Abstract
Atrial natriuretic peptide (ANP) exerts its biological effects by binding to guanylyl cyclase/natriuretic peptide receptor-A (GC-A/NPRA), which generates the second messenger cGMP. The molecular mechanism mediating Npr1 (coding for GC-A/NPRA) gene regulation and expression is not well understood. The objective of this study was to elucidate the mechanism by which Ets-1 contributes towards the regulation of Npr1 gene transcription and expression. Chromatin immunoprecipitation and gel shift assays confirmed the in vivo and in vitro binding of Ets-1 to Npr1 promoter. Overexpression of Ets-1 significantly enhanced NPRA mRNA levels, protein expression, guanylyl cyclase (GC) activity, and ANP-stimulated intracellular accumulation of cGMP levels in transfected cells. Depletion of endogenous Ets-1 by small interfering RNA (siRNA) dramatically decreased promoter activity by 80%. Moreover, methylation of the Npr1 promoter region −356 to +55 significantly reduced the promoter activity and hypermethylation around the Ets-1 binding sites directly reduced the Ets-1 binding to Npr1 promoter. Collectively, our present results demonstrate that the Npr1 gene transcription and GC activity of the receptor are critically controlled by Ets-1 in target cells.
Keywords: Atrial natriuretic peptide, guanylyl cyclase receptor, Npr1 gene promoter, gene transcription and expression, Ets-1
Introduction
Atrial natriuretic peptide (ANP) and brain natriuretic peptide (BNP) principally mediate natriuretic, diuretic, vasorelaxant, and antimitogenic responses, largely directed to reduce blood pressure, and to maintain fluid volume homeostasis [1–3]. ANP and BNP specifically bind to guanylyl cyclase-A/natriuretic peptide receptor-A (GC-A/NPRA), which produces the intracellular second messenger cGMP in response to hormone binding [4–7]. Several studies with Npr1 (coding for GC-A/NPRA) gene-disruption mouse models have revealed the hallmark significance of NPRA in lowering arterial pressure and protecting against renal and cardiac pathophysiological functions [8–11]. It has been reported that in Japanese individuals, genetic mutations in the human Npr1 gene promoter confer increased susceptibility to essential hypertension and left ventricular hypertrophy [12]. It has also been demonstrated that the human Npr1 deletion allele lacking eight nucleotides, which alters the binding sites of activator protein 2 (AP-2) and zeste, leading to decreased ability to bind transcription factor AP-2 and decreased promoter activity than the wild-type allele.
Relatively little is known about the regulation of Npr1 gene expression in target cells. Earlier studies have demonstrated that functional interaction of NF-Y with Sp1 is essential for optimal transcription of the Npr1 gene in vascular smooth muscle cells [13]. The complete genomic nucleotide sequence and promoter region analysis of murine Npr1 gene indicated that the core transcriptional machinery of the TATA-less Npr1 promoter contains three potential Sp1 binding sites, one inverted CCAAT box, and several putative cis-acting motifs for known transcription factors, including Ets-1, suggesting that they have a function in regulating Npr1 gene transcription [14]. Previous studies have shown that Npr1 basal promoter lies between the region −356 to +55 relative to transcription start site and its transcriptional activity is modulated by GATA-1, Ets-1, and LyF-1 transcription factors [15]. Ets proteins activate or repress the expression of various genes, including c-fos, c-myc, jun B, p53, and bcl-2 [16, 17]. Ets-1 protein, which is expressed in a variety of cell types including endothelial cells, mesangial cells, and vascular smooth muscle cells, regulates the transcription of several genes involved in angiogenesis and remodeling of the extracellular matrix [18, 19]. Previous studies have shown that Ets-1 protein is essential for normal coronary and myocardial development [17, 20]. Ets-1 also plays a role in kidney development by activating fork-head-related transcription factor detected during nephrogenesis [21].
Although Npr1 gene regulation is poorly understood, the activity and expression of NPRA, assessed primarily through ANP-stimulated cGMP accumulation, are regulated by various factors, including hormones such as endothelin, glucocorticoids, growth factors, and certain physiological and pathophysiological conditions [22–24]. Angiotensin II (Ang II) has been shown to repress Npr1 transcriptional activity by binding to its response element (RE), located in the promoter region −1346 to −916 base pairs (bp) from the transcription start site (TSS) [25, 26]. Previous studies have shown that NPRA expression is regulated by natriuretic peptides [27] as well as transcriptional repression by cGMP, which is mediated by cGMP-RE in the Npr1 promoter at position −1372 to −1354 bp from TSS [28]. Indeed, better understanding of the regulation of NPRA expression requires a more extensive functional characterization of its promoter region and the functional significance of the potential cis-elements in this region. The present study demonstrates that Ets-1 plays an integral role in the regulation of Npr1 transcription and GC activity of the receptor through its consensus binding sites present in the Npr1 gene promoter.
Materials and Methods
Materials
The pGL3-basic vector, pRL-TK, and dual luciferase assay system were purchased from Promega (Madison, WI). The plasmid isolation kit and RNeasy mini-kit for total RNA isolation were obtained from Qiagen (Valencia, CA). We purchased sequence-specific oligonucleotides from Midland Certified Reagent Company (Midland, TX). Cell culture media, fetal calf serum, ITS (insulin, transferrin, and sodium selenite), Lipofectamine-2000, and Superscript one-step RT-PCR kit were obtained from Invitrogen (Carlsbad, CA). Phenylmethyl sulfonyl fluoride (PMSF), aprotinin, and leupeptin were obtained from Sigma Chemical Co. (St. Louis, Mo). The HhaI, HpaII, and SssI methylases and restriction enzymes HhaI, HpaII, and BstUI were purchased from New England Biolabs (Ipswich, MA). We obtained the LightShift Chemiluminescent EMSA kit from Pierce (Rockford, IL) and the EZ ChIP kit from Upstate Biotechnology (Temecula, CA). The direct cyclic GMP correlate-EIA kit was purchased from Assay Designs (Ann Arbor, MI.). Antibodies; anti-Ets-1 (N-276) and anti-rabbit IgG, Ets-1 siRNA, control siRNA and protein A-agarose were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). NPRA antibody was obtained from Genway Biotech (San Diego, CA). The expression vectors for Ets-1 (pEVRF0-Ets-1) and its empty vector (pEVRF0) were kindly provided by Dr. Paul Brindle [29]. All other chemicals used were of molecular biology reagent grade.
Plasmids and promoter constructs
The promoter-luciferase reporter constructs were generated by cloning PCR-amplified DNA fragments of various lengths of murine Npr1 gene promoter upstream of the promoterless firefly luciferase gene in the vector pGL3-basic. The cloning of various deletion constructs of Npr1 promoter and constructs having mutations at Ets-1A, Ets-1B or both sites have been described earlier [15].
Cell Transfection and Luciferase Assay
Mouse mesangial cells (MMCs) were isolated and cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal calf serum and ITS as previously described [30]. The MMCs were seeded in 24-well plates and experiments were performed using cells between 4 to 18 passages. Cloned mouse Leydig tumor (MA-10) cells were cultured in modified Waymouth’s medium supplemented with 15% horse serum as previously described [31]. The cultures were maintained at 37°C in an atmosphere of 5% CO2 and 95% O2. The cells were transfected using Lipofectamine-2000 reagent with 1 µg of promoter-reporter construct and 300 ng of pRL-TK carrying the Renilla luciferase gene downstream of thymidine kinase promoter, which was used as an internal transfection control. For cotransfection experiments, expression plasmids of varying concentrations were transfected along with the promoter-reporter construct. Cells were lysed after 48 h and the lysate was used to measure firefly and Renilla luciferase activities with Promega dual luciferase assay kit using a TD 20/20 luminometer (Turner Designs). The results were normalized for the transfection efficiency as relative to light units per Renilla luciferase activity. In Ets-1 overexpression experiments, cells were transfected with expression vectors for Ets-1 (pEVRF0-Ets-1) or its empty vector (pEVRF0). The total DNA content was equalized by inclusion of pEVRF0 plasmid. To examine the transfection efficiency, cells were transfected with pCMV β-galactosidase control plasmid and transfection efficiency was assessed by using in situ β-galactosidase staining kit from Stratagene (La Jolla, CA) according to manufacturer’s protocol. In MMCs and MA-10 cells, the transfection efficiency was found to be 85% and 90%, respectively using Lipofectamine-2000.
Reverse Transcriptase–PCR Assay
Cells were transfected with appropriate expression vector and 48 h after transfection, total RNA was extracted using RNeasy mini-kit (Qiagen, Valencia, CA). One microgram of total RNA was reverse-transcribed using the Superscript one-step RT-PCR with platinum Taq system. Primer sequence for amplification of NPRA and β-actin, as well as PCR conditions, were used as described previously [25]. The amplified PCR product increased linearly up to 40 cycles (data not shown). Control experiments were performed with RNA samples but without reverse transcriptase. The specific primers for β-actin gene were included in the PCR reaction as an internal control. The expected sizes of the amplified NPRA and β-actin PCR products are 456 and 256 bp, respectively. After amplification, 15 µl of each PCR reaction mixture were electrophoresed through a 1.5% agarose gel with ethidium bromide (0.5 µg/ml). The gel was digitized and signal intensities of the corresponding bands were quantified using an Alpha Imaging System (Alpha Innotech, San Leandro, CA).
Whole Cell Lysate Preparation and Immunoblot Assay
Forty-eight hours after transfection, cells were lysed and whole cell lysate was prepared essentially as described earlier [15]. The protein concentration of the lysates was estimated using a Bradford protein detection kit (Bio-Rad). Whole cell lysate (50–80 µg) from each sample was mixed with sample loading buffer and separated by using 10% SDS-polyacrylamide gel electrophoresis (PAGE). Proteins were electrotransferred onto a polyvinylidene fluoride (PVDF) membrane. The membrane was blocked with 1X Tris-buffered saline-Tween 20 (TBST) containing 5% fat-free milk for 2 h at room temperature and incubated overnight at 4oC in TBST containing 3% fat-free milk with primary antibodies (1:1000 dilution). The membrane was then treated with corresponding secondary anti-rabbit, anti-mouse, or anti-chicken horseradish peroxidase (HRP)-conjugated antibodies (1:5000 dilutions). Protein bands were visualized by enhanced chemiluminescence (ECL) plus detection system.
Electrophoretic Mobility Shift Assay
Nuclear extract was prepared according to the method of Digman et al., [32]. Protein-DNA complexes were detected using 5' biotin end-labeled double-stranded DNA probes prepared by annealing complementary oligonucleotides. The forward sequences of the wild-type oligonucleotides for Ets-1A and Ets-1B were 5′-ccgcccgcctccggaacggccggag-3′ and 5′-tgggccagccggacgccccttctg-3′, respectively. The respective forward sequences for the mutant Ets-1A and Ets-1B binding sites are underlined; 5′-cccgcccgcctcattcacggccgg-3′ and 5′-gggccagcattccgccccttctg-3′. Briefly, MMCs nuclear extract (1.5 µg of protein) in binding buffer was incubated on ice for 5 min in a total volume of 20 µl before addition of biotin-labeled probe of Ets-1A (40 fmol) or Ets-1B (20 fmol). The reaction was allowed to incubate for an additional 25 min at room temperature. In competition experiments, the nuclear extract was preincubated with 200-fold molar excess of unlabeled double-stranded wild-type or mutant oligonucleotides for 20 min on ice. Protein-DNA complexes were separated on nondenaturing polyacrylamide gel and observed using the LightShift Chemiluminescent EMSA kit.
Chromatin Immunoprecipitation Assay
The chromatin immunoprecipitation (ChIP) assay was done using the EZ ChIP kit according to the manufacturer’s instructions. Briefly, 48 h after transfection, cells (1 × 106 ) were cross-linked in 1% formaldehyde for 10 min at room temperature and the reaction was quenched with 0.1 M glycine. Cells were scraped, pelleted, resuspended in 300 µl SDS lysis buffer on ice, and then sonicated 3 times for 10 sec each at 30% input producing fragments between 500 to 1,000 bp. The suspension was centrifuged at 10,000 × g for 10 min at 4°C. Supernatant was diluted 10-fold in ChIP dilution buffer. Immunoprecipitation was performed with protein G-agarose and 5 µg of the Ets-1 antibody or control IgG. The chromatin solution was precleared by adding protein G-agarose for 2 h at 4°C and incubated with antibodies overnight at 4°C with rotation. The DNA-protein-antibody complex was captured on protein G-agarose beads by incubating for 1 h at 4°C. Beads were pelleted and washed sequentially for 5 min each with low-salt buffer, high-salt buffer, lithium chloride wash buffer, and 1 X Tris-EDTA. Bound protein was eluted twice from the beads by gently rotating for 15 min in ChIP elution buffer at room temperature. After elution of protein/DNA complex from the beads, crosslinking was reversed at 65°C overnight to release DNA. Immunoprecipitated DNA was sequentially treated with RNase A and Proteinase K, and then purified. PCR of the Npr1 promoter region containing Ets-1 binding sites was carried out using purified DNA as a template. For PCR amplification, the forward primer was 5′-ctctcttgtcgccgaatctg-3′; the reverse primer was 5′- tctcgttctctcgctctccac −3′.
cGMP Assay
Forty-eight hours after transfection, cells were treated with ANP at 37°C for 20 min in the presence of 0.2 mM 3-isobutyl-1-methylxanthine. Cells were washed three times with PBS and scraped into 0.5 N HCl. Cell suspensions were subjected to 5 cycles of freeze and thaw, and then centrifuged at 10,000 rpm for 15 min. The supernatant thus collected was used for the cGMP assay using the direct cyclic GMP correlate-EIA kit according to the manufacturer’s protocol.
Plasma Membrane Preparation and Guanylyl Cyclase Activity Assay
The plasma membranes were prepared by suspending cell pellet in 5 volumes of sodium phosphate buffer (10 mM, pH 7.4) containing 250 mM sucrose, 150 mM NaCl, 1 mM PMSF, 5 mM benzamidine, 5 mM EDTA, and 10 µg/ml each of leupeptin and aprotinin, as described previously [26]. Briefly, cells were homogenized and centrifuged at 400 × g for 10 min at 4°C and the supernatant collected was recentrifuged at 80,000 × g for 1 h at 4°C. The resultant supernatant was discarded, the pellet was resuspended in 1 ml of HEPES buffer (50 mM, pH 7.4) containing 150 mM NaCl, 1 mM PMSF, 5 mM benzamidine, 5 mM EDTA, and 10 µg/ml each of leupeptin and aprotinin, and centrifuged at 80,000 × g for 1 h at 4°C. The final pellet was suspended in 200 µl of HEPES buffer (pH 7.4). GC activity was assayed as described by Leitman et al., [33], with some modifications [34]. Fifty microgram aliquot of plasma membrane was added to 100 µl GC assay buffer containing Tris-Cl buffer (50 mM, pH 7.6), 4 mM MnCl2, 2 mM IBMX, 1 mM BSA, 5 units of creatinine phosphokinase, 7.5 mM creatine phosphate, 0.5 mM GTP, and 1 µM ANP. The samples were incubated in a water bath at 37°C for 10 min. Reaction was stopped by adding 900 µl of sodium acetate (55 mM, pH 6.2) and sample tubes were placed in boiling water bath for 3 min and then on ice for 15 min to stop the reaction. Samples were then centrifuged at 13,000 × g for 5 min, supernatant was collected, and cGMP was determined.
Transfection of Small Inhibitory RNA
Cells were cultured to 70%–80% confluence in 10% FBS-supplemented antibiotic-free DMEM with ITS and transfected with Ets-1 small inhibitory RNA (siRNA) (a pool of 3 target-specific 20–25 nucleotide sequence siRNAs) using Lipofectamine-2000 reagent. A nontargeting 20–25 nucleotide sequence siRNA was used as a negative control. Four hours after transfection, fresh medium was added to the plates and, after 24 h, medium was replaced. After 48 h, cells were lysed and the clear supernatant was used to measure firefly and Renilla luciferase activities.
Immunofluorescence Assay
Cells were plated on chamber slides and were transiently transfected with Ets-1 expression vector or siRNA using Lipofectamine-2000 reagent. Twenty-four hours after transfection, cells were fixed for 10 min at room temperature in 4% paraformaldehyde in phosphate-buffered saline (PBS), washed in PBS, permeabilized with PBS/0.2% Triton X-100 for 5 min, and blocked in 2% (w/v) bovine serum albumin in PBS with 0.1% Tween 20 for 10 min. Cells were then incubated with a 1:200 dilution of Ets-1 polyclonal antibody at 4°C overnight, washed three times for 5 min with PBS/0.1%Tween 20, then incubated with 1:400 dilution of FITC-conjugated anti-rabbit secondary antibody at room temperature for 1 h. Slides were washed 3 times for 5 min each with PBS/0.1%Tween 20 before coverslips were mounted on the slides, using Vectashield mounting medium with 4',6-diamidino-2-phenylindole (DAPI) to visualize nuclei. Immunofluorescence was measured using an Olympus BX51TRF microscope at 40 x magnification. An integrated Magnafire SP Digital "Firewire" Camera System was used for image processing.
In Vitro Methylation of Npr1 Promoter
Plasmids pGL3-basic and −356/+55 Npr1 promoter construct were in vitro methylated by HhaI, HpaII, and SssI methyltransferase according to the procedures recommended by the manufacturer (New England Biolabs, Ispwich, MA). The completeness of methylation was checked by measuring the extent of protection from digestion by the restriction enzymes HhaI, HpaII, and BstUI, respectively. Transfection of the plasmids was performed as described above. The wild type Ets-1A and Ets-1B oligonucleotides used in EMSA were also in vitro methylated by SssI methylase as described above. For the unmethylated control, pGL3-basic, −356/+55 Npr1 promoter construct, and the Ets-1A and Ets-1B oligonucleotides were mixed with all components required for in vitro methylation except methylases.
Statistical analysis
The results are expressed as mean ± SE. The statistical significance was evaluated by one-way ANOVA, followed by Dunnett’s multiple comparison tests using PRISM computer software (GraphPad Software, San Diego, CA). A p value of < 0.05 was considered significant.
Results
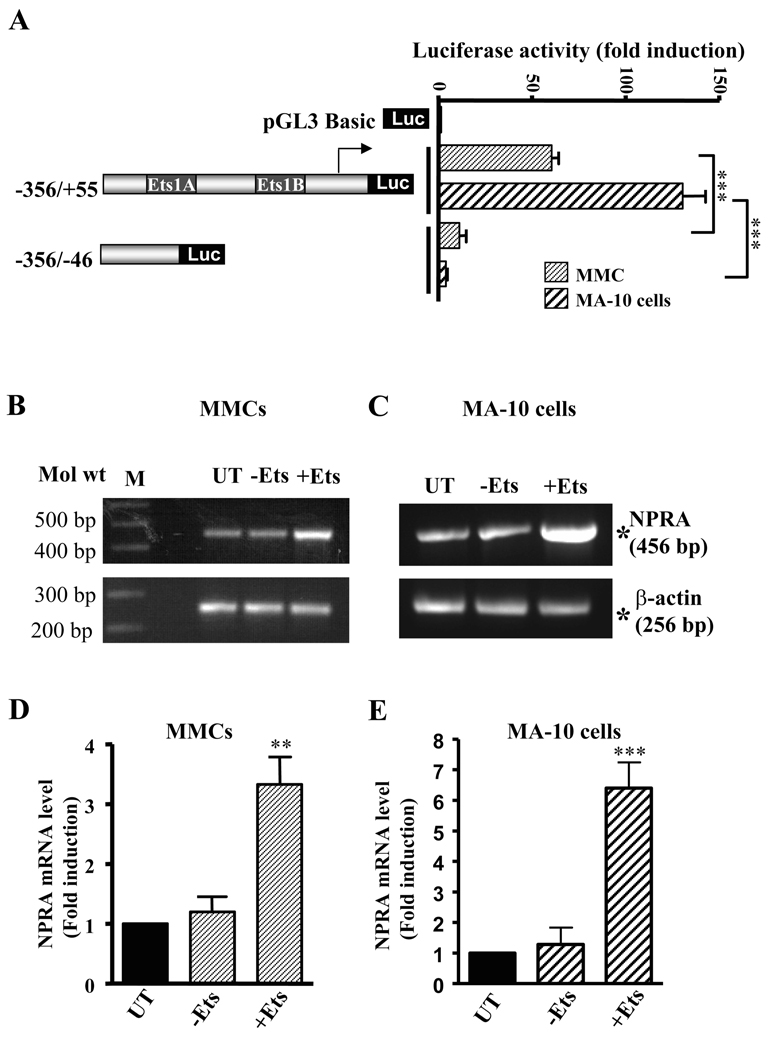
The Npr1 promoter construct (−356/+55) containing Ets-1 binding sites, exhibited 60- to 140-fold induction in luciferase activity when transfected into MMC and MA-10 cells, respectively, as compared with the pGL3-basic plasmid (Fig. 1A). Deletion of the region −46 to +55 containing Ets-1 binding sites in the construct −356/−46 significantly decreased the promoter activity in both cell lines. To investigate the effect of overexpression of Ets-1 on the endogenous Npr1 gene expression in MMCs and MA-10 cells, we analyzed the mRNA levels of NPRA by reverse transcription-PCR assay. Representative levels of Ets-1-enhanced NPRA mRNA in MMCs and MA-10 cells are shown in Fig. 1, B and C, respectively. There was 3.5- to 5-fold induction in NPRA mRNA levels in MMCs and MA-10 cells when transfected with Ets-1 expression plasmid as compared with untransfected controls (Fig. 1, D and E), respectively.
Fig. 1. Effect of Ets-1 on Npr1 gene transcription and expression.
A) Left panel shows the schematic representation of the deletion construct of the Npr1 promoter. Right panel shows the transcriptional activity of these constructs in MMCs and MA-10 cells. Values represent fold induction relative to pGL3-Basic vector. Representative mRNA levels of NPRA and β-actin in (B) MMCs and (C) MA-10 cells transfected with (+Ets-1) and without (−Ets-1) Ets-1 expression plasmid. β-actin was used as a control. M indicates DNA marker; UT indicates untransfected control. (D) and (E) shows the densitometry of NPRA mRNA levels in MMCs and MA-10 cells, respectively. Values represent fold induction relative to untransfected controls. Bars in A, D, and E represent the mean ± SE of three independent experiments in triplicates. **, p < 0.01; ***, p < 0.001.
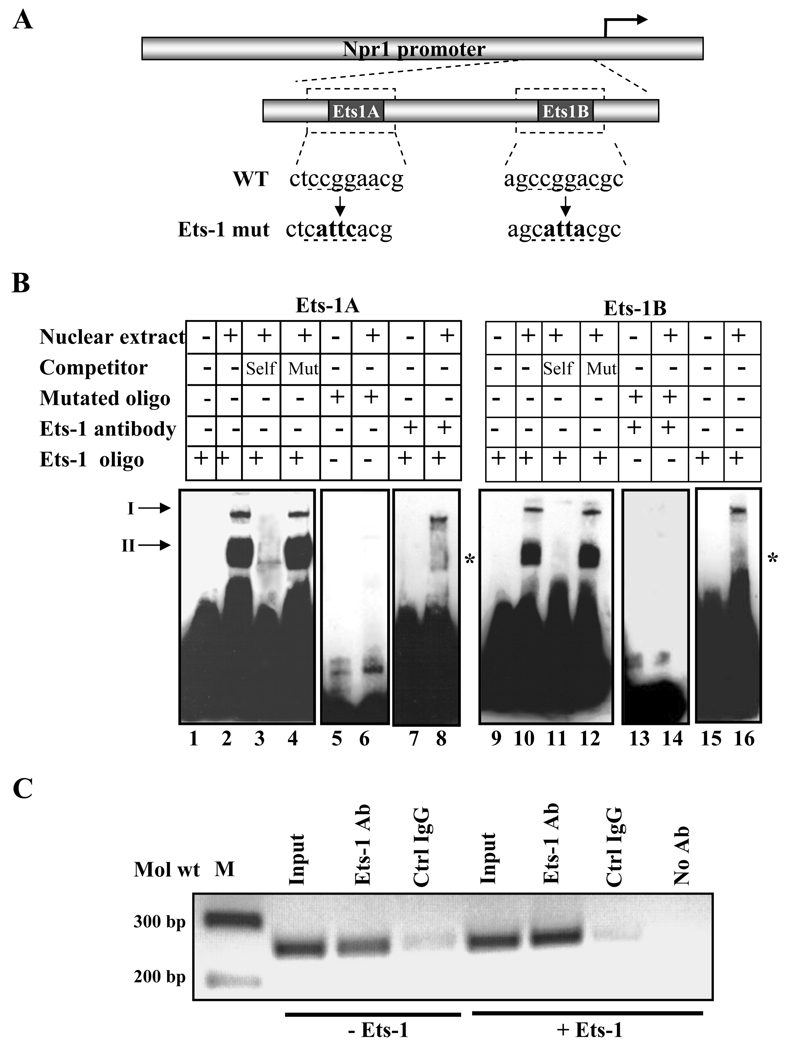
We further investigated the binding of endogenously expressed Ets-1 protein to each of the two Ets-1 binding sites present in the Npr1 promoter using a combination of electrophoretic mobility shift assay including; competition binding, mutant oligonucleotides, and immunoshift assay. Fig. 2A shows the schematic map of the wild-type and mutant Ets-1 binding sites. Incubation of nuclear extract with wild-type Ets-1A and Ets-1B oligonucleotides showed the formation of specific nucleoprotein complexes (Fig. 2B, lanes 2 and 10). These complexes were effectively competed with an addition of 200-fold molar excess of corresponding unlabeled wild-type probe (Fig. 2B, lanes 3 and 11). The addition of unlabeled mutant Ets-1A and Ets-1B site oligonucleotides created no competition for DNA-protein binding (Fig. 2B, lanes 4 and 12). Furthermore, the incubation of labeled Ets-1A and Ets-1B mutant probes with nuclear extract did not exhibit the binding of specific nucleoprotein (Fig. 2B, lanes 6 and 14). The specificity of the protein/DNA complex was confirmed by antibody supershift assays. Incubation of nuclear extract with Ets-1 antibody before the addition of probe disrupted the DNA-protein complex, thus ablating the specific band (Fig. 2B, lanes 8 and 16). In Fig. 2B, lanes 1, 5, 7, 9, 13, and 15 shows the mobility of the probe alone, which was taken as a negative control. We used ChIP assay and PCR analysis to determine whether Ets-1 interacts with Npr1 promoter in a natural chromosome configuration. PCR amplification of the Npr1 promoter region containing Ets-1 binding sites from the immunoprecipitated DNA demonstrated that endogenous Ets-1 binds to the Npr1 promoter in vivo (Fig. 2C). Overexpression of Ets-1 showed enhanced binding of Ets-1 to Npr1 promoter, but no amplification was detected with DNA immunoprecipitated in the absence of antibody.
Fig. 2. In vitro and in vivo binding of Ets-1.
A) Schematic diagram showing the sequence of the wild-type and mutated Ets-1 binding site in the Npr1 promoter. WT and mut indicate the wild-type and mutant sequences, respectively, and underlined nucleotides show the mutated sequence. B) The gel retardation assay in nuclear extract from MMCs using Ets-1A and Ets-1B oligonucleotides. Lanes 2 and 10 show the nuclear protein complex binding with Ets-1A and Ets-1B sites, respectively. Unlabeled competitor DNA was used in a 200-fold molar excess concentration in Lanes 3 and 11 (wild-type Ets-1A and Ets-1B) and Lanes 4 and 12 (mutant Ets-1A and Ets-1B). In lanes 6 and 14, labeled mutant Ets-1A and Ets-1B oligonucleotides, respectively, were used for binding reactions. Ets-1 antibody was used for a supershift assay in lanes 8 and 16. Arrows I and II indicate specific DNA-protein binding complex; asterisk shows the disruption of Ets-1 nuclear protein complex binding in the presence of the antibody. C) ChIP assay of the Npr1 gene promoter in Ets-1-transfected cells. PCR amplification of the immunoprecipitated DNA shows binding of Ets-1 to the Npr1 promoter region, which contains two Ets-1 binding sites. Samples immunoprecipitated with control IgG (Crtl IgG) showed a very faint band and in the absence of antibody showed no detectable signals after PCR amplification. -Ets indicates transfection with empty vector (pEVRF0) and +Ets indicates transfection with Ets-1 expression plasmid. Representative results of three experiments are shown.
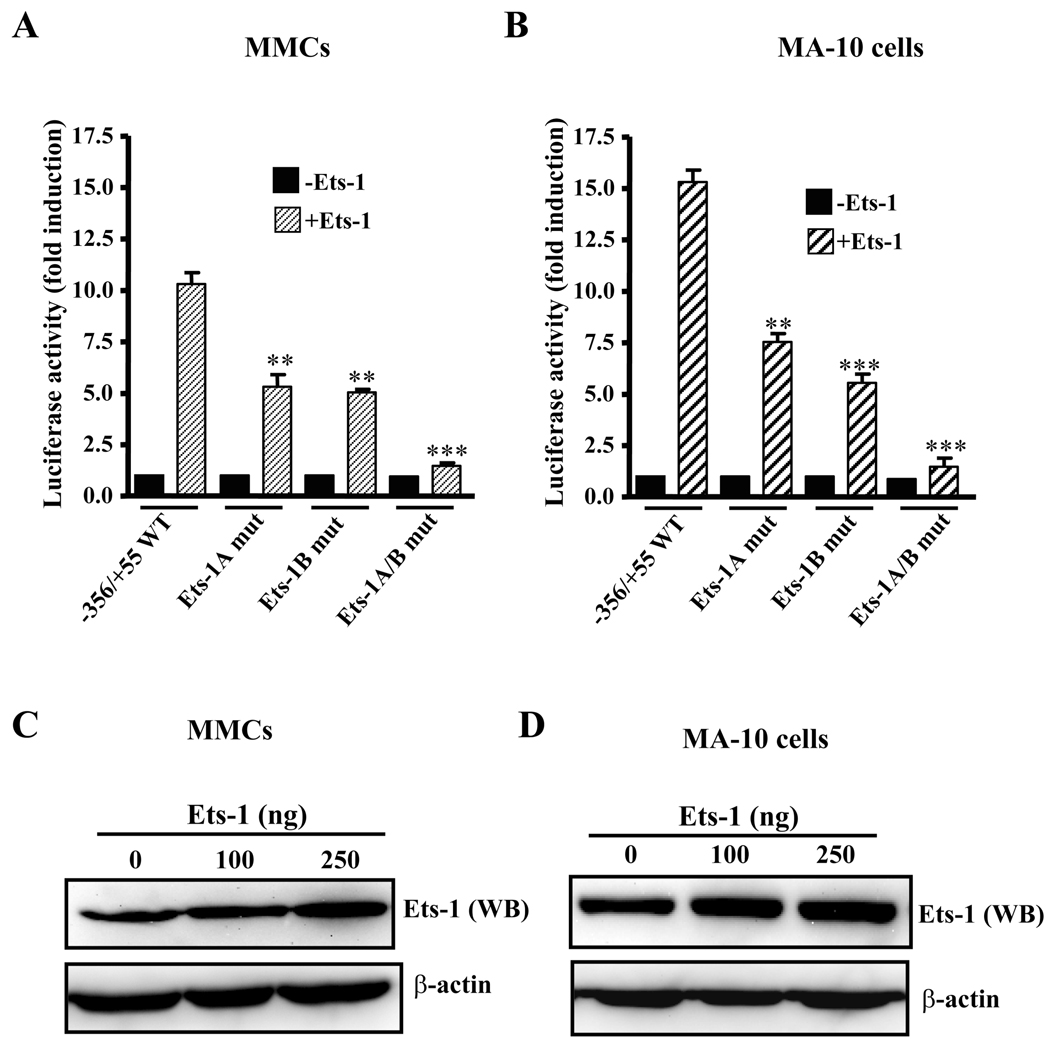
To demonstrate the effect of overexpression of Ets-1 protein on Ets-1-inducible Npr1 gene transcription, we used −356/+55 Npr1 promoter constructs having wild-type or mutants of Ets-1A or Ets-1B sites individually or together and transiently transfected with Ets-1 expression plasmid in MMCs and MA-10 cells (Fig. 3, A and B, respectively). Overexpression of Ets-1 with the wild-type construct increased the promoter activity by 11- and 15-fold in MMC and MA-10 cells, respectively. There was significant reduction of approximately 50% in Ets-1-induced promoter activity with either Ets-1 site mutated as compared with the wild-type −356/+55 construct in both the cell lines (Fig. 3, A and B). Simultaneous mutation of both the Ets-1 sites did not respond to Ets-1-induced Npr1 gene transcription in either cell line (Fig. 3, A and B). The increase in luciferase activity observed after cotransfection with Ets-1 was due to an increase in firefly luciferase activity, whereas there was no significant change in Renilla luciferase activity. The enhanced expression of Ets-1 protein in transfected MMCs and MA-10 cells was confirmed by Western blot analysis using Ets-1 polyclonal antibody (Fig. 3, C and D, respectively). β-actin was used as loading control.
Fig. 3. Effect of overexpression of Ets-1 on Npr1 promoter activity.
Luciferase activity of Npr1 promoter constructs having wild-type (WT) and mutant (mut) Ets-1A and Ets-1B binding sites when transfected in (A) MMCs and (B) MA-10 cells. Ets-1 expression plasmid (250 ng) or empty plasmid pEVRF0 (-Ets-1) was cotransfected along with Npr1 promoter construct. The results were normalized for the transfection efficiency as relative to light units per Renilla luciferase activity. Values represent fold induction as compared with empty vector (−Ets-1). Western blot (WB) analysis of Ets-1 in transfected (C) MMCs and (D) MA-10 cells along with β-actin as a control. Bars in A and B represent the mean ± SE of three to four independent experiments in triplicates. **, p< 0.01; ***, p < 0.001 vs Ets-1-transfected wild type construct.
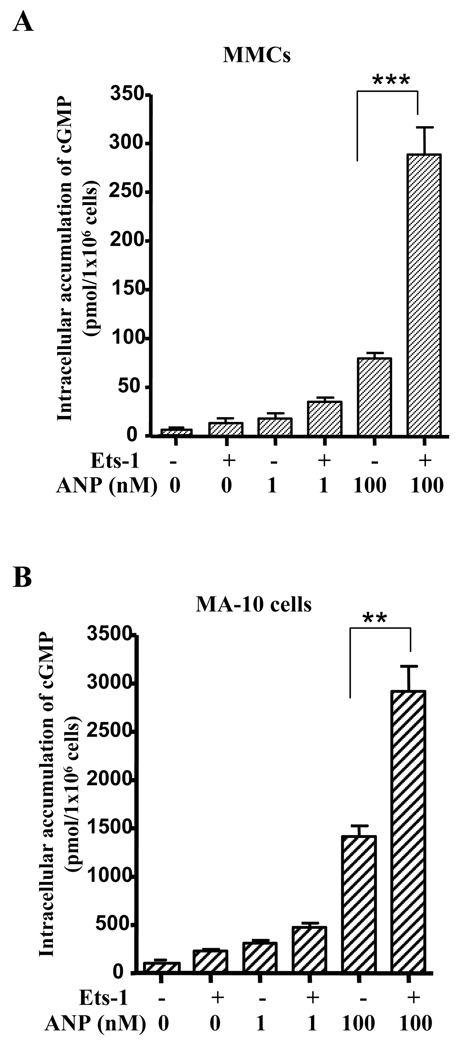
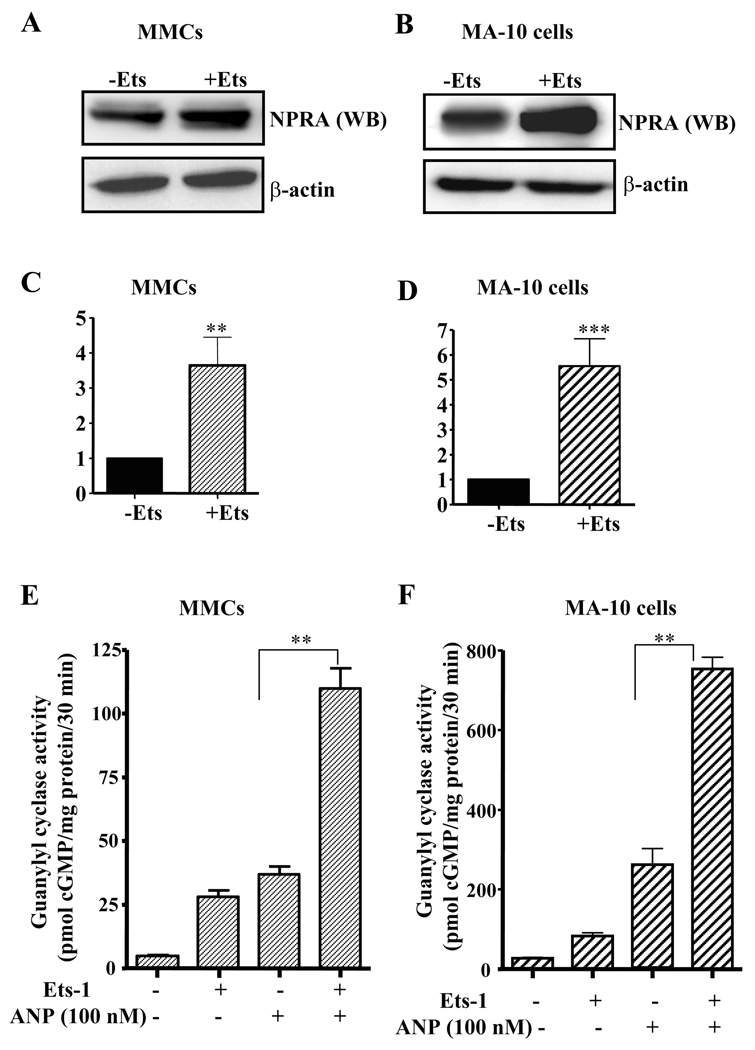
We examined the effect of Ets-1 on intracellular accumulation of cGMP in both MMCs and MA-10 cells. The treatment of Ets-1-transfected MA-10 cells and MMCs with 100 nM ANP showed an increase in the intracellular accumulation of cGMP levels by 2-fold and 3.5-fold, respectively, as compared with the untransfected control cells (Fig. 4, A and B). To investigate the effect of Ets-1 on expression and function of NPRA protein, we performed Western blot and GC activity assay. Fig. 5, A and B shows the representative levels of NPRA protein expression in MMCs and MA-10 cells, respectively. NPRA protein expression was induced by 3.5- and 5.5-fold in Ets-1 transfected MMCs and MA-10 cells, respectively, as compared with the empty vector-transfected control cells (Fig. 5, C and D). MA-10 cells showed higher expression of NPRA protein as compared with MMCs. The effect of Ets-1 on GC-A/NPRA signaling was measured by GC activity assay. Plasma membrane preparations of Ets-1-transfected MMCs and MA-10 cells showed significant increase in GC activity as compared with untransfected controls (Fig. 5, E and F). There was an almost 3-fold increase in GC activity in Ets-1-transfected MMCs when stimulated with ANP as compared with untransfected control cells. Similarly, Ets-1-stimulated GC activity was also enhanced by almost 3.5-fold in MA-10 cells as compared with unstimulated control cells (Fig. 5F).
Fig. 4. Effect of Ets-1 on intracellular accumulation of cGMP levels.
A) MMCs and B) MA-10 cells were transiently transfected with Ets-1 expression plasmid and treated with 0, 1, and 100 nM ANP. Intracellular accumulation of cGMP was quantitated by ELISA. Bars represent the mean ± SE of four independent experiments in triplicate. **, p < 0.01, ***, p < 0.001.
Fig. 5. Ets-1-mediated regulation of NPRA expression and GC activity in MMCs and MA-10 cells.
A) MMCs and B) MA-10 cells were transfected with and without Ets-1 expression plasmids. Forty-eight hours after transfection, cells were lysed and total protein was isolated and subjected to Western blot using antibodies directed against NPRA. β-actin was used as loading control. (C) and (D) shows the densitometry quantitation of NPRA mRNA levels in MMCs and MA-10 cells, respectively. Values represent the fold induction relative to empty vector-transfected controls. GC activity in the plasma membrane preparations of Ets-1 transfected (E) MMCs and (F) MA-10 cells was measured as described in the ‘Materials and Methods’ section. WB indicates Western blot, -Ets indicates transfection with empty vector (pEVRF0), and +Ets indicates transfection with Ets-1 expression plasmid. Representative results of Western blots from 3 experiments are shown. Bars in C, D, E, and F represent mean ± S.E. of three separate determinants in triplicates. **, p < 0.01; ***, p<0.001.
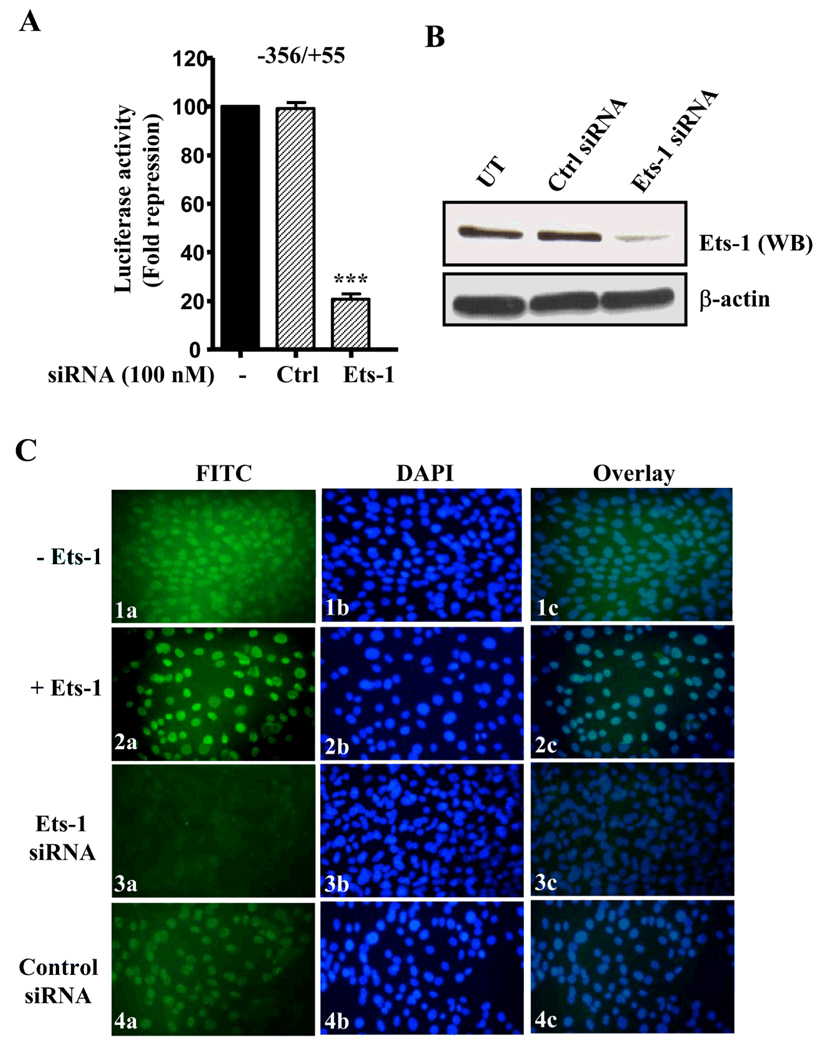
Cotransfection of Ets-1 siRNA along with Npr1 basal promoter reduced the luciferase activity by 80% as compared with cells transfected with Npr1 basal promoter alone, however, transfection of control siRNA showed no change in luciferase activity (Fig. 6A). We assessed the efficiency of siRNA knockdown by Western blot analysis using Ets-1 antibody. Ets-1 protein expression was markedly reduced in siRNA-transfected cells as compared with that in control siRNA-transfected cells (Fig. 6B). Immunofluorescent staining with Ets-1 antibody and FITC-labeled secondary antibody showed the expression of endogenous Ets-1 protein in the nuclei of cells transfected with empty vector (Fig. 6C, panel 1a). Ets-1 protein was found to be overexpressed in Ets-1-transfected cells; however, Ets-1 siRNA significantly reduced the expression of Ets-1 as compared with that in control siRNA-transfected cells (Fig. 6C, panels 2a, 3a, and 4a). Nuclei staining in the corresponding cells with DAPI are shown in Fig. 6C, panels 1b–4b. Overlay of Ets-1 protein with DAPI nuclear stain confirmed the nuclear localization of Ets-1 protein (Fig. 6C, Panels 1c–4c).
Fig. 6. Ets-1 gene silencing effect on Npr1 gene transcription and expression using fluorescence microscopy.
A) Cells were transiently transfected with Npr1 promoter construct, 356/+55 with and without Ets-1 siRNA. Forty-eight hours after transfection, cells were lysed and luciferase assay was performed. B) Western blot analysis of knockdown effect of Ets-1 siRNA and control siRNA (Ctrl siRNA) in transfected cells along with untransfected (UT) cells. β-actin was taken as loading control. C) Immunofluorescence staining of Ets-1 indicating cells transfected with 1) vector, 2) Ets-1 expression plasmid, 3) Ets-1 siRNA, and 4) control siRNA. Panels (1a–4a), Ets-1 protein expression in vivo; panels (1b–4b), nuclei staining of the corresponding cells with DAPI; panels (1c–4c), the merge image. Bars shown in (A) represent mean ± S.E. of three separate determinants in triplicates. ***, p < 0.001.
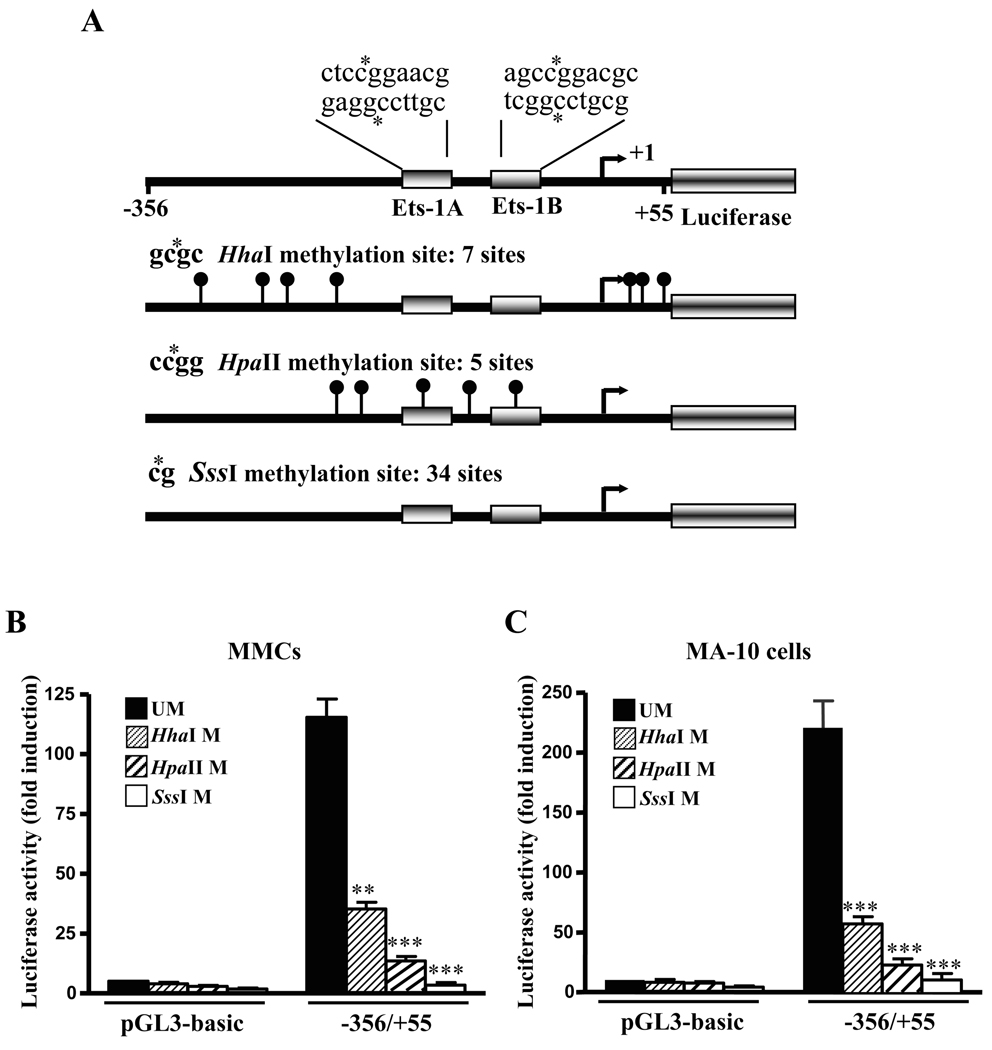
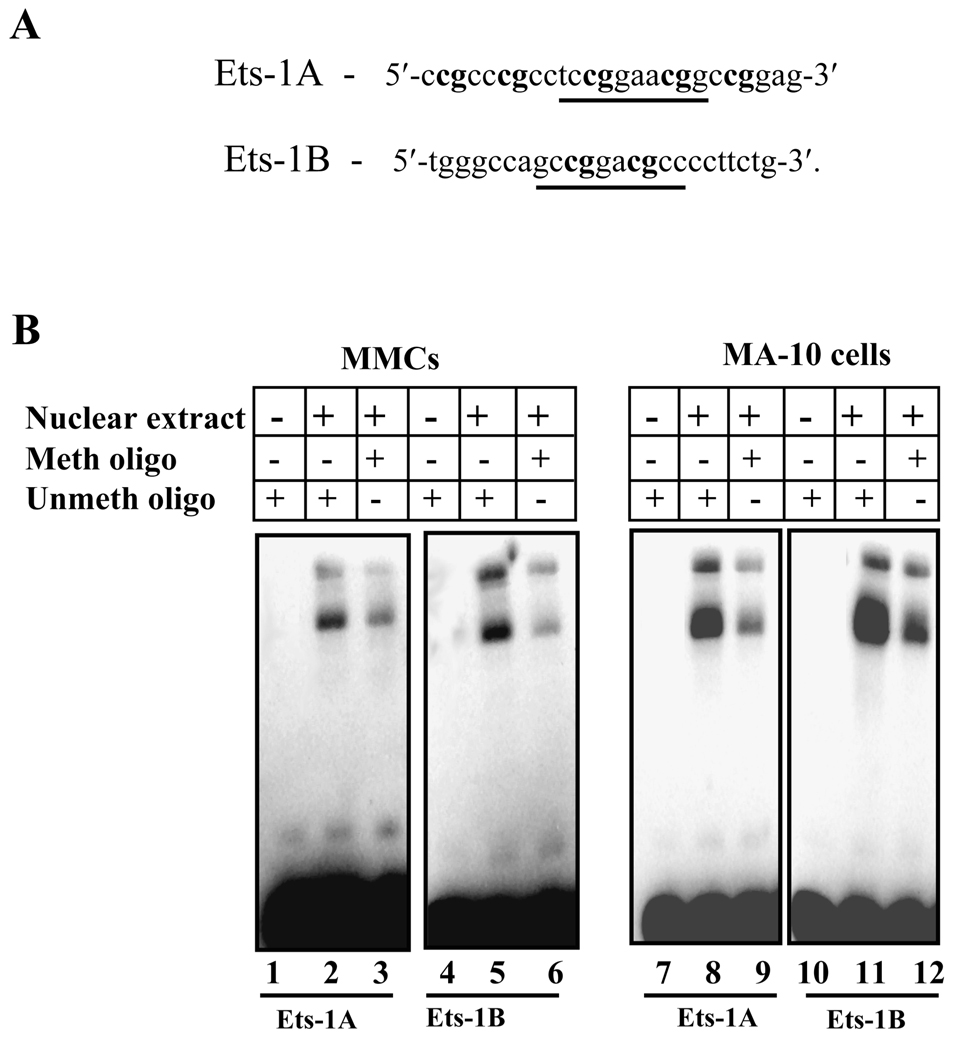
We further investigated the role of methylation in Npr1 gene transcription and its effect on Ets-1 binding to the Npr1 promoter. By analyzing the sequence of Npr1 promoter between positions −356 and +55 relative to the TSS, we noted that there are 34 CpG sites. To examine the effect of methylation on the Npr1 promoter activity both MMCs and MA-10 cells were transfected with in vitro methylated pGL3-basic and −356/+55 Npr1 promoter constructs. The schematic representation of −356/+55 Npr1 promoter constructs containing CG sites recognized by various methylases is shown in Fig. 7A. Luciferase activity of these plasmids was compared with the corresponding unmethylated plasmids. The luciferase activity of partially HhaI-methylated −356/55 construct was reduced by 70% and 75% in MMCs and MA-10 cells, respectively, exhibiting the partial effect of methylation on Npr1 gene transcription (Fig. 7, B and C). Similarly partial methylase HpaII reduced the luciferase activity of −356/+55 construct by almost 80% as compared with the unmethylated construct in both MMCs and MA-10 cells. On the other hand, the inhibition of Npr1 gene transcription by methylation was further confirmed by transfection of full methylase SssI-treated plasmids, which suppressed Npr1 promoter activity by 95% and 98% in MA-10 cells and MMCs, respectively, (Fig. 7, B and C). Subsequently, EMSA was also performed to investigate whether the methylation of Ets-1 consensus sequences influences binding of Ets-1 to its recognition sites in the Npr1 promoter. The Ets-1A and Ets-1B oligonucleotides sequence used for methylation are shown in Fig. 8A. The unmethylated Ets-1A and Ets-1B oligonucleotides were mixed with the nuclear proteins of MMCs and MA-10 cells, which exhibited the binding of Ets-1 to Npr1 promoter (Fig. 8B, lanes 2, 5, 8, and 11). Methylation of the wild-type probe at all the CpG sites present in the Ets-1 recognition site produced significantly weaker shifted bands than the unmethylated probes in both MMCs and MA-10 cells (Fig. 8B, lanes 3, 6, 9, and 12).
Fig. 7. Effect of methylation on Npr1 promoter activity.
A) Schematic representation of the potential CpG methylation island of −356/+55 in Npr1 promoter. The construct contains 411 bp between positions −356 to +55 relative to the TSS (bent arrow), which is indicated as +1. Boxes represent Ets-1A and Ets-1B binding sites. Location of methylation sites by HhaI, HpaII and SssI methylases are indicated as lollipop, together with the recognition sequences of the methylases. Asterisk indicates the position of methylated cytosine by each methylase. Luciferase activity of the in vitro methylated pGL3-basic and −356/+55 Npr1 promoter construct transfected in (B) MMCs and (C) MA-10 cells. The luciferase activity of the constructs was compared with those of corresponding unmethylated constructs. UM indicates unmethylated; M indicates methylase. Bars represent the mean ± SE of four independent experiments in triplicate. **, p < 0.01, ***, p < 0.001 versus unmethylated construct.
Fig. 8. Effect of methylation on in vitro Ets-1 binding in Npr1 promoter.
A) Schematic diagram showing the sequence of the wild-type Ets-1A and Ets-1B oligonucleotides used for EMSA. Sequences in bold show the CpG sites recognized by SssI methylase. Underlined sequences show the Ets-1A and Ets-1B consensus sites. B) The gel retardation assay in nuclear extract from MMCs and MA-10 cells using unmethylated and methylated Ets-1A and Ets-1B oligonucleotides. Lanes 1, 4, 7, and 10 contain free probe. Lanes 2 and 5 show MMCs nuclear protein complex binding with Ets-1A and Ets-1B sites, respectively, and lanes 8 and 11 show the MA-10 cells nuclear protein complex binding with Ets-1A and Ets-1B sites, respectively. Lanes 3 and 6 show the binding reaction of MMCs nuclear protein extract with methylated Ets-1A and Ets-1B probes, respectively, and lanes 9 and 12 show binding reactions with MA-10 nuclear protein. Meth oligo indicates methylated oligonucleotide and Unmeth oligo indicates unmethylated oligonucleotides.
Discussion
The results of the present study demonstrate that Npr1 promoter activity is regulated by Ets-1 and show that the endogenously expressed Ets-1 protein physically associates with Npr1 promoter in vivo and binds to its consensus motifs under in vitro conditions. Overexpression of Ets-1 greatly increased NPRA mRNA and protein levels and stimulated GC activity of the receptor in both MMCs and MA-10 cells. On the other hand, gene silencing of Ets-1 significantly reduced the basal promoter activity, indicating the critical role of Ets-1 in Npr1 gene transcription. Deletion of Ets motifs along with the TSS present in the region −46 to +55 showed significant decrease in luciferase activity in the construct −356/−46 (Fig. 1A). The decrease in luciferase activity may also be attributed to removal of initiation sequence CATACTCC present at the TSS in the region −46 to + 55. Therefore, to confirm the involvement of Ets sites in Npr1 gene regulation we performed in vitro site-directed mutagenesis experiments. The individual contribution of each Ets-1 motif appears to be equivalent because mutation of either element reduced Ets-1-induced promoter activity by almost 50% as compared with wild type construct thus emphasizing the importance of Ets sites in Npr1 gene transcription. Mutation of both the sites together abolished the Ets-1-dependent Npr1 promoter activity by almost 92% as compared with wild-type control plasmid, further confirming the stimulatory role of Ets-1 transcription factor in Npr1 gene transcription. A significantly higher Npr1 gene transcription and expression was observed in MA-10 cells as compared with MMCs. This could be explained by the fact that MA-10 cell line express NPRA at a very high density and has been used as a model system to investigate the receptor-mediated cellular and biochemical responses of ANP [35]. Ets factors share a winged helix-turn-helix DNA-binding domain and interact with the 5'-GGA (T/A)-3' motif [36]. It has been suggested that Ets family members have a primary function in formation of the initiation complex on core promoters lacking the TATA sequence and are frequently located in the vicinity of the TSS in various TATA-deficient gene promoters [37, 38]. The murine Npr1 gene can be added to this group of genes because it exhibits features typical of TATA-less GC-rich promoters and has two Ets binding motifs close to the TSS [14]. The Ets motifs present in the region −46 to +55 of the Npr1 promoter show high sequence homology with the Ets-1 consensus sequence PuCC/a-GGAA/T-GCPy, suggesting that this arrangement might serve as a core for recruiting and/or stabilizing transcriptional complexes [39].
The expression of cell type specific genes is tightly regulated by a hierarchical mechanism composed of genetic and epigenetic factors. Epigenetic changes influence gene expression by changes in chromatin structure via modification of DNA (methylation of CpG islands) and/or histones (e.g. methylation, acetylation, and phosphorylation) [40]. A strong correlation between promoter methylation and gene silencing has been extensively demonstrated [41, 42]. Our results show the involvement of methylation in Npr1 gene silencing. Thereby, in vitro methylated plasmid constructs exhibited a significant decrease in luciferase activities in the transfected cells. The extent of the decrease in luciferase activity was dependent on the density of methylation in the region −356 to +55 of the Npr1 promoter. Methylation of all 34 sites by SssI methylase dramatically decreased the promoter activity by almost 95- 98% in MMCs and MA-10 cells. However, the partial methylation by HhaI (7 sites) or HpaII (5 sites) methylases reduced the luciferase activity by 70% and 80%, respectively, in both MMCs and MA-10 cells. Methylation of HpaII sites reduced the promoter activity more significantly than HhaI sites, which can be explained by the fact that two HpaII sites are present within the Ets-1 binding sites that can inhibit the binding of Ets-1 to the promoter. Methylation of the binding sites of various transcription factors, such as AP-2, Sp3, and Sp1/Sp3, has previously been shown to have a direct influence on expression of downstream genes and is another mechanism for methylation-induced gene repression [43, 44]. We observed that in vitro methylation of Ets-1A and Ets-1B recognition sequence significantly decreased the binding of Ets-1 to these sites.
Recent studies have indicated that Ets transcription factors are involved in gene activation in response to vascular inflammation [45]. Ets-1 transcriptionally induces the expression of Caspase-1 [46], which plays a prominent role in the apoptotic induction of inflammatory response cells [47]. On the other hand, studies in Ets-1 null mutant mice have shown that Ets-1 acts as a transcriptional mediator for Ang II-induced vascular remodeling and generation of reactive oxygen species in hypertension and vascular diseases [48, 49]. Our recent studies have also shown that ablation of Npr1 gene provokes inflammatory response in Npr1 null mutant mice [50]. Since Ets-1 is stimulated by pro-inflammatory mediators and also enhances Npr1 gene transcription, it is implicated that it may have a dual function in cardiovascular disease states. On one hand, it plays a role in inflammatory responses downstream of Ang II signaling pathways, and on the other hand it stimulates Npr1 gene transcription, which seems to inhibit pro-inflammatory responses in hypertension and cardiovascular events.
In summary, the present results provide direct evidence that Ets-1 is essential for Npr1 gene transcription and mediates its effect by binding to its consensus sites present in the Npr1 promoter. The findings of the present study should prove important in elucidating the molecular mechanisms for the expression and regulation of members of the GC receptor family. The results of the present study will greatly enhance our understanding of the transcriptional regulation of Npr1 gene, an important locus in the control of hypertension and cardiovascular homeostasis.
Acknowledgements
The authors thank Mr. Edward Au for technical assistance and Mrs. Kamala Pandey for assistance during the preparation of this manuscript. Our special thanks are due to Dr. Susan L. Hamilton, Department of Molecular Physiology and Biophysics at Baylor College of Medicine, and to Dr. Bharat B. Aggarwal, Department of Experimental Therapeutics and Cytokine Research Laboratory at MD Anderson Cancer Center, for providing their facilities during our displacement period due to Hurricane Katrina. We sincerely thank Dr. Paul Brindle for the kind gift of expression vectors. This work was supported by the National Institutes of Health Grants (HL57531 and HL62147), and partial support was received from the developmental funds of the Tulane Cancer Center.
Abbreviations
- ANP
Atrial natriuretic peptide
- BNP
brain natriuretic peptide
- GC-A/NPRA
guanylyl cyclase/natriuretic peptide receptor-A
- MMCs
mouse mesangial cells
- TSS
transcription start site
- RE
response element
- RT-PCR
reverse transcription polymerase chain reaction
- EMSA
electrophoretic mobility shift assay
- siRNA
small inhibitory RNA
- TBST
Tris-buffered saline-tween-20
- PMSF
phenylmethylsulfonyl fluoride
- DMEM
Dulbecco’s modified Eagle’s medium
- PBS
phosphate buffered saline
- PVDF
Polyvinylidene fluoride
- HEPES
N-(2-Hydroxyethyl)piperazine-N′-(2-ethanesulfonic acid)
References
- 1.de Bold AJ, Borenstein HB, Veress AT, Sonnenberg H. A rapid and potent natriuretic response to intravenous injection of atrial myocardial extract in rats. Life Sci. 1981;28:89–94. doi: 10.1016/0024-3205(81)90370-2. [DOI] [PubMed] [Google Scholar]
- 2.de Bold AJ. Atrial natriuretic factor a hormone produced by the heart. Science. 1985;230:767–770. doi: 10.1126/science.2932797. [DOI] [PubMed] [Google Scholar]
- 3.Brenner BM, Ballermann BJ, Gunning ME, Zeidel ML. Diverse biological actions of atrial natriuretic peptide. Physiol. Rev. 1990;70:665–699. doi: 10.1152/physrev.1990.70.3.665. [DOI] [PubMed] [Google Scholar]
- 4.Chen YF. Atrial natriuretic peptide in hypoxia. Peptides. 2005;26:1068–1077. doi: 10.1016/j.peptides.2004.08.030. [DOI] [PubMed] [Google Scholar]
- 5.Levin ER, Gardner DG, Samson WK. Natriuretic peptides. N. Engl. J. Med. 1998;339:321–328. doi: 10.1056/NEJM199807303390507. [DOI] [PubMed] [Google Scholar]
- 6.Sharma RK. Evolution of the membrane guanylate cyclase transduction system. Mol. Cell. Biochem. 2002;230:3–30. [PubMed] [Google Scholar]
- 7.Pandey KN. Biology of natriuretic peptides and their receptors. Peptides. 2005;26:901–932. doi: 10.1016/j.peptides.2004.09.024. [DOI] [PubMed] [Google Scholar]
- 8.Holtwick R, van Eickels M, Skryabin BV, Baba HA, Bubikat A, Begrow F, Schneider MD, Garbers DL, Kuhn M. Pressure-independent cardiac hypertrophy in mice with cardiomyocyte-restricted inactivation of the atrial natriuretic peptide receptor guanylyl cyclase-A. J. Clin. Invest. 2003;111:1399–1407. doi: 10.1172/JCI17061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Misono KS, Ogawa H, Qiu Y, Ogata CM. Structural studies of the natriuretic peptide receptor: a novel hormone-induced rotation mechanism for transmembrane signal transduction. Peptides. 2005;26:957–968. doi: 10.1016/j.peptides.2004.12.021. [DOI] [PubMed] [Google Scholar]
- 10.Tokudome T, Horio T, Kishimoto I, Soeki T, Mori K, Kawano Y, Kohno M, Garbers DL, Nakao K, Kangawa K. Calcineurin-nuclear factor of activated T cells pathway-dependent cardiac remodeling in mice deficient in guanylyl cyclase A, a receptor for atrial and brain natriuretic peptides. Circulation. 2005;111:3095–3104. doi: 10.1161/CIRCULATIONAHA.104.510594. [DOI] [PubMed] [Google Scholar]
- 11.Vellaichamy E, Khurana ML, Fink J, Pandey KN. Involvement of the NF-kappa B/matrix metalloproteinase pathway in cardiac fibrosis of mice lacking guanylyl cyclase/natriuretic peptide receptor A. J. Biol. Chem. 2005;280:19230–19242. doi: 10.1074/jbc.M411373200. [DOI] [PubMed] [Google Scholar]
- 12.Nakayama T, Soma M, Takahashi Y, Rehemudula D, Kanmatsuse K, Furuya K. Functional deletion mutation of the 5'-flanking region of type A human natriuretic peptide receptor gene and its association with essential hypertension and left ventricular hypertrophy in the Japanese. Circ. Res. 2000;86:841–845. doi: 10.1161/01.res.86.8.841. [DOI] [PubMed] [Google Scholar]
- 13.Liang F, Schaufele F, Gardner DG. Functional interaction of NF-Y and Sp1 is required for type a natriuretic peptide receptor gene transcription. J. Biol. Chem. 2001;276:1516–1522. doi: 10.1074/jbc.M006350200. [DOI] [PubMed] [Google Scholar]
- 14.Garg R, Oliver PM, Maeda N, Pandey KN. Genomic structure, organization, and promoter region analysis of murine guanylyl cyclase/atrial natriuretic peptide receptor-A gene. Gene. 2002;291:123–133. doi: 10.1016/s0378-1119(02)00589-9. [DOI] [PubMed] [Google Scholar]
- 15.Kumar P, Arise KK, Pandey KN. Transcriptional regulation of guanylyl cyclase/natriuretic peptide receptor-A gene. Peptides. 2006;27:1762–1769. doi: 10.1016/j.peptides.2006.01.004. [DOI] [PubMed] [Google Scholar]
- 16.Sementchenko VI, Watson DK. Ets target genes: past, present and future. Oncogene. 2000;19:6533–6548. doi: 10.1038/sj.onc.1204034. [DOI] [PubMed] [Google Scholar]
- 17.Seth A, Watson DK. ETS transcription factors and their emerging roles in human cancer. Eur. J. Cancer. 2005;41:2462–2478. doi: 10.1016/j.ejca.2005.08.013. [DOI] [PubMed] [Google Scholar]
- 18.Mizui M, Isaka Y, Takabatake Y, Sato Y, Kawachi H, Shimizu F, Takahara S, Ito T, Imai E. Transcription factor Ets-1 is essential for mesangial matrix remodeling. Kidney Int. 2006;70:298–305. doi: 10.1038/sj.ki.5001541. [DOI] [PubMed] [Google Scholar]
- 19.Hultgardh-Nilsson A, Cercek B, Wang JW, Naito S, Lovdahl C, Sharifi B, Forrester JS, Fagin JA. Regulated expression of the ets-1 transcription factor in vascular smooth muscle cells in vivo and in vitro. Circ. Res. 1996;78:589–595. doi: 10.1161/01.res.78.4.589. [DOI] [PubMed] [Google Scholar]
- 20.Lie-Venema H, Gittenberger-de Groot AC, van Empel LJ, Boot MJ, Kerkdijk H, de Kant E, DeRuiter MC. Ets-1 and Ets-2 transcription factors are essential for normal coronary and myocardial development in chicken embryos. Circ. Res. 2003;92:749–756. doi: 10.1161/01.RES.0000066662.70010.DB. [DOI] [PubMed] [Google Scholar]
- 21.Cederberg A, Hulander M, Carlsson P, Enerback S. The kidney-expressed winged helix transcription factor FREAC-4 is regulated by Ets-1. A possible role in kidney development. J. Biol. Chem. 1999;274:165–169. doi: 10.1074/jbc.274.1.165. [DOI] [PubMed] [Google Scholar]
- 22.Lanier-Smith KL, Currie MG. Glucocorticoid regulation of atrial natriuretic peptide receptors on cultured endothelial cells. Endocrinology. 1991;129:2311–2317. doi: 10.1210/endo-129-5-2311. [DOI] [PubMed] [Google Scholar]
- 23.Fujio N, Gossard F, Bayard F, Tremblay J. Regulation of natriuretic peptide receptor A and B expression by transforming growth factor-beta 1 in cultured aortic smooth muscle cells. Hypertension. 1994;23:908–913. doi: 10.1161/01.hyp.23.6.908. [DOI] [PubMed] [Google Scholar]
- 24.Goto M, Itoh H, Tanaka I, Suga S, Ogawa Y, Kishimoto I, Nakagawa M, Sugawara A, Yoshimasa T, Mukoyama M, et al. Altered gene expression of natriuretic peptide receptor subtypes in the kidney of stroke-prone spontaneously hypertensive rats. Clin. Exp. Pharmacol. Physiol. 1995;22 Suppl:S177–S179. doi: 10.1111/j.1440-1681.1995.tb02871.x. [DOI] [PubMed] [Google Scholar]
- 25.Garg R, Pandey KN. Angiotensin II-mediated negative regulation of Npr1 promoter activity and gene transcription. Hypertension. 2003;41:730–736. doi: 10.1161/01.HYP.0000051890.68573.94. [DOI] [PubMed] [Google Scholar]
- 26.Arise KK, Pandey KN. Inhibition and down-regulation of gene transcription and guanylyl cyclase activity of NPRA by angiotensin II involving protein kinase C. Biochem. Biophy. Res. Commun. 2006;349:131–135. doi: 10.1016/j.bbrc.2006.08.003. [DOI] [PubMed] [Google Scholar]
- 27.Pandey KN, Nguyen HT, Sharma GD, Shi SJ, Kriegel AM. Ligand-regulated internalization, trafficking, and down-regulation of guanylyl cyclase/atrial natriuretic peptide receptor-A in human embryonic kidney 293 cells. J. Biol. Chem. 2002;277:4618–4627. doi: 10.1074/jbc.M106436200. [DOI] [PubMed] [Google Scholar]
- 28.Hum D, Besnard S, Sanchez R, Devost D, Gossard F, Hamet P, Tremblay J. Characterization of a cGMP-response element in the guanylyl cyclase/natriuretic peptide receptor A gene promoter. Hypertension. 2004;43:1270–1278. doi: 10.1161/01.HYP.0000126920.93207.53. [DOI] [PubMed] [Google Scholar]
- 29.Yang C, Shapiro LH, Rivera M, Kumar A, Brindle PK. A role for CREB binding protein and p300 transcriptional coactivators in Ets-1 transactivation functions. Mol. Cell. Biol. 1998;18:2218–2229. doi: 10.1128/mcb.18.4.2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pandey KN, Nguyen HT, Li M, Boyle JW. Natriuretic peptide receptor-A negatively regulates mitogen-activated protein kinase and proliferation of mesangial cells: role of cGMP-dependent protein kinase. Biochem. Biophys. Res. Commun. 2000;271:374–379. doi: 10.1006/bbrc.2000.2627. [DOI] [PubMed] [Google Scholar]
- 31.Pandey KN, Ascoli M, Inagami T. Induction of renin activity by gonadotropic hormones in cultured Leydig tumor cells. Endocrinology. 1985;117:2120–2126. doi: 10.1210/endo-117-5-2120. [DOI] [PubMed] [Google Scholar]
- 32.Dignam JD, Lebovitz RM, Roeder RG. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Leitman DC, Andresen JW, Catalano RM, Waldman SA, Tuan JJ, Murad F. Atrial natriuretic peptide binding, cross-linking, and stimulation of cyclic GMP accumulation and particulate guanylate cyclase activity in cultured cells. J. Biol. Chem. 1988;263:3720–3728. [PubMed] [Google Scholar]
- 34.Khurana ML, Pandey KN. Catalytic activation of guanylate cyclase/atrial natriuretic factor receptor by combined effects of ANF and GTP gamma S in plasma membranes of Leydig tumor cells: involvement of G-proteins. Arch. Biochem. Biophys. 1995;316:392–398. doi: 10.1006/abbi.1995.1052. [DOI] [PubMed] [Google Scholar]
- 35.Pandey KN, Singh S. Molecular cloning and expression of murine guanylate cyclase/atrial natriuretic factor receptor cDNA. J. Biol. Chem. 1990;265:12342–12348. [PubMed] [Google Scholar]
- 36.Donaldson LW, Petersen JM, Graves BJ, McIntosh LP. Solution structure of the ETS domain from murine Ets-1: a winged helix-turn-helix DNA binding motif. Embo. J. 1996;15:125–134. [PMC free article] [PubMed] [Google Scholar]
- 37.Rudge TL, Johnson LF. Synergistic activation of the TATA-less mouse thymidylate synthase promoter by the Ets transcription factor GABP and Sp1. Exp. Cell Res. 2002;274:45–55. doi: 10.1006/excr.2001.5451. [DOI] [PubMed] [Google Scholar]
- 38.Arman M, Calvo J, Trojanowska ME, Cockerill PN, Santana M, Lopez-Cabrera M, Vives J, Lozano F. Transcriptional regulation of human CD5: important role of Ets transcription factors in CD5 expression in T cells. J. Immunol. 2004;172:7519–7529. doi: 10.4049/jimmunol.172.12.7519. [DOI] [PubMed] [Google Scholar]
- 39.Dittmer J. The biology of the Ets1 proto-oncogene. Mol. Cancer. 2003;2:29. doi: 10.1186/1476-4598-2-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.D'Alessio AC, Weaver IC, Szyf M. Acetylation-induced transcription is required for active DNA demethylation in methylation-silenced genes. Mol. Cell. Biol. 2007;27:7462–7474. doi: 10.1128/MCB.01120-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 42.Plass C, Soloway PD. DNA methylation, imprinting and cancer. Eur. J. Hum. Genet. 2002;10:6–16. doi: 10.1038/sj.ejhg.5200768. [DOI] [PubMed] [Google Scholar]
- 43.Zhu WG, Srinivasan K, Dai Z, Duan W, Druhan LJ, Ding H, Yee L, Villalona-Calero MA, Plass C, Otterson GA. Methylation of adjacent CpG sites affects Sp1/Sp3 binding and activity in the p21(Cip1) promoter. Mol. Cell. Biol. 2003;23:4056–4065. doi: 10.1128/MCB.23.12.4056-4065.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Aoyama T, Okamoto T, Nagayama S, Nishijo K, Ishibe T, Yasura K, Nakayama T, Nakamura T, Toguchida J. Methylation in the core-promoter region of the chondromodulin-I gene determines the cell-specific expression by regulating the binding of transcriptional activator Sp3. J. Biol. Chem. 2004;279:28789–28797. doi: 10.1074/jbc.M401273200. [DOI] [PubMed] [Google Scholar]
- 45.Oettgen P. Regulation of vascular inflammation and remodeling by ETS factors. Circ. Res. 2006;99:1159–1166. doi: 10.1161/01.RES.0000251056.85990.db. [DOI] [PubMed] [Google Scholar]
- 46.Pei H, Li C, Adereth Y, Hsu T, Watson DK, Li R. Caspase-1 is a direct target gene of ETS1 and plays a role in ETS1-induced apoptosis. Cancer Res. 2005;65:7205–7213. doi: 10.1158/0008-5472.CAN-04-3566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rowe SJ, Allen L, Ridger VC, Hellewell PG, Whyte MK. Caspase-1-deficient mice have delayed neutrophil apoptosis and a prolonged inflammatory response to lipopolysaccharide-induced acute lung injury. J. Immunol. 2002;169:6401–6407. doi: 10.4049/jimmunol.169.11.6401. [DOI] [PubMed] [Google Scholar]
- 48.Zhan Y, Brown C, Maynard E, Anshelevich A, Ni W, Ho IC, Oettgen P. Ets-1 is a critical regulator of Ang II-mediated vascular inflammation and remodeling. J. Clin. Invest. 2005;115:2508–2516. doi: 10.1172/JCI24403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ni W, Zhan Y, He H, Maynard E, Balschi JA, Oettgen P. Ets-1 is a critical transcriptional regulator of reactive oxygen species and p47(phox) gene expression in response to angiotensin II. Circ. Res. 2007;101:985–994. doi: 10.1161/CIRCRESAHA.107.152439. [DOI] [PubMed] [Google Scholar]
- 50.Vellaichamy E, Zhao D, Somanna N, Pandey KN. Genetic disruption of guanylyl cyclase/natriuretic peptide receptor-A upregulates ACE and AT1 receptor gene expression and signaling: role in cardiac hypertrophy. Physiol. Genomics. 2007;31:193–202. doi: 10.1152/physiolgenomics.00079.2007. [DOI] [PubMed] [Google Scholar]