Abstract
Background
Aggressive periodontitis (AgP) is associated with impaired polymorphonuclear leukocyte (PMN) chemotaxis toward bacterial N-formylpeptides. Formylpeptide receptors (FPRs) play a major role in guiding PMNs to infection sites. Previous work revealed a significant association between FPR1 single nucleotide polymorphism (SNP) 348T>C and AgP in African Americans. We tested the hypothesis that 348T impairs PMN chemotaxis by decreasing FPR mRNA expression, thereby increasing susceptibility to AgP.
Methods
Blood samples were obtained from African American subjects (37 AgP cases and 38 controls). Chemotaxis to N-formyl-Met-Leu-Phe by freshly isolated PMNs was assayed in a modified Boyden chamber. RNA was isolated from PMNs and FPR1 gene expression was quantified by real time PCR. To detect FPR1 5’ SNPs, genomic DNA was isolated and four fragments spanning the FPR1 5’ region were PCR-amplified and sequenced. Haplotype associations between SNP 348T>C and 5’ SNPs were analyzed.
Results
The homozygous 348T genotype was only found in AgP cases (P=0.017, odds ratio=18.9). Subjects with this genotype exhibited a significantly lower PMN chemotactic response relative to controls and to subjects with the 348C/C or 348T/C genotypes (P<0.05). There were no significant differences in PMN FPR1 expression between subjects with the 348C/C, 348C/T and 348 T/T genotypes. Eleven FPR1 5’ SNPs were detected, but none of the predicted haplotypes reflected associations with AgP or with 348T.
Conclusions
Although the 348T/T genotype is relatively rare, it is associated with significantly impaired PMN chemotaxis and an increased risk of developing AgP in African Americans. These associations do not appear to be related to significant reductions in FPR1 transcripts in subjects expressing 348T.
Keywords: aggressive periodontitis, polymorphisms, leukocytes, FPR1 c.348T>C
INTRODUCTION
Aggressive periodontitis (AgP) is a rapidly progressive, multifactorial periodontal disease that occurs in localized and generalized forms and has a tendency toward familial aggregation. AgP is associated with invasive bacterial pathogens like A. actinomycetemcomitans and P. gingivalis.1,2 In addition, polymorphonuclear neutrophils (PMNs) from individuals affected by AgP exhibit reduced binding of N-formylpeptides and a significant reduction in chemotactic activity towards formylpeptides relative to PMNs from healthy control subjects.3,4,5,6
PMNs serve as the first line of defense against bacterial infections in the body. Upon stimulation, a complex signal transduction mechanism enables PMNs to migrate through endothelial cell walls into tissues. Bacterial formylpeptides are one of the major chemotactic stimuli that guide PMNs in their migration toward infection sites. PMNs typically express approximately 55,000 G-protein-coupled formylpeptide receptors (FPRs) per cell.7 Binding of formylpeptides to FPRs triggers a cascade of intracellular signals that coordinate cytoskeletal reorganization, formation of pseudopodia and migration up a chemotactic gradient.8 When PMNs arrive at an infection site, they attempt to kill bacteria by phagocytosis, production of reactive oxygen species and release of microbicidal enzymes.9,10 The defensive capabilities of PMNs can be undermined by aberrant FPR expression.11 FPR knockout mice exhibit an increased susceptibility to L. monocytogenes infection and their PMNs exhibit no chemotactic response to N-formyl-methionine-leucine-phenylalanine (fMLF).12
Previous analyses of FPR1 suggest that it is highly polymorphic.13,14,15,16,17 In some populations, AgP appears to be associated with specific single nucleotide polymorphisms (SNPs) in FPR1 (the gene encoding the FPR).13,14,15 FPR1 coding region SNPs located in crucial sites can potentially affect FPR function. The FPR second intracellular loop and carboxy-terminal tail serve as major G-protein contact sites.18,19 The amino-terminal domain can affect ligand binding to the FPR by functioning as a “lid” to the ligand-binding pocket.20,21,22 SNPs located in the 5’ region of the FPR1 can potentially affect gene expression. A recent study of Japanese subjects revealed that the -12915T variant of SNP -12915T>C in the FPR1 5’ region is associated with a decreased FPR1 transcription.15
Our previous work has shown that African Americans who are homozygous for the 348T variant of the synonymous SNP 348T>C have a significantly increased risk of developing AgP. The homozygous 348T genotype appears to occur only in individuals with AgP.23 Although 348T>C does not code for a change in the amino acid structure, synonymous coding region SNPs are occasionally associated with 5’ SNPs in promoter or enhancer regions that can alter gene expression. This phenomenon has been observed in the human β2 adrenergic receptor and the CC chemokine receptor.24,25 It is reasonable to hypothesize that the 348T FPR variant is associated with PMN FPR expression defects, PMN chemotaxis defects and increased susceptibility to AgP. It is also feasible that 348T is linked to one or more SNPs in the 5’ region of FPR1 that contribute to impaired FPR expression and increased susceptibility to AgP. To test these hypotheses, we assayed the chemotactic activity of PMNs obtained from African-Americans with AgP and from healthy ethnically matched controls to determine whether 348T is associated with reduced PMN chemotaxis. To ascertain the mechanism by which 348T could influence PMN chemotaxis, we examined the expression FPR1 transcripts and screened our subjects for FPR1 5’ SNPs that could potentially be associated with 348T.
MATERIALS AND METHODS
Subject recruitment
African American AgP cases and healthy control subjects were enrolled between January 2003 and July 2008 under a protocol approved by the Ohio State University Institutional Review Board. Written informed consent was obtained from each subject. The criteria for diagnosis of AgP were consistent with the 1999 International Workshop for Classification of Periodontal Diseases and Conditions.26 Individuals diagnosed with either localized aggressive periodontitis (LAgP, n=18) or generalized aggressive periodontitis (GAgP, n=19), and who were otherwise healthy, were included in the AgP case group. Cases with attachment loss involving no more than two permanent teeth other than the first molars and incisors were diagnosed with LAgP.26 These subjects had ≥4mm attachment loss involving at least two permanent first molars and incisors, including at least one first molar.26 Cases involving at least three teeth other than the first molars and incisors, with an attachment loss of ≥4mm were diagnosed with GAgP.26, 27 The control subjects (n=38) were periodontally healthy based on clinical and radiographic examinations. None of the controls exhibited clinical attachment loss of >3 mm or probing depths >4 mm.
PMN isolation and chemotaxis assays
Peripheral blood was drawn from cases and controls by venipuncture. PMNs were isolated by Ficoll-Hypaque density gradient centrifugation and dextran sedimentation as previously described.28 Freshly isolated PMNs were resuspended at a density of 106/ml in RPMI 1640 medium containing 0.3 % bovine serum albumin. Chemotaxis was assayed in a 48-well modified Boyden chamber.‡ Suspended PMNs were added to the upper wells and fMLF§ was added to the lower wells at concentrations of 0, 10 and 30 nM in RPMI 1640 medium containing 0.3% bovine serum albumin. Upper and lower wells were separated by a polycarbonate filter∥ (5 µm pore size). The chamber was incubated at 37° C for 30 minutes. Following incubation, the chamber was disassembled and the filter was carefully removed and washed with sterile PBS. PMNs that failed to migrate through the filter were wiped off the top surface of the filter. PMNs that had migrated through the filter were stained with a commercially available kit¶ and counted with an oil immersion microscope objective (400X, 5 fields per well). Chemotaxis was assayed with PMNs isolated from a total of 26 cases and 32 controls.
RNA isolation and cDNA synthesis
Total RNA was isolated from PMNs obtained from 26 AgP subjects and 37 healthy controls with TRIzol.# Freshly isolated PMN pellets were suspended in TRIzol (1ml per 107cells) and vortexed for 15 seconds. After addition of chloroform (0.2ml per 107 cells), the samples were vortexed for 15 seconds and placed on ice for 10 minutes. The samples were then centrifuged at 4900 × g and the aqueous phase containing RNA was aspirated and placed in new centrifuge tubes. Isopropanol (0.5 ml per ml of TRIzol) was added to the new tubes, which were then vortexed for 15 seconds and placed on ice for 10 minutes. The samples were centrifuged for 10 minutes at 4900 × g and the supernatant was discarded. The RNA pellets were washed in 70% isopropanol (1 ml per ml of TRIzol). The resultant RNA pellets were then air-dried for 5 minutes and resuspended in RNase free water. RNA concentrations were measured by spectrophotometry at 260 nm and 280 nm.
Reverse transcription of total RNA was performed with a kit** according to manufacturer’s instructions. For each sample, 2 µg of total RNA was added to a reaction mixture containing 5 mM MgCl2, 4 µl of 10 X reverse transcription buffer, 1 mM of each deoxynucleoside triphosphate, 1 µg of random primers, 40 U of recombinant ribonuclease inhibitor and 26.4 U of avian myeloblastosis virus reverse transcriptase (40 µl total volume). The samples were incubated at room temperature for 10 minutes, heated to 42° C for 1 hour and then placed in boiling water for 5 minutes. Immediately afterward, samples were cooled on ice for 5 minutes and 60 µl of RNase free water was added to bring the final volume of each sample to 100 µl. The resultant cDNA samples were stored at −80° C until the time of assay by real-time polymerase chain reaction (PCR).
Quantitative real-time PCR
All cDNA samples were subjected to real time PCR to quantify FPR1 expression. A known positive control sample expressing FPR1 was included in each plate at 1, 1:10, 1:100 and 1:1000 dilutions for obtaining standard curves and a baseline value. Human β-actin was used as the internal control. For each sample, reactions were run in duplicates on 96-well plates. To quantify FPR1 and β-actin, commercially available gene expression assays†† containing primer-probe combinations were used. To each reaction well, 1.25µl of primer-probe mix, 12.5 µl universal PCR master mix ‡‡, 2.5 µl of cDNA and RNase free water were added to give a final volume of 25 µl. Samples were subjected to 40 cycles of amplification, each consisting of 2 minutes at 50° C, 10 minutes at 95°, 15 seconds at 95°, and 1 minute at 60°. Real-time PCR analysis was performed with a dedicated software package. §§
Real time PCR data was analyzed by the comparative Ct (threshold cycle) method as previously described.29 The delta Ct (ΔCt = FPR1 Ct minus β-actin Ct) values calculated for each sample were used to make comparisons between groups. The ΔCt values are normally distributed and amenable to analysis by one way ANOVA, but are not inherently suitable for descriptive purposes. For presentation (Figure 1, Table 3 and Table 4), they were standardized and transformed to ΔΔCt (which represents the fold difference in gene expression relative to baseline). The transformation process results in positive skewing of the variability within samples. Since this skewing misrepresents the actual variability, standard errors were not included in the data presentation.
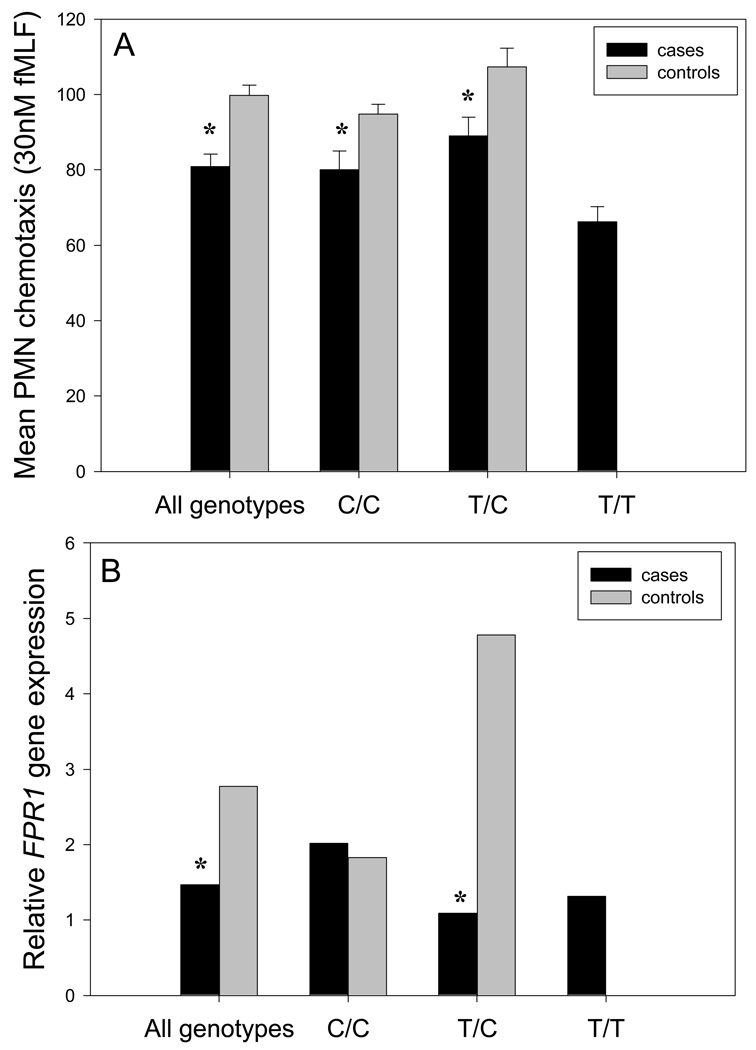
Figure 1.
Relationship of 348T>C genotype to PMN chemotaxis activity and PMN FPR1 expression. Statistically significant differences between cases and controls are denoted by * (P < 0.05, t test). Upper panel: Chemotaxis activity was assayed in the presence of 30 nM fMLF with PMNs obtained from 26 case subjects and 32 controls. Data are presented as mean ± SEM. Lower panel: FPR1 expression was assayed with RNA isolated from 26 case and 37 control subjects. Transformed values (ΔΔCt) represent n-fold change in FPR1 expression relative to baseline. Since the transformation skews the variation within samples, error bars were not included.
Table 3.
PMN Chemotactic Activity and Relative FPR1 Expression in Healthy Controls and AgP Cases of Various Genotypes*
| Group | Chemotaxis activity to 30 nM fMLF† | Fold increase in FPR1 expression¶ |
|---|---|---|
| All controls | 99.7 ± 2.8‡ § | 2.77# |
| (n = 32) | (n = 37) | |
| 348 C/C Cases | 80.0 ± 5.0§ | 2.02 |
| (n = 11) | (n = 11) | |
| 348 T/C Cases | 89.0 ± 5.0∥ | 1.09# |
| (n = 10) | (n = 10) | |
| 348 T/T Cases | 66.2 ± 4.0‡∥ | 1.32 |
| (n = 5) | (n = 5) | |
Differences among the groups in the columns are statistically significant (P ≤ 0.006, ANOVA). Within columns, like superscripts indicate values that are significant different (P<0.05, Holm-Sidak test)
Data are reported as mean ± SEM
Transformed values (ΔΔCt) represent n-fold change in FPR1 expression relative to baseline. Variability is not shown since the transformed data is positively skewed.
Table 4.
Relationship of 348T>C Genotype to PMN Chemotactic Activity and Relative FPR1 Expression in a Pool of All Case and Control Subjects
| Genotype Group | Chemotaxis activity to 30 nM fMLF* | Fold increase in FPR1 expression‡ |
|---|---|---|
| 348 C/C | 89.3 ± 2.8† | 1.92 |
| (n = 30) | (n = 32) | |
| 348 T/C | 99.3 ± 4.0† | 2.93 |
| (n = 23) | (n = 26) | |
| 348 T/T | 66.2 ± 4.0† | 1.32 |
| (n = 5) | (n = 5) | |
Data are reported as mean ± SEM. Differences among the groups in this column are statistically significant (P < 0.001, ANOVA). Within this column, like superscripts indicate values that are significantly different (P <0.05, Holm-Sidak test)
Transformed values (ΔΔCt) represent n-fold change in FPR1 expression relative to baseline. Variability is not shown since the transformed data is positively skewed and can be misleading. Differences among the groups in this column are not statistically significant (P =0.26, Kruskal-Wallis ANOVA on ranks).
PCR and sequencing of the FPR1 coding and 5’ regions
Genomic DNA was isolated from whole blood obtained from 37 cases and 38 controls with a commercially available kit∥∥. FPR1 SNP 348T>C and other coding region SNPs were detected by PCR amplification and sequencing of a 439 bp fragment of FPR1 as previously described.23 To detect 5’ SNPs, validated primer sets were used to amplify four fragments spanning approximately 10 kb of the FPR1 5’ region, of sizes 500 bp, 264 bp, 188 bp and 783 bp.15 PCR was carried out for 37 cycles in a reaction mixture containing 45 μl PCR supermix ¶¶, 1 µl of each of the two primers, 1.5 µl (18–30 pg) of the DNA sample and 1.5 µl water. Prior to initiating PCR, the reaction mixture was heated at 94° C for 5 minutes. The parameters for one PCR cycle included denaturation at 94° C for 30 seconds, annealing at 60° for 30 seconds and extension at 72° for 30 seconds. The cycles were followed by a final 7 minute extension step at 72°. PCR products were purified and sequenced with an automated DNA analyzer.## Differences in allele frequencies for each SNP were analyzed by the Fisher exact test.
Stability of mRNA structure
Genetic association software*** was used to predict the predominant FPR1 haplotypes and analyze potential associations of SNP 348T>C with 5’ SNPs. The secondary mRNA structures derived from FPR1 haplotypes were predicted with the mfold web server.30
RESULTS
Clinical Characteristics
The case group comprised 18 subjects diagnosed with LAgP and 19 subjects diagnosed with GAgP. No differences were observed between LAgP and GAgP subjects with respect to mean PMN chemotaxis and 348T allele association, so data presentation in the following sections does not distinguish between these clinical diagnoses. The clinical and demographic characteristics of the case and control groups are presented in Table 1. Clinical attachment loss in the control group was noted only at isolated sites and did not exceed 3 mm at any site. None of the control subjects exhibited radiographic evidence of bone loss.
Table 1.
Clinical and Demographic Characteristics of the Study Population*
| Cases † (n = 37) | Controls‡ (n = 38) | |
|---|---|---|
| Age | 24.1 ± 1.31 | 27.7 ± 1 |
| Gender composition | 62.2% Female | 63.2% Female |
| Number of teeth with Probing Depth ≥ 5 mm | 15.6 ± 1.14 | 0 |
| Depth of sites with Probing Depth ≥ 5 mm | 6.24 ± 0.11 mm | N/A |
| No. of teeth with Clinical Attachment Loss ≥ 4 mm | 16.5 ± 1.3 | 0 |
| AL of sites with Clinical Attachment Loss ≥ 4 mm | 5.38 ± 0.15 mm | N/A |
Data are presented as mean ± SEM
FPR1 5’ SNPs and 348T>C were analyzed in all 37 cases, but some subjects were lost to follow-up studies. Chemotaxis and FPR1 gene expression were assayed in a subset of 26 subjects. The characteristics of the subset were not significantly different from the full case population.
FPR1 5’ SNPs and 348T>C were analyzed in all 38 controls, but some subjects were lost to follow-up studies. Chemotaxis was assayed with 32 subjects and FPR1 gene expression was assessed with 37 subjects. The characteristics of these two subsets were not significantly different from the full control population.
Association of 348T with Aggressive Periodontitis
Apart from SNP 348T>C, four other previously identified FPR1 SNPs (301G>C, 546C>A, 568A>T and 576T>C>G) were detected in the fragment analyzed. None of these were associated with AgP (data not shown). Three different 348T>C genotypes (C/C, T/C and T/T) were detected in the AgP case group, while only two genotypes (C/C and C/T) were detected in controls. The 348T/T genotype was expressed by 7 of the 37 case subjects. There was a significant relationship between 348T/T and case/control status (P = 0.017; odds ratio = 18.9, Table 2).
Table 2.
Allele association of FPR1 SNP 348T>C with AgP cases in African-Americans*
| Genotype | AgP | Controls | ||
|---|---|---|---|---|
| n | % | n | % | |
| 348 C/C | 17 | (45.9) | 22 | (57.9) |
| 348 T/C | 13 | (35.1) | 16 | (42.1) |
| 348 T/T | 7 | (19) | 0 | (0) |
The relationship between genotype and AgP case/control status is significant (P=0.017, Fisher Exact test). Individuals with the 348T/T genotype are at greater risk for AgP than those with the 348T/C and 348C/C genotypes combined (odds ratio=18.9, 95% confidence interval=1.04–333).
PMN Chemotaxis and Relative FPR1 Expression
To examine the relationship between 348T, the PMN chemotactic response and chemotactic receptor expression, chemotaxis activity was assayed with PMNs obtained from a subset of 26 cases and 32 controls and relative FPR1 gene expression assayed in a subset of 26 AgP cases and 37 controls. In the presence of 10 nM fMLF, the mean chemotaxis of PMNs obtained from AgP cases was approximately 22% lower than in PMNs obtained from controls (P = 0.001, Mann-Whitney rank sum test, data not shown). The mean chemotactic response to 30 nM fMLF was 19% lower in cases than in control subjects (P < 0.001, t-test, Figure 1A). There was no significant difference between cases and controls with respect to random migration (data not shown). The overall level of PMN FPR1 expression in cases was significantly lower than in controls (P < 0.001, t-test, Figure 1B).
The mean chemotactic response to 30 nM fMLF was 34% lower in cases with the 348T/T genotype than the average response by controls (P < 0.05, Holm-Sidak test, Table 3), and chemotaxis in cases with the 348C/C genotype was 20% lower than in controls (P < 0.05, Holm-Sidak test). The level of FPR1 gene expression by PMNs from cases with the 348T/T genotype was not significantly different from the levels observed in controls or cases with the 348C/C or 348T/C genotypes (P = 0.26, Table 3). However, PMNs from cases with the 348T/C genotype expressed significantly lower levels of FPR1 transcripts than the mean level expressed by controls (P = 0.006, Holm-Sidak test).
Table 4 portrays the relationship between PMN chemotaxis activity, FPR1 expression levels and 348T>C genotype in a pool of case and control subjects. There were significant differences in chemotaxis activity between subjects with the 348C/C, 348T/C and 348T/T genotypes (P < 0.001, ANOVA). In particular, the mean chemotaxis activity observed with subjects with the 348T/T genotype was 26% lower than in subjects with the 348T/C genotype and 33% lower than those with the 348C/C genotype (P < 0.05, Holm-Sidak test). Although subjects with the 348T/T genotype exhibited the lowest observed level of FPR1 expression, no statistically significant differences in relative FPR1 expression were found between the three 348 genotype groups (P = 0.26, Kruskal-Wallis one way ANOVA).
Figure 1 compares chemotaxis activity and FPR1 gene expression between cases and controls with the 348C/C, 348T/C and 348T/T genotypes. The mean chemotaxis activity of PMNs from 348C/C cases was 16% lower than controls with the same genotype (P = 0.008, t-test, Figure 1A), while the mean chemotaxis activity in 348T/C cases was 17% lower than in 348T/C controls (P = 0.02). Cases with the 348T/T genotype exhibited the lowest chemotactic response. The level of FPR1 expression was not significantly different between cases and controls with the 348C/C genotype (P = 0.63, Figure 1B), but expression by 348T/C controls was significantly higher than observed in 348T/C cases (P < 0.001, t-test).
FPR1 5’ SNPs
Eleven 5’ SNPs were identified, including seven (-12915C>T, -10107A>G, -10056T>C, -8430A>G, -8318A>G, -5426G>A and -4949C>T) that have been previously described15 and four newly identified SNPs (-13045C>T>A, -12888T>A, -9997C>T and -8375G>T). None of the SNPs exhibited significant differences in allele frequencies between cases and controls (Table 5), but -13045C>T>A, -12915C>T and -8430A>G most closely approached statistical significance. Haplotype association of 348T>C with each of these three SNPs was analyzed. However, none of the predicted haplotypes containing 348T in combination with one or more of these three 5’ SNPs exhibited associations with AgP that were any more significant than found with 348T alone (data not shown).
Table 5.
Allele Frequencies of FPR1 5’ SNPs in Cases and Controls
| SNP | AgP Cases | Controls | P value* | |||
|---|---|---|---|---|---|---|
| n | % | n | % | |||
| −13045C>T>A | C | 58 | (78.4) | 68 | (89.5) | 0.11 |
| T | 3 | (4) | 3 | (3.9) | ||
| A | 13 | (17.6) | 5 | (6.6) | ||
| −12915C>T | C | 2 | (2.7) | 8 | (10.5) | 0.098 |
| T | 72 | (97.3) | 68 | (89.5) | ||
| −12888 T>A | T | 73 | (98.6) | 72 | (94.7) | 0.37 |
| A | 1 | (1.4) | 4 | (5.3) | ||
| −10107A>G | A | 61 | (82.4) | 63 | (82.9) | 1 |
| G | 13 | (17.6) | 13 | (17.1) | ||
| −10056T>C | T | 2 | (2.7) | 5 | (6.6) | 0.44 |
| C | 72 | (97.3) | 71 | (93.4) | ||
| −9997C>T | C | 72 | (97.3) | 72 | (94.7) | 0.68 |
| T | 2 | (2.7) | 4 | (5.3) | ||
| −8430A>G | A | 70 | (94.6) | 66 | (86.8) | 0.16 |
| G | 4 | (5.4) | 10 | (13.2) | ||
| −8318A>G | A | 55 | (74.3) | 56 | (73.7) | 1 |
| G | 19 | (25.7) | 20 | (26.3) | ||
| −8375G>T | G | 60 | (81.1) | 65 | (85.5) | 0.51 |
| T | 14 | (18.9) | 11 | (14.5) | ||
| −5426G>A | G | 62 | (83.8) | 67 | (88.2) | 0.49 |
| A | 12 | (16.2) | 9 | (11.8) | ||
| −4949C>T | C | 49 | (66.2) | 50 | (65.8) | 1 |
| T | 25 | (33.8) | 26 | (34.2) | ||
Determined by Fisher exact test
Stability of Predicted mRNA Secondary Structures associated with 348T and 348C
A total of thirteen haplotypes with frequencies ≤ 0.01 were predicted among AgP cases. Five pairs of haplotypes that differed only at 348T>C were identified. The free energy (dG) values of mRNA structures predicted for the 348T-containing haplotypes 301G.348T.546A.568A.576T, 301C.348T.546C.568A.576G, 301C.348T.546C.568A.576T, 301G.348T.546C.568A.576G and 301G.348T.546C.568A.576T were, on average, 2.73 ± 0.26 kcal/mole higher than those predicted for the 348C-containing haplotypes 301G.348C.546A.568A.576T, 301C.348C.546C.568A.576G, 301C.348C.546C.568A.576T, 301G.348C.546C.568A.576G and 301G.348C.546C.568A.576T (P < 0.001, paired t test). The mRNA associated with haplotypes containing 348T consistently exhibited a different secondary structure and higher free energy (lower stability) than those containing 348C (data not shown).
DISCUSSION
The present study extends our previous observation that FPR1 SNP 348T>C is associated with AgP to a larger population of African Americans. Subjects with the 348T/T genotype appear to have a significantly increased risk of developing AgP compared to those with the 348T/C or 348C/C genotypes (odds ratio = 18.9). Consistent with our hypothesis, subjects with the 348T/T genotype exhibit a significantly lower PMN chemotactic response to formylpeptides than observed with subjects with the 348T/C or 348C/C genotypes. However, our studies refute the hypothesis that 348T contributes to impaired chemotaxis through a mechanism involving reduction of FPR1 transcripts and provide no evidence that 348T is associated with SNPs in the 5’ region of FPR1.
Consistent with previous studies,3,4,5,6 PMNs from our AgP cases exhibited significantly decreased chemotaxis toward fMLF and reduced FPR1 expression relative to PMNs from controls. We observed a 19–22% decrease in chemotaxis activity, while previous investigations reported a decrease of approximately 20–50%. This discrepancy may be related to differences in chemotaxis assay techniques. Many past studies used blind-well Boyden chambers and measured average PMN migration at various levels of a thick filter. The present study used a modified Boyden chamber and assayed PMN migration though a thin filter, an approach that is perhaps less sensitive. Subjects with the 348T/T genotype, who were all AgP cases, exhibited the lowest level of PMN chemotactic activity observed in this study. This finding supports the hypothesis that 348T/T increases the risk of developing AgP through a mechanism that involves impairment of PMN chemotaxis.
Although 348T is a synonymous SNP that does not alter the FPR amino acid sequence, it is reasonable to hypothesize that it could be associated with other SNPs in the FPR1 promoter or enhancer regions. Linkages of this type have been observed in the human β2 adrenergic receptor24 and the CC chemokine receptor,25 in which specific haplotypes containing variants of synonymous coding region SNPs and non-synonymous promoter SNPs were associated with altered receptor expression. In the present study, subjects with the 348T/C genotype exhibited the highest level of relative FPR1 expression, while those with the 348T/T genotype presented the lowest level of expression. Interestingly, FPR1 expression was significantly higher in 348T/C controls than in cases with the same genotype. It is not unusual for genes to exhibit individual differences in allelic expression.31 While there were no statistically significant differences in FPR1 expression between the three 348T>C genotypes (Table 4), the relatively small number of subjects with the 348T/T genotype limited the power of statistical analysis.
Sequencing analysis of the FPR1 5’ region in our subjects revealed eleven SNPs, including the -4949C>T SNP that is located in the putative FPR1 promoter region. A previous study of Japanese AgP cases and controls reported a total of eight SNPs in this region.15 Seven of these SNPs were detected in the present study, along with four SNPs that had not been previously described. None of these 5’ SNPs occurred at a significantly different frequency in cases than in controls, although there were differences in the frequencies of -13045C>T>A, -12915C>T, and -8430A>G that approached statistical significance. In a previous study of Japanese subjects, -12915C>T and -8430A>G were associated with AgP, while -12915C>T was associated with decreased FPR1 expression and decreased transcriptional efficiency.15 Haplotype analysis was conducted with -13045C>T>A, -12915C>T, and -8430A>G to determine whether any of these 5’ SNPs are linked with 348T, but no association was found. Collectively, our findings suggest that the 348T allele is not associated with significantly decreased FPR1 expression.
While it is unclear why individuals with the 348T/T genotype exhibit a lower PMN chemotactic response to fMLF, it is possible that 348T could reduce the number of FPRs through a post-transcriptional regulatory mechanism. Consistent with this possibility, we recently reported that the mRNA associated with the haplotype 348T.568A exhibited higher free energy (lower stability) and a different secondary structure than that predicted for 348C.568A, which could potentially lower mRNA stability and decrease translational efficiency.23 Moreover, within five haplotype pairs differing only at 348T>C, the predicted secondary mRNA structures of haplotypes containing 348T were all associated with higher free energy (and lower stability) than those containing 348C. There is increasing evidence that synonymous SNPs that are associated with disease can affect mRNA stability. As an example, a single synonymous SNP 971T>C in the gene encoding corneodesmosin is associated with psoriasis and contributes to a 2-fold increase in mRNA stability. Transcripts associated with the 971T variant of this SNP have a decreased affinity for a cytoplasmic protein, which affects the rate of mRNA decay.32 Furthermore, the synonymous SNP 3435C>T in the gene encoding the multidrug resistance 1 protein contributes to a 2-fold decrease in mRNA levels. The 3435T variant of this SNP reportedly changes mRNA folding structure.33
AgP is a multifactorial disorder that involves a complex interplay of environmental, genetic and behavioral factors that could potentially affect individual susceptibility to disease. It often results in early tooth loss and significant functional and esthetic problems. The findings of this study suggest that African Americans who have a homozygous 348T genotype have an increased risk of developing AgP. This susceptibility may, in part, be related to impaired PMN chemotaxis to formylpeptides. Contrary to our hypothesis, 348T does not appear to be linked to FPR1 5’SNPs that could potentially regulate the level of FPR1 expression at the level of transcription. However, evidence from molecular modeling suggests that 348T could alter the folding of FPR mRNA in a manner that decreases its stability. It is uncertain whether these changes in folding decrease the half-life of FPR1 mRNA and exert a clinically significant post-transcriptional influence on the number of FPR expressed by PMNs. Further investigations are needed to fully understand the mechanism by which 348T is associated with decreased PMN chemotaxis.
Key finding
The homozygous FPR1 348T genotype is associated with an increased risk of Aggressive Periodontitis and significant impairment of PMN chemotaxis, but is not associated with a significant reduction in the level of FPR1 transcripts in PMNs.
ACKNOWLEDGMENTS
This investigation was supported by United States Public Health Service research grant R21 DE017178 from the National Institute of Dental and Craniofacial Research, National Institutes of Health, Bethesda, MD 20892, USA. The authors are grateful for the assistance of Dr. John Mills of Montana State University, Bozeman, MT. Drs. Maney and Walters report no conflicts of interest related to commercial products used in this study.
Footnotes
Supported by USPHS grant R21 DE017178 from the National Institute of Dental and Craniofacial Research, National Institutes of Health, Bethesda, MD 20892, USA
AP-48, Neuroprobe Inc, Gaithersburg, MD, USA
Sigma-Aldrich, St. Louis, MO
Neuroprobe Inc, Gaithersburg, MD, USA
Protocol Hema3® stain kit, Fisher Scientific, Pittsburgh, PA,USA
TRIzol® Reagent, Invitrogen, Carlsbad, CA, USA
Reverse Transcription System, Promega, Madison, WI., USA
Taqman® Gene Expression Assays, Applied Biosystems, Foster City, CA, USA
Taqman Universal PCR Master Mix Applied Biosystems, Foster City, CA, USA
Prism 7000 Sequence Detection System Software, Applied Biosystems, Foster City, CA, USA
QIAamp DNA Blood Mini Kit, Qiagen, Valencia, CA, USA
PCR SuperMix, Invitrogen, Carlsbad, CA, USA
3730 automated DNA analyzer, Applied Biosystems, Foster City, CA, USA
Golden Helix Inc., Bozeman, MT, USA. HelixTree® Software
REFERENCES
- 1.Lang N, Bartold PM, Cullinan M, et al. Consensus report: aggressive periodontitis. Ann Periodontol. 1999;4:53. [Google Scholar]
- 2.Holt SC, Kesavalu L, Walker S, Genco CA. Virulence factors of Porphyromonas gingivalis. Periodontology 2000. 1999;20:168–238. doi: 10.1111/j.1600-0757.1999.tb00162.x. [DOI] [PubMed] [Google Scholar]
- 3.Van Dyke TE, Levine MJ, Tabak LL, Genco RJ. Reduced chemotactic peptide binding in juvenile periodontitis: A model for neutrophil function. Biochem Biophys Res Commun. 1981;100:1278–1284. doi: 10.1016/0006-291x(81)91962-8. [DOI] [PubMed] [Google Scholar]
- 4.Sigusch B, Eick S, Pfister W, Klinger G, Glockmann E. Altered chemotactic behavior of crevicular PMNs in different forms of periodontitis. J Clin Periodontol. 2001;28:162–167. doi: 10.1034/j.1600-051x.2001.028002162.x. [DOI] [PubMed] [Google Scholar]
- 5.Perez HD, Kelly E, Elfman F, Armitage G, Winkler J. Defective polymorphonuclear leukocyte formyl peptide receptors in juvenile periodontitis. J Clin Invest. 1991;87:971–976. doi: 10.1172/JCI115105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tufano MA, Ianniello R, Sanges MR, Rossano F. Neutrophil function in rapidly progressive and adult periodontitis. Eur J Epidemiol. 1992;8:67–73. doi: 10.1007/BF03334974. [DOI] [PubMed] [Google Scholar]
- 7.Pike MC, Snyderman R. Leukocyte chemoattractant receptors. Meth Enzymol. 1988;162:236–245. doi: 10.1016/0076-6879(88)62080-5. [DOI] [PubMed] [Google Scholar]
- 8.Katanaev VL. Signal transduction in neutrophil chemotaxis (in English) Biochemistry (Moscow) 2001;66:351–368. doi: 10.1023/a:1010293809553. [DOI] [PubMed] [Google Scholar]
- 9.Miyasaki KT. The neutrophil-mechanisms of controlling periodontal bacteria. J Periodontol. 1991;62:761–774. doi: 10.1902/jop.1991.62.12.761. [DOI] [PubMed] [Google Scholar]
- 10.Smith JA. Neutrophils, host defense and inflammation: a double-edged sword. J Leukoc Biol. 1994;56:672–686. doi: 10.1002/jlb.56.6.672. [DOI] [PubMed] [Google Scholar]
- 11.Mills JS, Miettinen HM, Vlases MJ, Jesaitis AJ. The N-formyl Peptide Receptor: Structure, Signaling and Disease. In: Serhan CN, Ward PA, editors. Molecular Biology of Inflammation. New York: Humana Press; 1999. pp. 215–245. [Google Scholar]
- 12.Gao J, Lee EJ, Murphy PM. Impaired antibacterial host defense in mice lacking the N-formyl peptide receptor. J Exp Med. 1999;189:657–662. doi: 10.1084/jem.189.4.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gwinn MR, Sharma A, De Nardin E. Single nucleotide polymorphisms of the N-formyl peptide receptor in localized juvenile periodontitis. J Periodontol. 1999;70:1194–1201. doi: 10.1902/jop.1999.70.10.1194. [DOI] [PubMed] [Google Scholar]
- 14.Zhang Y, Syed R, Uygar C, et al. Evaluation of human leukocyte N-formylpeptide receptor (FPR1) SNPs in aggressive periodontitis patients. Genes Immun. 2003;4:22–29. doi: 10.1038/sj.gene.6363900. [DOI] [PubMed] [Google Scholar]
- 15.Gunji T, Onouchi Y, Nagasawa T, et al. Functional polymorphisms of the FPR1 gene and aggressive periodontitis in Japanese. Biochem Biophys Res Commun. 2007;364:7–13. doi: 10.1016/j.bbrc.2007.09.105. [DOI] [PubMed] [Google Scholar]
- 16.Sahagun-Ruiz A, Colla JS, Juhn J, Gao JL, Murphy PM, McDermott DH. Contrasting evolution of the human leukocyte N-formylpeptide receptor subtypes FPR and FPRL1R. Genes Immun. 2001;2:335–342. doi: 10.1038/sj.gene.6363787. [DOI] [PubMed] [Google Scholar]
- 17.Nibali L, Parkar M, Brett P, Knight J, Tonetti MS, Griffiths GS. NADPH oxidase (CYBA) and FcgammaR polymorphisms as risk factors for aggressive periodontitis: a case-control association study. J Clin Periodontol. 2006;33:n529–n539. doi: 10.1111/j.1600-051X.2006.00952.x. [DOI] [PubMed] [Google Scholar]
- 18.Schreiber RE, Prossnitz ER, Ye RD, Cochrane CG, Bokoch GM. Domains of the human neutrophil N-formyl peptide receptor involved with G protein coupling. J Biol Chem. 1994;269:326–331. [PubMed] [Google Scholar]
- 19.Wenzel-Seifert K, Seifert R. Functional differences between human formyl peptide receptor isoforms 26, 98 and G6. Naunyn-Schmiedeberg’s Arch Pharmacol. 2003;367:509–515. doi: 10.1007/s00210-003-0714-7. [DOI] [PubMed] [Google Scholar]
- 20.Perez HD, Holmes R, Vilander LR, et al. Formyl peptide receptor chimeras define domains involved in ligand binding. J Biol Chem. 1993;268:2292–2295. [PubMed] [Google Scholar]
- 21.Perez HD, Vilander LR, Andrews WH, Holmes R. Human formyl peptide receptor ligand binding domains. J Biol Chem. 1994;269:22485–22487. [PubMed] [Google Scholar]
- 22.Palczewski K, Kumasaka T, Hori T, et al. Crystal structure of rhodopsin: a G-protein-coupled receptor. Science. 2000;289:739–774. doi: 10.1126/science.289.5480.739. [DOI] [PubMed] [Google Scholar]
- 23.Maney P, Emecen P, Mills JS, Walters JD. Neutrophil Formylpeptide Receptor Single Nucleotide Polymorphism 348T>C in Aggressive Periodontitis. [Accessed December 27, 2008];Online J Periodontol (serial online) 2008 December 26; doi: 10.1902/jop.2009.080225. doc 225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Drysdale CM, McGraw DW, Stack CB, et al. Complex promoter and coding region β2-adrenergic receptor haplotypes alter receptor expression and predict in vivo responsiveness. Proc Natl Acad Sci USA. 2000;97:10483–10488. doi: 10.1073/pnas.97.19.10483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mummidi S, Banshad M, Ahuja SS, et al. Evolution of human and non-human primate CC chemokine receptor 5 gene and mRNA. J Biol Chem. 2000;275:18946–18961. doi: 10.1074/jbc.M000169200. [DOI] [PubMed] [Google Scholar]
- 26.Armitage GC. Development of a classification system for periodontal diseases and conditions. Ann Periodontol. 1999;4:1–6. doi: 10.1902/annals.1999.4.1.1. [DOI] [PubMed] [Google Scholar]
- 27.Tonetti MS, Mombelli A. Early-onset periodontitis. Ann Periodontol. 1999;4:39–52. doi: 10.1902/annals.1999.4.1.39. [DOI] [PubMed] [Google Scholar]
- 28.Boyum A. Isolation of mononuclear cells and granulocytes from human peripheral blood. Scand J Clin Lab Invest. 1968;21:77–89. [PubMed] [Google Scholar]
- 29.Bailey MT, Engler H, Powell ND, Padgett DA, Sheridan JF. Repeated social defeat increases the bactericidal activity of splenic macrophages through a Toll-like receptor-dependent pathway. Am J Physiol Regul Integr Comp Physiol. 2007;293:R1180–R1190. doi: 10.1152/ajpregu.00307.2007. [DOI] [PubMed] [Google Scholar]
- 30.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;3:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Buckland PR. Allele-specific gene expression differences in humans. Hum Mol Genet. 2004;13:R255–R260. doi: 10.1093/hmg/ddh227. Review. [DOI] [PubMed] [Google Scholar]
- 32.Capon F, Allen MH, Ameen M, et al. A synonymous SNP of the corneodesmosin gene leads to increased mRNA stability and demonstrates association with psoriasis across diverse ethnic groups. Hum Mol Genet. 2004;13:2361–2368. doi: 10.1093/hmg/ddh273. [DOI] [PubMed] [Google Scholar]
- 33.Wang D, Sadée W. Searching for polymorphisms that affect gene expression and mRNA processing: example ABCB1 (MDR1) AAPS J. 2006;8:E515–E520. doi: 10.1208/aapsj080361. Review. [DOI] [PMC free article] [PubMed] [Google Scholar]