Abstract
Caenorhabditis elegans is an important model organism for modern biologic research. An essential aspect of C. elegans research is the production of transgenic animals for study. These are often generated via microinjection, but biolistic bombardment has become increasingly popular. However, many of the plasmids previously generated for use in microinjection are not readily used for bombardment due to the lack of a convenient marker. The unc-119 gene is often used as a marker since unc-119 rescue can be observed at low magnification, allowing rescued animals to be easily distinguished from the larger number of non-rescued animals. Here we report the use of homologous recombination in E. coli as a method to insert a cassette containing the unc-119 gene into commonly used plasmids at the site of the ampicillin-resistance gene which is simpler than other methods like subcloning. These cassettes are flanked by regions homologous to the 5′ and 3′ ends of the ampicillin resistance gene and contain either the unc-119 gene and the kanamycin resistance gene or a unc-119:mCherry fusion gene and the kanamycin resistance gene. The resulting plasmids may be used for biolistic bombardment to yield animals that display unc-119 rescue, and also express the recipient plasmid transgene.
Keywords: C. elegans, transgenic animals, homologous recombination, plasmid, fluorescent protein, unc-119, transgene
1. Introduction
The non-parasitic nematode Caenorhabditis elegans has emerged as a useful and powerful model system to study a wide-range of biologic questions ranging from development to cell signaling to neurodegenerative diseases. An attractive aspect of C. elegans research is the ability to generate transgenic animals to create GFP reporter genes to visualize the timing or pattern of expression, to mis-express a gene to probe biologic function, or to generate mutant transgenes to probe the function of specific protein domains [1]. A set of vectors generated by the lab of Dr. Andrew Fire further advanced this approach by providing a set of modular vectors including features such as GFP optimized for use in C. elegans [1, 2].
Transgenic animals have often been generated by microinjection into the worm gonad with a glass pipette [1, 3, 4]. This approach is successful and used by many laboratories. However, the technique requires some time and effort to become proficient and requires specialized equipment. Further, the transgenic animals generated carry the transgene as an extrachromosomal array which can demonstrate either mosaic expression or changes during continued passage and also do not obey Mendelian genetics [4].
Recently, the use of biolistic bombardment has become increasingly common as a means of generating transgenic worms [5]. This technique involves bombarding worms with gold or tungsten particles coated with DNA. The advantages of this approach are the ability of lab members to successfully generate transgenic animals with little specialized training or practice, and the ability to obtain animals with both extrachromosomal arrays and integrated transgenes [5-7]. However, the range of co-injection marker genes is much more limited with bombardment in contrast to microinjection [5-7]. Further for bombardment it is preferable to have the marker gene in cis to the transgene, as while bombardment with multiple plasmids can lead to the identification of transgenic animals with all of the plasmids, it is also possible to only find animals with some but not all of the plasmids as has been found by our group and others [6]. This is particularly important with transgenes that cannot be easily visualized, such as those not linked to GFP or those with low levels of GFP or other fluorescent protein expression. To place the marker in cis, existing transgenes would need to either be subcloned into a vector with the marker or vice versa. This step could be challenging if the transgene possesses few unique sites or if multiple vectors are to be modified at once.
To facilitate the use of older vectors with bombardment, we explored whether homologous recombination in E. coli. could be used to insert the commonly used unc-119 gene onto the vector backbone [8]. We found that we could easily integrate a cassette consisting of the unc-119 marker gene and the kanamycin resistance gene into the site of the ampicillin resistance gene. We were then able to generate transgenic animals with the modified plasmid which are both unc-119(+) and express the transgene from the original plasmid. To further increase the utility of the technique we also created a second vector with the unc-119 gene fused to the monomeric red fluorescent protein mCherry which allows the transgene to be also tracked via the visualization of RFP [9].
Beyond the use in modifying C. elegans plasmids, this basic strategy could have general application as a means to quickly modify any of the large number of ampicillin resistant plasmids used in scientific research.
2. Materials and Methods
2.1 Plasmids
The pPD93.97 plasmid which expresses the myo-3p:GFP transgene was obtained from Addgene, Inc. (Cambridge, MA)(a gift of Dr. Andy Fire). The pmyo-2 mCherry plasmid which expresses the myo-2p:mCherry transgene was a gift of Drs. Cliff Luke and Gary Silverman (University of Pittsburgh, Pittsburgh, PA). The pDP#MM016b plasmid which carries C. elegans genomic DNA for the unc-119 gene was a gift from Drs. Denis Dupuy and Marc Vidal (Dana Farber, Boston, MA) [10].
punc-119 was constructed in the following manner. To create the recombination cassette, 50 nucleotide sequences (sequences available upon request) that were homologous to the 5′ and 3′ ends of the ampicillin resistance gene were inserted into a plasmid called pLoxP unc-119 [11]. The annealed 3′ oligo was ligated with XbaI/SbfI digested pLoxP unc-119. The resulting plasmid was cut with NarI and XhoI, and the annealed 5′ oligo was inserted. This plasmid contained a 5.7 kb unc-119 genomic DNA rescue fragment flanked by 50 nucleotide regions of homology to the ampicillin resistance gene. The kanamycin resistance gene was then excised from pKRP11 with XbaI and inserted into the XbaI site of this plasmid to give punc-119 [12]. This cassette was initially tested in recombination reactions, but was found to have a high background requiring gel purification of the cassette prior to use and to not consistently give high numbers of true positives. To address these limitations, we sought to minimize the size of the plasmid and similarity to other plasmids via substitution of the 2.1 kb unc-119p:unc-119cDNA rescue fragment instead of the genomic DNA, substitution of a kanamycin resistance gene PCR product, and substitution of a minimal R6K origin PCR product as described below.
punc-119c was constructed from punc-119 in the following manner. The unc-119 promoter, cDNA, and 3′ UTR were excised from pCG150 (a gift from Dr. Geraldine Seydoux and available at Addgene Inc., Cambridge, MA) with XhoI and XbaI, and subcloned into pBluescript II KS(+) (Stratagene, La Jolla, CA). A BamHI restriction site was then destroyed by fill-in. The resulting plasmid was digested with XhoI and XbaI and the unc-119 promoter, cDNA, and 3′ UTR were inserted into punc-119 digested with XhoI and XbaI. The resulting plasmid was called pMOD4 Amp unc-119c. The kanamycin resistance gene was then PCR amplified from pKRP11 (oligo sequences available upon request), digested with XbaI, and then cloned into pMOD4 Amp unc-119c to give pMOD4 Amp unc-119ck. The R6K origin of replication was then PCR amplified from pLoxP unc-119 (oligo sequences available upon request), digested with XmaI, and then inserted into pMOD4 Amp unc-119ck digested with XmaI. This PCR product eliminated the ampicillin resistance gene and other plasmid sequences common to many plasmids found in the pMOD4 vector (Epicentre Biotechnologies, Madison, WI) backbone which is used by punc-119. The resulting plasmid was named punc-119c.
To make the unc-119 marker visible for use in non-unc-119 mutant strains, we generated a unc-119cDNA:mCherry fusion gene in the cassette carried by punc-119cR. punc-119cR was constructed in the following manner. The mCherry monomeric red fluorescent protein and unc-54 3′ UTR were PCR amplified (oligos available upon request) from pPD95.79 mCherry and TOPO cloned (Invitrogen Corp., Carlsbad, CA) for sequencing. pPD95.79 mCherry contains a C. elegans optimized mCherry RFP cDNA instead of the GFP sequences in pPD95.79 (A. Fisher, unpublished) [9, 13]. Following sequencing, the mCherry:unc-54 3′ UTR fragment was excised with NsiI and XbaI and inserted into pMOD4 Amp unc-119c. As this construct contained the unc-54 3′ UTR which is used by many Fire lab vectors, we substituted the unc-119 3′ UTR by substituting a PCR product generated from pCG150 (oligo sequences available upon request) as a SpeI/XbaI fragment into this plasmid to give pMOD4 Amp unc-119cR3. The kanamycin resistance gene from punc-119c was then inserted as a XbaI fragment into pMOD4 Amp unc-119cR3 digested with XbaI. Finally, the R6K origin from punc-119c was substituted for the pMOD4 backbone by XmaI digestion to resulting in punc119cR.
Both plasmids are unable to replicate in standard lab E. coli strains due to the R6K replication origin which requires the trans-acting π protein for replication. These plasmids were grown in the EC100D pir-116 (F- mcrA △(mrr-hsdRMS-mcrBC) φ80d lacZ△M15 △lacX74 recA1 endA1 araD139 △(ara, leu)7697 galU galK λ-rpsL nupG pir-116(DHFR)) (Epicentre Biotechnologies, Madison, WI) which provides this factor.
These plasmids will be made available through Addgene Inc. (Cambridge, MA).
2.2 Homologous Recombination
For recombination, the pKD78 plasmid (a gift of Dr. Barry Wanner and available at The Coli Stock Center, Yale University, New Haven, CT) which expresses the λred recombination genes under the control of the araBAD promoter was transformed into DH5α (F- φ80lacZ△M15 △(lacZYA-argF)U169 recA1 endA1 hsdR17(rk−, mk+) phoA supE44 thi-1 gyrA96 relA1 λ-). This plasmid is similar to pKD46 except chloramphenicol resistance is used for selection [14]. The transformed DH5α bacteria were grown at 30°C due to the temperature sensitive replicon used by pKD78.
Bacteria carrying pKD78 were induced and made electrocompetent by inoculating 500 μL of an overnight culture into 500 mL SOB with 1.5% arabinose and 34 μg/mL chloramphenicol. This culture was grown at 30°C to an O.D. of 0.6-0.8. The cells were washed once in 500 mL and then twice in 50 mL of ice cold 10% glycerol. After the final wash, the pellet was brought to a final volume of approximately 5 mL with 10% glycerol, and 50μL aliquots were stored at −80°C. Comparable results were obtained even with bacteria frozen for weeks or months (not shown).
punc-119c or punc-119cR were digested with BamHI which digests the plasmid upstream and downstream of the regions of homology to the ampicillin resistance gene. The digested DNA was desalted and purified with the Zymo DNA Clean & Concentrator - 5 kit (Zymo Research, Orange, CA). Gel purification was not found to improve results. An Eppendorf 2510 electroporator (Eppendorf, Westbury, NY) set at 1350 volts was used with 0.1 cm gap cuvettes to electroporate 100 ng of the BamHI digested punc-119c or punc-119cR with 50 ng of the recipient pPD93.97 vector into 50 uL of DH5α carrying pKD78 prepared above. The cells were recovered in 1 mL LB, incubated with shaking for 2 hours at 37°C, and plated on LB plates with kanamycin (30 μg/mL). Incubation at 37°C was used to inhibit replication of the pKD78 plasmid. We routinely obtained 50-500 colonies. Electroporation with BamHI digested or non-digested punc-119c or punc-119cR alone produced no colonies.
From these plates, four colonies were selected and grown overnight at 37°C in LB with kanamycin. DNA was prepared for analysis with a Qiagen Quicklyse miniprep kit (Qiagen Inc., Valencia, CA). This DNA along with the parent vector was digested with XhoI, and run on an agarose gel to identify modified plasmids. XhoI digests the cassette twice and produces a 2329 bp fragment for punc-119c and a 3171 bp fragment for punc-119cR which were used to aid in identifying modified plasmids. We frequently observed bands that corresponded to both the parent plasmid, as well as the modified vector. In these cases it was necessary to destroy the parent plasmid by digesting the plasmid miniprep with ScaI, which cuts only in the ampicillin resistance gene in the parent plasmid. The digest was transformed into DH5α, and plated on LB plates containing 30 μg/mL kanamycin.
2.3 Transgenic Animals
The DP38 (unc-119(ed3)) strain was obtained from the Caenorhabditis elegans Genetics Center which is supported by NIH funding. DP38 was grown in mass culture on egg plates as previously described for bombardment [15, 16]. The worms were isolated from the plates by washing in S-basal buffer followed by gravity sedimentation to concentrate the adult animals [15]. The concentrated adult worms were bombarded with gold microparticles coated with 10 μg. of plasmid DNA, for transgenes prepared with punc-119c and punc-119cR, following a previously published protocol [15]. For the co-bombardment experiment, the gold microparticles were coated with a mixture of 10 μg. of pPD93.97, 10 μg. of pDP#MM016b, and 10 μg. of pmyo-2 mCherry using the same protocol. We used a Bio-Rad PDS-1000 system (Bio-Rad Laboratories, Hercules, CA) fitted with a hepta adapter, and we used a 1350 PSI rupture disk and vacuum of 27 inches Hg for each bombardment. Following bombardment, the worms were allowed to recover for 10 minutes at 20°C before being resuspended in S-basal and dispensed to 20 -10 cm. NGA plates for growth.
Transgenic animals were identified by rescue of the morphologic and mobility deficits of unc-119. For the co-bombardment experiment, we found that the myo-2:mCherry transgene was sufficiently bright that transgenic worms could also be identified by mCherry fluorescence. Single rescued animals were used to establish transgenic lines, and those lines with transmission of the transgene to the resulting progeny were kept for further study. Only one line was generated from each large NGA plate to ensure that all lines are distinct.
DP38 and transgenic worms were mounted for digital photography using an Olympus BX51 upright microscope (Olympus America, Center Valley, PA) as previously described [16]. Images were captured with Nomarski optics for transmitted light, a FITC filter set for GFP, and a Cy3 filter set for mCherry. All photos within a panel were taken on the same day with identical camera settings to allow direct comparison.
3. Results and Discussion
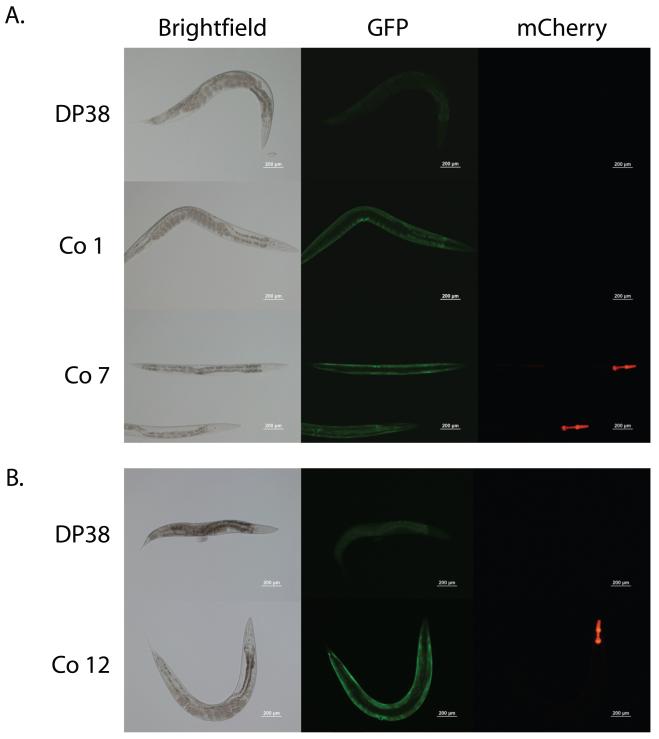
Previous work in C. elegans suggested that bombardment of worms with multiple plasmids could result in the identification of transgenic worms carrying some but not all of the plasmids [6]. However, the work of Wilm et. al. used a custom-made bombardment apparatus that may behave differently than the systems manufactured by Bio-Rad Laboratories (Bio-Rad Laboratories, Hercules, CA) that are in common use by our lab and others [7, 15]. To address whether this is routinely seen using this equipment and commonly used transgenes, we performed a co-bombardment using equal amounts of the pDP#MM016b plasmid, pPD93.97, and the pmyo-2 mCherry plasmids as this would represent a common scenario consisting of the unc-119 marker for bombardment, a GFP reporter, and then a secondary marker to facilitate identification of transgenic animals when crossed to unc-119(+) animals. We identified transgenic animals using either unc-119 rescue or the very bright pharyngeal mCherry expression provided by the pmyo-2 mCherry plasmid. We identified five stable transgenic lines and tested for the presence of each transgene by phenotype. Two of the lines showed clear unc-119 rescue and easily visible myo-3:GFP, which expresses in the cytoplasm of body wall muscles, and myo-2:mCherry, which expresses in the cytoplasm of the pharyngeal muscle cells, expression (Figure 1A, line Co 7, bottom row). However, one line showed unc-119 rescue and myo-3:GFP expression, but lacked myo-2:mCherry expression (Figure 1A, line Co 1, middle row). Further, two lines showed easily visible myo-3:GFP and myo-2:mCherry expression, but showed at best weak unc-119 rescue which might not have been identified using unc-119 rescue as the primary screen (Figure 1B, line Co 12, bottom row). Together, our results suggested that co-bombardment can often be successful and is probably not necessary with easily visible transgenes. However, for experiments using weak, conditional, mutated, or non-fluorescent transgenes, the co-bombardment approach could lead to the identification of transgenic animals lacking a transgene. Consequently, placing the transgenes in cis on the same plasmid is an important safeguard against this possible outcome.
Figure 1. Identification transgenic animals lacking individual transgenes following co-bombardment.
(A) Bombardment of the DP38 (unc-119(ed3)) strain (top row) with the pDP#MM016b, pPD93.97, and pmyo-2 mCherry plasmids can produce transgenic worms which are both unc-119(+) and express myo-3:GFP and myo-2:mCherry (line Co 7, bottom row). However, transgenic worms lacking the myo-2:mCherry transgene were also identified (line Co 1, middle row). (B) From the same co-bombardment, two stable transgenic lines which expressed both myo-3:GFP and myo-2:mCherry but had weak unc-119 rescue based on morphology and movement were also identified (line Co 12, bottom row).
To facilitate the addition of the unc-119 marker to ampicillin resistant plasmids used for the construction of transgenic C. elegans, we generated the punc-119c and punc-119cR plasmids which carry a cassette consisting of a unc-119 cDNA capable of rescuing the unc-119(ed3) mutation and a kanamycin resistance gene for selection in E. coli flanked by 50 nucleotide regions of homology to the ampicillin resistance gene. This cassette uses common sequences found in Fire lab vectors and others such as pBluescript (Stratagene Inc., La Jolla, CA) as a site of homologous recombination (Figure 2A).
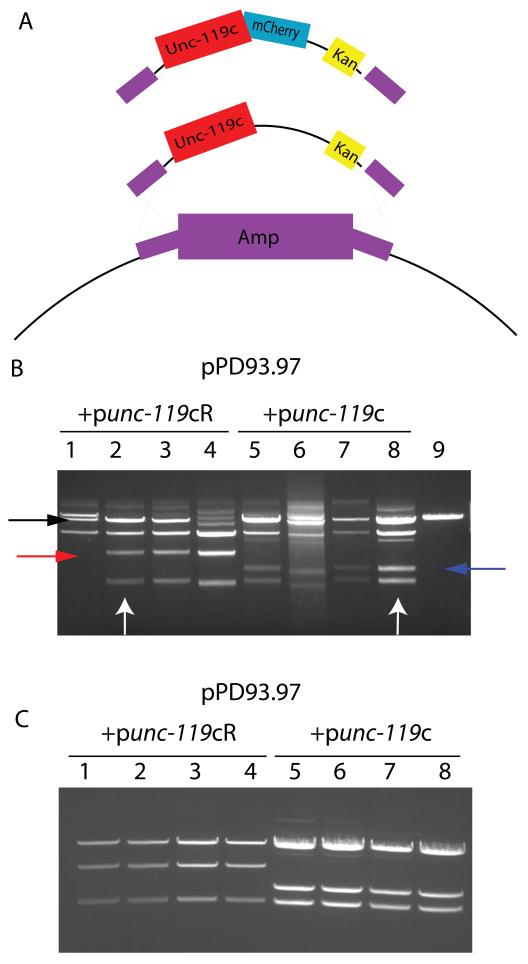
Figure 2. Generation of retrofitted plasmids by homologous recombination.
(A) Diagram of the flanking regions of ampicillin resistance gene homology (purple) , unc-119 promoter and unc-119 cDNA (unc-119c)(red), mCherry RFP fusion (punc-119cR only)(red and blue), and kanamycin resistance gene (yellow) carried by the recombination cassettes carried by the punc-119c and punc-119cR plasmids. These cassettes can be inserted into ampicillin-resistant plasmids by recombination using the 50 nucleotide homology regions at the 5′ and 3′ ends. (B) Mini-preps of colonies containing pPD93.97 retrofitted with punc-119c or punc-119cR. XhoI digest reveals both the parent plasmid and a second retrofitted plasmid (lane 9 and black arrow). This new plasmid is identified by a change in the restriction pattern compared with the parent vector and the appearance of new 2329 b.p. (punc-119c) (blue arrow) and 3171 b.p. (punc-119cR) (red arrow) fragments derived from the inserted cassette. (C) Elimination of the parent plasmid by ScaI digest. Mini-prep DNA from (B) indicated by white arrows was digested with ScaI and transformed into bacteria. Mini-preps of the resulting colonies reveal only the presence of only the retrofitted plasmid.
To evaluate whether punc-119c or punc-119cR could be integrated into a commonly used vector backbone, we electroporated induced DH5α bacteria transformed with the pKD78 plasmid with linearized punc-119c or punc-119cR and pPD93.97. pPD93.97 is from the Fire lab vector collection and consists of the myo-3 promoter driving GFP in the body wall musculature [14]. The transformed bacteria were then plated on LB-kanamycin plates to select for plasmids which have successfully undergone integration. Control transformations with punc-119c or punc-119cR alone or pPD93.97 alone did not result in any kanamycin resistant colonies (not shown). From the punc-119 and pPD93.97 transformations, we obtained many colonies and selected four for mini-prep to analyze the resulting plasmids. Following XhoI digestion, we found that 3 of 4 colonies from the punc-119c and 4 of 4 colonies from the punc-119cR electroporations contained plasmids consistent with both the unmodified and correctly modified pPD93.97 plasmids (Figure 2B).
Finding both plasmids inside the initial transformants is not unexpected as pPD93.97 is a multi-copy plasmid so even with out-growth and selection each bacterium could contain a random mixture of each plasmid [8]. To obtain a pure population of modified plasmid, we used the mini-prep DNA to either just retransform DH5α following dilution or retransform DH5α following brief digestion with ScaI. The ScaI site is located in the ampicillin resistance gene of all Fire lab vectors and pBluescript-derived vectors, and this site is eliminated in the vectors that have undergone recombination. The ScaI sequence also appears to be uncommon in C. elegans DNA (not shown). We found that retransformation without digestion resulted in colonies carrying both modified and unmodified plasmids, whereas digestion of the unmodified plasmids with ScaI produced colonies only carrying the modified vector (Figure 2C).
We then tested if the modified plasmids generated by recombination could be successfully used to generate transgenic animals. We bombarded the DP38(unc-119(ed3)) strain with both the pPD93.97 (unc-119c) and the pPD93.97 (unc-119cR) plasmids. We obtained multiple unc-119(+) lines from each bombardment which indicated that both the unc-119 promoter and cDNA and the unc-119 promoter and cDNA fused to mCherry could rescue the unc-119 mutation. As expected the majority of the obtained lines transmitted unc-119(+) consistent with an extrachromosomal array [15].
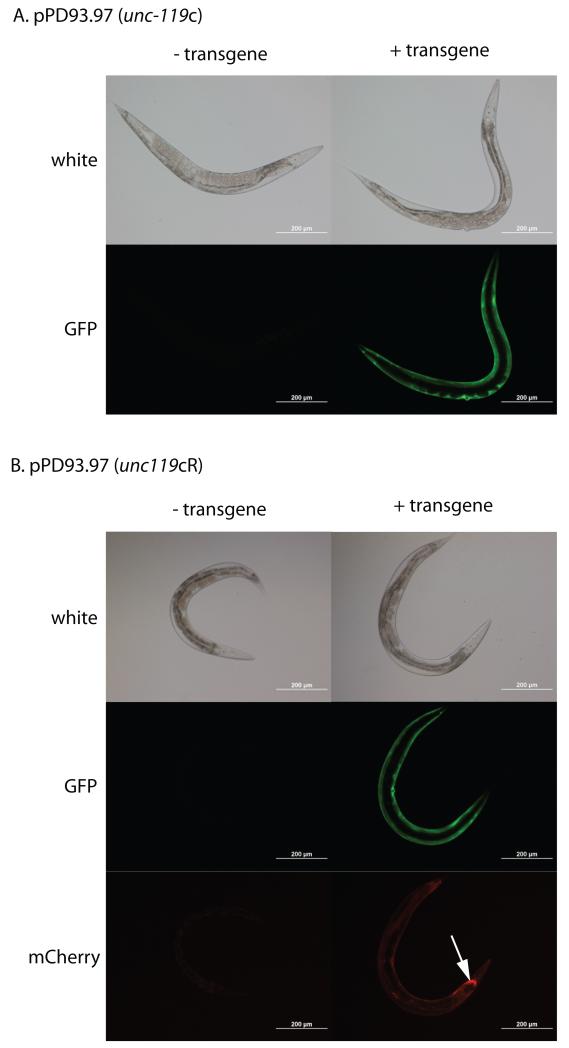
To assess whether the insertion of the unc-119c or unc-119cR cassette interfered with the expression of the existing transgene on the cassette, we examined the unc-119(+) transgenic animals via fluorescent microscopy. We found the myo-3::GFP transgene carried by pPD93.97 still expressed in the cytoplasm of body-wall muscles (Figure 3A and 3B). Further, we found that all of the stable unc-119(+) transgenic animals obtained detectable myo-3::GFP which demonstrated the utility of placing the unc-119 gene in cis to the transgene.
Figure 3. Generation of transgenic animals with retrofitted plasmids.
(A) Bombardment of the DP38 (unc-119(ed3)) strain with the unc-119c modified pPD93.97 plasmids produces transgenic worms which are both unc-119(+) and express myo-3:GFP. (B) Bombardment of the DP38 (unc-119(ed3)) strain with the unc-119cR modified pPD93.97 plasmids produces transgenic worms which are unc-119(+) and express both unc-119:mCherry (white arrow) and myo-3:GFP.
Finally, we assessed whether the unc-119::mCherry transgene encoded by unc-119cR gave rise to a fusion protein that could be detected by fluorescent microscopy. If visible, this transgene would then have a dual function of allowing rescue of the unc-119(ed3) mutant for initial selection after bombardment, and then later the transgene could serve as a visible marker if the transgene is crossed into worms lacking the unc-119 mutation. This feature would be particularly useful for transgenes lacking fluorescence, such as a TAP-tagged transgene, or for transgenes with low levels of fluorescence. We found that the unc-119:mCherry transgene is indeed visible and can be detected both by a stereomicroscope and also with a compound microscope (Figure 3B and not shown). The selection of mCherry also carries the advantage that the fluorescence from this protein is not visible under the standard FITC filter set used to visualize GFP (Figure 3B).
Acknowledgements
AAF and ALF were supported by grants from the NIH (AG028977 and AG029870) to ALF. AAF and ALF would like to thank the Caenorhabditis elegans Genetics Center, the Cloning Vector collection hosted at the National Institute of Genetics (Japan), Drs. Barry Wanner, Cliff Luke, Gary Silverman, Denis Dupuy, Marc Vidal, and Dr. Geraldine Seydoux for strains and plasmids.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Mello C, Fire A. DNA transformation. Methods Cell Biol. 1995;48:451–482. [PubMed] [Google Scholar]
- 2.Miller DM, 3rd, Desai NS, Hardin DC, Piston DW, Patterson GH, Fleenor J, Xu S, Fire A. Two-color GFP expression system for C. elegans. Biotechniques. 1999;26:914–8. doi: 10.2144/99265rr01. 920-1. [DOI] [PubMed] [Google Scholar]
- 3.Fire A. Integrative transformation of Caenorhabditis elegans. EMBO J. 1986;5:2673–2680. doi: 10.1002/j.1460-2075.1986.tb04550.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mello CC, Kramer JM, Stinchcomb D, Ambros V. Efficient gene transfer in C.elegans: extrachromosomal maintenance and integration of transforming sequences. EMBO J. 1991;10:3959–70. doi: 10.1002/j.1460-2075.1991.tb04966.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Praitis V, Casey E, Collar D, Austin J. Creation of low-copy integrated transgenic lines in Caenorhabditis elegans. Genetics. 2001;157:1217–1226. doi: 10.1093/genetics/157.3.1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wilm T, Demel P, Koop HU, Schnabel H, Schnabel R. Ballistic transformation of Caenorhabditis elegans. Gene. 1999;229:31–5. doi: 10.1016/s0378-1119(99)00043-8. [DOI] [PubMed] [Google Scholar]
- 7.Evans TC. Transformation and microinjection Wormbook. 2006:1–15. [Google Scholar]
- 8.Thomason LC, Costantino N, Shaw DV, Court DL. Multicopy plasmid modification with phage lambda Red recombineering. Plasmid. 2007;58:148–58. doi: 10.1016/j.plasmid.2007.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McNally K, Audhya A, Oegema K, McNally FJ. Katanin controls mitotic and meiotic spindle length. J Cell Biol. 2006;175:881–91. doi: 10.1083/jcb.200608117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maduro M, Pilgrim D. Identification and cloning of unc-119, a gene expressed in the Caenorhabditis elegans nervous system. Genetics. 1995;141:977–988. doi: 10.1093/genetics/141.3.977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang Y, Nash L, Fisher AL. A simplified, robust, and streamlined procedure for the production of C. elegans transgenes via recombineering. BMC Dev Biol. 2008;8:119. doi: 10.1186/1471-213X-8-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reece KS, Phillips GJ. New plasmids carrying antibiotic-resistance cassettes. Gene. 1995;165:141–2. doi: 10.1016/0378-1119(95)00529-f. [DOI] [PubMed] [Google Scholar]
- 13.Shaner NC, Campbell RE, Steinbach PA, Giepmans BN, Palmer AE, Tsien RY. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat Biotechnol. 2004;22:1567–72. doi: 10.1038/nbt1037. [DOI] [PubMed] [Google Scholar]
- 14.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc.Natl.Acad.Sci.U.S.A. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Berezikov E, Bargmann CI, Plasterk RH. Homologous gene targeting in Caenorhabditis elegans by biolistic transformation. Nucleic Acids Res. 2004;32:e40. doi: 10.1093/nar/gnh033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fisher AL, Page KE, Lithgow GJ, Nash L. The Caenorhabditis elegans K10C2.4 Gene Encodes a Member of the Fumarylacetoacetate Hydrolase Family: A CAENORHABDITIS ELEGANS MODEL OF TYPE I TYROSINEMIA. J Biol.Chem. 2008;283:9127–9135. doi: 10.1074/jbc.M708341200. [DOI] [PMC free article] [PubMed] [Google Scholar]