Structure
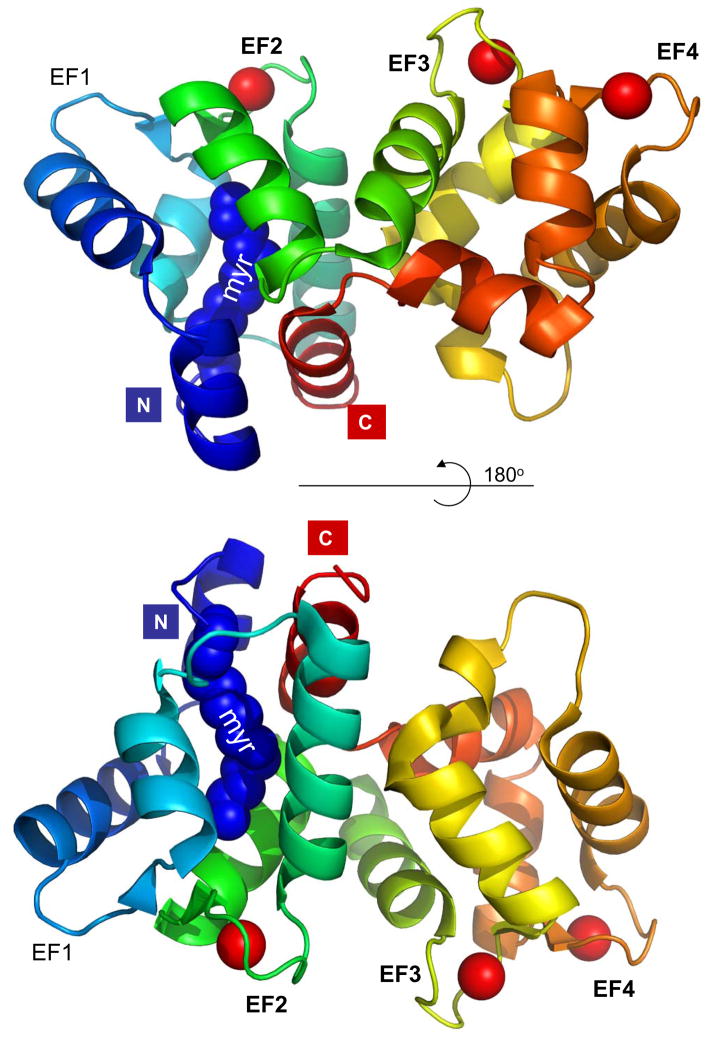
GCAPs are neuronal Ca2+-binding proteins of the calmodulin (CaM) superfamily expressed in photoreceptors (Palczewski et al., 1994). Whereas mammals express up to three GCAPs (GCAP1-3), as many as eight distinct GCAPs can be identified in the genomes of lower vertebrates. These proteins exhibit about 50% sequence similarity and identical domain structures; all are N-myristoylated or modified by other fatty acid groups. GCAPs are composed of four EF-hand motifs, two each in the N-terminal and C-terminal domains. Crystallization of chicken myrGCAP1 with Ca2+ bound reveals a compact structure (Fig. 1) (Stephen et al., 2007). In contrast to other neuronal Ca2+-binding proteins, the myristoyl group in GCAP1 is completely buried between the N- and C-terminal helices (green and blue in Fig. 1), and remains buried even after Ca2+ dissociation from the protein.
Figure 1.
Structure of Ca2+-bound, myristoylated GCAP1 (pdb 2R2I, generated in pymol). Helices are coded in rainbow colors; Ca2+ is represented by red balls. The EF-hand loops are identified by EF1–4. ‘myr’ denotes the N-terminal myristoyl group buried between the C-terminal (green) and the N-terminal helix (blue). The N- and C-terminal ends are indicated with colored boxes.
Hallmarks of GCAP structure are high affinity Ca2+-binding sites termed EF-hand motifs consisting of helix-loop-helix secondary elements. In the canonical EF-hand motif, the loop consists of a 12 amino acid-long sequence rich in acidic residues that provide oxygen groups for Ca2+ coordination. Residues adjacent to this loop are generally hydrophobic (I, L, Y, W), and the entire loop is flanked by α-helical structures. In GCAP1, EF-hand 1 (residues 17–42) and EF-hand 2 (residues 50–82) form the N-terminal domain, whereas EF-hand 3 (residues 87–118) and EF-hand 4 (residues 130–160) are contained in the C-terminal domain. EF hands 2–4 are fully functional, canonical EF-hand Ca2+-binding sites, whereas EF-hand 1 does not bind Ca2+ because the loop of this motif lacks the acidic side chains that provide oxygen for Ca2+ coordination. A short loop between EF-hand 2 and EF-hand 3 and an N-terminal helix fasten the N- and C-terminal domains together (Fig. 1) (Stephen et al., 2007). The conformational changes that take place when GCAP1 converts from a Ca2+ bound to Ca2+ free state have not yet been structurally defined.
Function
GCAP1 and GCAP2 are expressed in rod and cone cells and target membrane guanylate cyclases (GCs) located in retinal outer segment disk membranes (Baehr et al., 2007). In the dark-adapted photoreceptor, basal GC activity is balanced by the activity of phosphodiesterase 6 (PDE6) that adjusts the cytoplasmic [cGMP] to about 1–10 μM. In photoactivated photoreceptors, [Ca2+] decreases from about 500 nM (dark) to <50 nM and dissociates from GCAPs. The Ca2+-free GCAPs then activate GCs, increasing their catalytic activity about 10-fold. Provided that all of the major components of the phototransduction cascade (R*, G*, PDE*) are “silenced” by returning to their inactive dark-adapted states, activation of GC will restore [cGMP] to dark levels. It is assumed that the N-terminal region of GCAPs, including EF1, plays a key role in interacting with the target protein GC. GCAPs interact with an intracellular domain of GC, because deletion of the extracellular domain of GC has no effect on GCAP stimulation whereas deletion of the kinase-like domain diminishes the stimulation by GCAPs.
Disease Involvement
GCAP1 and GCAP2 genes (GUCA1A and GUCA1b, respectively) are organized in tail-to-tail arrays in mammals, presumably generated by gene duplication and inversion. In GCAP double knockout mouse photoreceptors, return to the dark-adapted state is delayed by a few hundred milliseconds, consistent with absence of the activator and a defect in GC stimulation. However, absence of GCAPs in photoreceptors does not cause recessive retinal degeneration. Transgenic GCAP1 can rescue the GCAP−/− phenotype in rods and cones, as tested by single rod cell recordings and electroretinography, even in the absence of GCAP2.
Pathogenic mutations of residues flanking EF-hand 3 and EF-hand 4, as well as several residues within the EF-hand 3 and the 4 loops of GCAP1 are associated with autosomal dominant cone or cone-rod dystrophy. These mutations are Y99C, N104K, I143NT, L151F, and E155G. Residue Y99 is located adjacent to the EF3-hand motif, and I143 is adjacent to the EF4 hand. Residue N104 is located in EF3, and E155G and L151F are situated in EF4 (Jiang et al., 2008). All these mutations disrupt co-ordination of Ca2+ to the mutant loop and decrease the Ca2+ sensitivity of GCAP1. As a result, mutant GCAPs are not fully inactivated in the dark, leading to persistent stimulation of GC1, elevated [cGMP] and [Ca2+] levels, and eventually to cell death.
Future Directions
The interface between GCAPs and guanylate cyclases is still poorly defined. Future experiments will attempt to crystallize the GC holoenzyme or its cytoplasmic domain with Ca2+-free/Ca2+-bound GCAP1 and GCAP2. Further, the presence of three GCAPs in human (two in mouse) retina and two guanylate cyclases is complicating our understanding of the GC/GCAP regulatory system. Generation of genetically engineered mice expressing single GCs and single GCAPs will likely shed light on the respective contributions of each GC and each GCAP to photoreceptor recovery, at least in mouse.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference List
- Baehr W, Karan S, Maeda T, Luo DG, Li S, Bronson JD, Watt CB, Yau KW, Frederick JM, Palczewski K. The function of guanylate cyclase 1 and guanylate cyclase 2 in rod and cone photoreceptors. J Biol Chem. 2007;282:8837–8847. doi: 10.1074/jbc.M610369200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L, Wheaton D, Bereta G, Zhang K, Palczewski K, Birch DG, Baehr W. A novel GCAP1(N104K) mutation in EF-hand 3 (EF3) linked to autosomal dominant cone dystrophy. Vision Res. 2008;48:2425–2432. doi: 10.1016/j.visres.2008.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palczewski K, Subbaraya I, Gorczyca WA, Helekar BS, Ruiz CC, Ohguro H, Huang J, Zhao X, Crabb JW, Johnson RS, Walsh KA, Gray-Keller MP, Detwiler PB, Baehr W. Molecular cloning and characterization of retinal photoreceptor guanylyl cyclase activating protein (GCAP) Neuron. 1994;13:395–404. doi: 10.1016/0896-6273(94)90355-7. [DOI] [PubMed] [Google Scholar]
- Stephen R, Bereta G, Golczak M, Palczewski K, Sousa MC. Stabilizing function for myristoyl group revealed by the crystal structure of a neuronal calcium sensor, guanylate cyclase-activating protein 1. Structure. 2007;15:1392–1402. doi: 10.1016/j.str.2007.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]