Abstract
The high incidence of resistance to DNA damaging chemotherapeutic drugs and severe side effects of chemotherapy have led to a search for biomarkers able to predict which patients are most likely to respond to therapy. ERCC1-XPF nuclease is required for nucleotide excision repair of helix-distorting DNA damage and the repair of DNA interstrand crosslinks. Thus it is essential for several pathways of repair of DNA damage by cisplatin and related drugs, which are widely used in treatment of non-small cell lung carcinoma and other late stage tumors. Consequently, there is tremendous interest in measuring ERCC1-XPF expression in tumor samples. Many immunohistochemistry studies have been performed, but the antibodies for ERCC1-XPF were not been rigorously tested for antigen specificity. Herein we survey a battery of antibodies raised against human ERCC1 or XPF for their specificity, using ERCC1-XPF deficient cells as a negative control. Antibodies were tested for the following applications: immunoblotting, immunoprecipitation from cell extracts, immunofluorescence detection in fixed cells, co-localization of ERCC1-XPF with UV radiation-induced DNA damage in fixed cells, and immunohistochemistry in paraffin-embedded samples. Although several commercially available antibodies are suitable for immunodetection of ERCC1-XPF in some applications, only a select subset is appropriate for detection of this repair complex in fixed specimens. The most commonly used antibody, 8F1, is not suitable for immunodetection in tissue. The results with validated antibodies reveal marked differences in ERCC1-XPF protein levels between samples and cell types.
Keywords: Nucleotide excision repair, interstrand crosslink repair, cisplatin, biomarker, chemotherapy
Introduction
It is important to identify molecular markers that help guide cancer treatment decisions to improve patient survival and quality of life in advanced stage disease. Non-small cell lung carcinoma (NSCLC), accounting for ~85% of all lung cancers in the United States, is a prime example. The overall five-year survival rate is only 17%, largely because NSCLC is rarely detected prior to dissemination and current methods of screening are ineffective (1, 2). Platinum-based chemotherapy can improve survival when used as adjuvant therapy after surgical resection of NSCLC (3, 4). For patients with stage IIIB or IV disease, platinum-based combination chemotherapy is the first line of treatment (5, 6). However, this is effective in only a subset of patients. Prediction of which patients are mostly likely to respond to platinum-based chemotherapy is therefore imperative.
Cisplatin [cis-diamminedichloroplatinum(II)] and related platinum-based drugs inhibit cell proliferation because they cause DNA damage, including adducts on single bases, intrastrand crosslinks between two bases, and interstrand crosslinks (ICLs) between DNA strands (7). The monoadducts and intrastrand crosslinks are repaired by nucleotide excision repair (NER), a DNA repair mechanism that excises helix-distorting lesions affecting one DNA strand. ICLs are repaired by mechanisms that remain poorly defined, but involve proteins from multiple DNA repair pathways including homologous recombination (8), Fanconi anemia (9, 10), translesion polymerases and NER (11). ERCC1-XPF is a structure-specific nuclease, that acts both in NER (12) and the repair of ICLs (8). Thus, ERCC1-XPF is uniquely important for protection against the cytotoxicity caused by cisplatin-induced DNA damage. If ERCC1-XPF levels vary between tumors, it would be an attractive target for use as a predictor of the efficacy of platinum-based chemotherapy.
XPF harbors the catalytic domain in the heterodimeric nuclease, (13) and ERCC1 is required for DNA binding (14). The levels of ERCC1 are reduced in XPF-deficient mammalian cells and vice versa, suggesting that the proteins stabilize one another in vivo (15). Due to the obligate nature of this partnership, it follows that cellular levels of the two proteins should be closely correlated and that either could potentially be a good biomarker of the response to platinum-based therapy.
There has been increasing interest in measuring the levels of ERCC1 in NSCLC specimens by immunohistochemistry (IHC) (16–19). ERCC1 expression, as measured by IHC, was reported to correlate with poor response to cisplatin chemotherapy and significantly decreased survival (17). On the other hand, it has also been reported that increased ERCC1 in tumours was associated with longer survival after surgical treatment of NSCLC (16, 19). Unfortunately, the antibody (8F1) used in almost all studies is not specific for ERCC1 (20). It recognizes human ERCC1, but also a second major nuclear antigen of unknown identity, and the antibody is unable to discriminate between cells expressing ERCC1-XPF and cells that do not (20).
To determine if ERCC1-XPF is a biomarker for prognosis and making therapeutic decisions in NSCLC or other types of cancer, the specificity of relevant antibodies must be critically evaluated. Here we examine a collection of antibodies raised against each subunit of the nuclease for their ability to specifically detect their respective antigens in several applications. This not only provides reliable tools for the detection of ERCC1 and XPF in clinical specimens, but also illustrates a strategy for the validation of other antibodies against other potential biomarkers that could stratify patients according to cancer risk, response to therapy and prognosis.
Material and methods
Cell lines, recombinant proteins and whole cell extract preparation
Primary human fibroblast cell lines C5RO (normal), XP51RO (XPF mutant) (21), 165TOR (ERCC1 mutant) (22) and XP25RO (XPA mutant) were cultured in Ham’s F-10 with 10% fetal bovine serum (FBS), antibiotics and non-essential amino acids. Transformed human fibroblasts XP2YO (XP-F) (23) were cultured in RPMI with 10% FBS, antibiotics and non-essential amino acids. CHO cell lines AA8 (wild-type) (24), UV47 (Xpf mutant) and 43-3B (Ercc1 mutant) (25) were cultured in D-MEM with 10% FBS, antibiotics and non-essential amino acids. Mouse embryonic stem (ES) cell lines IB10 (wild-type) and clone 49 (Ercc1−/−) (26) were cultured in a 1:1 mixture of DMEM and Buffalo rat liver (BRL) cell conditioned media with 10% FBS, antibiotics, non-essential amino acids, 0.1 mM 2-mercaptoethanol and leukemic inhibitory factor (1000 units/ml, Gibco).
Recombinant XPF with an N-terminal His tag and ERCC1 with a C-terminal His tag were expressed from a dicistronic construct in E. coli and purified, as previously described (27). For preparation of whole cell extracts (WCEs), cells were grown to 80–90% confluence in 10 cm dishes or 75 cm2 flasks, trypsinized and washed with PBS, and then lysed at 4°C for 30 min with NP-40 lysis buffer (1% NP-40, 10% Glycerol, 20 mM Tris, pH 7.4, 137 mM NaCl, 2 mM Na3V04 and protease inhibitor cocktail set III (Calbiochem).
Immunoblotting and immunoprecipitation
For immunoblotting, 50 µg of total protein from each WCE and 20 ng of recombinant HIS-tagged ERCC1 and XPF were boiled in 4× loading buffer (0.25 M Tris-HCl, pH 8.5, 8% SDS, 1.6 mM EDTA, 0.1 M DTT, 0.04% bromophenol blue and 40% glycerol), separated by SDS-PAGE (10% polyacrylamide gel), and transferred to a nitrocellulose membrane. Anti-ERCC1 and anti-XPF antibodies (listed in Supplemental Table 1) were tested for their ability to specifically detect the respective proteins.
For immunoprecipitation, WCEs of C5RO cells were pre-cleared for 30 min by incubation with protein A+G agarose beads (Calbiochem) and incubated at 4°C with 10 µl of each of the primary antibodies and 25 µl protein A+G beads with continuous mixing for 12 hr. The beads were collected by centrifugation at maximum speed (15 min), washed three times each with the NP-40 lysis buffer and PBS, boiled for 10 min in protein loading buffer, and eluted proteins separated by SDS-PAGE. Immunoprecipitated XPF was detected with anti-XPF antibody (Ab-1, Neomarkers, 1:1000) and AP conjugated goat anti-mouse secondary antibody (1:7500, Promega).
Immunofluorescence and UV-C-induced local damage
C5RO and XP2YO cells were plated at 75–80% confluence and labeled with 0.6 µm and 3 µm diameter latex beads (Sigma-Aldrich) respectively, as described (28, 29). After 24 hr, cells were washed three times with PBS, trypsinized and co-plated on coverslips. Twelve hr later, the cells were fixed with 2% paraformaldehyde (ICN Biomedicals) at 37°C for 15 min. Triton-X-100 (0.2% in PBS) was used to permeabilize the cells and 5% BSA in PBS was used for blocking. Primary antibodies were used as described in Supplemental Table 2. Goat anti-mouse IgG (1:1000), chicken anti-rabbit IgG (1:1000), goat anti-mouse IgG2b (1:500) or goat anti-mouse IgG1 (1:500) secondary antibodies conjugated either with Alexa fluor 488 or Alexa fluor 594 (Invitrogen) were used for visualization.
For co-localization of ERCC1-XPF with UVC radiation-induced cyclobutane pyrimidine dimers (CPD), experiments were performed as described (30, 31) with minor modifications. C5RO cells grown on glass coverslips were irradiated with 60 J/m2 UV through Isopore™ membrane filters (Millipore) with 8 µm pores to induce subnuclear domains of DNA damage. The cultures were then incubated in medium for 45 min to allow initiation of DNA repair, and then fixed with 2% paraformaldehyde with 0.15% Triton-×-100 for 15 min on ice. Samples were blocked with 5% BSA in PBS for 20 min at RT and incubated with anti-ERCC1 or anti-XPF antibodies at the same dilution as for immunofluorescence (Supplemental Table 2) for 90 min at RT. Secondary antibodies were used as described above. This was followed by fixation with 2% paraformaldehyde for 10 min, denaturation with 2N HCl for 5 sec and blocking with 5% BSA in PBS for 20 min, all at RT. Cells were stained secondarily for CPDs using mouse anti-thymine dimer antibody (1:200, Kamiya Biomedical) for 90 min at RT and either Alexa fluor 488 or Alexa fluor 594 conjugated goat anti-mouse secondary antibodies (1:500, Invitrogen).
Immunohistochemistry
C5RO, HeLa, XP42RO, XP51RO, 165TOR and XP2YO cells were grown to 75–80% confluence, fixed at room temperature with 10% neutral-buffered formalin for 15 min, and then collected by scraping. Cells were stored in neutral-buffered formalin at 4°C for at least 3 hr, then pelleted by centrifugation (1,200 rpm, 5 min) and washed twice with PBS. Cells were resuspended in 500 µl of 80% ethanol, transferred to Eppendorf tubes containing 300 µl of solidified 1% low melting point agarose in PBS, and re-pelleted. The ethanol was aspirated and the bottoms of the tubes cut off. The agarose plugs containing the cells were pushed and molded into the caps of Eppendorf tubes and snap frozen on dry ice. The solidified pellets were extruded from the caps and paraffin embedded according to standard methods for tissue.
For immunohistochemical staining of the cell plug and paraffin embedded tissue sections, antigen retrieval was carried out at pH 6 (Dako Target retrieval solution, Dako) at 95°C for 20 min. Samples were blocked with 20% swine serum for 30 min and then incubated with either FL297 (anti-ERCC1, rabbit polyclonal, 1:250) or 3F2 (anti-XPF, mouse monoclonal, 1:1000) for 60 min at room temperature. Primary antibody signal was then detected using biotinylated anti-rabbit or anti-mouse secondary antibody for 30 min and DAB+ for 5 min at room temperature (Vectastain ABC kit; Vector Laboratories). Haematoxylin was used for counterstaining.
Results
A panel of antibodies against human ERCC1 and XPF, including commercially available reagents and those from our laboratory (Supplemental Table I), were tested for their specificity in immunoblotting, immunoprecipitation, immunofluorescence and IHC.
Immunoblotting and Immunoprecipitation
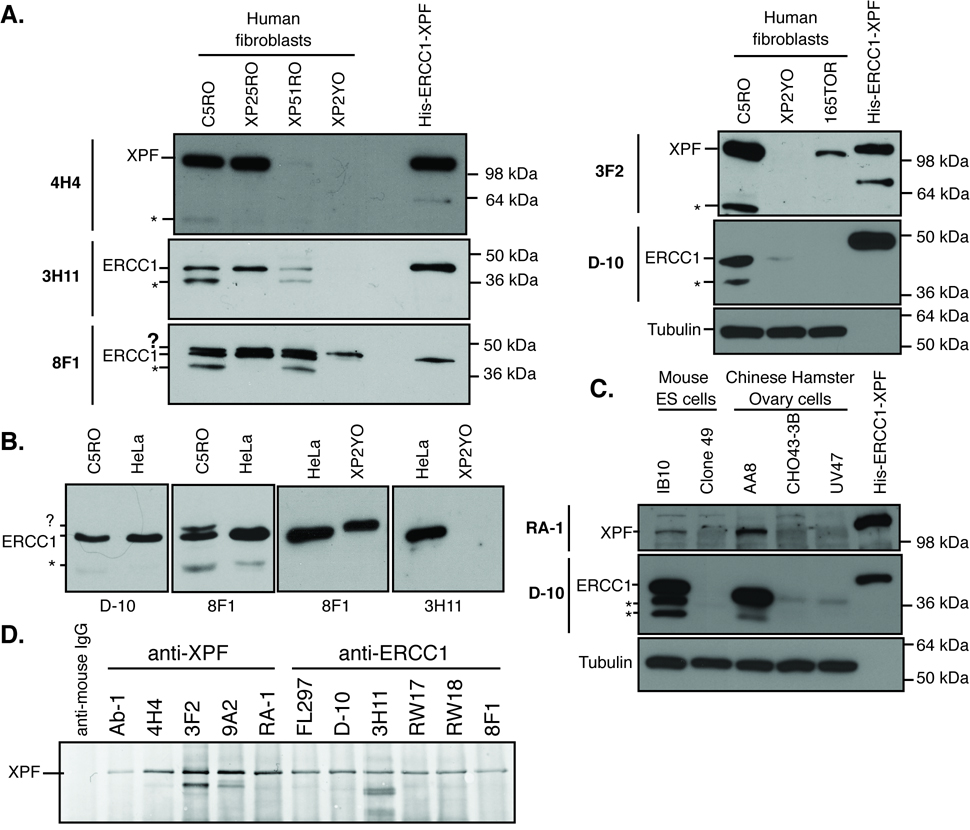
Antibodies were first screened for their specificity for ERCC1 or XPF by immunoblotting. Normal and ERCC1-XPF deficient human fibroblasts were lysed and immunoblotted along with recombinant His-tagged ERCC1 and XPF proteins (Fig. 1A). Antibodies were categorized as specific for their target protein if they: 1) detected a band of the appropriate molecular weight (~116 kDa for XPF and 37 kDa for ERCC1) in normal cells (C5RO) and in XPA mutant cells (XP25RO), 2) detected the recombinant protein (slightly retarded in its migration due to the HIS-tag), 3) showed significantly reduced or no signal in ERCC1 (165TOR) and XPF (XP2YO and XP51RO) mutant cell lines, and 4) did not detect additional bands of the incorrect molecular weight in both normal and mutant cells (results summarized in Supplemental Table 1). All of the antibodies screened were specific for ERCC1 or XPF on immunoblotting (e.g., 3H11 and D-10 for ERCC1; 4H4 and 3F2 for XPF in Fig. 1A) except for 8F1, which detected ERCC1 and an additional, spurious band of approximately 45 kDa. The spurious band is of equal intensity in normal and ERCC1-XPF deficient cells (Fig. 1A, bottom, left panel and (20)). Importantly, the spurious band picked-up by 8F1 is detected in extracts from fibroblast cell lines, but not HeLa cell extracts (Fig. 1B, indicated with a “?”), which were used previously as a negative control for immunodetection of ERCC1 (32). All of the antibodies also detected degradation products of XPF or ERCC1 in cell lines with abundant ERCC1-XPF expression (Fig. 1 A–C, bands marked by “*”). The optimal dilution of each antibody for immunoblotting was determined (Supplemental Table 2).
Figure 1.
Immunodetection of ERCC1-XPF in whole cell extracts. (A) Examples of how each of the test antibodies was screened for specificity for ERCC1 or XPF by immunoblot. WCEs were prepared from normal (C5RO), XPA mutant (XP25RO), XPF mutants (XP51RO and XP2YO) or ERCC1 mutant (165TOR) human fibroblasts. Proteins were separated by SDS-PAGE and identified on the blot by loading recombinant His-tagged ERCC1-XPF. Blots were immunostained with each of the test antibodies (e.g., anti-XPF: 4H4 and 3F2, and anti-ERCC1: 3H11, D-10 and 8F1). Tubulin was used as a loading control. An asterisk (*) is used to denote degradation products of ERCC1 or XPF detected by all antibodies and a question mark (?) is used to denote an unidentified spurious band detected only by the 8F1 antibody. (B) 8F1, but not D-10 or 3H11, detects a band that migrates more slowly than ERCC1 (denoted ?). This antigen is found in fibroblasts (C5RO and XP2YO), but not HeLa cells. (C) Two examples of screening the test antibodies for their ability to detect ERCC1 or XPF in mouse or hamster cell extracts. Lysates were prepared from wild-type (IB10) and Ercc1−/− mouse embryonic stem cells (clone 49) as well as wild-type (AA8), Ercc1 mutant (43-3B) and Xpf mutant (UV47) Chinese hamster ovary cells. These were run on SDS-PAGE along with recombinant His-tagged ERCC1-XPF. Blots were immunostained with each of the test antibodies (e.g., anti-XPF RA-1 and anti-ERCC1 D-10). Tubulin was used as a loading control. (D) Immunoprecipitation of ERCC1-XPF from WCEs from normal human fibroblasts with each of the test antibodies. Precipitates were separated by SDS-PAGE, blotted and immunostained with antibody Ab-1 to detect XPF protein.
To further test the utility of these antibodies, WCE from wild-type and ERCC1-XPF deficient mouse embryonic stem cells (IB10 and clone 49, respectively) and Chinese hamster ovary cells (AA8, CHO43-3B [Ercc1 mutant] and UV47 [Xpf mutant]) were also immunoblotted (Fig. 1C). In general, cross-reactivity for mouse and hamster proteins was poor (Supplemental Table I). The only commercially available antibody that detected rodent ERCC1 was D-10 (Fig. 1C).
Each of the ERCC1 and XPF antibodies were screened for their ability to immunoprecipitate their respective antigen from WCEs of normal cells (Fig. 1D). The precipitates were all probed for XPF using antibody Ab-1. XPF was detected in all samples precipitated with antibodies against XPF, as well as antibodies against its obligate binding partner ERCC1, as expected (20). Immunoblotting for XPF showed that all antibodies tested were able to immunoprecipitate the ERCC1-XPF nuclease, indicating that each is appropriate for this application (Supplemental Table 1).
Immunofluorescence
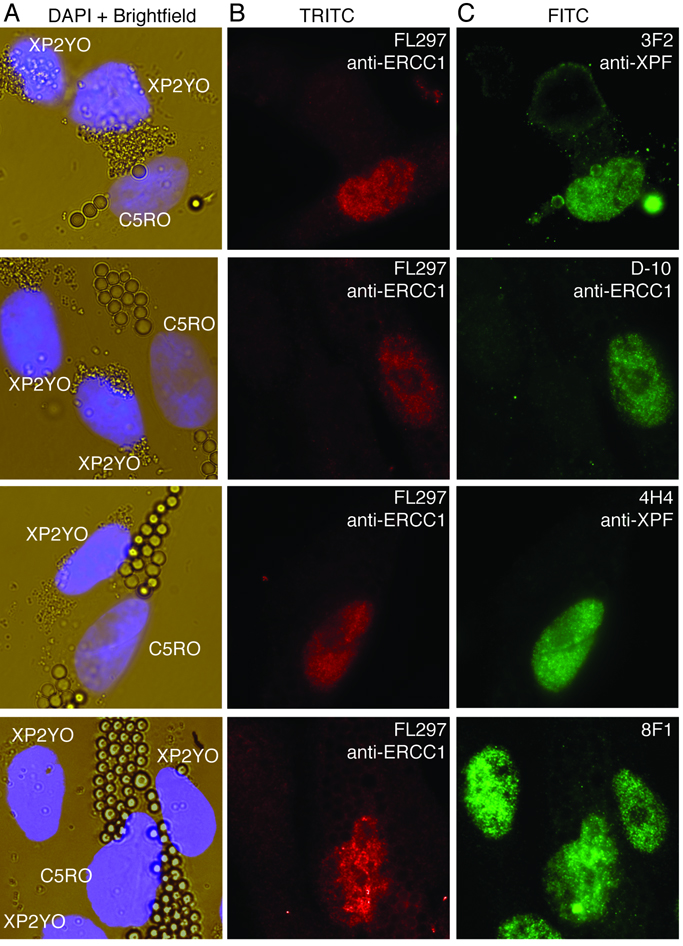
Antibodies that recognize denatured protein in immunoblots do not always detect their antigen in cells or tissues fixed with formaldehyde. Thus, each of the ERCC1 and XPF antibodies were screened for their ability to detect their respective antigen by immunofluorescence (IF) and immunohistochemistry (IHC). For IF, normal (C5RO) and XPF mutant human fibroblasts (XP2YO) were labeled with large (3 µm) and small (0.6 µm) latex beads, respectively, then co-cultured on glass coverslips to create a sample with an internal negative control (XP2YO cells). The coverslips were simultaneously immunostained with two antibodies (Fig. 2): the test antibody against ERCC1 or XPF, and FL297 an antibody against ERCC1, which we previously established specifically recognizes ERCC1-XPF positive cells in immunofluorescence (20). For the test antibody to be appropriate for IF application, it must stain the nuclei of normal cells only (those with large beads) and the same nuclei as FL297. Anti-XPF antibodies Ab-1, 4H4 and 3F2 and anti-ERCC1 antibody D-10 showed nuclear staining in normal cells (C5RO) and no staining in XPF mutant cells (XP2YO). Antibody 8F1 stained the nuclei of both cell types equally and therefore was unable to distinguish between normal and ERCC1-XPF deficient cells (Fig. 2, bottom panels and (20)). These data indicate that there are several commercially available antibodies (ERCC1: FL297 and D-10; XPF: Ab-1) appropriate for detecting ERCC1-XPF in human cells. Potential applications include fine needle aspirates and tumour biopsies.
Figure 2.
Immunofluorescence detection of ERCC1-XPF in cells. The cytoplasm of normal (C5RO) and ERCC1-XPF deficient XPF mutant human fibroblasts (XP2YO) was labeled with large and small latex beads, respectively. Then the cells were co-cultured on glass coverslips, fixed and stained. (A) Brightfield images reveal both cell types in a single field. The nuclei are stained with DAPI. (B) Immunostaining with antibody FL297 against ERCC1 distinguishes between the normal and ERCC1-XPF deficient cells. Nuclei of C5RO, but not XP2YO cells stain positively (red, Alexa-594). (C) Co-immunostaining with each of the test antibodies (3F2, D-10, 4H4 and 8F1; secondary antibody Alexa 488, in green) reveals which are specific for ERCC1-XPF. Only those nuclei that stain positively with FL-297 (red nuclei in B) should show green signal.
UV-C induced local damage
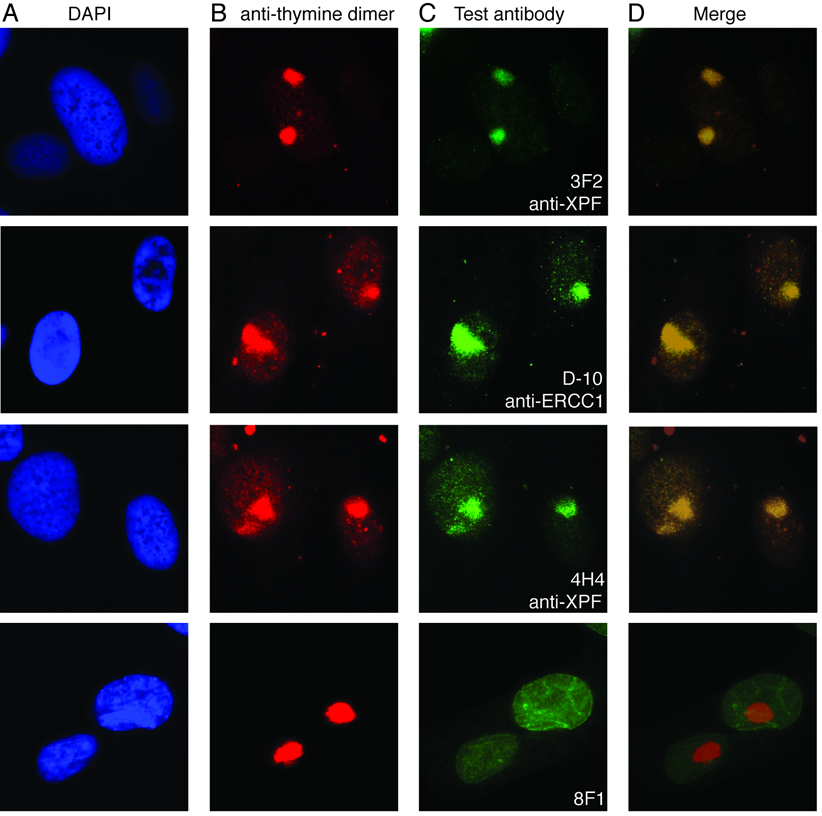
When cells are irradiated with UV-C through a polypropylene micropore filter, DNA damage is induced in regions of the nucleus corresponding to the filter pores (30). The sites of DNA damage can be identified with an antibody against UV radiation-induced cyclopyrimidine dimers (CPD) (30). ERCC1-XPF localizes to these sites of damage during NER and thus co-localizes with CPDs by immunofluorescence (30). Co-immunostaining of UV-C irradiated fibroblasts with anti-CPD antibody and test antibodies against ERCC1 or XPF revealed that the antibodies which discriminated between normal and ERCC1-XPF-deficient cells in IF also detected functional ERCC1-XPF in this local damage assay (Fig. 3). Antibody 8F1, which was unable to discriminate between normal and ERCC1-XPF deficient cells in IF, did not yield a signal that co-localized with CPDs (Fig. 3, bottom row). These results reinforce the IF data and indicate that several commercially available antibodies can be used to specifically detect ERCC1-XPF in fixed cells.
Figure 3.
Immunofluorescence detection of ERCC1-XPF at sites of UV-C induced DNA damage. C5RO fibroblasts were irradiated with UV-C through a filter to induce sub-nuclear domains of DNA damage. The cells were stained with (A) DAPI to identify the nuclei; (B) anti-CPD to identify the sites of DNA damage and (C) each of the test antibodies against ERCC1 or XPF. The merged images (D) reveal which antibodies give a signal that co-localizes with UV-induced DNA damage, indicating their specificity for ERCC1-XPF DNA repair endonuclease.
Immunohistochemistry
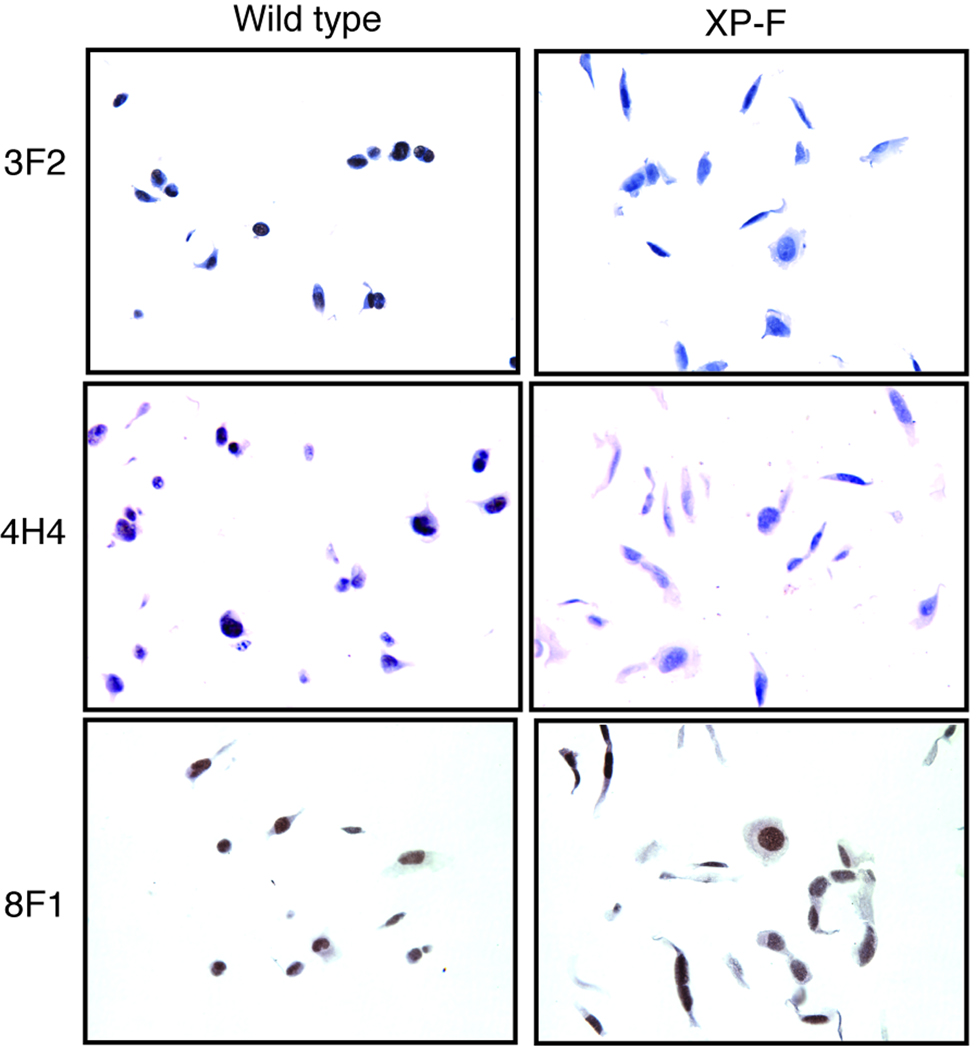
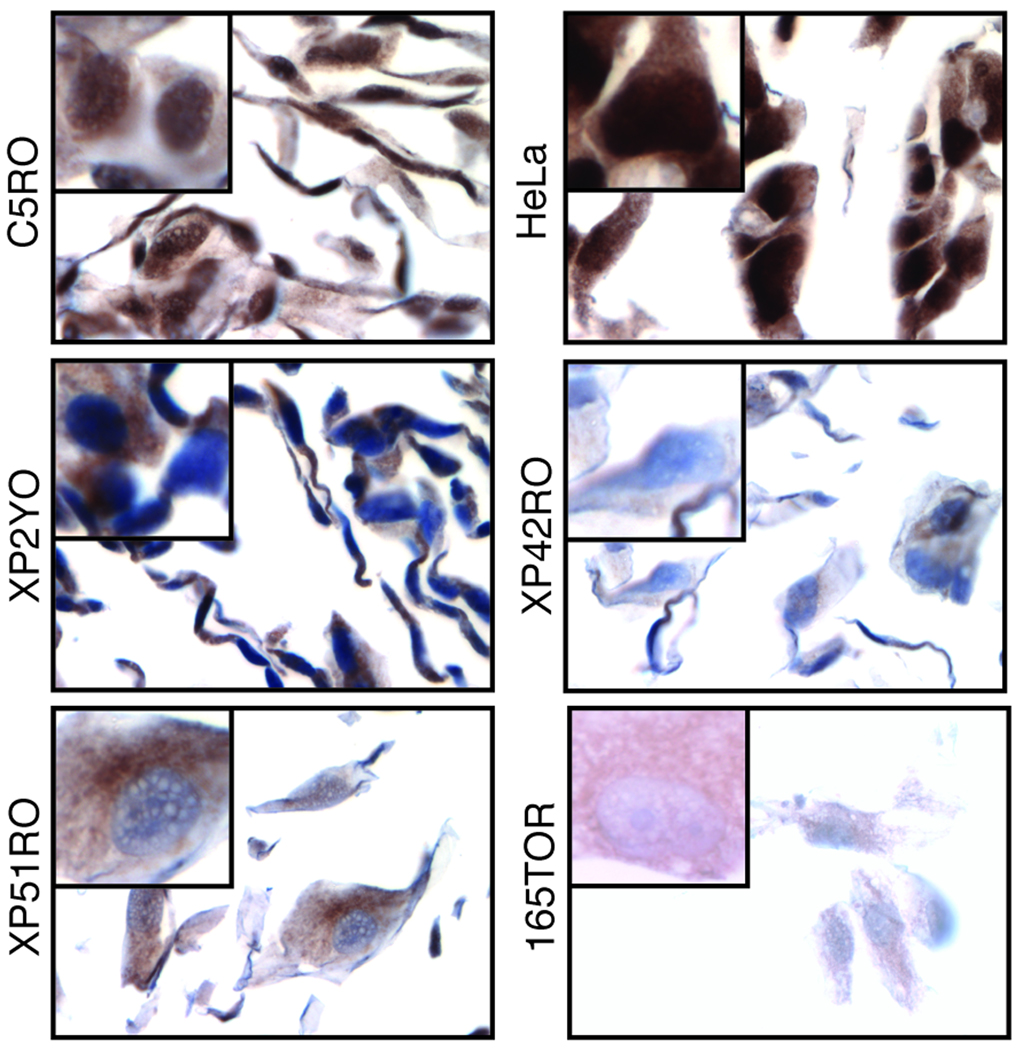
The most commonly used approach for measuring protein in tumors in clinical studies is IHC. However, for many essential proteins that are of interest as potential biomarkers in cancer prognosis, such as ERCC1 and XPF, true negative controls are not available. To circumvent this problem, we created paraffin-embedded blocks of normal and ERCC1-XPF deficient cell pellets for use as positive and negative controls to screen the test antibodies for their specificity in IHC. Normal and XPF mutant human fibroblasts were fixed in paraformaldehyde, pelleted in agarose, embedded in paraffin, sectioned and immunostained, according to the method used for solid tumors. Antibody 3F2, against XPF, stained the nuclei of paraffin-embedded normal but not ERCC1-XPF deficient cells by IHC (Fig. 4, first row). Similarly, antibody 4H4 against XPF (Fig. 4, second row) was able to discriminate between normal and ERCC1-XPF deficient cells, indicating that both of these antibodies are appropriate for detection of XPF by IHC (Supplemental Table 1). In contrast, antibody 8F1 against ERCC1 stained the nuclei of normal and ERCC1-XPF deficient cells equally (Fig. 4, third row), demonstrating its inability to discriminate between ERCC1 positive and negative specimens by IHC, as previously observed for other applications (20). Like 8F1, the other test antibodies were unable to discriminate between normal and ERCC1-XPF deficient cells by IHC (data not shown), with the exception of FL297. Antibody FL297 against ERCC1 was tested on six cell lines: two ERCC1-XPF proficient (C5RO and HeLa) and four ERCC1-XPF deficient (XP2YO, XP42RO, XP51RO and 165TOR). FL297 stained nuclei of both repair proficient cell lines (Fig. 5, C5RO and HeLa). All ERCC1-XPF deficient cells (Fig. 5, XP2YO, XP42RO, XP51RO and 165TOR) did not have nuclear staining, although there was a variable level of cytoplasmic staining in all cell lines. ERCC1-XPF functions exclusively as a nuclear complex. Thus the absence of nuclear staining indicates a negative result on IHC, as expected for these ERCC1-XPF deficient cell lines. HeLa cells showed a more intense nuclear staining than C5RO cells. This correlates with the relative expression of ERCC1 in these cell lines, indicating that FL297 can distinguish between different levels of ERCC1 on IHC. Therefore FL297 (anti-ERCC1), 4H4 (anti-XPF) and 3F2 (anti-XPF) are the only antibodies against ERCC1-XPF demonstrated to specifically detect this DNA repair nuclease by IHC.
Figure 4.
Immunohistochemical detection of ERCC1-XPF. Normal or ERCC1-XPF deficient human fibroblasts (XP2YO) were fixed in paraformaldehyde, pelleted in agarose, paraffin embedded, sectioned and stained as positive and negative controls, respectively, for IHC with antibodies against XPF (3F2 and 4H4) and ERCC1 (8F1). Notably, 8F1 was unable to discriminate between the normal and ERCC1-XPF deficient fibroblasts.
Figure 5.
Immunohistochemical detection of ERCC1 by antibody FL297. Six human cell lines were paraffin-embedded, sectioned and stained with FL297. Two were ERCC1-XPF proficient: C5RO and HeLa, and four were ERCC1-XPF deficient: XP2YO, XP42RO, XP51RO and 165TOR.
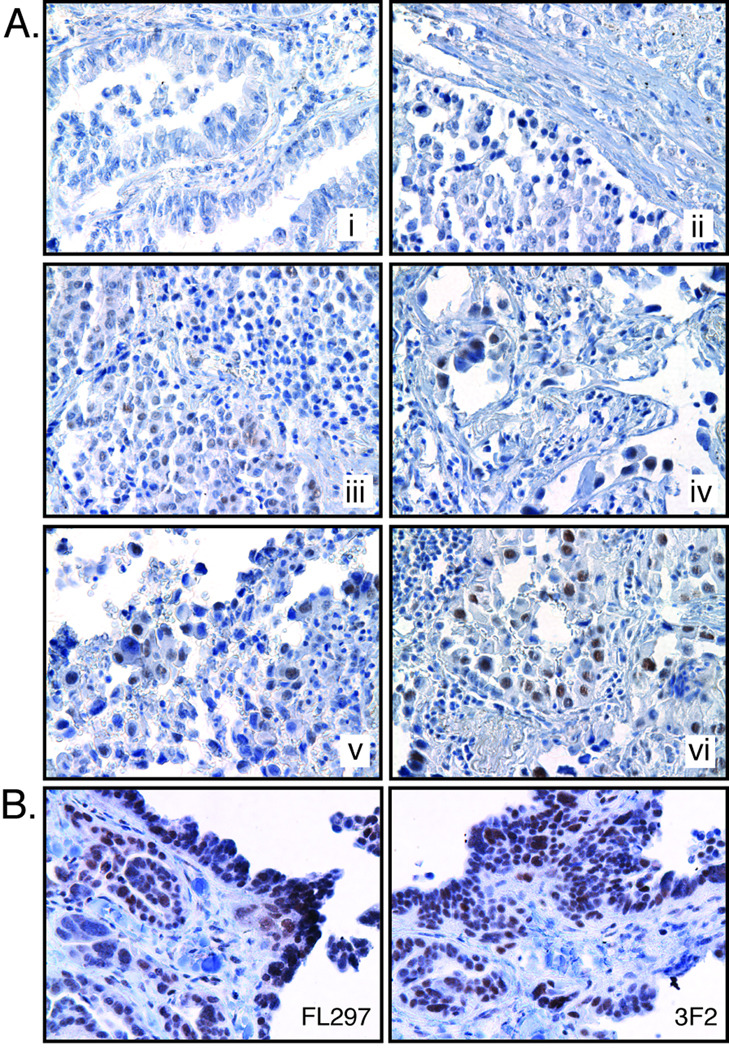
Lung tumor sections stained with antibody 3F2 display highly variable nuclear staining in different NSCLC samples (Fig. 6), suggesting that expression of XPF may vary in tumors. Sections from the same NSCLC tumor were immunostained with FL297 (anti-ERCC1) and 3F2 (anti-XPF) to determine if both proteins are detected in the same cells as expected (Fig. 6B). Both antibodies stained the same cell types with approximately the same intensity, confirming that ERCC1-XPF are co-expressed and both FL297 and 3F2 antibodies are appropriate for immunodetection of this repair complex in clinical specimens.
Figure 6.
Immunohistochemical staining of lung tumors for ERCC1-XPF. (A) Lung tumor sections were immunostained with 3F2. Representative sections have been arranged in increasing order of staining intensity (i–vi) showing the range of staining variability. (B) Cross-sections of the same tumor stained alternately with FL297 (anti-ERCC1) and 3F2 (ant-XPF) showing similarities in staining patterns.
Discussion
For several types of common cancers, e.g., lung cancer, patient survival rates have not improved in the last decade (33). Response to therapy is highly variable, and in some cases, e.g., ovarian cancer, does not improve overall survival (33–35). This is particularly true for platinum-based therapies to which patients frequently become refractory (36). Therefore, a major goal in oncology is to define biomarkers that can stratify cancer patients according to their likelihood of responding to chemotherapy. A highly active current focus of this effort is to measure expression of ERCC1 in tumors as a potential biomarker of DNA repair and therefore, resistance to genotoxic therapeutic agents (37–39). To infer conclusions about the mechanism of therapy failure, it is imperative to rigorously standardize methods to accurately measure biomarker expression.
Immunohistochemistry (IHC) is an extremely valuable method for measuring tumor biomarkers. Unlike other immunological methods, IHC is applied to fixed specimens, which are the most abundantly available from surgical resections of tumors, allowing the large-scale screens necessary to validate the biological significance of novel biomarkers. Small amounts of tumor tissue, such as those obtained by needle biopsy, are sufficient for a semi-quantitative measurement of the antigen of interest. IHC is therefore used in decisions regarding diagnosis, prognosis and therapy of malignancies. However, there are multiple variables in the processing of samples in IHC and data analysis that need to be addressed prior to the widespread use of IHC as a quantitative immunoassay (40). The staining intensity is affected significantly by the choice of fixative, the time of fixation (41), the extent of deparaffinization (42), thickness of the tissue section (41), the antigen retrieval technique (43), sensitivity and specificity of antibodies, and inter-observer inconsistencies in sample analysis (40). Furthermore, scoring of samples as positive or negative for a particular biomarker can be based on a subjective scale of staining intensity or percent of positively staining cells. Therefore, the importance of validating and standardizing every IHC protocol used in clinical trials cannot be overstated. In this manuscript we have critically analyzed the reagents available for measuring ERCC1 expression in tissue samples and developed a standardized method for ERCC1 IHC that could be applied to tumors.
In order to develop an IHC protocol, the first step is to identify an antibody that is specific for the target antigen. This is accomplished by immunoblotting using protein extracts from human cells or tissue and demonstrating that the antibody detects a single band of the appropriate molecular weight (44, 45). It is imperative to include positive and negative controls in which the antigen is known to be expressed or depleted (either genetically or by shRNA), respectively (Fig. 1). Another excellent positive control is recombinant tagged protein included as a molecular weight control on immunoblot or overexpressed in a cell line (Fig. 1). Of eleven screened antibodies that detect ERCC1-XPF, ten were specific for the repair complex (Supplemental Table 1).
The second step in developing an IHC protocol is to validate that an antibody retains its specificity for an antigen in fixed samples (44). This can be accomplished directly in tissue samples only if negative controls (tissues in which the antigen is not expressed) are available. For many of the tumor biomarkers this is not feasible because the antigens of interest are key regulators of cell cycle control and genome maintenance, and thus are ubiquitously expressed. Alternatively, the specificity of an antibody to detect a biomarker in fixed material can be validated using positive and negative control cell lines processed according to the IHC protocol (Fig. 4) (44, 46). In our experiments, normal human fibroblasts (C5RO) were used as a positive control and XP-F patient fibroblasts (XP2YO), deficient in ERCC1-XPF, were used as a negative control due to the absence of human tissue samples missing this protein (Fig. 4). Such internal controls must be included in every IHC analysis (41, 45) (47). One important step in validating proper controls is establishing that the antigen staining has a proper subcellular localization, for example, ERCC1-XPF is a nuclear antigen (Fig. 2). A second step that can provide internal validation of an immunostaining protocol is to differentially label the positive and negative control cell lines and co-culture them, creating a sample with both immunoreactive and unreactive cells (Fig. 2).
Using the above methods with stringent controls, we discovered that antibody 8F1 is not suitable for measurement of ERCC1 expression because it detects a second antigen (20). It has been reported that 8F1 could discriminate between HeLa cells and an isogenic strain in which ERCC1 expression was knocked down by siRNA (32). Based on this, it was argued that 8F1 is suitable for measurement of ERCC1 by IHC. However, HeLa cells do not have appreciable amount of the non-specific antigen (Fig. 1D) and are thus inappropriate for use as either positive or negative control for validating this antibody. In the present work, using formalin-fixed paraffin-embedded normal and ERCC1-XPF deficient cell lines, it was confirmed that 8F1 is unable to differentiate between normal and ERCC1 deficient cells by IHC (Fig. 4). Since antibody 8F1 is the most widely used antibody for IHC (16–18, 48–50), it is important to emphasize that it is not specific for ERCC1 and that validated alternative antibodies exist to reliably measure ERCC1 by IHC. The extensive literature on 8F1-IHC does however indicate that the antibody may have prognostic value. This warrants further investigation to identify the unknown antigen recognized by 8F1.
After testing a panel of antibodies raised against ERCC1 and XPF, antibodies suitable for a variety of immunodetection techniques were identified (Supplemental Table 1). Most of these are unsuitable for IHC, primarily because they do not discriminate between positive and negative controls in fixed material. Those that do work (FL297, 4H4, 3F2) should facilitate the intense interest in measuring ERCC1 expression in tumor samples by IHC. These are validated alternatives to 8F1 that can be used to reliably measure ERCC1 and XPF by IHC. FL297 and 3F2 were used to measure ERCC1 and XPF, respectively, in lung tumor sections (Fig. 6). Variable levels of cytoplasmic staining were seen in paraffin embedded cell lines (Fig. 5). This staining does not correlate with the level of ERCC1-XPF in cells, nor their sensitivity to genotoxic stress. It should therefore be noted that when using these antibodies, grading of expression levels should be based on the extent of nuclear staining only. The levels of both proteins vary between specimens, ranging from intense staining to virtually no staining, but parallel one another (Fig. 6). This indicates that either ERCC1 or XPF might serve as a biomarker of DNA repair in tumors. There was also differential expression within a tumor depending on the cell type. These results indicate that measuring ERCC1-XPF may be of value for stratifying patients for responsiveness to therapy.
Supplementary Material
Acknowledgements
This work was supported by NCI (UPCI SPORE in lung P50 CA090440). Additionally, L.J.N. and N.R.B. were supported by the Ellison Medical Foundation (AG-NS-0303-05) and NIEHS (R01 ES016114). We thank Maureen Biggerstaff (Cancer Research UK) and Melanie Hardman (Cancer Research Technology, Ltd) for discussion and assistance with antibody production.
References
- 1.Ries LAG MD, Krapcho M, Stinchcomb DG, Howlader N, Horner MJ, Mariotto A, Miller BA, Feuer EJ, Altekruse SF, Lewis DR, Clegg L, Eisner MP, Reichman M, Edwards BK, editors. SEER Cancer Statistics Review, 1975–2005. Bethesda, MD: National Cancer Institute; 2008. [Google Scholar]
- 2.Patz EF, Jr, Goodman PC, Bepler G. Screening for lung cancer. N Engl J Med. 2000;343:1627–1633. doi: 10.1056/NEJM200011303432208. [DOI] [PubMed] [Google Scholar]
- 3.Arriagada R, Bergman B, Dunant A, Le Chevalier T, Pignon JP, Vansteenkiste J. Cisplatin-based adjuvant chemotherapy in patients with completely resected non-small-cell lung cancer. N Engl J Med. 2004;350:351–360. doi: 10.1056/NEJMoa031644. [DOI] [PubMed] [Google Scholar]
- 4.Winton T, Livingston R, Johnson D, et al. Vinorelbine plus cisplatin vs. observation in resected non-small-cell lung cancer. N Engl J Med. 2005;352:2589–2597. doi: 10.1056/NEJMoa043623. [DOI] [PubMed] [Google Scholar]
- 5.Bulzebruck H, Bopp R, Drings P, et al. New aspects in the staging of lung cancer. Prospective validation of the International Union Against Cancer TNM classification. Cancer. 1992;70:1102–1110. doi: 10.1002/1097-0142(19920901)70:5<1102::aid-cncr2820700514>3.0.co;2-5. [DOI] [PubMed] [Google Scholar]
- 6.Bunn PA, Jr, Kelly K. New chemotherapeutic agents prolong survival and improve quality of life in non-small cell lung cancer: a review of the literature and future directions. Clin Cancer Res. 1998;4:1087–1100. [PubMed] [Google Scholar]
- 7.Eastman A. The formation, isolation and characterization of DNA adducts produced by anticancer platinum complexes. Pharmacol Ther. 1987;34:155–166. doi: 10.1016/0163-7258(87)90009-x. [DOI] [PubMed] [Google Scholar]
- 8.De Silva IU, McHugh PJ, Clingen PH, Hartley JA. Defining the roles of nucleotide excision repair and recombination in the repair of DNA interstrand cross-links in mammalian cells. Molecular and cellular biology. 2000;20:7980–7990. doi: 10.1128/mcb.20.21.7980-7990.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Niedernhofer LJ, Lalai AS, Hoeijmakers JH. Fanconi anemia (cross)linked to DNA repair. Cell. 2005;123:1191–1198. doi: 10.1016/j.cell.2005.12.009. [DOI] [PubMed] [Google Scholar]
- 10.Friedberg EC. DNA repair and mutagenesis. 2nd ed. Washington, D.C.: ASM Press; 2006. [Google Scholar]
- 11.Collins AR. Mutant rodent cell lines sensitive to ultraviolet light, ionizing radiation and cross-linking agents: a comprehensive survey of genetic and biochemical characteristics. Mutation research. 1993;293:99–118. doi: 10.1016/0921-8777(93)90062-l. [DOI] [PubMed] [Google Scholar]
- 12.Sijbers AM, de Laat WL, Ariza RR, et al. Xeroderma pigmentosum group F caused by a defect in a structure-specific DNA repair endonuclease. Cell. 1996;86:811–822. doi: 10.1016/s0092-8674(00)80155-5. [DOI] [PubMed] [Google Scholar]
- 13.Enzlin JH, Scharer OD. The active site of the DNA repair endonuclease XPF-ERCC1 forms a highly conserved nuclease motif. The EMBO journal. 2002;21:2045–2053. doi: 10.1093/emboj/21.8.2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tsodikov OV, Enzlin JH, Scharer OD, Ellenberger T. Crystal structure and DNA binding functions of ERCC1, a subunit of the DNA structure-specific endonuclease XPF-ERCC1. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:11236–11241. doi: 10.1073/pnas.0504341102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Biggerstaff M, Szymkowski DE, Wood RD. Co-correction of the ERCC1, ERCC4 and xeroderma pigmentosum group F DNA repair defects in vitro. The EMBO journal. 1993;12:3685–3692. doi: 10.1002/j.1460-2075.1993.tb06043.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee KH, Min HS, Han SW, et al. ERCC1 expression by immunohistochemistry and EGFR mutations in resected non-small cell lung cancer. Lung Cancer. 2008;60:401–407. doi: 10.1016/j.lungcan.2007.10.014. [DOI] [PubMed] [Google Scholar]
- 17.Olaussen KA, Dunant A, Fouret P, et al. DNA repair by ERCC1 in non-small-cell lung cancer and cisplatin-based adjuvant chemotherapy. The New England journal of medicine. 2006;355:983–991. doi: 10.1056/NEJMoa060570. [DOI] [PubMed] [Google Scholar]
- 18.Hwang IG, Ahn MJ, Park BB, et al. ERCC1 expression as a prognostic marker in N2(+) nonsmall-cell lung cancer patients treated with platinum-based neoadjuvant concurrent chemoradiotherapy. Cancer. 2008 doi: 10.1002/cncr.23693. [DOI] [PubMed] [Google Scholar]
- 19.Zheng Z, Chen T, Li X, Haura E, Sharma A, Bepler G. DNA synthesis and repair genes RRM1 and ERCC1 in lung cancer. The New England journal of medicine. 2007;356:800–808. doi: 10.1056/NEJMoa065411. [DOI] [PubMed] [Google Scholar]
- 20.Niedernhofer LJ, Bhagwat N, Wood RD. ERCC1 and non-small-cell lung cancer. The New England journal of medicine. 2007;356:2538–2540. doi: 10.1056/NEJMc070742. author reply 40-1. [DOI] [PubMed] [Google Scholar]
- 21.Niedernhofer LJ, Garinis GA, Raams A, et al. A new progeroid syndrome reveals that genotoxic stress suppresses the somatotroph axis. Nature. 2006;444:1038–1043. doi: 10.1038/nature05456. [DOI] [PubMed] [Google Scholar]
- 22.Jaspers NG, Raams A, Silengo MC, et al. First reported patient with human ERCC1 deficiency has cerebro-oculo-facio-skeletal syndrome with a mild defect in nucleotide excision repair and severe developmental failure. American journal of human genetics. 2007;80:457–466. doi: 10.1086/512486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yagi T, Tatsumi-Miyajima J, Sato M, Kraemer KH, Takebe H. Analysis of point mutations in an ultraviolet-irradiated shuttle vector plasmid propagated in cells from Japanese xeroderma pigmentosum patients in complementation groups A and F. Cancer research. 1991;51:3177–3182. [PubMed] [Google Scholar]
- 24.Thompson LH, Fong S, Brookman K. Validation of conditions for efficient detection of HPRT and APRT mutations in suspension-cultured Chinese hamster ovary cells. Mutation research. 1980;74:21–36. doi: 10.1016/0165-1161(80)90188-0. [DOI] [PubMed] [Google Scholar]
- 25.Wood RD, Burki HJ. Repair capability and the cellular age response for killing and mutation induction after UV. Mutation research. 1982;95:505–514. doi: 10.1016/0027-5107(82)90281-0. [DOI] [PubMed] [Google Scholar]
- 26.Niedernhofer LJ, Essers J, Weeda G, et al. The structure-specific endonuclease Ercc1-Xpf is required for targeted gene replacement in embryonic stem cells. The EMBO journal. 2001;20:6540–6549. doi: 10.1093/emboj/20.22.6540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kuraoka I, Kobertz WR, Ariza RR, Biggerstaff M, Essigmann JM, Wood RD. Repair of an interstrand DNA cross-link initiated by ERCC1-XPF repair/recombination nuclease. The Journal of biological chemistry. 2000;275:26632–26636. doi: 10.1074/jbc.C000337200. [DOI] [PubMed] [Google Scholar]
- 28.Vermeulen W, Bergmann E, Auriol J, et al. Sublimiting concentration of TFIIH transcription/DNA repair factor causes TTD-A trichothiodystrophy disorder. Nature genetics. 2000;26:307–313. doi: 10.1038/81603. [DOI] [PubMed] [Google Scholar]
- 29.Botta E, Nardo T, Lehmann AR, Egly JM, Pedrini AM, Stefanini M. Reduced level of the repair/transcription factor TFIIH in trichothiodystrophy. Human molecular genetics. 2002;11:2919–2928. doi: 10.1093/hmg/11.23.2919. [DOI] [PubMed] [Google Scholar]
- 30.Volker M, Mone MJ, Karmakar P, et al. Sequential assembly of the nucleotide excision repair factors in vivo. Molecular cell. 2001;8:213–224. doi: 10.1016/s1097-2765(01)00281-7. [DOI] [PubMed] [Google Scholar]
- 31.Mone MJ, Volker M, Nikaido O, et al. Local UV-induced DNA damage in cell nuclei results in local transcription inhibition. EMBO Rep. 2001;2:1013–1017. doi: 10.1093/embo-reports/kve224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Olaussen KA, Fouret P, Kroemer G. ERCC1-specific immunostaining in non-small-cell lung cancer. The New England journal of medicine. 2007;357:1559–1561. doi: 10.1056/NEJMc072007. [DOI] [PubMed] [Google Scholar]
- 33.Jemal A, Siegel R, Ward E, et al. Cancer statistics, 2008. CA Cancer J Clin. 2008;58:71–96. doi: 10.3322/CA.2007.0010. [DOI] [PubMed] [Google Scholar]
- 34.Armstrong DK. Relapsed ovarian cancer: challenges and management strategies for a chronic disease. Oncologist. 2002;7 Suppl 5:20–28. doi: 10.1634/theoncologist.7-suppl_5-20. [DOI] [PubMed] [Google Scholar]
- 35.McGuire V, Jesser CA, Whittemore AS. Survival among U.S. women with invasive epithelial ovarian cancer. Gynecol Oncol. 2002;84:399–403. doi: 10.1006/gyno.2001.6536. [DOI] [PubMed] [Google Scholar]
- 36.Modesitt SC, Jazaeri AA. Recurrent epithelial ovarian cancer: pharmacotherapy and novel therapeutics. Expert Opin Pharmacother. 2007;8:2293–2305. doi: 10.1517/14656566.8.14.2293. [DOI] [PubMed] [Google Scholar]
- 37.Vilmar A, Sorensen JB. Excision repair cross-complementation group 1 (ERCC1) in platinum-based treatment of non-small cell lung cancer with special emphasis on carboplatin: A review of current literature. Lung Cancer. 2008 doi: 10.1016/j.lungcan.2008.08.006. [DOI] [PubMed] [Google Scholar]
- 38.Simon GR, Ismail-Khan R, Bepler G. Nuclear excision repair-based personalized therapy for non-small cell lung cancer: From hypothesis to reality. Int J Biochem Cell Biol. 2007 doi: 10.1016/j.biocel.2007.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rosell R, Felip E, Paz-Ares L. How could pharmacogenomics help improve patient survival? Lung Cancer. 2007;57 Suppl 2:S35–S41. doi: 10.1016/S0169-5002(07)70426-9. [DOI] [PubMed] [Google Scholar]
- 40.Taylor CR, Levenson RM. Quantification of immunohistochemistry--issues concerning methods, utility and semiquantitative assessment II. Histopathology. 2006;49:411–424. doi: 10.1111/j.1365-2559.2006.02513.x. [DOI] [PubMed] [Google Scholar]
- 41.Grube D. Constants and variables in immunohistochemistry. Arch Histol Cytol. 2004;67:115–134. doi: 10.1679/aohc.67.115. [DOI] [PubMed] [Google Scholar]
- 42.Taylor CR. Quality assurance and standardization in immunohistochemistry. A proposal for the annual meeting of the Biological Stain Commission, June, 1991. Biotech Histochem. 1992;67:110–117. doi: 10.3109/10520299209110018. [DOI] [PubMed] [Google Scholar]
- 43.Shi SR, Imam SA, Young L, Cote RJ, Taylor CR. Antigen retrieval immunohistochemistry under the influence of pH using monoclonal antibodies. J Histochem Cytochem. 1995;43:193–201. doi: 10.1177/43.2.7822775. [DOI] [PubMed] [Google Scholar]
- 44.Holmseth S, Lehre KP, Danbolt NC. Specificity controls for immunocytochemistry. Anat Embryol (Berl) 2006;211:257–266. doi: 10.1007/s00429-005-0077-6. [DOI] [PubMed] [Google Scholar]
- 45.Pradidarcheep W, Labruyere WT, Dabhoiwala NF, Lamers WH. Lack of Specificity of Commercially Available Antisera: Better Specifications Needed. J Histochem Cytochem. 2008;56:1099–1111. doi: 10.1369/jhc.2008.952101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rhodes A, Jasani B, Couturier J, et al. A formalin-fixed, paraffin-processed cell line standard for quality control of immunohistochemical assay of HER-2/neu expression in breast cancer. Am J Clin Pathol. 2002;117:81–89. doi: 10.1309/4NCM-QJ9W-QM0J-6QJE. [DOI] [PubMed] [Google Scholar]
- 47.Leong AS. Quantitation in immunohistology: fact or fiction? A discussion of variables that influence results. Appl Immunohistochem Mol Morphol. 2004;12:1–7. doi: 10.1097/00129039-200403000-00001. [DOI] [PubMed] [Google Scholar]
- 48.Kwon HC, Roh MS, Oh SY, et al. Prognostic value of expression of ERCC1, thymidylate synthase, and glutathione S-transferase P1 for 5-fluorouracil/oxaliplatin chemotherapy in advanced gastric cancer. Ann Oncol. 2007;18:504–509. doi: 10.1093/annonc/mdl430. [DOI] [PubMed] [Google Scholar]
- 49.Steffensen KD, Waldstrom M, Jeppesen U, Brandslund I, Jakobsen A. Prediction of response to chemotherapy by ERCC1 immunohistochemistry and ERCC1 polymorphism in ovarian cancer. Int J Gynecol Cancer. 2007 doi: 10.1111/j.1525-1438.2007.01068.x. [DOI] [PubMed] [Google Scholar]
- 50.Kim MK, Cho KJ, Kwon GY, et al. Patients with ERCC1-negative locally advanced esophageal cancers may benefit from preoperative chemoradiotherapy. Clin Cancer Res. 2008;14:4225–4231. doi: 10.1158/1078-0432.CCR-07-4848. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.