Abstract
The aryl hydrocarbon receptor nuclear translocator (ARNT) is a promiscuous, basic helix-loop-helix Period/ARNT/Single-minded protein that forms dimeric transcriptional regulator complexes with other bHLH-PAS proteins to regulate various biological pathways. Intriguingly, the introduction of a single point mutation into the C-terminal PAS-B domain resulted in a protein that can simultaneously exist in two distinct conformations. The difference between these two structures is a +3 slip and inversion of a central Iβ-strand and an accompanying N448-P449 peptide bond isomerization in the preceding HI loop. Previous studies have indicated these two forms of Y456T interconvert on the approximate timescale of tens of minutes, allowing these two conformations to be separated by ion exchange chromatography. Here, we use time-resolved solution NMR spectroscopy to quantitatively characterize this rate and its temperature dependence, providing information into the transition state. When compared with fluorescence measurements of protein unfolding rates, we find data that suggest a linkage between interconversion and unfolding based on comparable temperature dependence and corresponding energetics of these processes. Notably, the N448-P449 peptide bond also plays a critical role for the interconversion between states, with a mutant unable to adopt a cis configuration at this bond (P449A/Y456T) being kinetically trapped under non-denaturing conditions. Taken together, these data provide information about a rare equilibrium model system for β-strand slippage.
The aryl hydrocarbon receptor nuclear translocator (ARNT) is a basic helix-loop-helix Period/ARNT/Single-minded (bHLH-PAS) protein that controls various biological pathways as part of dimeric transcriptional regulator complexes with other bHLH-PAS proteins1–4. These complexes utilize two PAS domains within ARNT, PAS-A and PAS-B, to mediate protein/protein interactions via residues located on their β-sheet surfaces5,6. Site-directed mutagenesis studies of ARNT PAS-B have demonstrated the plasticity of this domain to adopt different conformations7. In particular, one point mutation, Y456T, on the ARNT PAS-B solvent exposed β-sheet surface resulted in two conformations (“wt” and “+3”) that coexist in approximately equimolar concentrations. These conformations differ by a +3 shift in register and accompanying inversion of a central Iβ-strand along with an isomerization of the N448-P449 peptide bond in the preceding HI loop (Figure S1). Intriguingly, we found that the two conformations of Y456T interconvert slowly enough that the two forms could be separated by ion exchange chromatography7.
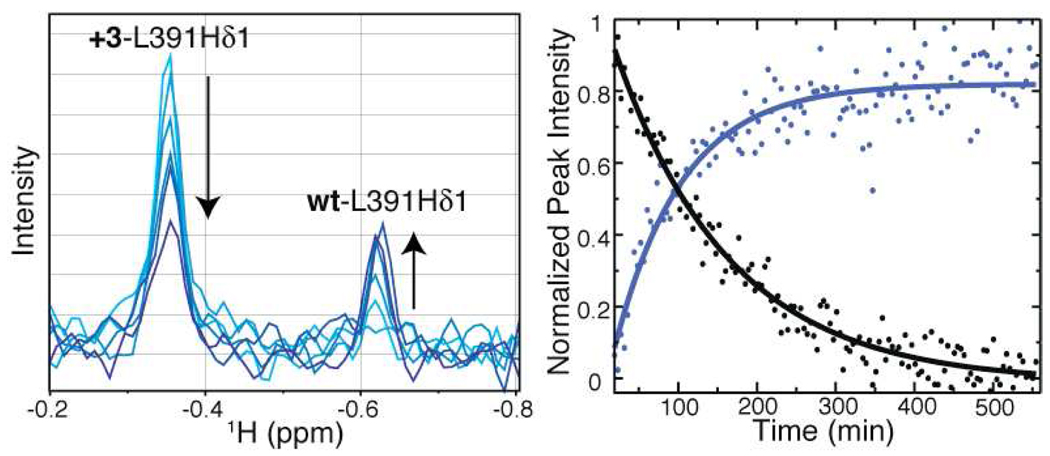
To measure the kinetics of this interconversion process for ARNT PAS-B Y456T, we recorded a series of sequential 13C-edited 1D 1H experiments on a sample enriched for the +3 conformation (12:88, wt:+3) as obtained by ion exchange chromatography of a Y456T sample (51:49, wt:+3). As expected, initial spectra in these series showed peaks resulting primarily from the +3 conformation, as illustrated by the peak at −0.348 ppm, which corresponds to the Leu391-δ1 methyl group in the +3 conformation (Figure 1). This same methyl group in the wt conformation is located at −0.628 ppm and was not observed in the initial spectra. By recording a series of spectra collected sequentially, approximately four minutes apart, we were able to monitor the process of interconversion between the two conformations as the system relaxed back to equilibrium. We fit the time-dependent peak intensity changes from these spectra to a single exponential decay, allowing the extraction of the kinetic rates. These analyses showed similar rates for the disappearance of +3 and concomitant formation of wt. In addition, we characterized the temperature dependence of this process between 278 K and 291 K and found a linear Eyring dependence, with rates between k = 5.49 × 10−5 s−1 and k = 2.13 × 10−4 s−1, respectively, Figure S2. Eyring analysis indicated that a large enthalpic barrier, 12.7 kcal/mol, must be crossed during interconversion (Table 1), most likely due to the breaking of backbone hydrogen bonds in the β-sheet. Therefore, we set out to determine the mechanism of interconversion, hypothesizing the domain may partially unfold, either locally around the Iβ-strand or more globally, in order to interconvert.
Figure 1.
Monitoring the rates of interconversion for ARNT PAS-B Y456T. Left: Peak intensity changes in 1D 1H NMR spectra are recorded as the protein re-establishes equilibrium (t=0 min, light blue; t=500 min, dark blue), as monitored by L391Hδ1. Right: Kinetic traces for the interconversion, corresponding to peak intensity changes for the wt (blue) and +3 (black) conformations.
Table 1.
Thermodynamic parameters for ARNT PAS-B Y456T.
| Parameter | Value | |
|---|---|---|
| Interconversion +3 → WT |
ΔG‡ | 18.0 kcal/mol |
| ΔH‡ | 12.7 kcal/mol | |
| ΔS‡ | −17.8 cal/mol·K (−5.3 kcal/mol @ 298 K) |
|
| Unfolding | ΔG‡ | 21.6 kcal/mol |
| ΔH‡ | 11.8 kcal/mol | |
| ΔS‡ | −32.9 cal/mol·K (−9.8 kcal/mol @ 298 K) |
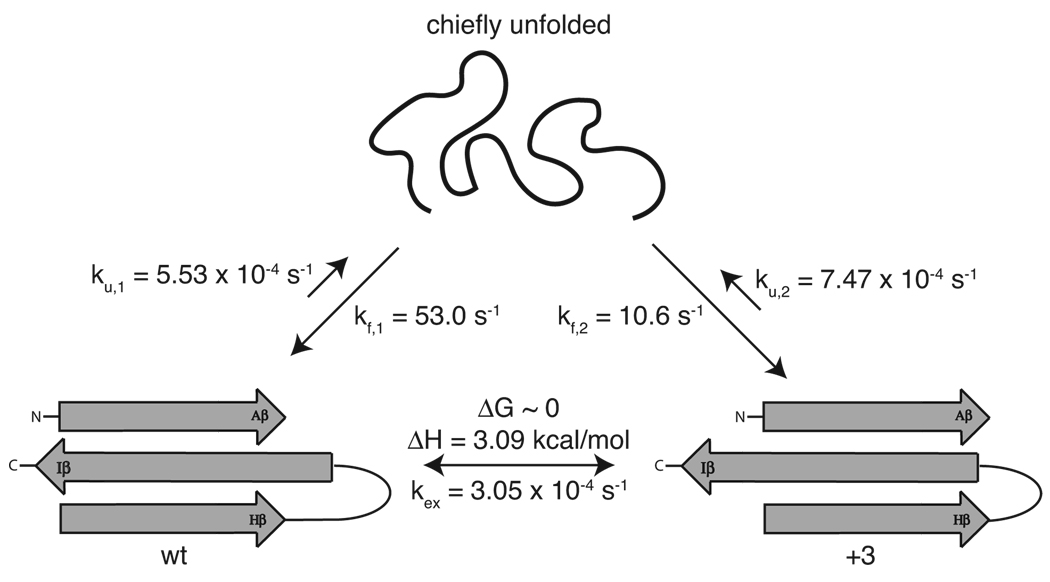
We considered two models of potential mechanisms of interconversion. The first model proposes local unfolding restricted to Iβ, while a second one alternatively involves global unfolding of the protein. In this latter case, the protein must unfold to a chiefly, unfolded state and that the sequence of the protein allows for equal probability of refolding for the Y456T point mutant, presumably with different probabilities for other variants7.
These models differ in their expected degree of protein unfolding, leading us to investigate the rates of global folding and unfolding using stopped-flow fluorescence in combination with Gdn·HCl. As expected, the rates of folding and unfolding at different denaturant concentrations from proteins stabilized in either conformation resulted in a chevron plot, Figure S3, with a denaturant midpoint (~2.5 M Gdn·HCl) that corresponded well to our previously determined equilibrium denaturation midpoint7. The wildtype (wt) and F444Q/F446A/Y456T (+3) proteins exhibited relatively slow unfolding kinetics, ku = 5.53 × 10−4 s−1 and ku = 7.47 × 10−4 s−1, respectively, comparable to dozens of other similarly sized proteins that have been reported with kinetics in the ku = 5.85 × 10−6 s−1 to 89 s−1 range8,9. Folding rates for the wt conformation (kf = 52.9 s−1) are slightly faster than for the +3 conformation (kf = 10.6 s−1) at 298 K and extracted to denaturant-free conditions, also corresponding well to reported folding rates of comparable proteins9. Repeating these experiments for ARNT PAS-B Y456T, which adopts both conformations, yields similar kinetics (ku = 6.76 × 10−4 s−1 and kf = 158 s−1) at 298 K with zero denaturant, indicating that Y456T folds and unfolds comparably to proteins locked into either conformation. Eyring analysis, Figure S4, for the unfolding rates of Y456T observed between 278 and 298 K resulted in a large enthalpic (11.8 kcal/mol) and entropic (−9.8 kcal/mol at 298 K) barriers to unfold (Table 1). The large entropic barrier is presumably due to a drop in water entropy upon exposure of non-polar core residues. Interestingly, these thermodynamic values for unfolding are very similar to those measured for the interconversion of Y456T.
Comparing the rates of interconversion and unfolding, we find support for interconversion proceeding through a chiefly unfolded transition state. Notably, the rates of unfolding are approximately two times faster than the rate of interconversion and that both interconversion and unfolding processes show similar temperature dependences and equivalently comparable enthalpic changes at the transition state. We interpret the different entropic changes among the two processes as reflecting the interconversion transition state as being less unfolded than the corresponding transition state on the unfolding pathway. These data are consistent with the protein unfolding to a chiefly disordered state as it interconverts between conformations (Figure 2). This also supports a model of global unfolding, as the unfolded state of ARNT PAS-B Y456T has an equal probability of refolding into either folded conformation, as established by the 51:49, wt:+3 equilibrium. This equal probability of refolding into either conformation makes the rates of interconversion appear to be twice as slow than if the interconversion was limited to a single direction, further supporting the idea that both processes (interconversion and unfolding) transition through the same chiefly, unfolded state.
Figure 2.
Summary of rates. The measured rates of interconversion are on a similar timescale as unfolding, implying the protein may undergo a global unfolding process in order to slip the central β-strand.
To examine a possible contributor to the slow interconversion kinetics observed in the Y456T protein, we investigated the N448-P449 peptide bond isomerization on the rates of interconversion. This peptide bond, found within the HI loop, was identified to exist in the trans and cis conformations for the wt and +3 proteins, respectively7. The trans/cis isomerization of X-Pro peptide bonds has been well documented as the rate-limiting step for various protein folding processes crucial for opening/closing membrane channels10, mediating conformer-specific ligand recognition11, and promoting phage infection of E. coli12, all of which occur on the second to minute timescale. The enthalpy of activation (ΔH‡) for interconversion is very close to the activation energy (Ea) of the trans/cis isomerization of X-Pro peptide bonds in proteins (13–20 kcal/mol)13,14. Coupled with the difference in configuration of the N448/P449 peptide bond we observed between wt and +37, we speculated that isomerization of this bond would be involved with the rate-limiting step for the interconversion.
Previous studies of P449A/Y456T showed the protein remains well folded with the N448/P449 peptide bond in the trans configuration, while the equilibrium (32:68, wt:+3) only slightly shifted compared to the Y456T mutant7. We again used ion exchange chromatography to purify a sample (>95%) in the +3 conformation and monitored interconversion using time-resolved NMR spectroscopy. Surprisingly, the enriched fraction did not return to equilibrium after over 300 hr, indicating that this conformation is kinetically trapped, Figure S5. Notably, these samples also started to slowly precipitate 100 hr post-separation. Despite this precipitation, total peak intensities in 15N/1H HSQC spectra decreased but the relative populations remained the same, indicating that the protein remained >95% in the +3 conformation. These data suggest once the protein folds into either conformation, it becomes kinetically trapped in that state. However, this can be escaped by denaturation in 3.5 M Gdn•HCl, letting the protein refold back to its initial equilibrium (28:72, wt:+3) upon its removal. To compare these interconversion results with unfolding kinetics, we used Gdn•HCl denaturation to probe the folding/unfolding rates for P449A/Y456T. Analysis of the chevron plot shows a two-fold increase in the folding rate (kf = 330 s−1) and a four-fold decrease in the unfolding rate (ku = 2.08 × 10−4 s−1) compared to Y456T. Taken together, these results indicated that isomerization of the N448/P449 peptide bond is critical for interconversion, but has only minor effects on unfolding.
We conclude from these data that ARNT PAS-B Y456T likely interconverts by unfolding to a chiefly disordered state, and refolds into both conformation with equal probability and thus establishing the 51:49 (wt:+3) equilibrium. In addition, the P449 residue located in the HI-loop critically affects the rate of interconversion, in contrast to the modest effects it has on the equilibrium distribution of this process. While the physiological relevance of this interconversion remains to be established, this system provides a useful equilibrium model for shifts in beta-strand register as observed in several biological systems15,16.
Supplementary Material
Acknowledgement
We thank John Richardson and Nicholas Malmquist for their technical assistance and Brian Zoltowski for his constructive comments and suggestions. This research was supported by a grant from the NIH (R01 GM081875) to K.H.G.
Footnotes
Supporting Information Available: Complete ref 9, protocols, and Figures S1–S6 are provided in Supporting Information. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Ema M, Morita M, Ikawa S, Tanaka M, Matsuda Y, Gotoh O, Saijoh Y, Fujii H, Hamada H, Kikuchi Y, Fujii-Kuriyama Y. Mol Cell Biol. 1996;16:5865–5875. doi: 10.1128/mcb.16.10.5865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Moffett P, Reece M, Pelletier J. Mol Cell Biol. 1997;17:4933–4947. doi: 10.1128/mcb.17.9.4933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lees MJ, Whitelaw ML. Mol Cell Biol. 1999;19:5811–5822. doi: 10.1128/mcb.19.8.5811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang GL, Jiang BH, Rue EA, Semenza GL. Proc Natl Acad Sci U S A. 1995;92:5510–5514. doi: 10.1073/pnas.92.12.5510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Card PB, Erbel PJ, Gardner KH. J Mol Biol. 2005;353:664–677. doi: 10.1016/j.jmb.2005.08.043. [DOI] [PubMed] [Google Scholar]
- 6.Scheuermann TH, Tomchick DR, Machius M, Guo Y, Bruick RK, Gardner KH. Proc Natl Acad Sci U S A. 2009;106:450–455. doi: 10.1073/pnas.0808092106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Evans MR, Card PB, Gardner KH. Proc Natl Acad Sci U S A. 2009;106:2617–2622. doi: 10.1073/pnas.0808270106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jackson SE. Fold Des. 1998;3:R81–R91. doi: 10.1016/S1359-0278(98)00033-9. [DOI] [PubMed] [Google Scholar]
- 9.Maxwell KL, et al. Protein Sci. 2005;14:602–616. doi: 10.1110/ps.041205405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lummis SC, Beene DL, Lee LW, Lester HA, Broadhurst RW, Dougherty DA. Nature. 2005;438:248–252. doi: 10.1038/nature04130. [DOI] [PubMed] [Google Scholar]
- 11.Mallis RJ, Brazin KN, Fulton DB, Andreotti AH. Nat Struct Biol. 2002;9:900–905. doi: 10.1038/nsb864. [DOI] [PubMed] [Google Scholar]
- 12.Eckert B, Martin A, Balbach J, Schmid FX. Nat Struct Mol Biol. 2005;12:619–623. doi: 10.1038/nsmb946. [DOI] [PubMed] [Google Scholar]
- 13.Schulz GD, Schirmer RH. Principles of Protein Structure. New York: Springer-Verlag; 1984. [Google Scholar]
- 14.Brandts JF, Halvorson HR, Brennan M. Biochemistry. 1975;14:4953–4963. doi: 10.1021/bi00693a026. [DOI] [PubMed] [Google Scholar]
- 15.Tuinstra RL, Peterson FC, Kutlesa S, Elgin ES, Kron MA, Volkman BF. Proc Natl Acad Sci U S A. 2008;105:5057–5062. doi: 10.1073/pnas.0709518105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goldberg J. Cell. 1998;95:237–248. doi: 10.1016/s0092-8674(00)81754-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.