Abstract
Unlike E. coli, Mycobacterium tuberculosis (Mt) expresses a Ku-like protein and an ATP-dependent DNA ligase that can perform non-homologous end-joining (NHEJ). We have expressed the Mt-Ku and Mt-Ligase D in E. coli using an arabinose-inducible promoter and expression vectors that integrate into specific sites in the E. coli chromosome. E. coli strains have been generated that express the Mt-Ku and Mt-Ligase D on a genetic background that is wild-type for repair, or deficient in either the RecA or RecB protein. Transformation of these strains with linearized plasmid DNA containing a 2-bp overhang has demonstrated that expression of both the Mt-Ku and Mt-Ligase D is required for DNA end-joining and that loss of RecA does not prevent this double-strand break repair. Analysis of the re-joined plasmid has shown that repair is predominantly inaccurate and results in the deletion of sequences. Loss of RecB did not prevent the formation of large deletions, but did increase the amount of end-joining. Sequencing the junctions has revealed that the majority of the ligations occurred at regions of micro-homology (1–4 bps), eliminating one copy of the homologous sequence at the junction. The Mt-Ku and Mt-Ligase D can therefore function in E. coli to re-circularize linear plasmid.
Keywords: Non-homologous end-joining, Mycobacterium tuberculosis, DNA repair, double strand break repair
1. Introduction
Non-homologous end-joining (NHEJ) is the process where two ends are directly joined using only base-pairing information at the DNA termini. NHEJ is the major double strand break-repair pathway in mammalian cells (for review see 1) and involves a number of proteins that bind the ends of the DNA (Ku 70, Ku80 and DNA protein kinase catalytic subunit), process (3′ phosphastases, nucleases and polymerases) and ligate (XRCC4 and DNA ligase IV) the termini. Only two proteins so far have been implicated in NHEJ in prokaryotes: Ku and an ATP-dependent ligase (2–4). The Ku and ligase are frequently expressed from a single operon. Such operons are known to exist in a number of prokaryotes (5) including Mycobacterium tuberculosis, Mycobacterium smegmatis and Bacillus subtilis, but not E. coli. It has been suggested that NHEJ may provide a selection advantage during dormant periods when homologous recombination cannot be used to repair DNA (5). Ku and ligase mutants of B. subtilis are sensitive to ionizing radiation during stationary phase, which implicates the prokaryote NHEJ proteins in double strand break repair (6).
M. tuberculosis has four DNA ligases: LigA is an NAD+-dependent ligase, while LigB, C and D are ATP-dependent ligases (7; 8). Only LigD (Mt-LigD) is expressed from an operon with a Ku protein (Mt-Ku). Mt-LigD is a multifunctional protein: It is a DNA-dependent DNA polymerase, DNA-dependent RNA primase, a 3′–5′ single stranded DNA exonuclease, and a terminal transferase as well as a DNA ligase (9). Mt-LigD has a polymerase domain at the N-terminus, a nuclease domain in the middle of the protein and a ligase domain at the C-terminus. The polymerase and the ligase domains can function as independent proteins; however, in vitro studies indicate that the Mt-Ku only interacts with the polymerase domain (10). It is highly likely therefore that the polymerase domain is essential for recruiting Mt-LigD to the damaged DNA after Mt-Ku has bound as a homodimer to the ends of the double strand break. Mt-Ku also specifically stimulates the end-joining activity of Mt-LigD (6). The termini processing activities of Mt-LigD suggest that auxiliary proteins are not required to process DNA breaks generated from ionizing radiation or damaging chemicals. In fact, the two proteins alone are able to perform non-homologous end-joining of DNA in a cell: expression of the Mt-Ku and Mt-LigD in yeast deficient in NHEJ corrected the yeast defect by ~50% (9).
Due to the slow growth and technical challenge of M. tuberculosis genetic manipulation, much of the work on Mt-Ku and Mt-LigD has been performed in vitro and the model cell systems available to study Mt- NHEJ are limited. The yeast expression system was able to show that Mt-Ku and Mt-LigD could perform NHEJ, but S. cerevisiae is a eukaryote that has NHEJ proteins. Even if mutants are used to disable yeast NHEJ, there are many proteins that are involved in this repair mechanism that could potentially interfere with or aid the Mt-NHEJ proteins. Work is emerging on the M. smegmatis and B. subtilis NHEJ proteins in cells, as these prokaryotes are easier to manipulate than M. tuberculosis. However, E. coli is the easiest prokaryote to grow and genetically manipulate, the DNA repair systems have been very well characterized and there are a multitude of plasmids available to study DNA repair and mutation frequency. Also E. coli does not have the capability to perform NHEJ and like replicating M. tuberculosis uses homologous recombination to repair double-strand breaks. E. coli and M. tuberculosis are also similar in other DNA repair pathways: they both express proteins for base excision repair, nucleotide excision repair and SOS repair, and have many of the same termini processing enzymes (11).
This work aimed to determine whether Mt-Ku and Mt-LigD could function in a model prokaryote organism that does not have any capability to perform NHEJ, and to produce an easy model system in which to study the Mt-NHEJ proteins. We created unique E. coli strains that express either Mt-Ku or Mt-LigD, or Mt-Ku and Mt-LigD using arabinose-inducible expression vectors that are integrated into specific sites on the E. coli chromosome. We have therefore generated E. coli strains in which NHEJ can be easily switched on or off by growth in arabinose, making it easy to identify the Mt-Ku and Mt-LigD repair products. None of the other model cell systems used to investigate prokaryote NHEJ have this ability. Since E. coli are easy to manipulate, we have established strains that express the NHEJ proteins in backgrounds that are wild-type for repair, or deficient in either RecA or RecB. This has allowed us to determine whether these proteins influence the end-joining process. Since this model system uses expression vectors integrated into the chromosome, assays can be performed to examine the repair of replicating extra-chromosomal plasmid DNA. The sequences of repair products can therefore be easily examined. We have assessed the repair of PacI or ClaI-linearized plasmid DNA and determined that the type of double strand break alters the fidelity of repair. Data presented in this paper indicates that this model will prove useful for examining how these NHEJ proteins interact with each other and possibly with other E. coli proteins during the repair of DNA. It will also allow us to assess whether this repair system can be beneficial to a replicating organism.
2. Materials and methods
2.1 Bacterial strains
Wild-type E. coli (strain BW35- Hfr KL16(PO-45) thi-1 relA1 spoT1 e14− λ−) were obtained from Dr. Susan S. Wallace (University of Vermont, Burlington, VT) and the isogenic strain deficient in RecA was BW386 (KL16 recA56 srlC300::Tn10). The recB mutation (recB268::Tn10) used in the transduction of strain BWKuLig#1 to generate BW1900 (also named in this paper as B−KuLig) was from strain C876 obtained from Dr. David Leach (University of Edinburgh, U.K.). BW23474 expresses the trans-acting II protein encoded by pir (12) and was used to propagate pAH143 and pLA2 plasmids and their derivatives. This strain was obtained from the Coli Genetics Stock Center (Yale University, New Haven, Connecticut).
2.2 Plasmids
The open reading frames used to generate the Mt-Ku and Mt-LigD expression vectors were from pET16b vectors containing the M. tuberculosis sequences RV0937c and RV0938, respectively (6). The CRIM vector system (12), which included pINT-ts, pLA2, pAH69 and pAH143 were obtained from Mary Berlyn from the Coli Genetics Stock Center (Yale University, New Haven, Connecticut). pINTts and pAH69 encode carbenicillin-resistance, while pLA2 and pAH143 encode kanamycin and gentamicin resistance, respectively. pACYC184 (New England Biolabs, Beverly, MA) is a low copy vector with a p15A origin of replication that encodes resistance to chloramphenicol (34 μg/ml). pBestluc (Promega, Madison, WI) is a high copy vector with a pUC origin of replication that encodes resistance to ampicilin or carbenicillin (50–100 μg/ml). pBestluc expresses the firefly luciferase open reading frame from a tac promoter. In our bacterial strains expression from the tac promoter was high without induction with IPTG, probably due to the high copy number of the vector compared to the expression of the lacI repressor. pBestluc does not contain the Chi recognition site (5′GCTGGTGG 3′) that is known to modulate the RecBCD exonuclease.
2.3 Oligodeoxyribonucleotides
Oligodeoxyribonucleotides (oligonucleotides) were purchased from Operon Technologies Inc. (Alameda, CA). The oligonucleotides His3, His4, MCS1 and MCS2 contained 5′ phosphates. The sequences for these oligonucleotides were as follow: His3: d(TATGGGCCATCATCATCATCATCATCATCATCATCACAGCAGCGGCCATATCGAAGGTCGTCA), His4: d(TATGACGACCTTCGATATGGCCGCTGCTGTGATGATGATGATGATGATGATGATGATGGCCCA), MCS1: d(TATGGCCATGGCTGCAGGATCCGGTAC), MCS2: d(CGGATCCTGCAGCCATGGCCA) Primers used to sequence the expression constructs were as follows: d(ATCCATAAGATTAGCGG), d(CGGATATTATCGTGAGG), d(CATAAACTGCCAGGCATC) The PCR primers used to check for integration of pLA-HisKu and pAH-HisLig were as follows: P1λ: d(GGCATCACGGCAATATAC), P4λ: d(TCTGGTCTGGTAGCAATG), P1HK: d(GGAATCAATGCCTGAGTG), P4HK: d(GGCATCAACAGCACATTC), P2: d(ACTTAACGGCTGACATGG), P3: d(ACGAGTATCGAGATGGCA). Primers used either for PCR to determine the size of deletions or to sequence repair products were as follows: Luc 1: d(TGGATGGCTACATTCTG), Luc 5: d(GCCTGGTATCTTTATAG), Hind 3: d(GAACGTGACGGACGTAAC), Luc 3: d(ATGTGGATTTCGAGTCGTCT), R: d(TCATCGTCTTTCCGTGCT).
2.4 Construction of the Mt-LigD and Mt-Ku expression constructs
pLA2 and pAH143 have the γ replication origin of R6K and hence cannot replicate in E. coli unless the cells express the trans-acting II protein, encoded by pir (12). For genetic manipulation of the vectors we used an E. coli strain that is pir+ (BW23474). pLA2 encodes kanamycin resistance and integrates at the λ phage attachment site in the E. coli chromosome, and pAH143 encodes gentamicin resistance and integrates at the HK022 phage site. pLA2 contains the araBp promoter upstream from the lacZ gene. We removed lacZ and inserted RV0937c between the NdeI and SmaI sites to generate pLA-Ku. To add the N-terminus tag (Met- Gly-[His]10 -Ser-Ser-Gly-His-Ile-Glu-Gly-Arg-His) encoded by the pET16b expression vector, we annealed two oligonucleotides (His3 and His 4) and the double-stranded oligonucleotide was inserted at the NdeI site upstream of the Mt-Ku coding sequence. This tag sequence does not disrupt the activity of the protein (6), but allows detection of the protein using an antibody to the tag sequence. The resulting construct, pLA-HisKu, was sequenced to confirm the oligonucleotide was inserted in the correct orientation. To insert the araBp promoter into pAH143, the PstI- KpnI fragment from pLA-HisKu was ligated between the PstI and KpnI sites of pAH143. Since this transferred the promoter and the HisKu coding sequence, the HisKu sequence was removed using NdeI and KpnI and a double-stranded oligonucleotide (created by annealing MCS1 and MCS2) was inserted to generate pAHMCS#6. The NcoI-BamHI fragment from pET16b RV0938 was then inserted downstream of araBp at the NcoI- BamHI sites of pAHMCS#6 to produce pAH-HisLigase. This construct expresses the Mt-LigD containing the tag sequence from pET16b plus two amino acids (Met and Ala) on the N-terminus of the protein. This construct was also sequenced to check the 5′ and 3′ sequence of the Mt-LigD open reading frame.
2.5 Integration of pLA-HisKu and pAH-HisLigase into the E. coli chromosome
Integration of the constructs was performed as described by Haldimann & Wanner (12). Briefly, bacteria were transformed by electroporation with pINTts or pAH69 for the integration of pLA-HisKu or pAH-HisLigase, respectively. pINTts and pAH69 are plasmids that express an integrase corresponding to the λ or HK022 phage attachment sites, and can only replicate at 30°C. Bacteria were therefore grown at 30°C on solid medium or in liquid culture during the preparation of electrocompetent bacteria. The strains containing either pINTts or pAH69 were transformed with 100 ng of the appropriate vector (pLA-HisKu or pAH-HisLigase) and grown at 37°C for 1 h and 42°C for 30 min. Growth at these temperatures activates the integrase and also prevents the replication of pINTts and pAH69. The culture was then grown on solid medium containing either 10 μg/ml kanamycin (for pLA-HisKu) or 5 μg/ml gentamicin (for pAH-HisLigase) and incubated overnight at 37°C. The colonies were then grown on solid medium without antibiotic, before re-plating on the appropriate antibiotic medium. This tested whether the antibiotic resistance gene was integrated into the chromosome. The bacteria were also checked to make sure they were no longer carbenicillin-resistant, which indicated they were “cured” of pINTts or pAH69. The resultant strains were then tested by PCR to determine whether the vector had integrated at the appropriate site. Strains no longer needed to be grown in kanamycin or gentamicin to maintain the integrated plasmid.
2.6 PCR from bacterial colonies to test for vector integration or deletions in repair products
A small amount of the bacterial colony was resuspended in 20 μl water. To test for the integration of pLA-HisKu or pAH-HisLigase, a 25-μl reaction was prepared containing 5 μl bacteria, 0.4 μM of each primer P1, P2, P3 and P4, 6.4 mM MgCl2, 0.2 mM of each of dGTP, dATP, TTP, dCTP, 10 mM Tris-HCl (pH 9), 50 mM KCl, 0.1% Triton® X-100 and 0.5 units Taq DNA polymerase (Promega, Madison, WI). For integration of pLA-HisKu at the λ site, P1λ and P4λ were used with an annealing temperature of 63°C, while P1HK and P4HK were used with an annealing temperature of 59°C to check for integration of pAH-HisLigase at the HK022 site. After 28 PCR cycles, the products were separated by electrophoresis through a 1.4% agarose gel.
To determine the presence and size of deletions in end-joining products, a 25-μl reaction was set up as described above containing 5 μl bacteria and two primers. The primer pairs were Hind 3 and Luc 5 with an annealing temperature 50°C, Luc 1 and R with an annealing temperature 55°C, or Luc 3 and Luc 5 with an annealing temperature 47°C. The predicted full length PCR products for pBestluc were 2.6 kb, 236 bp and 1.6 kb, respectively.
2.7 Construction of the RecB-deficient strain expressing Mt-Ku and Mt-LigD
Transduction with bacteriophage P1 was used to transfer the recB268::Tn10 mutation from strain C876 into BWKuLig#1 and was performed as previously described (13). Bacteria were selected using 15 μg/ml tetracycline and the loss of recB was confirmed by ultraviolet sensitivity.
2.8 Western analysis to examine expression of Mt-Ku and Mt-LigD
Bacteria were grown in a 5-ml LB culture containing the appropriate antibiotic overnight at 37°C at 250 rpm. The stationary phase culture was diluted 100 times in LB and antibiotic, and grown at 37°C, 300 rpm, for 30 min for B−KuLig and A−KuLig, or for 1.5 h for the wild-type strains, prior to the addition of L-arabinose to a final concentration of 0.2%. After a total growing time of 2.5 h, the bacteria were in exponential phase at an OD600 of ~0.6 and were harvested by centrifugation at 4°C. Bacteria were resuspended in 0.5 ml lysis buffer (0.1 mg/ml lysozyme, 45 mM HEPES pH 7.8, 80 mM KCl, 1 mM DTT, 1 mM PMSF, 2 μg/ml pepstatin A) and sonicated prior to centrifugation (4°C, 13,000rpm, 10 h) to remove cell debris. The protein concentration in the cell-free extract was determined according to Bradford (14), utilizing the Bio-Rad Protein Assay reagent (Bio-Rad, Hercules, CA). Protein (50–75 μg) was electrophoresed through a tris-glycine 4–20% gradient SDS polyacrylamide gel and transferred by electroblotting to 0.2 μm nitrocellulose. The membrane was probed using a His•Tag monoclonal antibody (final concentration 0.2 μg/ml; Novagen, Madison, WI,) according to manufacturer’s recommendations. This antibody recognizes five consecutive histidines. Bound His•Tag antibody was visualized using autoradiography following incubation with a secondary horse radish peroxidase antibody (diluted 1:3000; Amersham, Piscataway, NJ) and chemiluminescent substrate (ECL-plus substrate, Amersham, Piscataway, NJ).
2.9 End-joining assay
PacI or ClaI-linearized pBestluc were chosen as substrates for the end-joining assay. PacI and ClaI are unique restriction sites situated at the 3′ end of the firefly luciferase coding region and loss/alteration of sequence in this part of the protein frequently results in loss of luciferase activity (15). pBestluc was digested with PacI or ClaI and purified twice through 0.7% agarose gels. The linear DNA was isolated from the gel using the Qiaquick Gel Extraction kit (Qiagen Inc., Valencia, CA) and the concentration determined using an Agilent 2100 Bioanalyzer (Agilent Technologies, Wilmington, DE). By comparing the number of colonies obtained with the linear plasmid to that obtained for supercoiled pBestluc in DH5α bacteria, we determined that 100 ng of linear plasmid contained ~0.16 pg of contaminating circular DNA. 99–100% of these colonies expressed active firefly luciferase.
For the end-joining assay, bacteria were grown in LB or LB supplemented with 0.2% L-arabinose. Growth of bacteria for the preparation of electrocompetent cells was exactly as described above for the western analysis, with the arabinose being added for only the last 1 h (for wild-type strains) or 2 h (for RecA and RecB-deficient strains). Bacteria were then harvested and washed with water to prepare for electroporation as described by Seidman et al (16). Linearized pBestluc (1–100 ng) and 0.1 ng pACYC184 were co-transformed into the bacteria, which were then incubated in 1.5 ml LB, 1 mM IPTG at 37°C, 250 rpm for 1.5 h. A portion of the culture was then grown on triplicate plates of solid medium containing either 100 μg/ml carbenicillin or 34 μg/ml chloramphenicol. After overnight growth at 37°C, the colonies were counted and a ratio of the carbenicillin-resistant (CarbR) colonies/chloramphenicol-resistant (CmR) colonies calculated for each transformation. This was designated the ratio for the total number of CarbR colonies. Since pACYC184 encodes resistance to chloramphenicol, the calculation of this ratio normalized each sample for transformation efficiency. The CarbR colonies were then transferred to nylon and sprayed with firefly luciferase substrate and the number of colonies expressing active and inactive firefly luciferase was determined. A ratio of CarbR/CmR colonies was calculated for each transformation for the CarbR colonies expressing active (Luc+) or inactive luciferase (Luc−).
To examine the repair products, the culture from the transformation was also grown on extra plates of solid medium containing 100 μg/ml carbenicillin. The colonies were re-streaked and checked for luciferase activity and those that did not express activity were subjected to PCR as described above. To identify the sequence across the junctions of the repair products, plasmid was isolated from bacteria using the Wizard Plus Miniprep DNA purification system (Promega, Madison, WI) and the DNA was sequenced by the DNA Facility at the University of Iowa (Ames, IA).
2.10 Statistical analysis
The CarbR/CmR ratios obtained from the end-joining assay were compared for different growth conditions using the Instat3 program and the unpaired t test or the Dunn’s Multiple Comparisons test.
3. Results
3.1 Generation of bacteria containing pLA-HisKu and pAH-HisLigase integrated into the E. coli chromosome
Assays to examine the repair products of double-strand breaks frequently employ replicating plasmid DNA. It was therefore not practical to express the Mt-Ku and Mt-LigD from an episomal replicating plasmid. The CRIM vector system, developed by Haldimann & Wanner (12), uses plasmids that cannot replicate in E. coli because they have the γ replication origin of R6K. The vectors are designed to integrate into the bacterial chromosome at the different phage attachment sites. Multiple plasmids at different sites can be integrated without the risk of recombination between vectors, because the sequences between the phage attachment sites are essential for bacterial survival. Proteins are expressed from the araB promoter, which is repressed in the presence of glucose and induced in the presence of arabinose. We have generated two vectors using this system: pLA-HisKu can integrate into the λ phage attachment site and expresses Mt-Ku, and pAH-HisLigase can integrate into the HK022 attachment site and expresses Mt-LigD. Both the Mt-Ku and Mt-LigD were engineered to contain N-terminal histidine tag sequences to allow easy detection of the proteins by western analysis. This tag sequence does not interfere with protein function (6).
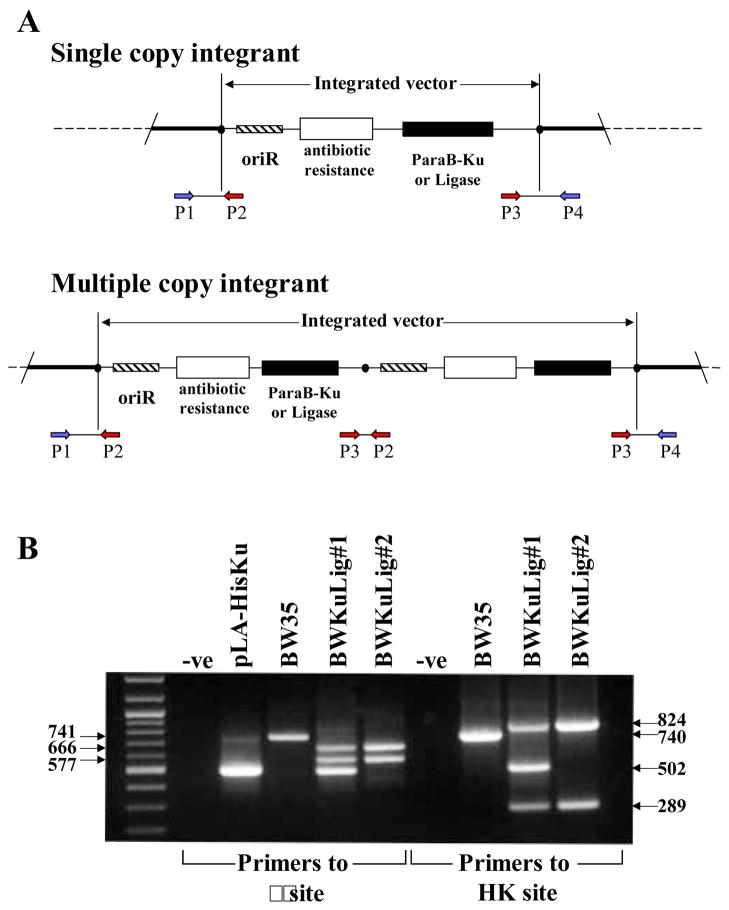
pLA-HisKu and pAH-HisLigase were individually integrated into the KL16 E. coli strain (BW35) to generate BWKu and BWLig, respectively (see Materials and Method section for details). pLA-HisKu was then integrated into BWLig to generate two strains BWKuLig#1 and BWKuLig#2. PCR, as described in Haldimann & Wanner (12), was used to screen colonies for integration of the plasmids. Four primers were used in the PCR reaction: P1-P4 (Figure 1A). P2 and P3 are primers common to the pLA-HisKu and pAH-HisLigase vectors, and generate a 502 bp band from pLA-HisKu and a 373 bp band from pAH-HisKu. Two types of P1 and P4 are required for screening since these primers are specific for the integration site: P1λ and P4λ for the reaction to check the λ site, and P1HK and P4HK to check the HK022 site. If there is not a vector integrated at the phage attachment site, one band (generated from P1 and P4) is seen on an agarose gel: 741 bp for the λ site and 740 bp for the HK022 site (Figure 1B, BW35). If a single copy integrates at the site two bands are seen: 577 bp and 666 bp for the λ site, and 289 bp and 824 bp for the HK022 site. BWKuLig#2 (Figure 1B) is an example of a strain which has a single copy of pLA-HisKu integrated at the λ site and a single copy of the pAH-HisLigase at the HK022 site. The 577 bp and 666 bp bands were seen for the BWKu strain when P1λ, P2, P3 and P4λ primers were used in the PCR, but only a 740 bp band was amplified when P1HK, P2, P3, and P4HK primers were used (data not shown). This indicated that the pLA-HisKu had integrated at the λ site, and that a vector was not integrated at the HK022 site. All strains were checked at both integration sites to confirm the correct integration pattern. Multiple copy integration results in 3 bands since an extra band is generated that corresponds to a product for P2 and P3 from the vector (Figure 1A). A bacterial strain (BWKuLig#1) was isolated during the integration of pLA-HisKu into BWLig that had multiple copies of pLA-HisKu integrated into the genome. This was seen from the amplification of the extra 502 bp band (Figure 1B). This 502 bp band was also generated during the PCR reaction to check the HK022 site, since the P2 and P3 primers are in both reactions (Figure 1B).
Figure 1. PCR was used to confirm the integration of the CRIM expression vectors into the specific phage attachment sites in the E. coli genome.
A small amount of bacteria and four primers are used in the PCR reaction: two primers (P1 and P4) are specific for the phage attachment site and two (P2 and P3) recognize CRIM vector sequences. In the schematic explanation of the PCR (A, adapted from 12), the black circle represents the integration site (λ or HK022) in the E. coli genome. An example of the PCR for strains BW35 and BWKuLig#1 and #2 is shown in B using primers specific for the λ site and the HK022 site. When a single copy of the vector integrates into the appropriate site, two products are amplified from primer sets P1-P2 and P3-P4 (B, see BWKuLig#2). When multiple copies are integrated an extra product is amplified from P2 and P3 (B, see BWKuLig#1). The size of this product is 502 bps for the Mt-Ku expression vector and 373 bps for the Mt-LigD expression vector. If integration has not occurred at the attachment site only one product is seen (B, BW35), which is generated by P1-P4 amplification of the region spanning the site.
The same procedure was performed using BW386 to generate A−KuLig, except the pLA-HisKu was integrated first and a strain was selected for the integration of pAH-HisLigase that contained multiple copies of the Mt-Ku expressing vector. A−KuLig is therefore deficient in RecA and contains multiple copies of both the Mt-Ku and the Mt-LigD expression vectors in the genome (data not shown).
To generate a strain deficient in RecB (B−KuLig), P1 transduction was used to transfer the recB268::Tn10 mutation from strain C876 into BWKuLig#1. The presence of the insertion at recB was checked by sensitivity to ultra-violet radiation and PCR was used to re-confirm the presence of the Mt-Ku and Mt-LigD expression plasmids. It is possible that the insertion at recB also disrupted RecD protein production since recB and recD exist in an operon, with recD downstream from recB. The promoter for the operon is upstream of recB and a weaker internal promoter exists for recD. Previously generated insertion mutations of recB (recB21) resulted in an 80% reduction of RecD protein (17).
3.2 Expression of Mt-Ku and Mt-LigD in E. coli
A monoclonal antibody to the histidine tag was used for western analysis to confirm expression of the proteins in cell-free extracts. It was necessary to examine expression of the Mt-Ku and Mt-LigD in bacteria in exponential phase to mimic the conditions used to prepare electrocompetent cells. This allowed the optimization of expression for the end-joining assay, which requires the proteins to be expressed at the time that the linear DNA enters the cell. An initial time course indicated that optimal induction with 0.2 % L-arabinose was achieved by addition of the arabinose for the last 1 h of growth for BWKuLig#1 and the last 2 h of growth for the RecA and RecB-deficient strains (data not shown). Cell-free extracts were prepared from all the strains after growth with and without arabinose for western analysis. As can be seen from Figure 2A, BWKu, BWLig and BWKuLig#2 only expressed proteins that cross-reacted with the anti-his tag antibody when arabinose was added to the culture. Two cross-reacting bands at sizes of ~35 kDa and ~90 kDa were detected, which corresponded to the expected sizes for the Mt-Ku and Mt-LigD, respectively. Only BWKu and BWKuLig expressed the ~35 kDa band, while the ~90kDa band was detected in BWLig and BwKuLig#2, as expected. It was evident that the expression of the Mt-Ku and Mt-LigD was not equal in strain BWKuLig#2; the ratio was ~1:3 for Ku:Ligase. However, expression of Ku:Ligase was ~1.5:1 in strains BWKuLig#1, A−KuLig and B−KuLig (Figure 2B), all of which contain multiple integrated copies of pLA-HisKu.
Figure 2. Expression of Mt-Ku and Mt-LigD requires growth in 0.2% L-arabinose.
Western analyses were performed on cell-free extracts prepared from exponential phase cultures grown in LB or LB supplemented with 0.2% arabinose (Ara+). Membranes were probed using a His•Tag monoclonal antibody that recognizes proteins containing five consecutive histidines. Expression was examined in wild-type strains that had integrated the expression constructs for Mt-Ku and/or Mt-LigD (A). Extracts from strains (wild-type, RecA− and RecB−) that had integrated both expression constructs were analyzed to compare the expression level of Mt-Ku and Mt-LigD following arabinose induction (B).
3.3 DNA End-joining activity of PacI-linearized DNA in wild-type E. coli
The end-joining assay uses a plasmid (pBestluc) that expresses firefly luciferase. Digestion with PacI linearizes the plasmid at the 3′ end of the coding region, leaving a 2-bp 3′ overhang (5′-AT-3′). If the double-strand break is accurately repaired, the plasmid is re-circularized resulting in a bacterial colony that is CarbR and that expresses firefly luciferase activity (Luc+). We have previously shown that alteration of sequence in this part of the protein frequently results in loss of luciferase activity (15). Hence inaccurate repair resulting in deletion or insertion of ≥1 bp at the PacI site would be expected to result in a frameshift mutation and hence a CarbR bacterial colony that does not express luciferase activity (Luc−). Expression of luciferase activity in bacteria can easily be determined by transferring the colonies to a nylon membrane and spraying the colonies with firefly luciferase substrate (15). Colonies emit light if active luciferase is expressed. This end-joining assay is therefore quick and easy to perform, and provides data not just about the ability of the bacteria to re-join linear DNA, but provides a measure of accurate and inaccurate repair.
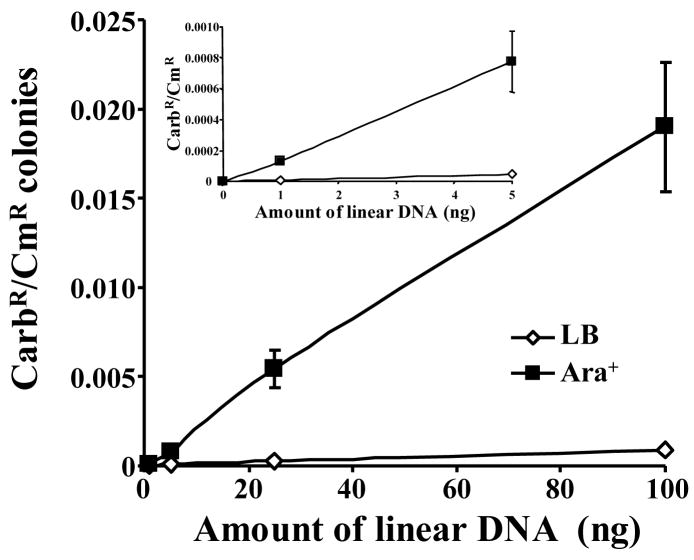
To determine whether the Mt-Ku and Mt-LigD could function in E. coli, BWKuLig#1 was grown in the presence or absence of arabinose and co-transformed with 0–100 ng of PacI-linearized pBestluc and 0.1 ng pACYC184. After 1.5 h at 37°C and 250 rpm, the culture was grown overnight on solid medium containing carbenicillin or chloramphenicol. The CarbR and the CmR colonies were then counted and the CarbR/CmR ratio calculated to normalize each sample for transformation efficiency. As can be seen from Figure 3, growth in arabinose (Ara+) significantly increased the amount of re-circularized pBestluc. Even with 1 ng of DNA, the average ratio increased from 8.25 × 10−6 to 1.3 × 10−4 by the addition of arabinose. This indicates that pBestluc was re-circularizied by Mt-Ku and Mt-LigD. It is also evident from Figure 3 that as the amount of transformed DNA increased, there was a linear increase in the CarbR/CmR ratio.
Figure 3. Re-circularization of plasmid DNA is linear over a range of 1–100ng in E. coli expressing Mt-Ku and Mt-LigD.
BWKuLig#1 bacteria were grown in LB or LB supplemented with 0.2% L-arabinose prior to the preparation of electrocompetent bacteria. Bacteria were then co-transformed with PacI-linearized pBestluc (1–100ng) and 0.1 ng pACYC184. The ratio of CarbR to CmR colonies was calculated for each transformation. At least three transformations were performed and the average ratio and standard deviation is shown. The inset graph shows the results for 1 and 5 ng DNA on an enlarged scale.
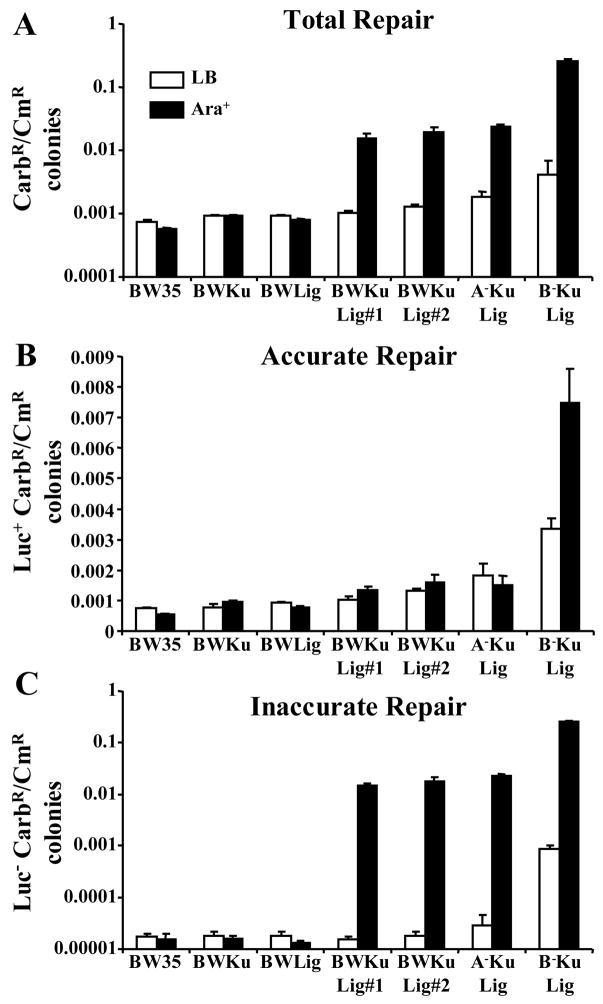
To confirm that repair of pBestluc required the expression of both Mt-Ku and Mt-LigD, the end-joining assay was repeated using BW35, BWKu, BWLig, BWKuLig#1 and BWKuLig#2 using 100 ng of PacI-digested DNA. From these bacterial strains only BWKuLig#1 and BWKuLig#2 (p<0.005, Figure 4A) showed a significant increase in the CarbR/CmR ratio when arabinose was added to the medium. However, there was not a significant difference between BWKuLig#1 and BWKuLig#2, even though BWKuLig#1 expresses a higher level of Mt-Ku than BWKuLig#2 (Figure 2B). The CarbR/CmR ratios for these two strains were also significantly higher than wild-type bacteria (p<0.001) under Ara+ conditions, where as the strains expressing only Mt-Ku (BWKu Ara+) or only Mt-LigD (BWLig Ara+) were similar (p>0.05) to wild-type (BW35 Ara+).
Figure 4. E. coli show enhanced RecA and RecB-independent inaccurate end-joining of linear DNA when both Mt-Ku and Mt-LigD are expressed.
Wild-type (BW35) bacteria and strains capable of expressing Mt-Ku (BWKu), Mt-LigD (BWLig) or Mt-Ku and Mt-LigD (BWKuLig#1 and #2, A−KuLig and B−KuLig) were grown in LB or LB supplemented with 0.2% L-arabinose prior to the preparation of electrocompetent bacteria. Bacteria were then co-transformed with 100 ng PacI-linearized pBestluc and 0.1 ng pACYC184. A ratio of CarbR to CmR colonies was calculated for each transformation for the total number of CarbR colonies (A – Total Repair), as well as the CarbR colonies expressing active luciferase (B – Accurate Repair) or inactive luciferase (C – Inaccurate Repair). At least seven transformations were performed and the average and standard error are shown.
To determine whether repair was accurate or inaccurate, the CarbR colonies were sprayed with luciferase substrate and a CarbR/CmR ratio calculated for the Luc+ and Luc− colonies. If accurate repair occurred, the CarbR/CmR for the Luc+ colonies was expected to increase under Ara+ conditions compared to when arabinose was absent (Ara−). It is evident from Figure 4B that there was very little difference between the Ara+ and Ara− conditions for BWKuLig#1 and #2 for Luc+ colonies. However, the proportion of CarbR colonies that were negative for luciferase activity increased by ~1000 times (Figure 4C) when Mt-Ku and Mt-LigD were expressed in these strains (Ara+). This indicates that DNA end-joining by the Mt-Ku and Mt-LigD proteins was predominantly inaccurate. As expected there was no significant increase in Luc+ or Luc− CarbR colonies for strains BW35, BWKu or BWLig when bacteria were grown in arabinose (Figures 4B and C), confirming that inaccurate repair required expression of both NHEJ proteins.
3.4 The products of inaccurate end-joining in wild-type E. coli
A further advantage to this end-joining assay is that colonies containing inaccurately rejoined repair products are easily identified and the product of repair can be analyzed for deletions and insertions. An initial PCR analysis was performed to determine whether sequence changes had occurred at the repair junction. From an experiment with BWKuLig#1 (Ara+), 104 Luc− colonies were examined and only 2 colonies generated the same size PCR product as wild-type pBestluc. We have previously found that a small percentage of colonies containing the correct sequence across this region do not express luciferase activity, and this can be attributed to a mutation in the promoter region. From the 102 colonies that contained plasmid with deletions, 99 had deletions ranging in size from ~ 20 bp to ~2 kb, and 3 contained deletions >2.6 kb. This latter deletion was defined by a lack of product from a PCR reaction using the Hind3 and Luc 5 primers. Plasmid DNA was isolated from 14 of the colonies and the sequence across the junction determined. In all but 1 of the repair products, the ligation had occurred at termini flanked by a region of microhomology (1–4 bp). Six of the sequences are shown in Table 1, and examination of the sequence before and after repair reveals that one of the microhomology sequences was deleted during repair. Deletions were not biased in a particular direction and the size of the microhomology region did not correlate with the size of the deletion.
Table 1.
Sequence at the repair junctions
| Type of bacteria |
Deletion size |
Sequence before repair | Sequence after repair |
Micro- homology region |
|---|---|---|---|---|
| BWKuLig #1 | 19 bp | TTGAAG-------AAGGATA | TTGAAGGATA | AAG |
| 48 bp | TGGGAC---------ACAAA | TGGGACAAA | AC | |
| 58 bp | GAAGA----------TGGCCC | GAAGATGGCCC | NONE | |
| 863 bp | GGAAGCG----------AGCGCTC | GGAAGCGCTC | AGCG | |
| 1251 bp | CAGATGC------------CCAACA | CAGATGCCAACA | C | |
| 2032 bp | GCTGAGC--------------GCAGAT | GCTGAGCAGAT | GC | |
| A−KuLig | 28 bp | TTGAAGT-------GTGGCCC | TTGAAGTGGCC | GT |
| 175 bp | GAAGCGAC----------CGACGCG | GAAGCGACGCG | CGAC | |
| 321 bp | ACAAGGATAT--AAGGATATCA | ACAAGGATATCA | AAGGATAT | |
| 484 bp | TAGTACC-----------CCCCCGCT | TAGTACCCCCGC | CC | |
| 868 bp | AAACGGAT-----------GGATATC | AAACGGATATC | GGAT | |
| 1368 bp | GAGCGGATA-------GGATATCA | GAGCGGATATCA | GGATA | |
| B−KuLig | 19 bp | CCGCTTGAAG------AAGGATA | CCGCTTGAAGGATA | AAG |
| 45bp | TTGAAG--------GGAAT | TTGAAGGAAT | G | |
| 569 bp | TTGAAGT------GTTATCA | TTGAAGTTATCA | GT | |
| 680 bp | GATTGAC--------TCTGCCA | GATTGACTCTGCCA | NONE | |
| 1637 bp | TAACAAT------------AATCAG | TAACAATCAG | AAT | |
| 1886 bp | GATAC--------------ACCCAGT | GATACCCAGT | AC |
After transformation of PacI-linearized DNA, the CarbR colonies were tested for firefly luciferase activity. PCR was used to test for plasmid containing deletions in Luc− colonies. Plasmid was then isolated and sequenced to identify the repair junction. Representative examples are shown. Microhomology sequences are in bold. The junction is underlined for samples that did not contain microhomology sequences.
3.5 DNA End-joining activity of PacI-linearized DNA in RecA-deficient E. coli
To eliminate the possibility that homologous recombination was interfering with the activities of Mt-Ku and Mt-LigD, a strain deficient in RecA that expressed the NHEJ proteins (A−KuLig) was examined for DNA end-joining activity using the same PacI-digested DNA as the wild-type. As can be seen from Figure 4, inaccurate DNA end-joining was not prevented by the lack of RecA protein.
PCR was performed on 115 Luc− colonies and all of these contained plasmids with deletions: 17 had deletions > 2.6 kb and 98 had deletions ranging in size ~ 10 bp to 2.3 kb. From these colonies, 14 were sequenced and six of the sequences are shown in Table 1. As found for BWKuLig#1, there was not a bias for the direction in which the deletions had occurred and the junction had been formed at a region of microhomology, eliminating one copy of the repeat sequence. The size of the microhomology region was from 1 to 6 bps. This analysis suggests that junctions made in the RecA-deficient bacteria tended to occur at sequences with a greater number of base pairs of microhomology compared to wild-type bacteria.
3.6 DNA End-joining activity of PacI-linearized DNA in RecB-deficient E. coli
Mt-LigD does possess a nuclease domain with 3′→5′ single stranded DNA exonuclease activity (9). However, the RecBCD nuclease is known to be involved in the degradation of foreign DNA entering the cell (for review of RecBCD see 18). It was therefore possible that RecBCD degraded the linear DNA prior to DNA end-joining by Mt-Ku and Mt-LigD. The RecB protein associated with RecC has DNA helicase activity but no nuclease activity (18), while RecD is essential for the nuclease activity (17). Mutations in either recB, recC or recD result in the loss of an ATP-dependent nuclease activity that is present in wild-type and recA mutant cells (19–22). Since plasmid DNA is unstable in recD mutants (23) and recB mutants do not have the RecBCD ATP-dependent nuclease activity, we chose to introduce a recB insertion mutation (recB268::Tn10) into BWKuLig#1 to test whether RecBCD was generating the deletions in the pBestluc repair products. The Chi sequence (5′ GCTGGTGG 3′) is known to cause the RecBCD protein to pause and to reduce the nuclease activity of the protein. The termini of deletions in the repair products may therefore have been expected to occur near this sequence. However, examination of the pBestluc sequence determined that this plasmid does not contain the Chi recognition sequence.
Disruption of recB did not prevent Mt-Ku and Mt-LigD from re-circularizing the DNA (Figure 4A, Ara+ condition). Again the repair was predominantly inaccurate: A comparison of the Ara+ and Ara− conditions showed that with arabinose there was a 287 times increase in the number of Luc− colonies. However, addition of arabinose also resulted in a significant increase (~2.2 times) in the number of Luc+ colonies (Figure 4B). This indicates that in the absence of RecB, the NHEJ proteins were able to perform a small amount of accurate repair.
By examining the Ara+/Ara− ratios the amount of repair is normalized for the background repair within the strain. For B−KuLig (Figure 4A) the ratio for total repair was 61.5, which was higher than BWKuLig#1 and 2 and A−KuLig where the ratios were 15 (wild-type strains) and 13 (RecA-deficient strain). Although it is possible that this increase in repair was due to fluctuations in protein expression, it may also be an indication that without RecB the linear DNA has a longer half-life and a greater chance of repair by Mt-Ku and Mt-LigD. This possible longer half-life and increased chance of repair was also seen to a smaller extent in the absence of arabinose. When the CarbR/CmR ratios were compared for all the strains for the Ara− condition (Figure 4A), it was found that there was a significant increase (2–4 times) in the total number of background colonies for the B−KuLig strain compared to BWKuLig#1 and #2, and A−KuLig. This was due to an increase in Luc+ and Luc− colonies (Figure 4B and 4C). Therefore in the absence of RecB, the bacteria were able to re-circularize a small amount of pBestluc even when the NHEJ proteins were not expressed.
PCR analysis was performed on 111 Luc− colonies that were generated from B−KuLig under Ara+ conditions. All of these colonies contained plasmids with deletions: 10 had deletions > 2.6 kb and 101 had deletions with the size ranging from ~10 bp to 2.2 kb. These results were therefore very similar to the PCR analysis of colonies from BWKuLig#1 and A−KuLig. Plasmid DNA was isolated from 19 colonies and sequenced across the repair junction. Six sequences are shown in Table 1. As found in the other bacterial strains, deletions were not biased to a particular direction and junctions occurred at regions of microhomology. The most frequently used size of homology was 2 bp. Only 1 sequence did not contain a region of microhomology at the junction (Table 1).
Since B−KuLig produced a significant number of Luc− colonies when electrocompetent bacteria were grown only in LB (Ara− condition), PCR analysis was performed on 34 of these background colonies. Approximately 44% of these colonies had deletions > 2.6 kb. Eleven samples were sequenced: Four samples contained deletions where the junction had a 1 or 2 bp microhomology region, 1 sample was found to contain no alteration in the sequence over the PacI region, 1 contained a deletion but no microhomology at the junction and 5 appeared to contain massive deletions/rearrangements that disrupted the carbenicillin resistance gene. Since the PCR and sequencing suggested that 5 contained rearrangements, we attempted to re-prepare the DNA for further analysis, but were unsuccessful. This suggested the plasmid was unstable. Only 1 sample from all the Ara+ samples was previously found to contain a plasmid similar to these 5, and this sample was from the B−KuLig strain. This suggests that this product was not joined by Mt-Ku and Mt-LigD.
3.7 DNA End-joining activity of ClaI-linearized DNA in wild-type E. coli
Results using PacI-digested DNA indicated that repair was inaccurate. In order to determine whether the type of DNA termini at the double strand break altered the fidelity of repair, the DNA end-joining assay was repeated using 100 ng of ClaI- digested pBestluc and BWKuLig#1. ClaI cleaves DNA to produce a 2-bp 5′ verhang with the sequence 5′ CG 3′ and therefore differs from PacI in that the overhang is 5′ instead of 3′ and the bases in the overhang have a greater hydrogen bonding capacity (i.e. CG compared to AT).
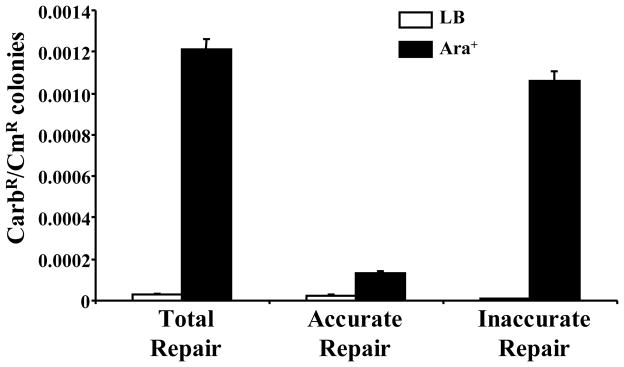
ClaI-digested DNA was re-circularized when Mt-Ku and Mt-LigD was induced by arabinose (Figure 5): The Ara+/Ara− ratio was 40 when the total number of CarbR colonies (Total Repair) was considered. Interestingly, when the colonies were checked for luciferase activity the CarbR colonies expressing active luciferase significantly increased when the cells were grown in arabinose (p< 0.0001). The majority of repair was however inaccurate: the Ara+/Ara− ratio was 5.2 for Luc+ colonies (Accurate repair) compared to 129 for Luc− colonies (Inaccurate repair).
Figure 5. DNA end-joining of ClaI-digested DNA by Mt-Ku and Mt-LigD in wild-type E. coli.
BWKuLig#1 was grown in LB or LB supplemented with 0.2% L-arabinose prior to the preparation of electrocompetent bacteria. Bacteria were co-transformed with 100 ng ClaI-linearized pBestluc and 0.1 ng pACYC184. A ratio of CarbR to CmR colonies was calculated for each transformation for the total number of CarbR colonies (Total Repair), as well as the CarbR colonies expressing active luciferase (Accurate Repair) or inactive luciferase (Inaccurate Repair). Nine transformations were performed and the average and standard error are shown.
PCR analysis of 103 Luc− colonies and XmnI digestion of isolated plasmid revealed that all 103 colonies contained pBestluc with deletions ranging in size from ~20bp to 2.3 kb. Two samples were sequenced across the junctions and both samples were joined at regions of microhomology (3 and 4 bp), with the deletion of one of the copies of the microhomology. The sequence of the junction for one sample suggested that the 3′ recessed end created by ClaI had been “filled in” to create a blunt end and the other terminus was deleted until a 4 bp microhomology sequence was located. Alignment and ligation then resulted in the deletion of one of the 4 bp regions of overlap.
4. Discussion
By integrating arabinose-inducible Mt-Ku and Mt-LigD expression vectors into the E. coli chromosome, we have generated unique prokaryote strains that can be switched from being unable to perform NHEJ to strains that are capable of NHEJ. This can be done simply by growth in arabinose. End-joining activity in E. coli required both Mt-Ku and Mt-LigD. This is in agreement with previous studies of the M. smegmatis NHEJ system (24; 25) and the expression studies of the M. tuberculosis NHEJ proteins in Saccharomyces cerevisiae (9). The M. tuberculosis end-joining activity in E. coli was not dependent on RecA. This implies that RecA is also not important for NHEJ in M. tuberculosis. In this respect the M. tuberculosis NHEJ system is similar to the M. smegmatis system, as a recA− M. smegmatis strain was capable of recircularizing plasmid with BamHI or EcoRI-EcoRV termini, albeit at a reduced frequency compared to the wild-type (25). The M. smegmatis study only examined plasmid re-circulariztion and not the accuracy of repair or the sequence of repair products. However, our work demonstrated that repair products generated in the absence of RecA were very similar to those generated in wild-type bacteria.
In E. coli, the Mt-NHEJ system predominantly repaired the PacI-digested DNA inaccurately, resulting in recircularized plasmid that contained deletions of 19 – 2300 bp. RecBCD nuclease is known to degrade invading foreign DNA in E. coli. It was therefore possible that the large deletions in the repair products were generated by RecBCD and not the end-trimming activities of Mt-LigD. To answer this, the DNA-end-joining assay was performed in a recB mutant expressing Mt-Ku and Mt-LigD. It was found that the majority of the repair was still inaccurate and the products still contained deletions. It is therefore unlikely that RecBCD generated the majority of the deletions in the repair products in the wild-type strain. It is possible that another exonuclease is interfering with the NHEJ proteins in E. coli. Future studies are planned with E. coli deficient in RecJ, which degrades single-stranded DNA in a 5′ to 3′ direction and has no known homologue in M. tuberculosis (11). In vitro studies do suggest, however, that PacI or ClaI termini would not be good substrates for RecJ since in vitro activity was low for substrates with 5′ single-stranded ends of 6 or less nucleotides (26). We hypothesize that the deletions identified in this work were predominantly made by Mt-LigD and not by other processing enzymes. Future work with this system will be aimed at answering this question and will include examining repair in mutants deficient in RecJ, the helicase UvrD and SbcCD, all of which could contribute to deletion formation.
Examination of the sequence of the repair products revealed that the junctions occurred at regions of microhomology (1–4 bp for wild-type bacteria and recB−), with one copy of the homology sequence deleted upon ligation. This type of junction is very similar to that found for the yeast (27; 28) and mammalian NHEJ systems (29), and it has been proposed that the DNA ends are brought in close proximity by protein-protein interactions and are then aligned prior to processing and ligation (30). It has recently been reported that the polymerase domain (PolDom) of M. tuberculosis and M. smegmatis LigD has potent template dislocation and realignment capacity (31), and these properties have been implicated in microhomology-mediated end-joining. Similar activities have also been shown for the eukaryote NHEJ polymerase, Polμ (32; 33). Our data supports the idea that Mt-LigD performs end-joining using microhomology sequences. Both Polμ and Polλ (a second eukaryote NHEJ polymerase) are also capable of bridging two 3′ protruding DNA ends, depending on the level of microhomology (34–36), and it has been proposed that PolDom may also have the capacity to align ends in the absence of significant base pairing (31). Our model E. coli system will prove useful in addressing questions concerning whether the Mt-NHEJ proteins can ligate DNA termini containing non-complementary sequences and whether the size of the microhomology region varies with the different types of termini.
When Mt-Ku and Mt-LigD were expressed in yeast the repair was found to be predominantly accurate (9), which is different from what we found in E. coli. The inaccurately repaired junctions in the yeast system contained insertions as well as deletions, but the deletions were small. The DNA breaks examined in the yeast studies contained 4-bp 3′ overhangs (5′ ATAA) generated from I-Sce I. PacI and ClaI-digested DNA used for this study contained complementary 2-bp overhangs with 3′ overhangs (5′ AT 3′) for PacI and 5′ overhangs (5′ CG 3′) for ClaI, and previous studies in M. smegmatis (24; 25) used linear DNA with 5′ overhangs or blunt termini. The M. smegmatis NHEJ system was also found to perform ~60% inaccurate repair resulting in junctions with small insertions, as well as deletions (8–1043 bp depending on the type of termini) at small regions of microhomology (24). In our studies the fidelity of repair was influenced by the type of DNA double strand break. The ClaI-digested DNA was repaired with greater accuracy than the PacI-digested DNA. We therefore hypothesize that the fidelity of repair is influenced by the type of complementary overhang (5′ or 3′) and/or by the strength of hydrogen bonding between the complementary overhangs (CG compared to AT). It is beyond the scope of this study to determine how different sizes of termini/hydrogen bonding capacity of cohesive DNA ends alter the fidelity of repair, although future studies are planned to try to answer this question. Double strand breaks introduced by DNA damaging agents, such as ionizing radiation, usually contain 5′ and 3′ termini that are not readily ligated. This model system will also prove useful in determining whether Mt-NHEJ can process such damaged termini or whether other prokaryote proteins are needed to complete the process.
This model system will prove useful for the study of how Mt-Ku and Mt-LigD interact with each other. It has the advantage that it can be switched on and off and does not contain a possible interfering NHEJ background end-joining system. By comparing the similarities and differences of this unique E. coli system with the M. smegmatis studies, a great deal can be learned about the mechanism of prokaryote NHEJ and whether there are E. coli proteins that can interfere/play a role in the Mt-NHEJ. Work is required using substrates with the same DNA termini to determine whether the products of repair are identical in the two organisms.
In summary, we have generated E. coli strains that can be induced to express the two M. tuberculosis NHEJ proteins: Mt-Ku and Mt-LigD. These proteins can perform DNA end-joining activity in E. coli and use regions of microhomology to align DNA termini. Repair is predominantly inaccurate, although the fidelity of repair varies depending on the type of cohesive double-strand break. The end-joining activity is not RecA or RecB-dependent. We suggest that this is also the scenario in M. tuberculosis, which expresses both RecA and RecB.
Future studies are planned to determine whether expression of the Mt-NHEJ proteins can enhance survival of E. coli after a DNA damaging challenge. It is possible that this inaccurate repair system may be detrimental to rapidly dividing cells, but beneficial to stationary phase cultures, as previously speculated by other groups (5).
Acknowledgments
This work was supported by the NIH grant CA 85693.
Abbreviations
- Mt
Mycobacterium tuberculosis
- NHEJ
non-homologous end-joining
- Mt-LigD
Mt-Ligase D
- oligonucleotide
oligodeoxyribonucleotide
- bp
base pair
- CarbR
carbenicillin resistant
- CmR
chloramphenicol resistant
- Luc+
expresses active firefly luciferase
- Luc−
expresses inactive firefly luciferase
- Ara+
grown in LB and 0.2% L-arabinose
- Ara−
grown in LB
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Weterings E, van Gent DC. The mechanism of non-homologous end-joining: a synopsis of synapsis. DNA Repair. 2004;3:1425–1435. doi: 10.1016/j.dnarep.2004.06.003. [DOI] [PubMed] [Google Scholar]
- 2.Doherty AJ, Jackson SP, Weller GR. Identification of bacterial homologues of the Ku DNA repair proteins. FEBS Letters. 2001;500:186–188. doi: 10.1016/s0014-5793(01)02589-3. [DOI] [PubMed] [Google Scholar]
- 3.Weller GR, Doherty AJ. A family of DNA repair ligases in bacteria? FEBS Letters. 2001;505:340–342. doi: 10.1016/s0014-5793(01)02831-9. [DOI] [PubMed] [Google Scholar]
- 4.Aravind L, Koonin EV. Prokaryotic homologs of the eukaryotic DNA-end-binding protein, Ku, novel domains in the Ku protein and prediction of a prokaryotic double-strand break repair system. Genome Research. 2001;11:1365–1374. doi: 10.1101/gr.181001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wilson TE, Topper LM, Palmbos PL. Non-homologous end-joining: bacteria join the chromosome breakdance. Trends in Biochemical Sciences. 2003;28:62–66. doi: 10.1016/S0968-0004(03)00005-7. [DOI] [PubMed] [Google Scholar]
- 6.Weller GR, Kysela B, Roy R, Tonkin LM, Scanlan E, Della M, Devine SK, Day JP, Wilkinson A, d’A di Fagagna F, Devine KM, Bowater RP, Jeggo PA, Jackson SP, Doherty AJ. Identification of a DNA nonhomologous end-joining complex in bacteria. Science. 2002;297:1686–1689. doi: 10.1126/science.1074584. [DOI] [PubMed] [Google Scholar]
- 7.Gong C, Martins A, Bongiorno P, Glickman M, Shuman S. Biochemical and genetic analysis of the four DNA ligases of mycobacteria. J Biol Chem. 2004;279:20594–20606. doi: 10.1074/jbc.M401841200. [DOI] [PubMed] [Google Scholar]
- 8.Bowater R, Doherty AJ. Making ends meet: Repairing breaks in bacterial DNA by non-homologous end-joining. PLoS Genetics. 2006;2:93–99. doi: 10.1371/journal.pgen.0020008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Della M, Palmbos PL, Tseng HM, Tonkin LM, Daley JM, Topper LM, Pitcher RS, Tomkinson AE, Wilson TE, Doherty AJ. Mycobacterial Ku and ligase proteins constitute a two-component NHEJ repair machine. Science. 2004;306:683–685. doi: 10.1126/science.1099824. [DOI] [PubMed] [Google Scholar]
- 10.Pitcher RS, Tonkin LM, Green AJ, Doherty AJ. Domain structure of a NHEJ DNA repair ligase from Mycobacterium tuberculosis. J Mol Biol. 2005;351:531–544. doi: 10.1016/j.jmb.2005.06.038. [DOI] [PubMed] [Google Scholar]
- 11.Mizrahi V, Andersen SJ. DNA repair in Mycobacterium tuberculosis. What have we learnt from the genome sequence? Molecular Microbiology. 1998;29:1331–1339. doi: 10.1046/j.1365-2958.1998.01038.x. [DOI] [PubMed] [Google Scholar]
- 12.Haldimann A, Wanner BL. Conditional-replication, integration, excision, and retrieval plasmid-host systems for gene structure-function studies of bacteria. Journal of Bacteriology. 2001;183:6384–6393. doi: 10.1128/JB.183.21.6384-6393.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sternberg NL, Maurer R. Bacteriophage-mediated generalized transduction. Methods Enzymol. 1991;204:18–43. doi: 10.1016/0076-6879(91)04004-8. [DOI] [PubMed] [Google Scholar]
- 14.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 15.Malyarchuk S, Youngblood R, Landry AM, Quillin E, Harrison L. The mutation frequency of 8-oxo-7,8 dihydroguanine (8-oxodG) situated in a multiply damaged site: Comparison of a single and two closely opposed 8-oxodG in Escherichia coli. DNA Repair. 2003;2:695–705. doi: 10.1016/s1568-7864(03)00040-5. [DOI] [PubMed] [Google Scholar]
- 16.Seidman CE, Struhl K, Sheen J, Jessen T. Introduction of plasmid DNA into cells. Current Protocols in Molecular Biology. 1997;137(supplement):1.8.4–1.8.5. doi: 10.1002/0471142727.mb0108s37. [DOI] [PubMed] [Google Scholar]
- 17.Amundsen SK, Taylor AF, Chaudhury AM, Smith GR. recD: The gene for an essential subunit of exonuclease V. PNAS USA. 1986;83:5558–5562. doi: 10.1073/pnas.83.15.5558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kowalczykowski SC, Dixon DA, Eggleston AK, Lauder SD, Rehrauer WM. Biochemistry of homologous recombination in Escherichia coli. Microbiological Reviews. 1994;58:401–465. doi: 10.1128/mr.58.3.401-465.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Emmerson PT. Recombination-deficient mutants of Escherichia coli K-12 that map between thyA and argA. Genetics. 1968;60:19–30. doi: 10.1093/genetics/60.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Oishi M. An ATP-dependent deoxyribonuclease from E. coli with a possible role in genetic recombination. PNAS USA. 1969;64:1292–1299. doi: 10.1073/pnas.64.4.1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goldmark PJ, Linn S. An endonuclease activity from Escherichia coli absent from certain rec− strains. PNAS USA. 1970;67:434–441. doi: 10.1073/pnas.67.1.434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chaudhury AM, Smith GR. A new class of Escherichia coli recBC mutants: Implications for the role of RecBC enzyme in homologous recombination. PNAS USA. 1984;81:7850–7854. doi: 10.1073/pnas.81.24.7850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Biek DP, Cohen SN. Identification and characterization of recD, a gene affecting plasmid maintenance and recombination in Escherichia coli. J Bacteriol. 1986;167:594–603. doi: 10.1128/jb.167.2.594-603.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gong C, Bongiorno P, Martins A, Stephanou NC, Zhu H, Shuman S, Glickman MS. Mechanisms of nonhomologous end-joining in mycobacteria: a low-fidelity repair system driven by Ku, ligase D and ligase C. Nature Structural & Molecular Biology. 2005;12:304–312. doi: 10.1038/nsmb915. [DOI] [PubMed] [Google Scholar]
- 25.Korycka-Machala M, Brzostek A, Rozalska S, Rumijowska-Galewicz A, Dziedzic R, Bowater R, Dziadek J. Distinct DNA repair pathways involving RecA and nonhomologous end joining in Mycobacterium smegmatis. FEMS Microbiol Lett. 2006;258:83–91. doi: 10.1111/j.1574-6968.2006.00199.x. [DOI] [PubMed] [Google Scholar]
- 26.Han ES, Cooper DL, Persky NS, Sutera VA, Jr, Whitaker R, Montello ML, Lovett ST. Rec J exonuclease: substrates, products and interaction with SSB. Nucleic Acids Res. 2006;34:1084–1091. doi: 10.1093/nar/gkj503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kramer KM, Brock JA, Bloom K, Moore JK, Haber JE. Two different types of double-strand breaks in Saccharomyces cerevisiae are repaired by similar RAD52-independent, non-homologous recombination events. Mol Cell Biol. 1994;14:1293–1301. doi: 10.1128/mcb.14.2.1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moore JK, Haber JE. Cell cycle and genetic requirements of two pathways of non-homologous end-joining repair of double strand breaks in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:2164–2173. doi: 10.1128/mcb.16.5.2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Roth DB, Wilson JH. Non-homologous recombination in mammalian cells: role for short sequence homologies in the joining reaction. Mol Cell Biol. 1986;6:4295–4304. doi: 10.1128/mcb.6.12.4295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hefferin ML, Tomkinson AE. Mechanisms of DNA double-strand break repair by non-homologous end joining. DNA Repair. 2005;4:639–648. doi: 10.1016/j.dnarep.2004.12.005. [DOI] [PubMed] [Google Scholar]
- 31.Pitcher RS, Brissett NC, Picher AJ, Andrade P, Juarez R, Thompson D, Fox GC, Blanco L, Doherty AJ. Structure and Function of a Mycobacterial NHEJ DNA repair polymerase. J Mol Biol. 2006 doi: 10.1016/j.jmb.2006.10.046. in press. [DOI] [PubMed] [Google Scholar]
- 32.Zhang Y, Wu X, Yuan F, Xie Z, Wang Z. Highly frequent frameshift DNA synthesis by human DNA polymerase mu. Mol Cell Biol. 2001;21:7995–8006. doi: 10.1128/MCB.21.23.7995-8006.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ruiz JF, Lucas D, Garcia-Palomero E, Saez AI, Gonzalez MA, Piris MA, Bernad A, Blanco L. Overexpression of human DNA polymerase mu (Pol mu) in a Burkitt’s lymphoma cell line affects the somatic hypermutation rate. Nucleic Acids Res. 2004;32:5861–5873. doi: 10.1093/nar/gkh929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ma Y, Lu H, Tippin B, Goodman MF, Shimazaki N, Koiwai O, Hsieh CL, Schwarz K, Lieber MR. A biochemically defined system for mammalian nonhomologous DNA end joining. Mol Cell. 2004;16:701–713. doi: 10.1016/j.molcel.2004.11.017. [DOI] [PubMed] [Google Scholar]
- 35.Nick McElhinny SA, Havener JM, Garcia-Diaz M, Juarez R, Bebenek K, Kee BL, Blanco L, Kunkel TA, Ramsden DA. A gradient of template dependence defines distinct biological roles for family X polymerases in nonhomologous end joining. Mol Cell. 2005;19:1–10. doi: 10.1016/j.molcel.2005.06.012. [DOI] [PubMed] [Google Scholar]
- 36.Daley JM, Laan RL, Suresh A, Wilson TE. DNA joint dependence of pol X family polymerase action in nonhomologous end joining. J Biol Chem. 2005;280:29030–2937. doi: 10.1074/jbc.M505277200. [DOI] [PubMed] [Google Scholar]