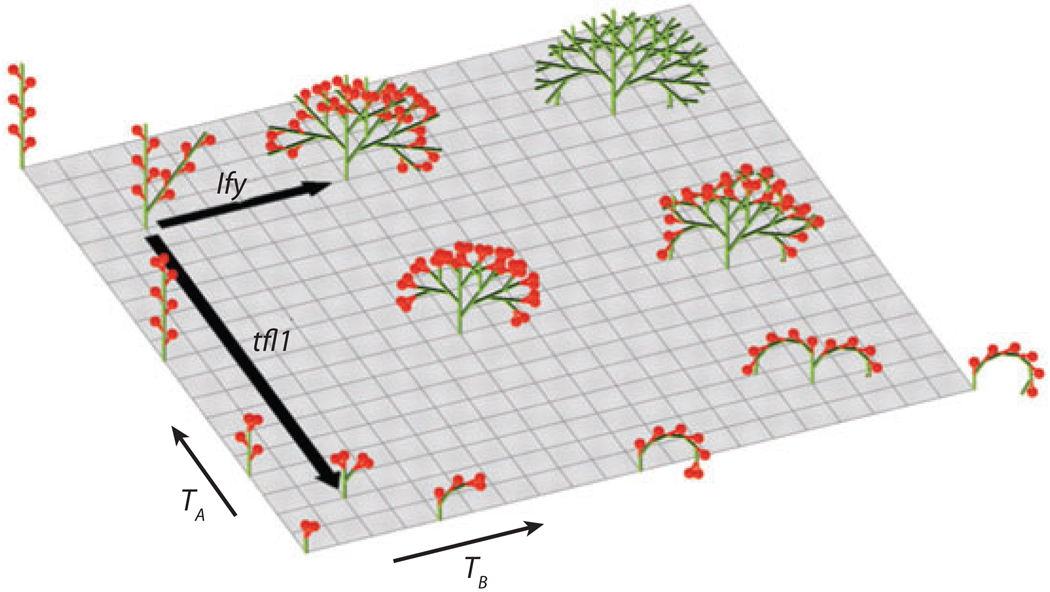
In a similar manner, a conceptual framework presented to understand inflorescence architecture incorporated previously characterized genes into its regulatory framework. Although a variety of inflorescence architectures are theoretically possible, only three general inflorescence classes are observed (Prusinkiewicz & Lindenmayer 1990, Prusinkiewicz et al. 2007, Weberling 1992). Prusinkiewicz et al. (2007) proposed a single developmental model that incorporates theoretical and molecular genetic studies and that accounts for the restricted range of architectures observed. In nature, the three inflorescence architecture types are (a) panicles, in which a series of branching axes ends in terminal flowers; (b) racemes, in which axes bear flowers in lateral positions or in a lateral axis that reiterates the pattern; and (c) cymes, with an axis bearing a terminal flower in a lateral position, and a lateral axis that reiterates this pattern (Figure 1). Prusinkiewicz and others proposed that a meristem can give rise to shoots or flowers and that this decision is dependent on the vegetativeness (veg) variable (Prusinkiewicz et al. 2007). High levels of veg will produce shoot meristem identity, whereas low levels will produce flower meristem identity. A meristem can be in one of two internal states: A, which is characterized by a high level of veg at time TA, or a transient state, B, which attains low levels of veg at time TB. Every lateral meristem initially formed is in state B, during which the lateral meristem can proceed to one of two possible fates, each dependent on veg levels. If veg is low, the meristem will become a flower, whereas if veg is high, the meristem enters state A and produces a branch. This model can generate cymes, racemes, and panicles by changing the TA and TB parameters (TA < TB, TA > TB, and TA = TB respectively) and have been represented in a single-parameter morphospace, that is, all possible morphologies given TA and TB parameters (Prusinkiewicz et al. 2007).
Figure 1.
Morphospace for the transient model. Different phenotypes are generated by varying the times TA and TB at which flowers begin to form. Values along each axis range from 0 to 10 plastochrons. Black arrows, pointing away from the wild-type architecture of Arabidopsis, indicate the effect of terminal flower 1 (tfl1) and leafy (lfy) mutations (under inductive conditions). Inflorescences are shown at six plastochrons; a plastochron is a time interval during which a meristem may either switch to floral identity or continue to produce further meristems. From Prusinkiewicz et al. (2007). Reprinted with permission from AAAS.
TERMINAL FLOWER1 (TFL1) and LEAFY (LFY) are two genes involved in regulating inflorescence architecture and proposed to increase and decrease veg levels, respectively. Wild-type inflorescence architecture results from high TFL1, low LFY, and therefore high veg in state A meristems, whereas high LFY and low TFL1 activity results in low veg and flowers in state B meristems. These different architectures were correlated with their fitness and modeled in two-dimensional fitness landscapes (Prusinkiewicz et al. 2007). The ability of these architectures to generate adaptive solutions in response to variable season length and environmental conditions was further explored in an additional dimension within these fitness landscapes. The path to move between racemes and cymes within this morphospace is quite large, and this model therefore suggested that this move would require multiple genetic changes, which may represent why racemes and cymes are rarely found within the same genera (Prusinkiewicz et al. 2007). This theoretical framework can be used to provide a mechanism to explain how the evolution of inflorescence architecture can be constrained by the nature of the developmental genetic mechanisms involved and the interaction between organism and environment. Both of these examples of theoretical modeling of networks can serve as frameworks upon which gene interaction data can be superimposed and that can be used further to analyze properties of gene regulatory networks.
Integrating Data Sources to Elucidate Networks
A brief search of the literature will reveal an astounding amount of information about various components of plant regulatory networks. However, only when these components have been integrated in a cohesive manner can one begin to generate models that predict how a network functions. In a recent study, researchers used a systems approach to identify protein kinases that integrate transcriptional regulation with stress and signaling (Baena-Gonzalez et al. 2007). Baena-Gonzalez et al. (2007) initially found a set of dark-inducible genes that were repressed by protein kinase inhibitors. On the basis of these data, these investigators screened for protein kinases that may play a role in stress response and found two, KIN10 and KIN11. Characterization of the promoter of one of the dark-inducible genes, DIN6, revealed a G-box regulatory element in its promoter. Surprisingly, KIN10 and KIN11 act synergistically with C-group bZIP transcription factors to bind the G-box in these promoters and to transcriptionally activate genes in major catabolic pathways (Baena-Gonzalez et al. 2007). Although future efforts include elucidating the relationship between these protein kinases and transcriptional regulators, studies such as these are a first step toward more comprehensive efforts to integrate two types of components within a system.
In addition to integrating signaling and regulatory networks, researchers have also begun to combine their understanding of miRNAs with that of signaling networks. Zhou et al. (2007) developed a novel computational approach in which miRNA targets were predicted on the basis of upregulation of miRNA expression due to UV-B exposure, the presence of common cis-regulatory elements, and whether a miRNA target’s expression was anticorrelated with expression of the target’s miRNA (Zhou et al. 2007). On the basis of this analysis, Zhou et al. were able to identify 21 putative UV-B-responsive miRNAs as well as their targets, of which the majority were transcription factors involved in auxin signaling. This further validated the regulatory role of miRNAs in a host of important biological processes.
Several recent studies have also combined transcriptomics and metabalomics. Hirai et al. (2007) compared condition-independent expression profiles from 1388 microarray experiments with those generated under sulfur-deficiency conditions. Using Pearson correlation, these researchers were able to detect coexpressed gene clusters, one of which contained two uncharacterized MYB transcription factors, MYB28 and MYB29. Further analysis of these genes, using loss-of-function and gain-of-function mutants, revealed that MYB28 activates expression of the aliphatic glucosinolate biosynthetic gene, but not indole or aromatic glucosinolate biosynthetic genes. Hirai et al. (2007) hypothesized that MYB29 functions in the induction of aliphatic glucosinolate biosynthetic genes only in response to methyl jasmonate signaling.
Other labs have integrated numerous systems components to develop and test regulatory networks under other stress conditions. For example, Coruzzi’s lab developed VirtualPlant, a multinetwork tool that integrates known and predicted information about Arabidopsis metabolic pathways as well as protein-protein, protein-DNA, and miRNA-target interactions (Gutierrez et al. 2007). After transcriptionally profiling Arabidopsis roots, using a systematic experimental space of carbon/nitrogen treatments, Coruzzi and coworkers applied these data to VirtualPlant (http://virtualplant.bio.nyu.edu/cgi-bin/vpweb/virtualplant.cgi). These investigators developed a qualitative multinetwork model for the carbon and nitrogen response, with 7635 nodes and more than 200,000 edges indicating putative regulatory interactions. In a subsequent study, Coruzzi and coworkers experimentally validated a subnetwork from this study, the proposed interaction between an miRNA, miR167, and its target, ARF8. Gifford et al. (2008) found that ARF8 is up-regulated in pericycle cells that are precursors to lateral roots and that miR167 is downregulated in these same cells under nitrogen stress conditions. These data, along with the finding that overexpression of miR176 results in complete loss of nitrogen-inducible lateral root emergence, led to the conclusion that miR167 targets ARF8 to control lateral root production (Gifford et al. 2008).
Emergent Properties: Utilization of High-Throughput Data
Above we describe components of plant regulatory networks and approaches for modeling these networks and show how researchers are beginning to integrate information about different systems components. As mentioned above, the goal of systems biology is the identification of emergent properties that arise from the interactions of different components within a system. Therefore, a comprehensive view of how each component interacts within a system is required. To that end we now describe recent efforts to generate gene regulatory network models from high-throughput data and emergent properties revealed by analysis of these networks.
In a comprehensive study, Cooper et al. (2003) integrated data from yeast-two-hybrid analysis, transcriptional profiling of different plant tissues under 11 different stress conditions, quantitative trait loci (QTL) analysis, and analysis of T-DNA insertion mutants to develop a comprehensive gene network of rice genes that play a role in stress responses. Through integration of these data sets, these researchers developed an interaction map that included protein-phosphatase interactions, transcription factor–target interactions, and interactions between signaling molecules such as 14-3-3 proteins and their targets. The modeling of this network allowed function to be ascribed to more than 200 genes, and five uncharacterized proteins were determined to play a role in disease resistance (Cooper et al. 2003).
Analyzing the NPR1-mediated gene regulatory network has revealed a complex set of regulatory interactions that influence systemic acquired resistance (SAR). Conditional induction of NPR1, a transcription cofactor that helps regulate SAR, in conjunction with microarray analysis, identified direct NPR1 targets (Wang et al. 2006). Of 64 putative targets, 8 were WRKY transcription factors. Pathogenicity analysis compared with microarray analyses of wild-type and npr1, single wrky, and double wrky mutants before and after SAR induction indicated that one of the factors, WRKY18, is a positive regulator of SAR, whereas another factor, WRKY58, is a negative regulator of spurious activation of SAR. Furthermore, a third factor, WRKY70, along with its functional homolog, appeared to negatively regulate SAR while positively mediating salicylic acid–mediated signaling (Wang et al. 2006). The emergent properties revealed by this study not only have enabled researchers to begin to understand the complex regulation of SAR but also can be used to identify further downstream SAR factors.
In addition to using high-throughput data to elucidate gene regulatory networks involved in stress response, investigators are also attempting to study plant developmental processes. A comprehensive plant protein-protein interaction map of more than 100 MADS box proteins in Arabidopsis revealed interactions between subgroups of MADS box proteins with similar biological functions. This study also revealed a surprising emergent property of floral development, namely, interactions between floral induction and floral organ identity proteins, suggesting feedback loops that regulate gene expression in these processes (de Folter et al. 2005).
High-resolution sampling and microarray expression analysis of developmental time points along the axis of the Arabidopsis root, in combination with the data from nearly all cell types in the Arabidopsis root, have yielded a large amount of spatiotemporal transcriptional information. Employing a computational pipeline that exploits methods used to identify strongly coexpressed groups of genes, Brady et al. (2007) obtained a set of distinct, dominant, transcriptional programs in both space and time. An example of an emergent property revealed by this systematic profiling of root development is a set of surprising expression patterns that underlie development. Nearly 50% of expression patterns fluctuate in developmental time, suggesting the need to separate biological processes along this temporal axis. Brady et al. (2007) used these high-resolution coexpression groups to infer a diverse set of transcriptional regulatory modules, one of which had direct support in experimental data. This suggests that, at least for the Arabidopsis root, we now have sufficient information to begin to elucidate transcriptional regulatory networks. These modules can be tested for their experimental validity and can then be used in modeling studies to understand higher-order properties of transcriptional regulatory networks.
Emergent Properties: Modeling Small Gene Regulatory Networks
A number of small gene regulatory networks, whose interactions have not necessarily been inferred by the use of high-throughput, genome-scale data, have greatly benefited from a systems biology framework. Modeling and testing of flower development, the circadian clock, and auxin flux regulatory networks have revealed a number of emergent network properties. These properties include novel regulatory component predictions and, strikingly, in all cases, the robustness of these networks in the face of stochastic biological fluctuations. These examples are intended to demonstrate the power of a systems approach as a framework to understand regulatory processes. As data obtained from genome-wide technologies are integrated, these model regulatory networks show how modeling can aid in elucidating the dynamics and properties of regulatory processes that give rise to plant form and function.
The logic of flower development
The Arabidopsis flower consists of four floral organs. From the outside of the flower to the inside, they include sepals, petals, stamens, and carpels, which are found in concentric whorls. In Arabidopsis thaliana and other plant species, the expression of overlapping transcription factors specifies floral organ development. The ABC model, which was one of the first plant gene regulatory networks modeled, described interactions among three classes of transcription factors that regulate floral pattern formation across plant species (Coen & Meyerowitz 1991). Additional studies identified genes that act prior to floral organ specification, in the vegetative-to-reproductive transition, as well as other modifiers of ABC transcription factor expression. These genes are involved in a variety of molecular processes from chromatin remodeling to signaling and protein turnover. Alvarez-Buylla and others have further modeled the genetic interactions controlling flower development, using Boolean logic (Espinosa-Soto et al. 2004, Mendoza & Alvarez-Buylla 1998, Mendoza et al. 1999). The main goal of their most recent model, which describes a small network of 15 genes, was to move beyond the ABC model to a dynamical explanation of how the steady-state pattern of gene expression characteristics in flowers is attained and maintained through the use of both ABC and non-ABC genes and logical rules in Arabidopsis and petunia (Espinosa-Soto et al. 2004). This analysis elucidated the robustness of the floral regulatory network and pointed to testable hypotheses concerning the existence of other regulators.
The directed interactions and logical rules were first assigned to 15 Arabidopsis nodes (genes) through the use of available experimental data. Eight nodes were assigned a binary activity state, that is, on or off, whereas seven nodes were assigned ternary activity states. In several cases, to maintain the coherence of the available experimental data, some interactions were inferred. One of the advantages of using a systems approach to understand network architecture or dynamics is that when a series of interactions is synthesized into a predictive model, gaps in available experimental data are revealed. One can use experimental data to predict the logical rules of these interactions, and these predictions can then be tested experimentally and incorporated into the model. Because the model describes the temporal dynamics of floral development, an inflorescence state exists, and one should make rules that describe the activation of each gene upon floral induction. As an example, the three SEPALLATA genes (SEP1–3) are expressed after floral induction, but no known activator exists in the inflorescence state. In this model, TFL1 was inferred to repress SEP1–3 expression prior to floral induction. An additional gap in the current data included a factor responsible for maintaining expression of PISTILLATA (PI) in the inner three whorls of the flower.
Transcriptional regulatory networks are generally recognized for their properties of robustness or their ability to function nearly independently of biochemical parameters that tend to fluctuate from cell to cell (Alon 2007). Robustness of this floral network was tested by simulating alterations in outputs of the logical rule tables for randomly selected nodes. Only a very small number of alterations gave rise to a steady-state condition that was not observed in the wild type, demonstrating the robustness of the floral development network.
The evolution of the circadian clock model: from single to multiple loops
The circadian clock is an internal timing mechanism that allows the temporal synchronization of physiology with environment to ensure successful growth and development (Más 2005). Circadian timing mechanisms entrain to rhythmic cues of light and dark, and these rhythms are generated by gene expression acting in positive and negative feedback loops. Although a single feedback loop can generate an oscillatory expression pattern, mathematical modeling of the Arabidopsis circadian clock, using differential equations, has revealed that this clock is generated through multiple interconnecting loops (Locke et al. 2005a,b, 2006; Zeilinger et al. 2006). Modeling is usually an iterative process, and through both simulation and experimental validation of these models, novel components have been predicted, tested, and continuously incorporated, illuminating a complex set of interactions acting to generate this robust, internal timing mechanism.
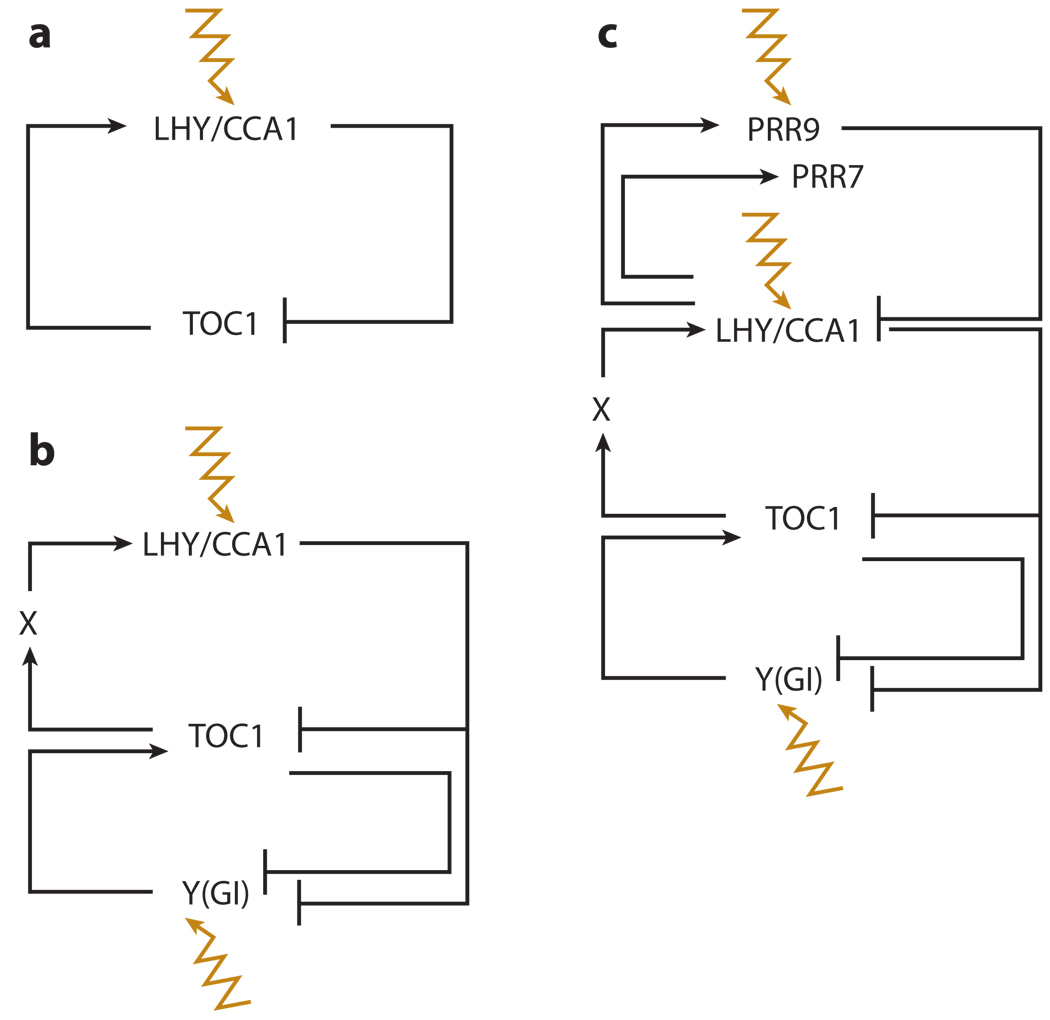
A first-generation model in which the partially redundant LATE ELONGATED HYPOCOTYL (LHY) and CIRCADIAN CLOCK ASSOCIATED 1 (CCA 1) genes repress expression of their activator, TIMING OF CAB EXPRESSION1 (TOC1), in a single feedback loop was modeled through the use of seven coupled differential equations, Michaelis-Menten kinetics for protein degradation, Hill functions to describe transcriptional activation or repression, and gene product concentrations including mRNA, cytoplasmic, and nuclear protein species (Figure 2a) (Locke et al. 2005a). In cases in which experimental data were not available, an entire set of parameter values was analyzed and an optimal set selected that best accounted for clock characteristics (Locke et al. 2005a). Although this model was able to simulate robust oscillations over a 24-h period, it was not able to validate all available experimental data. The primary failures were in its inability to recapitulate experimental observations of an abrupt reduction in TOC1 transcription after dusk during long-day conditions, a discrepancy in the accumulation of TOC1 protein at dawn when it should be activating LHY transcription maximally, and the inability of TOC1 to respond to photoperiod.
Figure 2.
The Arabidopsis circadian clock has been elucidated by the use of systems approaches. In particular, modeling and iterative inclusion of novel parameters have revealed that the circadian clock is composed of three interconnecting loops. (a) The first-generation Arabidopsis circadian clock model. Light-activated (orange line) LHY/CCA1 (LATE ELONGATED HYPOCOTYL and CIRCADIAN CLOCK ASSOCIATED 1, respectively) repress the expression of their activator, TIMING OF CAB EXPRESSION1 (TOC1), in a single feedback loop. (b) The second-generation circadian clock model. To resolve experimental observations, two hypothetical components X and Y were included. TOC1 protein activates transcription of X. X protein then activates LHY/CCA1 transcription, and LHY/CCA1 protein represses TOC1 transcription. In a second feedback loop, TOC1 protein represses Y transcription, and Y protein activates TOC1 transcription. A second light input was added so that in addition to the acute light responsiveness of LHY/CCA1 transcription, light is also able to activate Y transcription. Y is proposed to encode the GIGANTEA (GI) protein. (c) In the third-generation model, new network components, PSEUDO-RESPONSE REGULATOR 9 (PRR9) and PSEUDO-RESPONSE REGULATOR 7 (PRR7), were incorporated, resulting in a clock that can also sense the photoperiod. (Figure derived from Locke et al. 2005a,b, 2006; Zeilinger et al. 2006.)
Because the observed experimental values did not match with those predicted by the model, the model was refined and novel components incorporated within this same mathematical framework (Locke et al. 2005b). Two hypothetical components, X and Y, were predicted also to act within this oscillator and to comprise interlocking feedback loops (Figure 2b). TOC1 protein would activate transcription of X, X protein would activate LHY transcription, and then LHY protein would repress TOC1 transcription in a central loop. A second feedback loop would also exist: TOC1 protein would repress Y transcription, and Y protein would activate TOC1 transcription. An additional light input was added, so that in addition to an acute light responsiveness of LHY transcription, light would also be able to activate Y transcription. This model was able to account for a larger range of experimental data, primarily the appropriate timing of TOC1 transcription and protein expression, than could the previous model. Predicted expression values of Y were used to query genes that affect the circadian clock for similar expression values in the correct time period of the clock. GIGANTEA (GI) expression matched these predictions quite well, with a significant but transient light response in wild type and a strong light response in the cca1;lhy mutant. As postulated in a systems biology framework, modeling should inform further experiments. For example, if GI is the valid Y component within this system, and all components are accounted for, then a gi;cca1;lhy triple mutant should disrupt both oscillating loops.
On the basis of these predictions and further experiments that identified additional feedback regulators of LHY/CCA1 expression, PSEUDO-RESPONSE REGULATOR7 (PRR7) and PSEUDO-RESPONSE REGULA-TOR9 (PRR9), new network components were incorporated within the circadian clock model, resulting in a clock that also contains a photoperiod sensing mechanism (Figure 2c) (Locke et al. 2006, Zeilinger et al. 2006). Two slightly different models were proposed, with both models composed of three loops—a core LHY/CCA1-TOC1-X loop that is coupled with an evening-expressed TOC1-Y(GI) loop, and a novel morning-expressed PRR7/9-LHY/CCA1 loop. Of particular interest, Locke et al. (2006) biologically tested the identity of GI as the predicted Y component, using a gi;cca1;lhy triple mutant. These experimental observations performed very well compared with the model simulations, suggesting that GI is a good candidate for Y.
Locke et al. (2006) considered PRR7 and PRR9 as a single component; in contrast, on the basis of the partial redundancy of PRR7 and PRR9 in regulating LHY/CCA1, Zeilinger et al. (2006) incorporated PRR7 and PRR9 as separate components on the basis of their subtle transcriptional differences in response to light. This two-parameter inclusion revealed an emergent property of this network—that is, separate and distinct mechanisms of clock regulation. The magnitude of the PRR9 response to light is much higher than that of PRR7 and was accounted for by incorporating an acute light induction of PRR9 by a component P that is both active and unstable in light. Zeilinger et al. (2006) found that the acute peak of Y expression controls TOC1 expression, whereas the circadian peak modulates the fine-tuning response to day length, suggesting two distinct mechanisms of clock regulation. Zeilinger et al. (2006) also used sensitivity analysis to capture areas of network robustness. Many parameters indicated low sensitivity, a reflection of the robust nature of this oscillatory network. However, parameters with high sensitivity were identified; these most likely indicate the need for further experimentation. As an example, CCA1/LHY parameters showed low sensitivity relative to TOC1-Y. However, experimentally, cca1;lhy mutants also show an extreme phenotype, and therefore further experimentation regarding LHY/CCA1 regulation is needed.
Modeling auxin flux and regulatory networks in plant development
The plant hormone auxin is a potent regulatory molecule that acts in many aspects of plant growth and development. Auxin has been implicated as a morphogen, whereby its graded accumulation in cell types and tissues acts instructively to determine cell fate specification, division, and elongation. A systems approach has been used to model the dynamics of auxin flux in both the shoot and the root. Although the models we describe do not deal with gene regulatory networks directly, these modeling efforts describe how the flux of this hormone is established and maintained to generate complex patterns. Auxin gradients are dynamically and robustly controlled by the subcellular localization of auxin transporters, allowing auxin to act as a capacitor in morphogenesis. Dynamically controlled auxin flux is translated into gene regulatory networks, and therefore understanding the properties of auxin flux within the plant system is essential to comprehending auxin-mediated gene regulatory networks.
Modeling auxin flux in the shoot
Computer simulations of PIN-FORMED1 (PIN1), an auxin efflux facilitator, in the shoot apex were able to simulate successfully both a spiral and decussate phyllotaxis (de Reuille et al. 2005). These phyllotactic arrangements are dependent on the size of the central zone in the meristem. Surprisingly, however, these simulations also revealed an emergent property of this network—an auxin maximum that exists at the summit of the meristem (de Reuille et al. 2005). This was biologically tested, and indeed a free auxin maximum was found to exist in the center of the shoot apical meristem (SAM) (de Reuille et al. 2005). The simulations further predicted that this central auxin maximum is essential for the initiation of organ primordia but is not necessary for primordia maintenance, which remains to be experimentally tested. If biologically valid, this prediction suggests that the mechanisms by which primordia are maintained are remarkably robust.
The lab of Elliot Meyerowitz has established technology to collect time-lapse, single-cell, live images of key regulators of SAM formation (Reddy et al. 2004). This technology has revolutionized the ability to collect data on the dynamics of auxin flux and its role in establishing phyllotaxis and primordium development and has been used both to validate experimentally existing models and to infer novel models. One example of experimental validation involves models that describe floral primordium development. Live imaging of a developing inflorescence primordium confirmed model predictions that auxin accumulation and depletion are mediated by reversal of PIN1 localization in incipient primordia and that this reversal guides proper floral primordium development (Heisler et al. 2005). Genes known to be involved in floral primordium formation were analyzed to assess the temporal nature of this regulatory network in reference to this auxin flux. This regulatory network has also been analyzed by live-cell imaging during somatic embryogenesis (Gordon et al. 2007). Dynamic PIN1 concentrations were quantified and used to formulate a model based on the assumption that PIN1 polarity is regulated by relative auxin concentrations in neighboring cells within a two-dimensional space like the epidermis. This model also incorporated information regarding cellular growth and mechanics within the shoot apex (Jonsson et al. 2006). Simulations correctly recapitulated observations on polarized auxin flow and phyllotactic patterning. Future directions entail assessing the robustness of this network regulating phyllotaxis in response to mechanical disturbance or stress.
Modeling auxin flux in the root
The Arabidopsis root is composed of different cell types arranged in concentric cylinders around the root’s radial axis. Along the root’s longitudinal axis, moving upward from the tip, all cell types are further organized in zones characterized by rapid cell division (meristematic), elongation, and, finally, maturation. An auxin response maximum is required for cell-type specification at the root tip. However, auxin is also required for cell division and expansion in the meristematic and elongation zones. The molecular mechanism by which auxin acts to regulate pattern formation, division, and elongation was previously unclear. Through a systems biology approach, two models have been developed to describe auxin distribution within the root and its regulation of root development (Grieneisen et al. 2007, Likhoshvai et al. 2007). Both models take into account root length and different cell types within the root, incorporate auxin derived from the shoot, and exclude auxin synthesis within the root. The model proposed by Grieneisen et al. (2007) additionally incorporated the cell-type and subcellular location and activity of auxin transporters, in addition to the shape and local neighborhood of different cell types. This modeling and experimental testing have revealed a minimum regulatory mechanism by which auxin can specify and maintain root patterning (Grieneisen et al. 2007, Likhoshvai et al. 2007).
In the Grieneisen et al. (2007) model, cells were characterized by appropriately scaled sizes and shapes to understand the interaction of local neighborhoods in auxin flux, and four generalized tissue types were considered—epidermal tissue, vascular tissue, a border tissue between the epidermal tissue and vascular tissue, and distal cap tissue. Furthermore, auxin diffusion and facilitated transport were considered as independent parameters, and auxin synthesis was not considered implicitly. The goal of modeling this aspect of auxin regulation was to determine if a minimal amount of information could provide an accurate estimate of auxin flux dynamics in cellular growth, and to provide testable hypotheses regarding the influence of each component on growth and the robustness of flux within the root.
First, the critical features of transport in this system were examined in a static growth phase (Grieneisen et al. 2007). Auxin was simulated as coming from the uppermost vascular region, and a steady state was quickly achieved with a local maximum in cells above the cap region. This steady state is quite robust and is insensitive to ubiquitous auxin synthesis or to the position of the auxin source. The root maximum can also be maintained for a relatively long period of time after removal of the shoot. This prediction of auxin capacitance was tested experimentally and validated. The most important parameter in establishing this maximum magnitude is the density of lateral auxin efflux transporters (PINs) in the border and epidermal files. This lateral density is critical in establishing reflux directed through the vascular tissues, as experimentally tested with mutations in three PIN loci. The model also predicts that the key parameter for determining the position of the auxin maximum is the efflux carriers in the root cap. Interestingly, the model predicted that stabilization of the auxin maximum–associated gradient takes much longer than formation of the maximum itself. This maintenance state was termed a quasi-steady state: Auxin levels increase, but the auxin distribution profile maintains its overall shape.
The model then incorporated root growth and explored the relationship between auxin dynamics and the different growth zones within a root (Grieneisen et al. 2007). A graded response function that was equal for all cells and that specified growth, division, or elongation was implemented. The auxin maximum was maintained in growing roots, and emerging zones that displayed meristematic and elongation properties were rapidly realized. In addition, the model predicted increased mitotic activity in vascular regions relative to other cell types, which has also been observed experimentally. These zones become sharper over time owing to feedback between cell growth dynamics and the auxin gradient. The model simulated a cut within the root and predicted a shift in the boundary between the meristematic and elongation zones, with a much longer elongation zone. This was experimentally validated with a localized application of an auxin transport inhibitor. These rounds of modeling and experimentation have demonstrated a further emergent property of the auxin-mediated regulatory network: that the formation of a meristematic zone and an elongation zone is due to the self-organizing property of the auxin response threshold that governs a variety of growth processes.
CONCLUSIONS AND FUTURE DIRECTIONS
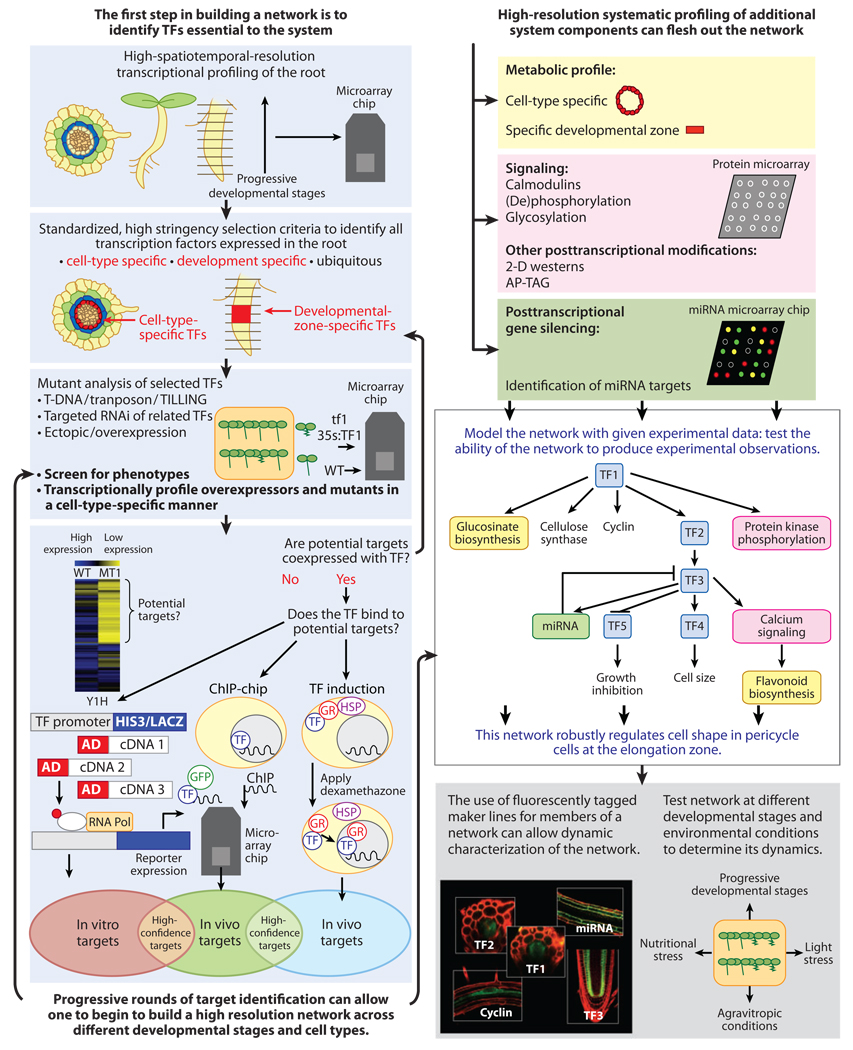
As we stand at the cusp of a major paradigm shift in how we approach science, some ask if we are really learning anything new. Initial indications are that moving from a reductionist to a systems approach can provide a clearer understanding of how biological systems are regulated, allowing one to predict their behavior. At the moment, plant biologists are collecting high-throughput data and identifying components acting in systems of interest. Researchers are using this information to model new gene regulatory networks and to expand upon older networks. However, when generating models that predict interactions between components, researchers are still testing models in a reductionist manner by perturbing a single component at a time in their respective network of interest. As we continue to use systems approaches, an open question is whether it is possible to test models in a nonreductionist manner. This will require the acquisition of dynamic, high-resolution, spatiotemporal expression and interaction data for all genes in a system. One area that seems tractable for this type of approach is transcriptional networks. Integration of expression data, interaction of transcription factors with DNA and with each other—in wild-type and mutant backgrounds, at the resolution of cell types and developmental stages, and in different environmental contexts—could generate high-confidence, dynamic gene regulatory models (Figure 3). However, this will require massive efforts from many labs. A potential problem is the temptation to focus on a few interactions for validation and further testing based on the current interests of a particular lab. However, the network would be expanded more rapidly if all targets were tested systematically. To address this issue, it may be possible to view systems biology endeavors as a group effort with two objectives: The first is to build a framework for understanding the inherent biology of plant systems of interest, and the second is to allow individual researchers to develop fine-tuned experiments that can elucidate missing aspects of a model.
Figure 3.
Systems approaches to studying root transcriptional regulatory networks regulating root development. Transcription factors (TFs) of interest are first identified by cell-type-specific and development-specific transcriptional profiling. Wild-type (WT) and mutants of cell-type-specific and development-specific TFs are analyzed and transcriptionally profiled to detect potential downstream targets of the initial TFs selected. One can validate physical interaction between the TFs and their potential targets first by determining that the TFs and their potential targets are coexpressed, using their transcriptional profile, then by in vitro tests such as yeast-one-hybrid (Y1H) assays, or in vivo by ChIP-chip or by conditional TF induction coupled with microarray analysis. High-confidence TF targets are validated by multiple methods of identification. Subsequently, those targets that are TFs themselves can be further analyzed to detect their targets. Subsequent rounds of TF/target identification can allow one to generate a model of a TF network. Other aspects of the network, such as the metabolic output, signaling, and posttranscriptional gene silencing, should also be characterized in a high-resolution, systematic manner. Combining all these results can allow researchers to generate a detailed, high-resolution, gene regulatory network model of root development. This model should then be tested experimentally to determine its ability to recapitulate experimental observations and for its robustness. The dynamics of these networks can then be monitored by measuring the expression of fluorescently tagged genes within the network and by testing the network under a range of conditions. Chip, microarray chip; ChIP, chromatin immunoprecipitation; GR, glucocorticoid receptor; HSP, heat shock protein.
ACKNOWLEDGMENTS
Work in P.N.B.’s lab on systems biology is funded by grants from the NIH, NSF, and DARPA. T.A.L. is funded by an NSF Minority Postdoctoral Research Fellowship. S.M.B. is funded by an NSERC postdoctoral fellowship. We thank Jalean Petricka, Rosangela Sozzani, Anjali Iyer Pascuzzi, and Jee Jung for critical reading of the manuscript.
Footnotes
DISCLOSURE STATEMENT
P.N.B. is a cofounder of a company, GrassRoots Biotechnology.
LITERATURE CITED
- The Arabidopsis Initiative. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408:796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- Alon U. An Introduction to Systems Biology: Design Principles of Biological Circuits. Boca Raton: Chapman and Hall/CRC; 2007. [Google Scholar]
- Alonso JM, Ecker JR. Moving forward in reverse: genetic technologies to enable genome-wide phenomic screens in Arabidopsis. Nat. Rev. Genet. 2006;7:524–536. doi: 10.1038/nrg1893. [DOI] [PubMed] [Google Scholar]
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, et al. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science. 2003;301:653–657. doi: 10.1126/science.1086391. [DOI] [PubMed] [Google Scholar]
- Baena-Gonzalez E, Rolland F, Thevelein JM, Sheen J. A central integrator of transcription networks in plant stress and energy signalling. Nature. 2007;448:938–942. doi: 10.1038/nature06069. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Bertalanffy LV. General Systems Theory. Harmondsworth: Penguin; 1973. [Google Scholar]
- Birnbaum K, Jung JW, Wang JY, Lambert GM, Hirst JA, et al. Cell type-specific expression profiling in plants via cell sorting of protoplasts from fluorescent reporter lines. Nat. Methods. 2005;2:615–619. doi: 10.1038/nmeth0805-615. [DOI] [PubMed] [Google Scholar]
- Birnbaum K, Shasha DE, Wang JY, Jung JW, Lambert GM, et al. A gene expression map of the Arabidopsis root. Science. 2003;302:1956–1960. doi: 10.1126/science.1090022. [DOI] [PubMed] [Google Scholar]
- Borevitz JO, Hazen SP, Michael TP, Morris GP, Baxter IR, et al. Genome-wide patterns of single-feature polymorphism in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA. 2007;104:12057–12062. doi: 10.1073/pnas.0705323104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady SM, Benfey PN. A systems approach to understanding root development. Can. J. Bot. 2006;84:695–701. [Google Scholar]
- Brady SM, Long TA, Benfey PN. Unraveling the dynamic transcriptome. Plant Cell. 2006;18:2101–2111. doi: 10.1105/tpc.105.037572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady SM, Orlando DA, Lee J-Y, Wang JY, Koch J, et al. A high-resolution root spatiotemporal map reveals dominant expression patterns. Science. 2007;318:801–806. doi: 10.1126/science.1146265. [DOI] [PubMed] [Google Scholar]
- Busch W, Lohmann JU. Profiling a plant: expression analysis in Arabidopsis. Curr. Opin. Plant Biol. 2007;10:136–141. doi: 10.1016/j.pbi.2007.01.002. [DOI] [PubMed] [Google Scholar]
- Chen S, Jin W, Wang M, Zhang F, Zhou J, et al. Distribution and characterization of over 1000 T-DNA tags in rice genome. Plant J. 2003;36:105–113. doi: 10.1046/j.1365-313x.2003.01860.x. [DOI] [PubMed] [Google Scholar]
- Clark RM, Schweikert G, Toomajian C, Ossowski S, Zeller G, et al. Common sequence polymorphisms shaping genetic diversity in Arabidopsis thaliana. Science. 2007;317:338–342. doi: 10.1126/science.1138632. [DOI] [PubMed] [Google Scholar]
- Coen ES, Meyerowitz EM. The war of the whorls: genetic interactions controlling flower development. Nature. 1991;353:31–37. doi: 10.1038/353031a0. [DOI] [PubMed] [Google Scholar]
- Cooper B, Clarke JD, Budworth P, Kreps J, Hutchison D, et al. A network of rice genes associated with stress response and seed development. Proc. Natl. Acad. Sci. USA. 2003;100:4945–4950. doi: 10.1073/pnas.0737574100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui H, Levesque MP, Vernoux T, Jung JW, Paquette AJ, et al. An evolutionarily conserved mechanism delimiting SHR movement defines a single layer of endodermis in plants. Science. 2007;316:421–425. doi: 10.1126/science.1139531. [DOI] [PubMed] [Google Scholar]
- Davidson EH, Rast JP, Oliveri P, Ransick A, Calestani C, et al. A genomic regulatory network for development. Science. 2002;295:1669–1678. doi: 10.1126/science.1069883. [DOI] [PubMed] [Google Scholar]
- de Folter S, Immink RGH, Kieffer M, Parenicova L, Henz SR, et al. Comprehensive interaction map of the Arabidopsis MADS box transcription factors. Plant Cell. 2005;17:1424–1433. doi: 10.1105/tpc.105.031831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Reuille PB, Bohn-Courseau I, Ljung K, Morin H, Carraro N, et al. Computer simulations reveal properties of the cell-cell signaling network at the shoot apex in Arabidopsis. Proc. Natl. Acad. Sci. USA. 2005;103:1627–1632. doi: 10.1073/pnas.0510130103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dembinsky D, Woll K, Saleem M, Liu Y, Fu Y, et al. Transcriptomic and proteomic analyses of pericycle cells of the maize primary root. Plant Physiol. 2007;145:575–588. doi: 10.1104/pp.107.106203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deplancke B, Dupuy D, Vidal M, Walhout AJM. A gateway-compatible yeast one-hybrid system. Genome Res. 2004;14:2093–2101. doi: 10.1101/gr.2445504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dequeant M-L, Glynn E, Gaudenz K, Wahl M, Chen J, et al. A complex oscillating network of signaling genes underlies the mouse segmentation clock. Science. 2006;314:1595–1598. doi: 10.1126/science.1133141. [DOI] [PubMed] [Google Scholar]
- Espinosa-Soto C, Padilla-Longoria P, Alvarez-Buylla ER. A gene regulatory network model for cell-fate determination during Arabidopsis thaliana flower development that is robust and recovers experimental gene expression profiles. Plant Cell. 2004;16:2923–2939. doi: 10.1105/tpc.104.021725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gifford ML, Dean A, Gutierrez RA, Coruzzi GM, Birnbaum KD. Cell-specific nitrogen responses mediate developmental plasticity. Proc. Natl. Acad. Sci. USA. 2008;105:803–808. doi: 10.1073/pnas.0709559105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilchrist EJ, Haughn GW. TILLING without a plough: a new method with applications for reverse genetics. Curr. Opin. Plant Biol. 2005;8:211–215. doi: 10.1016/j.pbi.2005.01.004. [DOI] [PubMed] [Google Scholar]
- Gordon SP, Heisler MG, Reddy GV, Ohno C, Das P, Meyerowitz EM. Pattern formation during de novo assembly of the Arabidopsis shoot meristem. Development. 2007;134:3539–3548. doi: 10.1242/dev.010298. [DOI] [PubMed] [Google Scholar]
- Grieneisen VA, Xu J, Maree AFM, Hogeweg P, Scheres B. Auxin transport is sufficient to generate a maximum and gradient guiding root growth. Nature. 2007;449:1008–1013. doi: 10.1038/nature06215. [DOI] [PubMed] [Google Scholar]
- Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34:D140–D144. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groover A, Fontana JR, Dupper G, Ma C, Martienssen R, et al. Gene and enhancer trap tagging of vascular-expressed genes in poplar trees. Plant Physiol. 2004;134:1742–1751. doi: 10.1104/pp.103.034330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez R, Lejay L, Dean A, Chiaromonte F, Shasha D, Coruzzi G. Qualitative network models and genome-wide expression data define carbon/nitrogen-responsive molecular machines in Arabidopsis. Genome Biol. 2007;8:R7. doi: 10.1186/gb-2007-8-1-r7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez RA, Shasha DE, Coruzzi GM. Systems biology for the virtual plant. Plant Physiol. 2005;138:550–554. doi: 10.1104/pp.104.900150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harbison CT, Gordon DB, Lee TI, Rinaldi NJ, Macisaac KD, et al. Transcriptional regulatory code of a eukaryotic genome. Nature. 2004;431:99–104. doi: 10.1038/nature02800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heisler MG, Ohno C, Das P, Sieber P, Reddy GV, et al. Patterns of auxin transport and gene expression during primordium development revealed by live imaging of the Arabidopsis inflorescence meristem. Curr. Biol. 2005;15:1899–1911. doi: 10.1016/j.cub.2005.09.052. [DOI] [PubMed] [Google Scholar]
- Hilson P, Allemeersch J, Altmann T, Aubourg S, Avon A, et al. Versatile gene-specific sequence tags for Arabidopsis functional genomics: transcript profiling and reverse genetics applications. Genome Res. 2004;14:2176–2189. doi: 10.1101/gr.2544504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirai MY, Sugiyama K, Sawada Y, Tohge T, Obayashi T, et al. Omics-based identification of Arabidopsis Myb transcription factors regulating aliphatic glucosinolate biosynthesis. Proc. Natl. Acad. Sci. USA. 2007;104:6478–6483. doi: 10.1073/pnas.0611629104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsing Y-I, Chern C-G, Fan M-J, Lu P-C, Chen K-T, et al. A rice gene activation/knockout mutant resource for high throughput functional genomics. Plant Mol. Biol. 2007;63:351–364. doi: 10.1007/s11103-006-9093-z. [DOI] [PubMed] [Google Scholar]
- Ideker T, Galitski T, Hood L. A new approach to decoding life: systems biology. Annu. Rev. Genomics Hum. Genet. 2001;2:343–372. doi: 10.1146/annurev.genom.2.1.343. [DOI] [PubMed] [Google Scholar]
- Jeong D-H, An S, Park S, Kang H-G, Park G-G, et al. Generation of a flanking sequence-tag database for activation-tagging lines in japonica rice. Plant J. 2006;45:123–132. doi: 10.1111/j.1365-313X.2005.02610.x. [DOI] [PubMed] [Google Scholar]
- Johnson C, Bowman L, Adai AT, Vance V, Sundaresan V. CSRDB: a small RNA integrated database and browser resource for cereals. Nucleic Acids Res. 2007;35:D829–D833. doi: 10.1093/nar/gkl991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones-Rhoades MW, Bartel DP, Bartel B. MicroRNAs and their regulatory roles in plants. Annu. Rev. Plant Biol. 2006;57:19–53. doi: 10.1146/annurev.arplant.57.032905.105218. [DOI] [PubMed] [Google Scholar]
- Jonsson H, Heisler MG, Shapiro BE, Meyerowitz EM, Mjolsness E. An auxin-driven polarized transport model for phyllotaxis. Proc. Natl. Acad. Sci. USA. 2006;103:1633–1638. doi: 10.1073/pnas.0509839103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kheradpour P, Stark A, Roy S, Kellis M. Reliable prediction of regulator targets using 12 Drosophila genomes. Genome Res. 2007;17:1919–1931. doi: 10.1101/gr.7090407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilian J, Whitehead D, Horak J, Wanke D, Weinl S, et al. The AtGenExpress global stress expression data set: protocols, evaluation and model data analysis of UV-B light, drought and cold stress responses. Plant J. 2007;50:347–363. doi: 10.1111/j.1365-313X.2007.03052.x. [DOI] [PubMed] [Google Scholar]
- Kim SY, Chung H-J, Thomas TL. Isolation of a novel class of bZIP transcription factors that interact with ABA-responsive and embryo-specification elements in the Dc3 promoter using a modified yeast one-hybrid system. Plant J. 1997;11:1237–1251. doi: 10.1046/j.1365-313x.1997.11061237.x. [DOI] [PubMed] [Google Scholar]
- Lalonde S, Ehrhardt DW, Frommer WB. Shining light on signaling and metabolic networks by genetically encoded biosensors. Curr. Opin. Plant Biol. 2005;8:574–581. doi: 10.1016/j.pbi.2005.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Last RL, Jones AD, Shachar-Hill Y. Towards the plant metabolome and beyond. Nat. Rev. Mol. Cell Biol. 2007;8:167–174. doi: 10.1038/nrm2098. [DOI] [PubMed] [Google Scholar]
- Lee J, He K, Stolc V, Lee H, Figueroa P, et al. Analysis of transcription factor HY5 genomic binding sites revealed its hierarchical role in light regulation of development. Plant Cell. 2007;19:731–749. doi: 10.1105/tpc.106.047688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levesque MP, Vernoux T, Busch W, Cui H, Wang JY, et al. Whole-genome analysis of the SHORT-ROOT developmental pathway in Arabidopsis. PLoS Biol. 2006;4:e143. doi: 10.1371/journal.pbio.0040143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Likhoshvai VA, Omel’yanchuk NA, Mironova VV, Fadeev SI, Mojolsness ED, Kolchanov NA. Mathematical model of auxin distribution in the plant root. Russ. J. Dev. Biol. 2007;38:374–382. [PubMed] [Google Scholar]
- Lin X, Kaul S, Rounsley S, Shea TP, Benito M-I, et al. Sequence and analysis of chromosome 2 of the plant Arabidopsis thaliana. Nature. 1999;402:761–768. doi: 10.1038/45471. [DOI] [PubMed] [Google Scholar]
- Locke JCW, Kozma-Bognár L, Gould PD, Féher B, Kevei E, et al. Experimental validation of a predicted feedback loop in the multi-oscillator clock of Arabidopsis thaliana. Mol. Syst. Biol. 2006;2:59. doi: 10.1038/msb4100102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locke JCW, Millar AJ, Turner MS. Modelling genetic networks with noisy and varied experimental data: the circadian clock in Arabidopsis thaliana. J. Theor. Biol. 2005a;234:383–393. doi: 10.1016/j.jtbi.2004.11.038. [DOI] [PubMed] [Google Scholar]
- Locke JCW, Southern MM, Kozma-Bognár L, Hibberd V, Brown PE, et al. Extension of a genetic network model by iterative experimentation and mathematical analysis. Mol. Syst. Biol. 2005b;1:2005.0013. doi: 10.1038/msb4100018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma S, Gong Q, Bohnert HJ. An Arabidopsis gene network based on the graphical Gaussian model. Genome Res. 2007;17:1614–1625. doi: 10.1101/gr.6911207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Más P. Circadian clock signaling in Arabidopsis thaliana: from gene expression to physiology and development. Int. J. Dev. Biol. 2005;49:491–500. doi: 10.1387/ijdb.041968pm. [DOI] [PubMed] [Google Scholar]
- Mayer K, Schuller C, Wambutt R, Murphy G, Volckaert G, et al. Sequence and analysis of chromosome 4 of the plant Arabidopsis thaliana. Nature. 1999;402:769–777. doi: 10.1038/47134. [DOI] [PubMed] [Google Scholar]
- Mendoza L, Alvarez-Buylla ER. Dynamics of the genetic regulatory network for Arabidopsis thaliana flower morphogenesis. J. Theor. Biol. 1998;193:307–319. doi: 10.1006/jtbi.1998.0701. [DOI] [PubMed] [Google Scholar]
- Mendoza L, Thieffry D, Alvarez-Buylla ER. Genetic control of flower morphogenesis in Arabidopsis thaliana: a logical analysis. Bioinformatics. 1999;15:593–606. doi: 10.1093/bioinformatics/15.7.593. [DOI] [PubMed] [Google Scholar]
- Morgenthal K, Weckwerth W, Steuer R. Metabolomic networks in plants: transitions from pattern recognition to biological interpretation. Biosystems. 2006;83:108–117. doi: 10.1016/j.biosystems.2005.05.017. [DOI] [PubMed] [Google Scholar]
- Mundermann L, Erasmus Y, Lane B, Coen E, Prusinkiewicz P. Quantitative modeling of Arabidopsis development. Plant Physiol. 2005;139:960–968. doi: 10.1104/pp.105.060483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakazono M, Qiu F, Borsuk LA, Schnable PS. Laser-capture microdissection, a tool for the global analysis of gene expression in specific plant cell types: identification of genes expressed differentially in epidermal cells or vascular tissues of maize. Plant Cell. 2003;15:583–596. doi: 10.1105/tpc.008102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawy T, Lee J-Y, Colinas J, Wang JY, Thongrod SC, et al. Transcriptional profile of the Arabidopsis root quiescent center. Plant Cell. 2005;17:1908–1925. doi: 10.1105/tpc.105.031724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson T, Tausta SL, Gandotra N, Liu T. Laser microdissection of plant tissue: What you see is what you get. Annu. Rev. Plant Biol. 2006;57:181–201. doi: 10.1146/annurev.arplant.56.032604.144138. [DOI] [PubMed] [Google Scholar]
- Orlando V. Mapping chromosomal proteins in vivo by formaldehyde-crosslinked-chromatin immunoprecipitation. Trends Biochem. Sci. 2000;25:99–104. doi: 10.1016/s0968-0004(99)01535-2. [DOI] [PubMed] [Google Scholar]
- Pe’er D. Bayesian network analysis of signaling networks: a primer. Sci. STKE. 2005;281:L4. doi: 10.1126/stke.2812005pl4. [DOI] [PubMed] [Google Scholar]
- Pennisi E. Genome sequencing: the greening of plant genomics. Science. 2007;317:317. doi: 10.1126/science.317.5836.317. [DOI] [PubMed] [Google Scholar]
- Popescu SC, Popescu GV, Bachan S, Zhang Z, Seay M, et al. Differential binding of calmodulin-related proteins to their targets revealed through high-density Arabidopsis protein microarrays. Proc. Natl. Acad. Sci. USA. 2007;104:4730–4735. doi: 10.1073/pnas.0611615104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price ND, Shmulevich I. Biochemical and statistical network models for systems biology. Curr. Opin. Biotechnol. 2007;18:365–370. doi: 10.1016/j.copbio.2007.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritsker M, Liu Y-C, Beer MA, Tavazoie S. Whole-genome discovery of transcription factor binding sites by network-level conservation. Genome Res. 2004;14:99–108. doi: 10.1101/gr.1739204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prusinkiewicz P, Erasmus Y, Lane B, Harder LD, Coen E. Evolution and development of inflorescence architectures. Science. 2007;316:1452–1456. doi: 10.1126/science.1140429. [DOI] [PubMed] [Google Scholar]
- Prusinkiewicz P, Lindenmayer A. The Algorithmic Botany of Plants. New York: Springer-Verlag; 1990. [Google Scholar]
- Ratcliffe RG, Shachar-Hill Y. Measuring multiple fluxes through plant metabolic networks. Plant J. 2006;45:490–511. doi: 10.1111/j.1365-313X.2005.02649.x. [DOI] [PubMed] [Google Scholar]
- Reddy GV, Heisler MG, Ehrhardt DW, Meyerowitz EM. Real-time lineage analysis reveals oriented cell divisions associated with morphogenesis at the shoot apex of Arabidopsis thaliana. Development. 2004;131:4225–4237. doi: 10.1242/dev.01261. [DOI] [PubMed] [Google Scholar]
- Rios-Estepa R, Lange BM. Experimental and mathematical approaches to modeling plant metabolic networks. Phytochemistry. 2007;68:2351–2374. doi: 10.1016/j.phytochem.2007.04.021. [DOI] [PubMed] [Google Scholar]
- Rohila JS, Chen M, Chen S, Chen J, Cerny R, et al. Protein-protein interactions of tandem affinity purification-tagged protein kinases in rice. Plant J. 2006;46:1–13. doi: 10.1111/j.1365-313X.2006.02671.x. [DOI] [PubMed] [Google Scholar]
- Rosso MG, Li Y, Strizhov N, Reiss B, Dekker K, Weisshaar B. An Arabidopsis thaliana T-DNA mutagenized population (GABI-Kat) for flanking sequence tag-based reverse genetics. Plant Mol. Biol. 2003;53:247–259. doi: 10.1023/B:PLAN.0000009297.37235.4a. [DOI] [PubMed] [Google Scholar]
- Sallaud C, Gay C, Larmande P, Bes M, Piffanelli P, et al. High throughput T-DNA insertion mutagenesis in rice: a first step towards in silico reverse genetics. Plant J. 2004;39:450–464. doi: 10.1111/j.1365-313X.2004.02145.x. [DOI] [PubMed] [Google Scholar]
- Saunders P, editor. Collected Works of A.M. Turing: Morphogenesis. Amsterdam: North-Holland; 1992. [Google Scholar]
- Schwab R, Palatnik JF, Riester M, Schommer C, Schmid M, Weigel D. Specific effects of microRNAs on the plant transcriptome. Dev. Cell. 2005;8:517–527. doi: 10.1016/j.devcel.2005.01.018. [DOI] [PubMed] [Google Scholar]
- Int. Rice Genome Seq. Proj. The map-based sequence of the rice genome. Nature. 2005;436:793–800. doi: 10.1038/nature03895. [DOI] [PubMed] [Google Scholar]
- Sessions A, Burke E, Presting G, Aux G, McElver J, et al. A high-throughput Arabidopsis reverse genetics system. Plant Cell. 2002;14:2985–2994. doi: 10.1105/tpc.004630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sethupathy P, Corda B, Hatzigeorgiou AG. TarBase: a comprehensive database of experimentally supported animal microRNA targets. RNA. 2006;12:192–197. doi: 10.1261/rna.2239606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer MW, Casson SA, Lindsey K. Transcriptional profiling of the Arabidopsis embryo. Plant Physiol. 2007;143:924–940. doi: 10.1104/pp.106.087668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T, Eiguchi M, Kumamaru T, Satoh H, Matsusaka H, et al. MNU-induced mutant pools and high performance TILLING enable finding of any gene mutation in rice. Mol. Genet. Genomics. 2008;279:213–223. doi: 10.1007/s00438-007-0293-2. [DOI] [PubMed] [Google Scholar]
- Thelen JJ, Peck SC. Quantitative proteomics in plants: choices in abundance. Plant Cell. 2007;19:3339–3346. doi: 10.1105/tpc.107.053991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theologis A, Ecker JR, Palm CJ, Federspiel NA, Kaul S, et al. Sequence and analysis of chromosome 1 of the plant Arabidopsis thaliana. Nature. 2000;408:816–820. doi: 10.1038/35048500. [DOI] [PubMed] [Google Scholar]
- Thibaud-Nissen F, Wu H, Richmond T, Redman JC, Johnson C, et al. Development of Arabidopsis whole-genome microarrays and their application to the discovery of binding sites for the TGA2 transcription factor in salicylic acid-treated plants. Plant J. 2006;47:152–162. doi: 10.1111/j.1365-313X.2006.02770.x. [DOI] [PubMed] [Google Scholar]
- Till B, Cooper J, Tai T, Colowit P, Greene E, et al. Discovery of chemically induced mutations in rice by TILLING. BMC Plant Biol. 2007;7:19. doi: 10.1186/1471-2229-7-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Till B, Reynolds S, Weil C, Springer N, Burtner C, et al. Discovery of induced point mutations in maize genes by TILLING. BMC Plant Biol. 2004;4:12. doi: 10.1186/1471-2229-4-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuskan GA, DiFazio S, Jansson S, Bohlmann J, Grigoriev I, et al. The genome of black cottonwood, Populus trichocarpa (Torr. & Gray) Science. 2006;313:1596–1604. doi: 10.1126/science.1128691. [DOI] [PubMed] [Google Scholar]
- Uno Y, Furihata T, Abe H, Yoshida R, Shinozaki K, Yamaguchi-Shinozaki K. Arabidopsis basic leucine zipper transcription factors involved in an abscisic acid-dependent signal transduction pathway under drought and high-salinity conditions. Proc. Natl. Acad. Sci. USA. 2000;97:11632–11637. doi: 10.1073/pnas.190309197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Leene J, Stals H, Eeckhout D, Persiau G, Van De Slijke E, et al. A tandem affinity purification-based technology platform to study the cell cycle interactome in Arabidopsis thaliana. Mol. Cell Proteomics. 2007;6:1226–1238. doi: 10.1074/mcp.M700078-MCP200. [DOI] [PubMed] [Google Scholar]
- Wang D, Amornsiripanitch N, Dong X. A genomic approach to identify regulatory nodes in the transcriptional network of systemic acquired resistance in plants. PLoS Pathog. 2006;2:e123. doi: 10.1371/journal.ppat.0020123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Kwak S-H, Zeng Q, Ellis BE, Chen X-Y, et al. TRICHOMELESS1 regulates trichome patterning by suppressing GLABRA1 in Arabidopsis. Development. 2007;134:3873–3882. doi: 10.1242/dev.009597. [DOI] [PubMed] [Google Scholar]
- Ward JL, Baker JM, Beale MH. Recent applications of NMR spectroscopy in plant metabolomics. FEBS J. 2007;274:1126–1131. doi: 10.1111/j.1742-4658.2007.05675.x. [DOI] [PubMed] [Google Scholar]
- Weberling F. Morphology of Flowers and Inflorescences. Cambridge, UK: Cambridge Univ. Press; 1992. [Google Scholar]
- Willmann MR, Poethig RS. Conservation and evolution of miRNA regulatory programs in plant development. Curr. Opin. Plant Biol. 2007;10:503–511. doi: 10.1016/j.pbi.2007.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woll K, Borsuk LA, Stransky H, Nettleton D, Schnable PS, Hochholdinger F. Isolation, characterization, and pericycle-specific transcriptome analyses of the novel maize lateral and seminal root initiation mutant rum1. Plant Physiol. 2005;139:1255–1267. doi: 10.1104/pp.105.067330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Soutto M, Xie Q, Servick S, Subramanian C, et al. Imaging protein interactions with bioluminescence resonance energy transfer (BRET) in plant and mammalian cells and tissues. Proc. Natl. Acad. Sci. USA. 2007;104:10264–10269. doi: 10.1073/pnas.0701987104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yazaki J, Gregory BD, Ecker JR. Mapping the genome landscape using tiling array technology. Curr. Opin. Plant Biol. 2007;10:534–542. doi: 10.1016/j.pbi.2007.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeilinger MN, Farré EM, Taylor SR, Kay SA, Doyle FJ. A novel computational model of the circadian clock in Arabidopsis that incorporates PRR7 and PRR9. Mol. Syst. Biol. 2006;2:58. doi: 10.1038/msb4100101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zelazny E, Borst JW, Muylaert M, Batoko H, Hemminga MA, Chaumont F. FRET imaging in living maize cells reveals that plasma membrane aquaporins interact to regulate their subcellular localization. Proc. Natl. Acad. Sci. USA. 2007;104:12359–12364. doi: 10.1073/pnas.0701180104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zentella R, Zhang Z-L, Park M, Thomas SG, Endo A, et al. Global analysis of DELLA direct targets in early gibberellin signaling in Arabidopsis. Plant Cell. 2007;19:3037–3057. doi: 10.1105/tpc.107.054999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Pan X, Cobb GP, Anderson TA. Plant microRNA: a small regulatory molecule with big impact. Dev. Biol. 2006;289:3–16. doi: 10.1016/j.ydbio.2005.10.036. [DOI] [PubMed] [Google Scholar]
- Zhang J, Li C, Wu C, Xiong L, Chen G, et al. RMD: a rice mutant database for functional analysis of the rice genome. Nucleic Acids Res. 2006;34:D745–D748. doi: 10.1093/nar/gkj016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Clarenz O, Cokus S, Bernatavichute YV, Pellegrini M, et al. Whole-genome analysis of histone H3 lysine 27 trimethylation in Arabidopsis. PLoS Biol. 2007;5:e129. doi: 10.1371/journal.pbio.0050129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Yazaki J, Sundaresan A, Cokus S, Chan SWL, et al. Genome-wide high-resolution mapping and functional analysis of DNA methylation in Arabidopsis. Cell. 2006;126:1189–1201. doi: 10.1016/j.cell.2006.08.003. [DOI] [PubMed] [Google Scholar]
- Zhou X, Wang G, Zhang W. UV-B responsive microRNA genes in Arabidopsis thaliana. Mol. Syst. Biol. 2007;3:103. doi: 10.1038/msb4100143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zilberman D, Gehring M, Tran RK, Ballinger T, Henikoff S. Genome-wide analysis of Arabidopsis thaliana DNA methylation uncovers an interdependence between methylation and transcription. Nat. Genet. 2007;39:61–69. doi: 10.1038/ng1929. [DOI] [PubMed] [Google Scholar]
- Zou M, Conzen SD. A new dynamic Bayesian network (DBN) approach for identifying gene regulatory networks from time course microarray data. Bioinformatics. 2005;21:71–79. doi: 10.1093/bioinformatics/bth463. [DOI] [PubMed] [Google Scholar]