Abstract
DNA alkylation or adduct formation occurs at nucleophilic sites in DNA, mainly the N7-position of guanine. Ever since identification of the first N7-guanine adduct, several hundred studies on DNA adducts have been reported. Major issues addressed include the relationships between N7-guanine adducts and exposure, mutagenesis, and other biological endpoints. It became quickly apparent that N7-guanine adducts are frequently formed, but may have minimal biological relevance, since they are chemically unstable and do not participate in Watson Crick base pairing. However, N7-guanine adducts have been shown to be excellent biomarkers for internal exposure to direct acting and metabolically activated carcinogens. Questions arise, however, regarding the biological significance for N7-guanine adducts that are readily formed, do not persist, and are not likely to be mutagenic. Thus, we set out to review the current literature to evaluate their formation and the mechanistic evidence for the involvement of N7-guanine adducts in mutagenesis or other biological processes. It was concluded that there is insufficient evidence that N7-guanine adducts can be used beyond confirmation of exposure to the target tissue and demonstration of the molecular dose. There is little to no evidence that N7-guanine adducts or their depurination product, apurinic sites, are the cause of mutations in cells and tissues, since increases in AP sites have not been shown unless toxicity is extant. However, more research is needed to define the extent of chemical depurination versus removal by DNA repair proteins. Interestingly, N7-guanine adducts are clearly present as endogenous background adducts and the endogenous background amounts appear to increase with age. Furthermore, the N7-guanine adducts have been shown to convert to ring opened lesions (FAPy), which are much more persistent and have higher mutagenic potency. Studies in humans are limited in sample size and differences between controls and study groups are small. Future investigations should involve human studies with larger numbers of individuals and analysis should include the corresponding ring opened FAPy derivatives.
Keywords: N7-guanine adducts, mutagenesis, biomarkers, exposure assessment, Dose-response
1 Introduction
DNA alkylation or adduct formation occurs at nucleophilic sites in DNA. Of these nucleophilic sites, the N7-position of guanine is the most reactive [1]. A PubMed search on “N7-guanine adducts” resulted in over 300 publications with 9 out of 10 focusing on basic characterization of chemical or biochemical properties of N7-guanine adducts alone or in DNA. In addition, N7-guanine adducts are classified as non promutagenic since they are chemically unstable and the N7-position does not participate in Watson Crick base pairing [2]. Ever since identification of the first N7-guanine adduct [1], several hundred studies on DNA adducts have been reported and reviewed from different perspectives [3-16]. Consequently, many studies sought to establish the relationship between DNA adduct formation and other biological endpoints (mutations, DNA strand breaks, etc.). Technical limitations, however, did not permit integration into large molecular epidemiological studies during this era of lesion characterization. Despite superior sensitivity of the 32P-postlabeling assay, insufficient chemical specificity made it impossible to identify the chemical source of damage, and chemical depurination of N7-guanine adducts during sample preparation was a major concern. Almost all studies started with in vitro proof of concept experiments demonstrating covalent binding of the compounds of interest or their metabolites to DNA. Surprisingly, the identification and development of sensitive analytical methods remain a primary focus of many DNA adduct studies, even 50+ years later.
N7-guanine adducts appear to be good biomarkers of internal exposure because of their higher abundance compared to other DNA alkylations. Questions arise, however, regarding the biological significance for N7-guanine adducts that are readily formed, do not persist, and are not likely to be mutagenic. Thus, we set out to review the current literature to evaluate their formation and the mechanistic evidence for the involvement of N7-guanine adduts in mutagenesis or other biological processes.
1.1 Formation of DNA adducts
Miller and Miller pioneered the field of chemical carcinogens and were the first to demonstrate covalent binding of chemical carcinogens to macromolecules in vivo [17,18]. The first evidence for binding of chemical carcinogens or their metabolites to nucleic acid was reported by Wheeler and Skipper [19]. It quickly became apparent that carcinogens comprise a diverse group of chemicals. Some of them were from endogenous sources or natural products, while others arise from synthetic products of modern human life. These chemicals are able to react with nucleophilic sites (electron rich, S, N, and O), in DNA and proteins. Subsequent in vitro and in vivo studies quickly demonstrated that under physiological conditions (pH 7.4, 37°C), alkylation of DNA primarily occurred at the N7-position of guanine (Table 1) [20]. The distribution of methylation and ethylation adducts in DNA was studied in in vitro reactions, in bacterial or mammalian cell cultures, and in several tissues of mice and rats (reviewed by Beranek [8]). Overall, it confirmed the notion that the relative distribution of alkylation in DNA is similar in vivo and in vitro [21]. However, as technology advanced and allowed examination of lower exposures distinct differences in adduct distribution were established (see Section 2). Binding was shown to mainly occur via monomolecular (SN1, e.g., nitrogen mustards) or bimolecular nucleophilic (SN2, e.g., sulfonyl esters) substitutions [22-24]. SN2 reactions are heavily dependent on steric accessibility while SN1 reactions generally follow first-order kinetics. In DNA, the ring nitrogens and the exocyclic oxygens are the preferred sites for alkylation. Although the N7-position is the major site of alkylation, the electrophilic species formed by the N-nitroso compounds for example, following SN1 kinetics, will have a greater preference for reaction at the exocyclic oxygens than will the alkanesulfonates, which are limited to SN2 reactions. The larger the alkyl group is, the stronger will be its preference for reaction at the O6-position of guanine. Hence, N-ethyl-N-nitrosourea (ENU) binds more efficiently to the O6-position than does N-methyl-N-nitrosourea (MNU) (Table 1)[8, 25, 26]. The important difference in alkylation agents undergoing SN1 or SN2 reactions is that agents capable of SN1 reactions react more frequently at the O6-position of guanine, thus producing more mutagenic O6-guanine adducts, compared to agents that solely react via the SN2 mechanism.
Table 1. Relative reactivity [%] of nucleophilic sites in DNA.
| Chemical | N7-Gua | O6-Gua | N2-Gua | N3-Ade | N1Ade/N6 Ade | N3-Cyt | N4-Cyt | Reference |
|---|---|---|---|---|---|---|---|---|
| PO | 100 | 0.5 | - | 4.4/10 | 3.5 | 2 | - | [269, 270] |
| EO | 100 | - | - | 10 | 10 | 1 | - | [125] |
| AGE | 100 | - | - | 11 | 14 | 6 | - | [271] |
| ECH | 100 | 9 | 4 | [125] | ||||
| SO | 100 | 4.5 | 10 | 16 | 5 | 2 | [125] | |
| EB | 100 | 15 | 25 | 1.5 | [153, 272, 273] | |||
| MNU | 100 | 10 | - | 12 | 1.2 | 0.8 | - | [274, 275] |
| MMS | 100 | 3.7 | 12 | 2 | - | - | [274, 275] | |
| ENU | 100 | 66 | 30 | 2 | 2 | - | [274, 275] | |
| EMS | 100 | 3.4 | 7 | 3 | 0.6 | [274, 275] |
AGE; allyl glycidyl ether, ECH; epichlorohydrin
These early binding experiments in DNA, cell culture and animal studies also showed that some carcinogens required metabolic activation to gain their ability to form DNA adducts and to exhibit their mutagenic and carcinogenic effects. Consequently, compounds were classified as “direct-acting” or “metabolically activated” carcinogens. The latter type is also termed a pro-carcinogen. In addition to mono adducts, compounds with multiple reactive groups were shown to have the ability to form protein-protein, DNA-DNA or protein-DNA cross-links [20]. The decades following have produced a better understanding of the relationship between carcinogen exposures, DNA adduct formation, mutagenesis, and carcinogenesis [4-7, 10, 12, 27]. Various technologies have been applied to animal and human exposure studies for routine analysis of N7-guanine adducts and other DNA adducts. These studies have increased our understanding of formation and persistence of DNA adducts, and their relationship to carcinogenesis. It has become clear that cancer is a complex, multi-step process that varies with types of exposure, site of tumor induction, and species. Understanding the implications of N7-guanine adducts has significantly contributed to identification of the mode of action in chemically-induced mutagenesis and carcinogenesis. These findings have subsequently led to a better understanding of the role of DNA adducts in mutagenesis and mechanism-based risk assessment [27].
1.2 Stability of N7-guanine adducts
Compared to many other DNA adducts, N7-guanine adducts are chemically unstable, with half lives in double-stranded DNA (dsDNA) ranging from as little as 2 h to 150 h. The instability of N7-guanine adducts is created by the formal placement of an additional positive charge on the guanine ring system. In general, larger alkyl groups promote depurination in dsDNA. This has been demonstrated under physiological conditions (pH 7.4, 37°C), where the half-lives for N7-methyl-guanine (N7-Me-Gua), N7-(2-hydroxy-3-butene)-guanine (N7-HB-Gua) and N7-(trihydroxy-benzo[a]pyreneyl) guanine are 150 h, 50 h, and 3 h, respectively [28-31]. In addition, N7-guanine adducts accumulate in DNA with continuous exposure or treatment and usually reach a plateau (steady state) after ∼7-10 days [15, 32-34]. Steady state is reached when the rate of N7-guanine adducts formed is equal to the rate of adducts lost. In contrast, adducts that are more persistent, such as O4-ethyl-thymidine (O4-Et-Thy), accumulate over a period of 4 weeks [35], and O6-methyl-guanine (O6-Me-Gua) in the brain continue to accumulate over 6 weeks of dosing [36]. The formal placement of an additional positive charge on the guanine ring system also promotes further reactions that have been reviewed by Gates et al. [37]. Relative to guanine, N7-Me-Gua depurinates 106 times more rapidely at pH7, 37 °C [37]. Reactions characteristic for N7-guanine adducts are: (i) loss of C-8 proton, (ii) depurination, (iii) ring opening to yield 5-N-alkyl-2,6,-diamino-4-hydroxyformamidopyrimidine (alkyl-FAPy), (iv) hydrolysis of the N7-alkyl bond, and (v) rearrangement to C8 adducts. For details of the chemical reactivity of N7-guanine adducts, the reader is referred to the thorough review by Gates [37].
1.3 Methods for detection of N7-guanine adducts
During the past 50 years many technologies have been used for analysis of N7-guanine adducts. These technologies have been applied to rodent and human exposure studies for routine analysis of N7-guanine adducts and other DNA adducts. In the earliest studies, radiolabeled carcinogens were administered to rodents and binding to protein, RNA, and DNA was assessed by scintillation counting of the corresponding cellular fractions [38]. After DNA isolation and hydrolytic treatments, individual DNA adducts could be purified and quantified by basic column chromatography. This approach allowed analysis of one sample per day, with a detection limit of 1 adduct per 106 normal nucleotides (nnt) using ≥5 mg DNA [29, 39]. Longer exposure regimes were laborious and expensive, due to the requirement of radiolabeled carcinogen for these studies. Consequently, most studies employed single exposures [40, 41].
By the 1980's, HPLC with fluorescence detection, radioimmunoassay, or enzyme-linked immuno sorbent assay were commonly used for the analysis of DNA adducts. These approaches significantly increased throughput, reduced cost via elimination of custom radioisotope synthesis, and allowed application to study designs that included multiple exposure protocols. The extended exposure protocols provided information on the steady-state concentrations of DNA adducts and demonstrated that what had previously appeared to be minor adducts following single exposures could actually become major adducts if they were poorly repaired and accumulated with extended exposure [42]. These methods, however, had limited sensitivity compared to present day technology and some of the immunoassays were prone to false positive results due to cross-reactivity.
A major break though in methodology occurred in 1982, when Randerath and colleagues developed 32P-postlabeling methods for DNA adducts [43]. The limit of detection for the early 32P-postlabeling assays was 1 adduct per 108 nnt using as little as 1-2 μg DNA [43, 44]. Subsequently, combinations of 32P-postlabeling with HPLC or immunoaffinity permitted larger amounts of DNA to be analyzed and improved the sensitivity by one or more orders of magnitude. The major problems associated with this methodology include the lack of chemical-specific identity and poor reproducibility [45, 46]. The 32P-postlabeling method was most suitable for more stable DNA adducts, such as etheno adducts [47-49] and N2-guanine adducts derived from polycyclic aromatic hydrocarbons (PAH) [50], and less so for N7-guanine adducts, due to their instability.
More recently, advances in mass spectrometry have lowered the limit of detection for this chemical-specific and quantitative technology, making it the method of choice in contemporary investigations. Earlier studies applied gas chromatography - negative ion chemical ionization - mass spectrometry (GC-MS), after hydrolysis and derivatization, to the analysis of DNA adducts [51-56]. Presently, however, the vast majority of quantitative analysis of DNA adducts is performed with liquid chromatography tandem mass spectrometry (LC-MS/MS). The application of mass spectrometry for DNA adducts has been recently reviewed by Singh and Farmer [57] and others [15, 16, 58-63]. Major advances in instrumentation for both mass spectrometry and chromatography have increased the detection limits for DNA adducts up to 100-fold, making it possible to routinely measure 1 adduct per 108 nnt. A major advantage of GC- and LC-MS/MS methods is the utilization of chemically identical stable isotope labeled internal standards for accurate quantitation.
The greatest sensitivity for measuring DNA adducts, however, is achieved with accelerator mass spectrometry (AMS), which can quantitate down to 3 adduct per 1011 nucleosides using 1 μg DNA [64]. While this method is extremely sensitive, it is limited to the following radioisotopes (3H, 14C, 26Al, 41Ca, 10Be, 36Cl, 59Ni, 63Ni, and 129I), of which 14C and 3H are the most commonly used in biomedical research. Therefore, specific chemical syntheses are required to either obtain radioisotope-labeled test compounds or for chemical labeling of compounds of interest (postlabeling, derivatization). Unfortunately, access to AMS is limited worldwide (only 5 instruments as of 2007), mainly due to the expense of the mostly custom-made instruments [65].
2 Formation of N7-guanine adducts in animal models
Several investigators have successfully utilized N7-guanine adducts as biomarkers to answer important toxicology questions in rodent models. These studies used multi-dose exposure protocols to generate comprehensive dose-response curves. Data from studies in mice and rats are presented in supplemental materials (Table S1 and Table S2). Adduct formation was compared to other biological endpoints such as unscheduled DNA synthesis, mutation frequency, micronucleus, apurinic sites (AP sites), gene expression, and others [7, 10, 12, 66]. In the following sections we will describe studies demonstrating factors influencing the formation and persistence of N7-guanine adducts. First, metabolic activation and subsequent formation of N7-guanine adducts can be species, strain, tissue, and cell dependant [32, 67-69]. Second, chemical stability and DNA repair influence the ratios of N7-guanine adducts to other DNA adducts [36, 40]. Lastly, there is some evidence for accumulation and increased tolerance to, and higher formation rates of N7-guanine adducts in DNA at later ages compared to young ages [70, 71].
2.1 Nitrosourea compounds
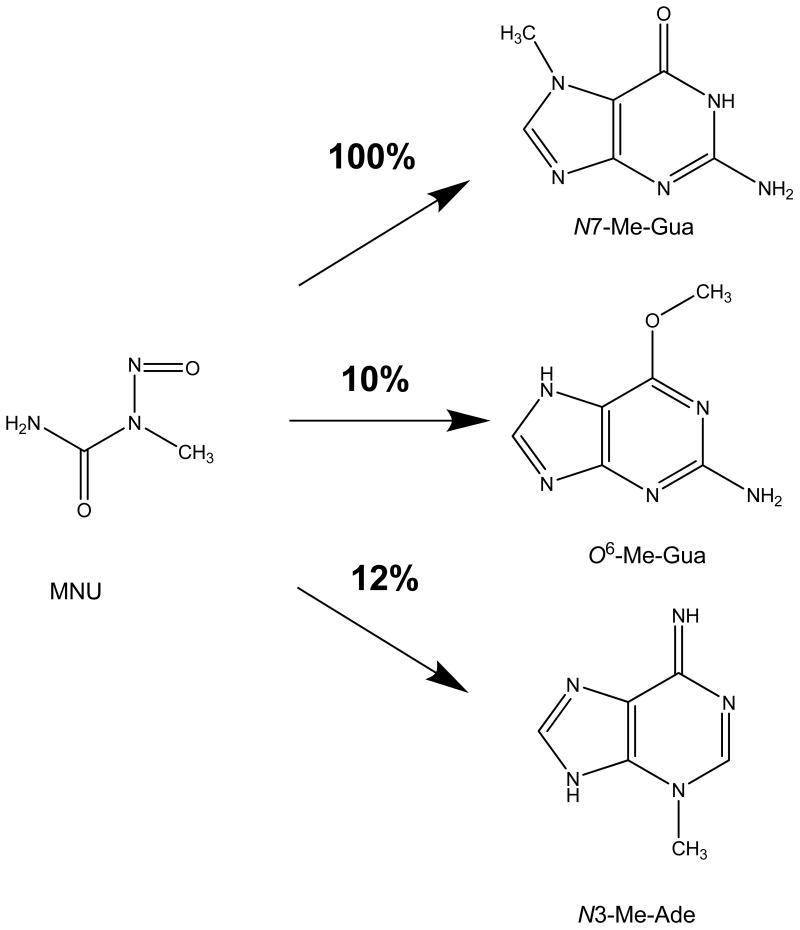
N-methyl-N-nitrosourea (MNU) primarily induced tumors in the nervous system in rats and lymphoid tumors in mice [72-74]. Subsequent analysis of N7-Me-Gua in several tissues of MNU-treated A/J and C3H3B/FeJ mice showed the highest formation of N7-Me-Gua and O6-methyl-guanine (O6-Me-Gua) in liver, kidney, lung, and brain, suggesting that adduct formation might be involved in carcinogenesis (Figure 1) [75]. Comparison of the ratio of N7-guanine adducts to O6-guanine adducts in brain and liver, assuming no active repair for N7-Me-Gua, suggested somewhat slower repair of O6-Me-Gua in the more susceptible C3H3B/FeJ strain than in A/J mice [75]. Persistence of N7-Me-Gua was similar in liver (non-target tissue) and brain (target tissue), suggesting that N7-Me-Gua adducts are most likely not causally linked to mutagenesis and carcinogenesis. O6-Me-Gua adducts, however, were removed much faster in liver than in brain, demonstrating active removal in the liver and suggesting involvement of O6-Me-Gua in mutagenesis and carcinogenesis [76]. In contrast, the persistence and amounts of O6-Me-Gua were similar in brain of both rats and mice, although these species differ markedly in their susceptibility to brain tumorigenesis, with the rat being a much more susceptible species [73, 77]. It was concluded that organotropic carcinogenic effects of methylating carcinogens do not solely depend on DNA adduct formation and persistence, since formation and persistence correlated only in certain cases with tumor formation [75, 78, 79].
Figure 1.
Guanine and adenine methylation by MNU relative to N7-Me-Gua formation (100%).
From the earliest studies on N7-guanine adducts, it quickly became apparent that certain adducts exist in control DNA and the notion of endogenous DNA damage was established. Amounts of N7-Me-Gua were 2-fold higher in liver DNA of rats that were 24 months old compared to 6 month-old animals, suggesting accumulation with age [71]. Similarly, endogenous amounts of N7-Me-Gua were twice as high in 29 month-old C57BL/6NNia mice, compared to 11 month-old mice [70]. More importantly, not only was the endogenous amount higher in older mice (29 month), they also formed twice as many N7-Me-Gua adducts as younger mice after 25 or 50 mg/kg MNU-treatment (11 months) [70]. These two reports demonstrate that endogenous background amounts and adduct formation may be significantly different at different ages. Unfortunately, to our knowledge age dependence of N7-guanine adduct formation has not been systematically investigated. Future studies should include establishing endogenous background amounts of N7-guanine adducts and their formation in target tissues at different ages in mice, rats, and humans.
N-Ethyl-N-nitrosourea (ENU), similar to MNU, is a potent carcinogen inducing mainly tumors of nervous system and forming N7-ethyl-guanine (N7-Et-Gua) and O6-ethyl-guanine (O6-Et-Gua) adducts. The persistence of N7-Et-Gua adducts is dependent upon their chemical stability and possible elimination by active DNA repair enzymes. It has been shown that DNA repair capacity is tissue-specific and influences accumulation of promutagenic O6-Et-Gua, O4-Et-Thy and O2-ethyl-thymidine (O2-Et-Thy) adducts, but not N7-Et-Gua adducts [40]. Tissue-specific DNA repair can greatly affect dose-response of DNA adducts as exemplified with O6-Me-Gua and O6-Et-Gua in brain and liver of MNU- or ENU-treated mice or rats [36, 40, 41]. O6-Me-Gua and O6-Et-Gua adducts are actively repaired in liver but not in the brain, the target tissue of MNU and ENU tumorigenesis in rats [80]. After a single exposure, O6-guanine adducts are readily removed in the repair-proficient hepatic tissue but remain relatively persistent in the repair-deficient brain tissue. In contrast, the chemically less stable N7-Et-Gua and N3-Et-adenine adducts have similar elimination rates in both tissues, suggesting that these adducts are lost due to spontaneous depurination [40, 41]. Consequently, the ratio of N7-Et-Gua to O6-Et-Gua decreases in target tissues with time and duration of exposure, and the presence of N7-guanine adducts does not predict a fixed amount of O6-guanine adducts. Most importantly, the presence of N7-guanine adducts cannot serve as quantitative indicator for the existence of other mutagenic or genotoxic lesions, without reliable knowledge of the half-lives in tissue and time since exposure. Although ratios of formation of related adducts can be expected to following in vitro chemistry, tissue-specific differences in repair and stability of the individual adducts will alter adduct-distribution and this dynamic must be considered in subsequent interpretations.
2.2 Nitrosamines
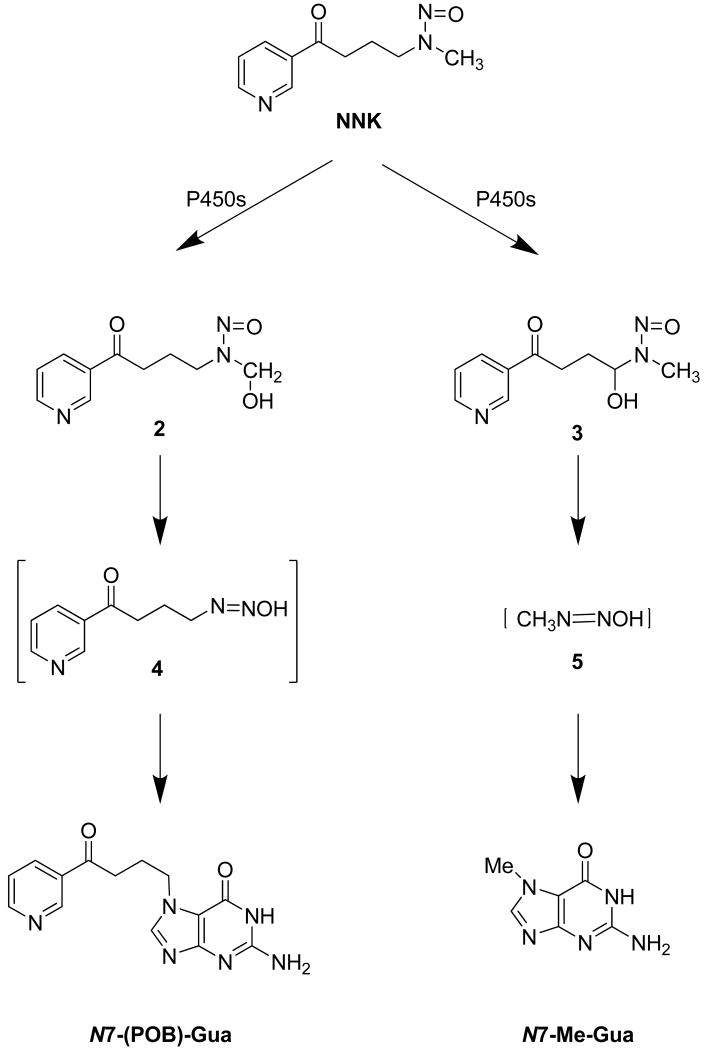
Nitrosamines are a class of chemical compounds that were first described in the chemical literature over 100 years ago, but not until 1956, did they receive much attention. During a routine screening of chemicals that were being proposed for use as solvents in the dry cleaning industry, John Barnes and Peter Magee, reported that dimethylnitrosamine (DMN) produced liver tumors in rats [81]. Magee and Barnes' landmark discovery caused scientists around the world to investigate the carcinogenic properties of other nitrosamines and N-nitroso compounds. Approximately 300 of these compounds have been tested, and 90% of them have been found to be carcinogenic in a wide variety of experimental animals, with many of them exhibiting organ specificity in their carcinogenicity [74]. For instance, DMN causes liver, kidney, and lung cancer in experimental animals, and some of the tobacco-specific nitrosamines are very potent pulmonary carcinogens [82-85]. N-nitrosamines require metabolic activation to exhibit their mutagenic and carcinogenic effects. Metabolic activation of nitrosamines is catalyzed by various forms of P450 enzymes in reactions that in general form a highly reactive diazonium ions and aldehydes [86]. Both the diazonium ions and the aldehydes form DNA adducts, including N7-guanine adducts (Figure 2).
Figure 2.
Formation of N7-guanine adducts form diazonium ion metabolites of NNK. NNK is metabolized by P450–catalyzed α-hydroxylation, producing hydroxymethyl NNK (2) and α-methylenehydroxy NNK (3), which spontaneously decompose to their corresponding diazonium ions (4,5), and keto aldehyde and formaldehyde, respectively. The highly reactive diazonium ions (4,5) subsequently form DNA adducts including N7-guanine adducts [82].
N,N-Dimethylnitrosamine (DMN) is potent rodent carcinogen inducing liver, kidney, and lung tumors, with mice being more susceptible than rats [81, 87]. To investigate potential mechanisms responsible for species differences, the formation of N7-Me-Gua and O6-Me-Gua were determined in mice and rats treated with DMN [88]. The experimental results of this study indicated distinct differences in rat and mouse hepatocytes for repair of the promutagenic DNA lesion, O6-Me-Gua. The ratio of N7-Me-Gua to O6-Me-Gua indicated similar repair of O6-Me-Gua at low DMN exposures in rats and mice. However, the ratios of N7-Me-Gua to O6-Me-Gua changed with increasing DMN dose, suggesting differential ability of these 2 species to repair increasing amounts of O6-Me-Gua. This difference was due to readily inducible O6-methylguanine DNA-methyltransferase (O6MT), found in the rat liver, but not the mouse. Pegg and Hui [89] first observed that the dose-response curves for O6-Me-Gua induced by DMN were different from that of N7-Me-Gua. Studying DNA adducts derived from DMN, it was noticed that the amounts of O6-Me-Gua were much lower relative to N7-Me-Gua at low exposures than at high exposures. The ratio of N7-Me-Gua to O6-Me-Gua at high doses was approximately 10, while at lower exposure the amounts of N7-Me-Gua were about 100-fold higher than that of O6-Me-Gua [89]. The change in ratio of N7-Me-Gua to O6-Me-Gua was later shown to be a result of depletion of O6MT at high doses. O6MT is a highly efficient and specific DNA repair protein that selectively repairs O6-Me-Gua. O6MT removes the alkyl groups from the O6-guanine position by transferring them to the protein itself, effectively restoring the guanine base in DNA, and thereby inactivating the protein [90]. Thus at low exposures O6-Me-Gua adducts are effectively repaired by O6MT producing a higher N7-Me-Gua to O6-Me-Gua ratio. As exposure increases, this O6-Me-Gua specific DNA repair system is depleted and amounts of O6-Me-Gua start to accumulate at a rate similar to their chemical formation. This may explain why early studies of adduct distribution with relative high exposures suggested that the adduct distribution in vivo is similar to the reaction of alkylating compounds with isolated DNA in vitro [8]. However as methodology improved, measurement at lower exposure concentrations were possible and differences in adduct profiles depending on exposures and tissue types were observed. Subsequent advancements in DNA repair research enabled correlation of specific DNA adduct profiles with repair capacities. In general, the dose response for N7-Me-Gua is linear, while it is sub-linear for O6-Me-Gua in O6MT repair proficient tissues. The slope of the upper portion of such a sub-linear dose response is similar to that of N7-Me-Gua and represents the chemical formation rate, since proportionately fewer adducts are removed by repair enzymes like O6MT. At low exposures O6-alkylguanine adducts are readily repaired in most tissues. Consequently, at higher exposures where DNA repair activity is saturated the number of DNA adducts formed increases per unit dose of carcinogen, producing a non-linear dose-response.
4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone (NNK), a tobacco-specific nitrosamine is a potent pulmonary carcinogen inducing lung tumors in experimental animals independent of route of exposure [83]. NNK is metabolically activated in several tissues by an P450-catalyzed hydroxylation of the methylene or methyl carbon adjacent to the N-nitroso group (α-hydroxylation, reviewed by Jalas et al. [91]). These hydroxylation reactions form unstable intermediates that spontaneously decompose to reactive diazonium ions, which can form DNA adducts including N7-guanine adducts, and formaldehyde (Figure 2)[92-100], as reviewed by Hecht [82]. The formation of NNK-derived N7-Me-Gua and O6-Me-Gua has been observed in a number of tissues including placenta, esophagus, larynx, liver, and white blood cells (WBC) [101]. The dose-responses for N7-Me-Gua and O6-Me-Gua in NNK-treated rat lung and liver tissues are similar in shape to the ones observed after DMN treatment discussed above. The dose-responses for N7-Me-Gua and O6-Me-Gua are supra-linear in the lung and liver of NNK-treated rats due to saturation of the α-hydroxylation step in metabolic activation [102]. Furthermore, in lung the persistence of O6-Me-Gua was greatest in Clara cells, the progenitor cells for nitrosamine-induced lung tumors [67, 103, 104]. NNK metabolism was shown to be greatest in Clara cells followed by alveolar macrophages, type II cells and minor in alveolar small cells (300, 220, 100, <10 mol/106cells/1h, respectively), demonstrating cell specific activation [69, 69, 103]. In addition, O6MT activity was 2-fold greater in macrophages and Type II cells than in alveolar small cells or Clara cells [69, 69, 103]. Therefore, the Clara cell-specific accumulation of O6-Me-Gua in rats was attributed to reduced O6MT activity coupled with a high capacity for NNK activation.
2.3 Hydrazines
Similar to nitrosamines, hydrazine compounds have been studied both as potential anticancer drugs and as cancer-causing agents. Early studies of hydrazines, including hydrazine sulfate, were conducted and these compounds were found to induce tumors in laboratory animals [105-107]. Hydrazines require metabolic activation to exhibit their carcinogenic effects and experiments with rat liver microsomes suggested involvement of P450s [108]. Treatment of mice, rats or hamsters with hydrazines substantially increases the incidence of several tumor types [107]. When administered by gavage, 1,2-dimethylhydrazine (SDMH) increased the incidence of lung tumors in female mice. When administered in drinking water, SDMH induced high incidences of angiosarcomas in various organs and tumors of the kidneys, lungs, and liver in mice of both sexes [107]. The same route of administration induced liver carcinomas and angiosarcomas in rats.
Herron and Shank [109] investigated the time course of N7-Me-Gua in liver and kidney of rats over a 25-week period, during which the animals received 21 mg/kg SDMH s.c., once every week during the first 14 weeks. During the treatment period, N7-Me-Gua adducts accumulated in liver and kidney and then were below the limit of detection of 156 N7-Me-Gua/ 107 nnt six or 11 weeks post treatment. No adducts were found in non target tissues (lung or pancreas) [109]. Unfortunately, the limit of detection at these early studies was not sufficient to determine potential endogenous amounts of N7-Me-Gua, which are discussed in detail below. This study clearly demonstrated that metabolic activation, and subsequent adduct formation was tissue-specific, and that reactive hydrazine metabolites were not stable enough to diffuse to distant sites.
Surprisingly, in rats no adduct accumulation was found in colon, another target site for SDMH tumorigenesis [110-112]. In colon DNA, N7-Me-Gua and O6-Me-Gua have the same half-life, ∼30 h, which is much lower than the ∼150 h determined in other tissues [29]. Therefore, the removal of N7-Me-Gua and other DNA adducts in colon is to a greater extent dependant on cell division and sloughing compared to liver, where DNA repair and spontaneous depurination are responsible for adduct removal [113]. O6-Me-Gua accumulated in kidney, while in liver it was only detected after the first SDMH treatment, adding support to the inducibility of O6MT in rat liver, but not in kidney, one of the main target organ of SDMH-induced tumorigenesis [89, 114].
Furthermore, metabolic activation, detoxication, and DNA repair can be different among cell types. The first evidence for different DNA adduct profiles between cells within a target organ was demonstrated in rats exposed to single [32] or chronic [115] doses of SDMH. Swenberg and colleagues investigated the formation and persistence of N7-Me-Gua and O6-Me-Gua in individual liver cell types. SDMH induces mainly malignant liver angiosarcoma [116-118]. Analysis of N7-Me-Gua in purified non-parenchymal cells (NPC), the origin of angiosarcomas, compared with hepatocytes demonstrated similar increases in both cell types, confirming even distribution of exposure between cell types. The corresponding amounts of O6-Me-Gua were significantly different, with amounts in hepatocytes peaking one day after exposure and then declining to 1/30 of the initial amounts, while in NPC cells, amounts of O6-Me-Gua steadily accumulated for 8 days [119]. The significant decline of O6-Me-Gua in hepatocytes was due to both constitutive and an additional 4-fold enhancement of O6MT activity during continuous exposure, an activity that is much lower and remained constant in NPC cells [119].
Summary
Even though O6-Me-Gua was identified as the potentially promutagenic adduct in the examples described above, it would have been difficult to come to such a conclusion without normalization of the data to N7-Me-Gua. These and other studies with pro-carcinogens clearly established that metabolic activation of carcinogens and DNA alkylation can be species-, strain-, tissue-, and cell-type specific. In addition, evidence has been reported that adduct formation rates and endogenous background levels could be age-dependent, and are modified by DNA repair. Consequently, these studies have greatly advanced our understanding in the origin and molecular mechanisms of tissue specificity of these model carcinogens. Unfortunately, as has been described above, the ratio of N7-guanine adducts to other DNA adducts (e.g., O6-guanine adducts) is not constant and is heavily dependent on time since exposure, dose, species, tissue, and cell type. Therefore, to use N7-guanine adducts as a surrogate biomarker for other DNA adducts, it is essential to have detailed knowledge of the rate of formation and persistence of N7-guanine adducts, and the corresponding adducts of interest.
2.4 Olefins: ethylene, propylene, butadiene, and their metabolites
Olefins are characterized by one or more double bonds, with ethylene (ET), propylene, and 1,3-butadiene (BD) being the best studied in respect to DNA adduct formation. BD and the ET metabolite, ethylene oxide (EO), have been classified as human carcinogens [120]. Occupational exposure to butadiene has been associated with increased risk for leukemia in workers exposed to BD in synthetic rubber production [121-123] and an increase in lymphohematopoietic cancers in BD monomer production workers that was not exposure related [124]. Olefins are mainly metabolized by P450s to the corresponding epoxides that are known to form DNA adducts, including N7-guanine adducts via SN2 reactions (Figure 3). These olefin epoxides are more stable than the reactive metabolites discussed above (diazonium ion, hydrazine, etc.) and they circulate freely throughout the body. Consequently, DNA adducts are formed in similar amounts in target and non target tissues [68, 125-127]). The main adducts of olefin epoxides are the corresponding N7-guanine adducts, and N3-adenine adducts in addition to other minor adducts. The relative reactivities of nucleophilic sites in DNA toward some olefin epoxides are shown in Table 1.
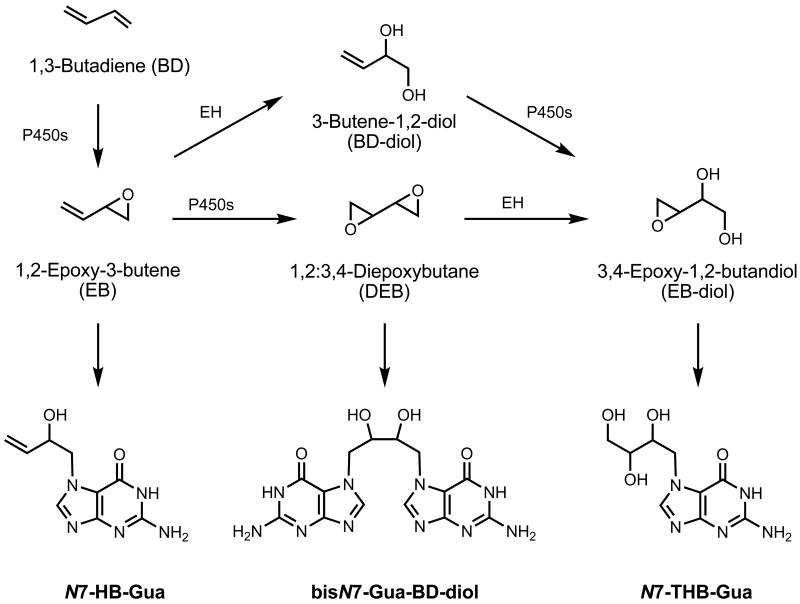
Figure 3.
Example of olefin metabolism and formation of N7-guanine adducts illustrated on BD. BD is metabolized by P450s to several epoxides that form DNA adducts including N7-guanine adducts. Shown are N7-HB-Gua, bisN7-Gua-BD-diol and THB-Gua as representative N7-guanine adducts formed from EB, DEB and EB-diol, respectively.
Ethylene and ethylene oxide
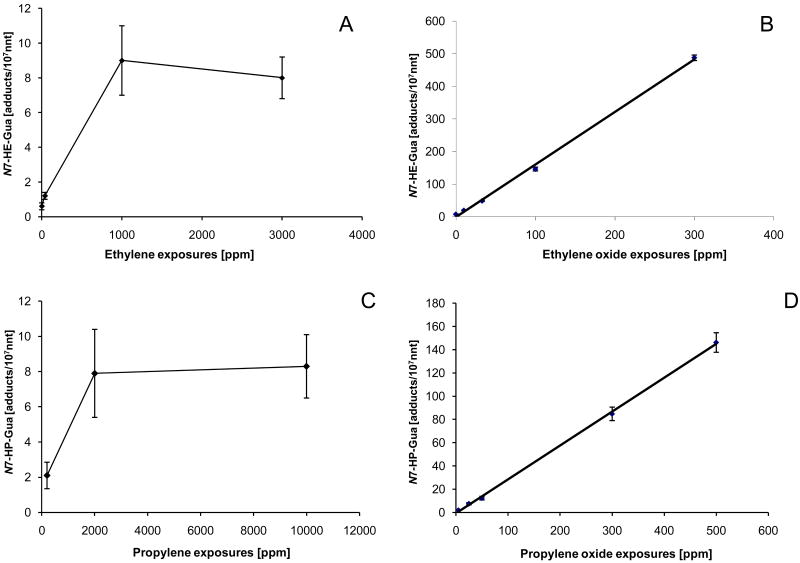
ET is metabolically activated by P450s to ethylene oxide (EO) and forms N7-hydroxyethyl-guanine (N7-HE-Gua) as its major DNA adduct. Inhalation studies with ET in mice and rats established supra-linear dose-responses over the exposure concentrations studied, suggesting saturation of the metabolic activation [128, 129]. The metabolism of ET to EO in mice saturates at ∼1000 ppm ET [130, 131], producing in liver ∼3.5 N7-HE-Gua/ 107 nnt (Table 2, Figure 4) [33, 132, 133]. In contrast, a linear dose-response for N7-HE-Gua has been found in experiments with mice or rats exposed to EO over a wide range of exposures (Figure 4)[133, 134]. The molecular dose of N7-HE-Gua can be orders of magnitude greater for exposures to EO than can ever be achieved by ET. In subacute or chronic exposures to EO, N7-HE-Gua adducts increase daily until they reach a steady state after 7-10 days [33]. This supra-linear response over time is attributed to the chemical instability of N7-HE-Gua. At steady state, the number of N7-HE-Gua adducts formed is equal to the number of adducts lost due chemical depurination or cell death.
Table 2. Dose response of some N7-guanine adducts in rats exposed to representative olefins in rat liver.
| Compound | Dose response Max adduct (adducts / 107) | Adduct | Reference |
|---|---|---|---|
| Ethylene | Saturates at ∼7 adduct/ 107 nnt | N7-HE-Gua | [33] |
| Ethylene oxide | Linear up to 463.8 adducts / 107 nnt | N7-HE-Gua | [129, 133] |
| Propylene | Saturates at 6.5 adducts / 107 nnt | N7-HP-Gua | [149] |
| Propylene oxide | Linear up to 915 adducts / 107 nnt | N7-HP-Gua | [127] |
| Butadiene | Saturates at 23 adducts / 107 nnt | N7-THB-Gua | [68] |
Figure 4.
Comparison of the dose-responses for formation of N7-hydroxyalkyl-Gua adducts following repeated exposure to the olefins ethylene (A) and propylene (C), and their epoxy metabolites EO (B) and PO (C), respectively. Formation of N7-guanine adducts is much more efficient for the epoxide metabolite compared to the parent olefin. Data are from rat lung after 20 days inhalation exposures to olefins and their epoxides [33, 127, 133, 149].
All animals and humans are constantly exposed to ET and EO produced by numerous endogenous metabolic processes and by conversion of ET to EO by the liver [135, 136]. Additional EO exposures are from numerous food products that can contain up to <0.05 to 1800 μg ET/g [137]. The endogenous formation and steady exposure via the diet produces a background level of N7-HE-Gua that has been observed in mice, rats, and humans [128, 138-143]. Yong et al. reported N7-HE-Gua in granulocytes from humans that were not exposed to known sources of ET or EO [143]. The presence of endogenous EO exposure in humans has been confirmed by detection of hydroxyethyl-valine hemoglobin adducts, another EO-specific biomarker [144, 145]. Endogenous or background amounts in mice and rats were in the range of 1-18 N7-HE-Gua adducts/107 nnt [132, 133, 146-148]. The existence of endogenous N7-guanine adducts has to be accounted for in mechanism-based risk assessments and evaluations of low exposures [27].
Propylene and propylene oxide
Propylene is metabolically activated by P450s to propylene oxide (PO) and forms N7-hydroxypropyl-guanine (N7-HP-Gua). The formation of N7-HP-Gua in rat liver saturates at exposures greater than 2000 ppm propylene and produces about 6.5 adducts /107 nnt (Figure 4) [149]. In contrast, exposures to the activated oxide, PO, produces almost 200-fold greater amounts of N7-HP-Gua and the dose-response is linear (Figure 4). Comparing the adduct amounts formed after propylene and PO exposures, it can be estimated that a maximum of 0.5% of the propylene dose is activated to PO and associated with DNA adduct formation. The fact that the adduct formation appears linear for PO-derived N7-HP-Gua and that exposures to PO produce much higher amounts of these adducts demonstrates that adduct formation is not limited by available binding sites in DNA or induction of specific DNA repair systems. Therefore, the saturation observed with propylene is due to saturation of the metabolic activation and/or induction of detoxification pathways. Similar data were obtained from a variety of chemicals and are summarized in Table 2.
Butadiene and butadiene-derived epoxides
1,3-Butadiene (BD) is an olefin of special interest because it is metabolized to several reactive epoxides (Figure 3)[150]. All BD-derived epoxides are known to form DNA and protein adducts. Althought the different oxidation reactions are catalyzed by the same enzymes, mainly by P450 2E1, 2A6 and 3A4, the dose-responses of their internal formation are vastly different in mice, rats, and humans [151, 152]. BD is first oxidized to 1,2-epoxy-3-butene (EB), a metabolite known to form 2-hydroxy-3-butenyl DNA and protein adducts. To date, the N7-(2-hydroxy-3-butenyl)-guanine and N7-(1-hydroxy-3-butenyl)-guanine (N7-HB-Gua)* are the only EB-specific N7-guanine adducts found in mice and rats exposed to BD (Figure 3)[68]. The dose-response for N7-HB-Gua has been shown to be linear in mice and rats from the lowest exposure studied (20 ppm BD 4 weeks) to high exposures known to induce tumorigenesis [68]. The presence of a minor adduct, N1-HB-Ade, has been reported in rat liver after 5 days of exposure to 300 ppm BD [153] and its formation was about 3-fold lower than the N7-HB-Gua.
In vitro formation of N3-HB-adenine, N6-HB-adenine, and N1-HB-inosine adducts have been identified, but their existence in vivo has not been shown [154, 155]. Of these theoretical adducts, N1-HB-inosine is of special interest, since it has the highest mutagenic potency (>95% per replication cycle) [156, 157]. The mutagenic potencies were significantly lower (<1%) for N2-HB-guanine and N6-HB-adenine [158]. Unfortunately, the N7-HB-Gua adduct is not suitable for site-directed mutagenesis studies because of its chemical instability that leads to spontaneous depurination. Therefore, the specific mutagenic potency of N7-HB-Gua remains to be assessed, although mutagenic potency may be very low given that N7-guanine adducts are relative unstable and do not participate in hydrogen bonding in the DNA double helix [2].
EB can undergo a second oxidation reaction, catalized by P450s 2E1, 2C9, and 2A6, producing the 1,2;3,4-diepoxybutane (DEB) [159], which is a bi-functional carcinogen that can form DNA-DNA [160-162] and DNA-protein crosslinks (Figure 3)[163, 164]. In vivo, the presence of N7-guanine-N7-guanine [1,4-bis(guan-7-yl)-2,3-butanediol (bisN7-Gua-BD-diol)] and N7-guanine-N1-adenine [1-(guan-7-yl)-4-(aden-1-yl)-2,3-butanediol (N7-Gua-N1-Ade-BD-diol)] were observed in liver and lung of mice exposed by inhalation to 625 ppm BD [160, 161]. The bisN7-Gua-BD-diol is a weak mutagen, producing miscoding in less than 1% of all replication cycles, compared to the more mutagenic N6,N6-adenine intrastrand cross-links, which produce miscoding in 8% of the replication cycles, [156, 157]. The dose-responses of bisN7-Gua-BD-diol and N7-Gua-N1-Ade-BD-diol cross links were recently reported in mice and rats [165]. It was shown that mice form much more DEB than rats and that adduct formation was higher in lung and liver compared to kidney, brain and thymus. The tissue difference appears to be due to the high reactivity of the bi-functional DEB compared to the mono-functional EB or EB-diol and suggests DEB formation in liver and lung tissue. Further, formation of DEB seemed to saturate in rats at exposures above 62.6 ppm BD and showed a decrease in slope in mice at exposures above 200 ppm. This dose-response for DEB derived adducts have been confirmed by the analysis of N,N-(2,3-dihydroxy-1,4-butadiyl)-valine (pyr-Val), the corresponding DEB-specific globin adduct, in the same animals (Georgieva et al in preparation).
The epoxides EB and DEB can be hydrolyzed by epoxide hydrolase (EH) producing 3-butene-1,2-diol (BD-diol) and 1,2-epoxy-3,4-butanediol (EB-diol), respectively [151, 166-171]. The latter, EB-diol, can also be formed by a second oxidation of BD-diol [167, 171]. Analysis of the EB-diol-derived N7-(trihydroxybutanyl)-guanine (N7-THB-Gua) adducts demonstrated that EB-diol is the main BD-derived epoxide in mice and rats (Figure 3)[68]. Most interestingly, while the dose-response for N7-HB-Gua (EB-derived) is linear, the dose-response for N7-THB-Gua (EB-Diol derived) shows saturation at exposures greater then 62.5 ppm BD in rats after 20 days of exposure, and its slope of formation is reduced with higher exposures in mice [68]. In addition, using N7-HB-Gua and N7-THB-Gua, it was shown that mice form much more EB and EB-diol than rats, a finding that is consistent with the higher susceptibility of mice to BD induced tumorigenesis.
BD species-dependent tumorigenesis is attributed to species-specific differences in BD metabolism. The formation of N7-guanine adducts support the hypothesis that mice are more susceptible because they are more efficient at BD oxidation. Molecular modeling of P450 2E1 also suggested significant species differences between mouse, rat, and human in BD oxidation [172]. The species differences in formation of N7-HB-Gua, DEB-derived cross-link adducts and N7-THB-Gua has been confirmed using the corresponding globin adducts hydroxybutenly-valine (HB-Val), pyr-Val and trihydroxy-butanly–valine (THB-Val), suggesting that the corresponding hemoglobin adducts are a good biomarker for formation of these reactive metabolites [173, 174]. Lastly it was shown that EB can bind to human P450 2E1 itself [175], and may alter catalytic activity in a similar fashion as the mechanism-based inhibition by other P450 substrates including ET [176-186]. Thus, monitoring N7-HB-Gua may provide a biomarker for functional changes in the metabolic pathways, possibly due to alkylating inactivation of important enzymes.
In summary, the studies with olefins described above clearly demonstrate that direct alkylating agents produce a linear dose-response for N7-guanine adducts vs dose, confirming the validity of N7-guanine adducts as a biomarker for internal dose. In contrast, adducts resulting from metabolites of metabolically activated compounds show strong species differences and usually exhibit supra-linear dose-response curves. This is primarily due to saturation of metabolic activation or changes in detoxification pathways, which means that the N7-guanine adducts are better dosimeters for internal dose than administered dose, especially if quantified in tissues of interest. In chronic exposure situations, steady state levels are typically reached after 7-10 days for both olefins and their epoxide metabolites.
3 Formation of N7-guanine adducts in human specimens
Despite the ubiquitous nature of N7-guanine adducts, their application as a biomarker of exposure in larger molecular epidemiology studies is not common practice. A review of the literature demonstrated limited numbers of studies using N7-guanine adducts as biomarkers for exposure to environmental or occupational pollutants. Furthermore, reported data are not extensive and mostly contain small numbers of individuals per group.
Similar to the data from animal studies, the presence of N7-Me-Gua and other N7-guanine adducts has been demonstrated ubiquitously in DNA from humans not known to be exposed to alkylating agents (Table 3) [146, 187-196]. Most of these reports investigated DNA from blood samples; either white blood cells (WBC) or individual blood cell types, and measured N7-Me-Gua by 32P-postlabeling or HPLC-ECD. While this seems to be sufficient evidence for the presence of N7-Me-Gua from endogenous sources in human DNA, it needs to be mentioned that the numbers of subjects per group in most of these studies were relatively small (≤20). The amount of N7-Me-Gua in controls, representing endogenous or background adducts, were about half of the amounts reported for the exposed groups, which were primarily smokers. Consequently, the statistical power of these studies is reduced because of the small number of subjects and the limited difference between controls and exposed indviduals. Furthermore, it is not possible to differentiate endogenous adducts from background adducts, e.g., formed from potential environmental sources due to their indistinguishable chemical identity. Interestingly, N7-Me-Gua amounts were 3-fold and 5-fold higher in tumor tissue compared to normal tissues from cervical or bladder tissues, respectively [187, 194]. In contrast, N7-Me-Gua amounts were lower in colon tumor DNA compared to normal colon tissue [192]. The amounts of N7-Me-Gua in all controls (representing endogenous background amounts) range from 0.8 to 13.5 adducts/107 nnt compared to 3.9 to 23.6 adducts/107 nnt in the exposed subjects from the corresponding study groups. Evaluation of N7-Me-Gua amounts in lymphocyte DNA and solid tissues suggest slightly higher amounts in solid tissues, however sample numbers are small (Table 3).
Table 3. Amounts of N7-guanine adducts in human DNA.
| Adduct/ Tissue | Group | Method | N7-Gua adducts/107 nnt (n) | Ref |
|---|---|---|---|---|
| N7-Me-Gua | ||||
| WBC | Age<50 | 32P-HPLC | 1.0 ± 0.9 (14) | [188] |
| WBC | Age >80 | 32P-HPLC | 0.8 ± 0.4 (20) | [188] |
| WBC | NS | 32P-HPLC | 2.5 | [191] |
| WBC | NS | 32P-AEC | 2.3 (6) | [190] |
| WBC | S | 32P-AEC | 11.5 (11) | [190] |
| WBC | NS | 32P-AEC | 3.4 (10) | [189] |
| WBC | S | 32P-AEC | 6.9 (10) | [189] |
| Granulocytes | NS | 32P-AEC | 2.8 (10) | [189] |
| Granulocytes | S | 32P-AEC | 4.7 (10) | [189] |
| Lymphocytes | NS | 32P-AEC | 13.5 (10) | [189] |
| Lymphocytes | S | 32P-AEC | 23.6 (10) | [189] |
| Lymphocytes | 32P-HPLC | 6.94 (5) | [142] | |
| Lung | Lung cancer patients | 32P-HPLC | 6.3 ± 1.9 (5) | [146] |
| Bronchial tissue | Lung cancer patients | 32P-HPLC | 6.1 ± 1.5 (5) | [146] |
| Lymphocytes | Lung cancer patients | 32P-HPLC | 3.3 ± 0.9 (5) | [146] |
| Lung | 32P-HPLC | 39.4 (5) | [276] | |
| Lung | 32P-HPLC | 2.7 (10) | [142] | |
| Bronchial tissue | NS | 32P-HPLC | 4.7 (6) | [190] |
| Bronchial tissue | S | 32P-HPLC | 17.3 (11) | [190] |
| Cervical cytology | NS | ISB | 0.82b (17) | [187] |
| Cervical cytology | S | ISB | 2.5b (22) | [187] |
| lymphocytes | NS | HPLC-ECD | 4.5 (8) | [193] |
| lymphocytes | S | HPLC-ECD | 3.9 (14) | [193] |
| Colon mucosa | ISB & 32P | 1.7b (6) | [192] | |
| Rectal mucosa | ISB & 32P | 0.7b (5) | [192] | |
| Bronchial Lavage | 11.9 | [277] | ||
| Bronchial Lavage | Ex-smoker | 32P-HPLC | 9.99 ± 20.3 (10) | [195] |
| Bronchial Lavage | Ex-Smokers | 32P-HPLC | 5.59 ± 15.6 (28) | [195] |
| Bronchial Lavage | Never smokers | 32P-HPLC | 0.58 ± 0.5 (6) | [195] |
| Bladder | Tumor | 0.1 | [194] | |
| 0.5 | [194] | |||
| Lung | 32P | 2.5 (80) | [278] | |
| Lung | 32P-HPLC | 2.74 (10) | [142] | |
| Lung | 32P | 2.11 (90) | [142] | |
| N7-alkyl-Gua | ||||
| Larynx tumor | NS | 32P | 11.3 23.2 (5) | [196] |
| Larynx tumor | FS | 32P | 23.2 ± 24.1(34) | [196] |
| Larynx tumor | S | 32P | 61.8 ± 84.5 (5) | [196] |
| Larynx non-tumor | NS | 32P | 8.3 ± 6.0 (3) | [196] |
| Larynx non-tumor | FS | 32P | 21.1 ±18.7 (25) | [196] |
| Larynx non-tumor | S | 32P | 36.3 ± 20.2 (6) | [196] |
| N7-Me-Gua N7-HE-Gua | ||||
| WBC | NS | 32P-HPLC | 2.9 ± 0.9 (8) | [146] |
| WBC | S | 32P-HPLC | 3.9 ± 0.8 (11) | [146] |
| Lung | NS | 32P-HPLC | 4.0 [3.6, 4.4](2) | [146] |
| Lung | S | 32P-HPLC | 6.5 ± 2.2 (7) | [146] |
| N7-Et-Gua | ||||
| Liver | 0.084(25) | [279] | ||
| Lymphocytes | 32P-HPLC | 1.0 (5) | [142] | |
| Lung | 32P-HPLC | 1.46 (10) | [142] | |
| Lung | 32P | 1.6 (75) | [278] | |
| N7-HE-Gua | ||||
| WBC | Controls subjects | HPLC-UV | 10.5 ± 4.8a (5) | [141] |
| WBC | Immuno blot | 0.65 | [280] | |
| WBC | S | Immuno blot | 0.11 | [280] |
| WBC | NS | Immuno blot | 0.095 | [280] |
| lymphocytes | Control subjects | GC-HRMS | 4.85± 3.1b (23) | [128] |
| Granolocytes | Hospital Workers | GC-MS | 9.6 ± 5.8 (6) | [143] |
| Granolocytes | Non exposed | |||
| Granolocytes | Hospital Workers | GC-MS | 13.6 ± 3.9(38) | [143] |
| Granolocytes | <32 ppm-hr ET | |||
| Granolocytes | Hospital Workers | GC-MS | 22.7 ± 11.7 (20) | [143] |
| Granolocytes | >32 ppm-hr Et | |||
| Lung | Lung cancer patients | 32P-HPLC | 0.8 ± 0.3 (5) | [146] |
| Bronchial tissue | Lung cancer patients | 32P-HPLC | 1.0 ± 0.8 (5) | [146] |
| Lymphocytes | Lung cancer patients | 32P-HPLC | 0.6 ± 0.2 (5) | [146] |
| N7-HOEt-Gua | ||||
| brain tissue | Non treated | HPLC-ECD | 1.15 ± 0.63 (6)b | [281] |
| Brain tissues distal | DTI-015 treated | HPLC-ECD | 1.6 (6)b | [281] |
| Brain tissues medial | DTI-015 treated | HPLC-ECD | 5.1 (6)b | [281] |
| brain tissues adjacent | DTI-015 treated | HPLC-ECD | 721 (6)b | [281] |
| RAL | Non cancer | 32P | 2.9 (0.6-11.5) | [282] |
| RAL | Cholangiocarcinoma | 32P | 7.2 (1.8-48.4) | [282] |
| RAL | Tumor adjacent | 32P | 8.6 (1.2-51.6) | [282] |
Adduct unit was converted using following calculation: 1 fmol/ μg DNA = 3.25 adducts/ 107 nnt
Adduct unit was converted using following calculation: 1 pmol/ μmol Guanine = 1.95 adducts/ 107 nnt
NS; non smokers, S; smokers, ISB; Immunoslot blot, 32P; 32P-postlabeling, 32P-AEC; 32P-postlabeling anion exchange chromatography
Wu et al. reported mean endogenous amounts of 4.8 ± 3.1 N7-HE-Gua adducts / 107 nnt in lymphocytes (n=23) of unexposed humans [128]. More recently, Yong et al. [143] demonstrated the presence of N7-HE-Gua in granulocytes of hospital workers not known to be exposed to sources of ET or EO [143]. The mean amount of N7-HE-Gua was 9.6 ± 5.8 adducts/ 107 nnt in subjects categorized as unexposed controls. The range of these amounts is similar as reported previously for five control subjects [141]. These N7-HE-Gua background amounts are believed to stem from EO that forms endogenously from ET derived from environmental exposures such as vegetation, urban air, smoking, and various endogenous metabolic processes [137, 197-199]. Tompkins et al. report in this special issue evidence of increased N7-HE-Gua in rat tissue due to oxidative stress [200]. H2O2 may catalize the formation of ET from methionine, similar to the ET production in fruits and vegetables [201, 202]. The confirmed presence of background DNA alkylation (N7-Me-Gua and N7-HE-Gua) from environmental and endogenous sources in humans is relevant for consideration in risk assessments used to make regulatory decisions. No evidence for endogenous N7-HP-Gua and N7-THB-Gua have been reported, although presence of hydroxypropyl-valine (HP-Val) and THB-Val, the corresponding PO- and EB-diol-specific globin adducts, have been reported in subjects or animals not exposed to known sources of propylene, BD or their epoxide metabolites, PO and EB-diol, respectively [126, 203, 204]. This may suggest that additional unknown endogenous N7-guanine adducts may exist that need to be considered.
4 Evidence for mutations resulting from N7-guanine adducts
4.1 N7-Me-Gua and N7-Et-Gua and mutagenesis in mammalian cells
Early efforts aimed to compare DNA alkylation with mutation frequency (MF) and mutation spectra to identify adducts involved in mutagenesis. Beranek et al. reported a good correlation between DNA methylation (N7-Me-Gua and O6-Me-Gua) and mutation frequency (MF) in the HPRT gene in CHO cells after treatment with MMS or MNU [205]. In contrast, the formation of N7-Et-Gua did not correlate with mutation frequency in HPRT or Na-K-ATPase genes in CHO cells treated with diethylsulfate (DES), EMS, or ENU [206]. The formation of N7-Et-Gua was linear with exposures for each ethylating agent, but the slopes were significantly different. In contrast to the N7-Et-Gua, the amounts of O6-Et-Gua adducts induced by all three ethylating agents correlated with MF in the HPRT gene, while the MF in the Na-K-ATPase gene correlated only with EMS and ENU, and not with DES [206]. This suggests that such correlation studies may be inadequate to analyze multi-component phenomena like mutagenesis, and that DES induces mutations by a mechanism not involving N7-Et-Gua adducts.
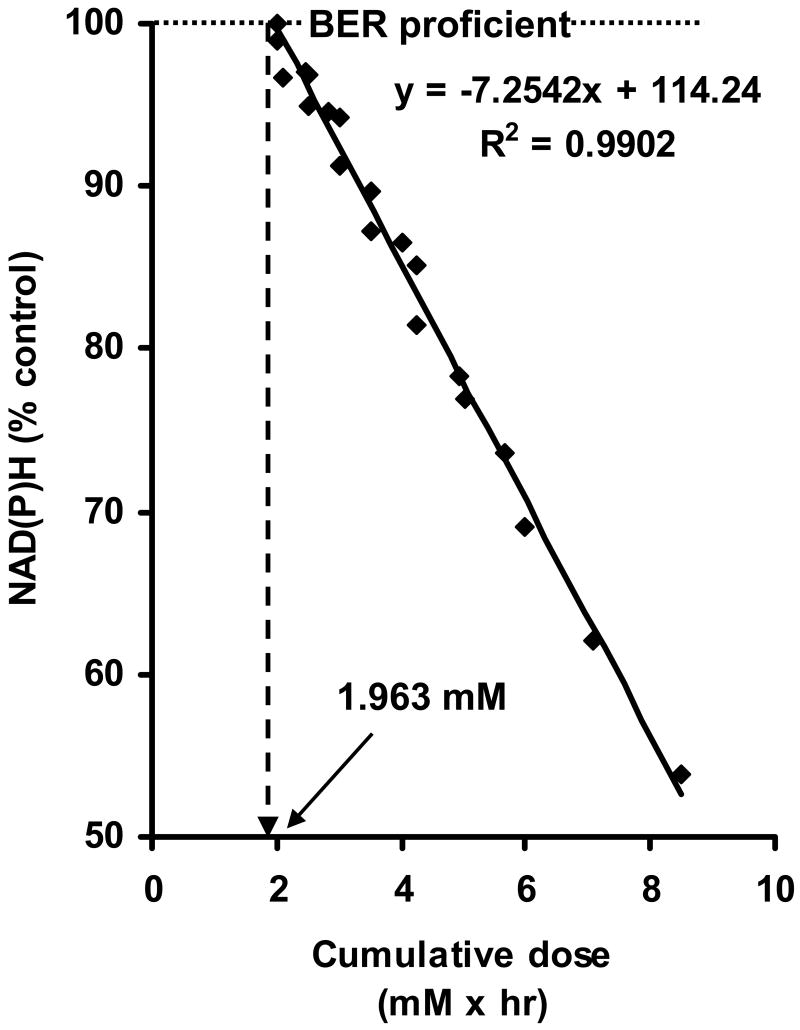
Methyl methanesulfonate (MMS) predominantly produces N7-Me-Gua (∼92%) in DNA and a very low percentage of mutagenic O6-Me-Gua (∼0.3%) [21]. In Chinese hamster ovary (CHO) cells exposed to MMS at 0.75 mM for 1 hour, the majority of HPRT mutations were GC to AT transitions [207], with no increase in other types of mutations. This type of mutation is also found in other Chinese hamster cell lines following exposure to MMS either at 2 mM for 30 minutes or at 1 mM for 1 hour [208, 209]. In these three studies, MMS appears to introduce 750 to 1000 N7-Me-Gua/107 nnt, 0.8 to 1.0 N3-Me-dAde/107 nnt, and 0.027-0.036 O6-Me-Gua/ 107 nnt roughly estimated from our data and from the ratio of N7-Me-Gua/N3-Me-Ade/O6-Me-Gua = 83/8.7/0.3 [21]. It was concluded that this specific GC to AT transition is most likely caused by mispairing of O6-Me-Gua with thymine during replication and cell division [210, 211]. These results strongly suggest that AP sites likely formed by spontaneous depurination of N7-Me-Gua do not contribute to the induction of HPRT mutations in these MMS-exposed cells. Interestingly, preliminary results from our lab show that an imbalance of base excision repair (BER) repair in CHO AA8 cells may start at exposure to 1.9 mM MMS for 1 hour as determined by the extent of depletion of intracellular NAD(P)H in cells (Figure 5) (Pachkowski, unpublished data). Therefore, if the N7-Me-Gua are 1000 lesions/107 nnt or less and N3-Me-Ade are 100 lesions/107 nnt or less, the increase in HPRT mutations in cells exposed to MMS can all be accounted for by induction of O6-Me-Gua (0.036 lesions/107 nnt or less) and are not due to formation of AP sites as BER intermediates.
Figure 5.
Determination of the number of N7-Me-Gua adducts present with an imbalance in BER. NAD(P)H values for CHO AA8 cells were plotted against the corresponding cumulative dose (the product of mM MMS and exposure duration). The X value corresponding to the intersection of the resulting linear regression line and y = 100 (i.e., 100% NAD(P)H relative to controls) was determined to be the start of an imbalance in BER. In the case of AA8 cells, exposure to 1.96 mM MMS for 1 hr initiated the imbalance of BER (unpublished results from Pachkowski et al).
4.2 N7-HE-Gua and N7-HP-Gua and mutagenesis
The formation of N7-HP-Gua saturates at levels of 6.5 adducts /107 nnt in rats after 20 days of exposure to propylene (Table 2, Figure 4) [149]. This amount is 15-fold lower than the molecular dose associated with induction of mutations in Drosophila [212]. In that study, male Drosophila exposed to PO for 24 h did not show a significant increase in mutations until the internal dose, measured as N7-HP-Gua adducts, reached approximately 100 adducts /107 nnt. Since no other adducts were measured, these data do not demonstrate that N7-HP-Gua was the causal adduct. Together, these reports provide mechanistic evidence that propylene is not mutagenic. Pottenger et al. [149] demonstrated that metabolic activation of propylene does not produce sufficient numbers of DNA adducts in mice or rats, compared to that associated with mutagenesis in Drosophila [212]. Furthermore, the Drosophila system most likely over estimates the mutagenic potency of PO since cell proliferation is much faster in the Drosophila gonadocytes, leaving less time for DNA repair compared to mammalian systems.
The range of N7-guanine adducts of 0.8-23 adducts /107 nnt in humans, summarized in Table 3, are significantly below the N7-HP-Gua adducts shown to correlate with mutations in Drosophila. In contrast, significant increases in mutations in Drosophila after EO exposure were accompanied by 30 N7-HE-Gua/ 107 nnt. However NER repair decreased the EO-induced mutation response in Drosophila, suggesting involvement of adducts other then N7-HE-Gua in mutagenesis [212]. This adduct amount is close to the amounts observed in hospital workers exposed to EO as a sterilant and only 3 to 6-times higher than the background level of endogenous N7-HE-Gua [143]. These data may suggest that the large background of endogenous DNA adducts, derived from endogenous metabolites arising from oxidative stress and endogenous ET may significantly contribute to background mutagenesis [200, 213]. This is an area that clearly needs additional research.
4.3 AP sites derived from N7-guanine adducts
Apurinic sites (AP) are the most common form of endogenous DNA damage and are the product of spontaneous depurination, oxidative damage, and the result of DNA repair. The notion that N7-guanine adducts convert to AP sites and subsequently cause mutations was investigated by Rusyn et al. [129]. It was demonstrated that AP sites are not increased following repeated exposure to ET or EO; the presence of N7-HE-Gua indicates that AP sites would be formed by spontaneous depurinations, so they must be efficiently repaired so that they do not cause any increase over the endogenous level [129]. In addition, the HPRT mutation frequency in mice and rats, and micronuclei in polychromatic or normochromatic erythrocytes (MNPCE) in rats, were not increased after repeated exposures to 3000 ppm ET, while the N7-HE-Gua adducts significantly increased over endogenous levels and their formation saturated at exposures of 3000 ppm ET. This exposure resulted in 6 adducts /107 nnt (Table 2) [33]. In contrast, exposures of 100 ppm EO significantly increased HPRT mutation frequency and MNPCE [33, 214], but still did not result in accumulation of AP sites [129]. The number of AP sites ranged from 30 to 70 AP sites /107 nnt for controls, ET-, and EO-exposed rats, while the number of N7-HE-Gua increased more than 10-fold from ∼6 to 64 /107 nnt in rats exposed to 0 or 100 ppm EO for 4 weeks, respectively [129]. The authors concluded that the mutagenesis from EO exposures most likely resulted from minor promutagenic adducts, rather than AP sites or N7-HE-Gua. Measurement of AP sites after neutral thermal hydrolysis of DNA confirmed that EO produced heat labile lesions as a result of enhanced depurination. Heat-treated spleen DNA had a 5-fold increase in AP sites after EO exposure [129].
Similarly, PO exposures did not increase AP sites in nasal epithelium, the target tissue for PO carcinogenesis, despite the high internal dose of N7-HP-Gua, with up to ∼1000 N7-HP-Gua /107 nnt in rats exposed to 500 ppm PO for 4 weeks [215]. While the number of adducts was much lower than that found after high PO exposures, 4 week inhalation exposures to 10,000 ppm propylene did not increase the mutation frequency in the HPRT gene in spleenocytes [149].
AP sites are also intermediates of BER, however, formation from enzymatic removal of N7-guanine adducts is minimal, compared to those arising from spontaneous depurination. This area is not fully understood, however, enzymatic removal has been shown to reduce the half-lives of some N7-guanine adducts to minutes instead of hours [216-218]. The bacterial enzyme AlkA has been shown to remove a wide variety of damaged purine and pyrimidine bases, including N7-Me-Gua [219-221]. Mammalian cells do not possess a homologue of AlkA, but AAG, a member of the N-methylpurine DNA glycosylase family, removes a broad spectrum of modified purines from DNA including N7-Me-Gua [222, 223]. Additional studies on knockout animals will be needed to demonstrate that DNA repair plays a significant role in removal of N7-guanine adducts, compared to chemical depurination.
4.4 Mutations induced by BER intermediates in mammalian cells
To characterize the mutagenic potential of AP sites, a BER intermediate in mammalian cells, an SV-40-derived shuttle vector with a single lesion of either an abasic (AP) site or 5′-deoxyribose-5-phosphate (5′dRp: 5′-cleaved AP sites), at a defined position, was transfected into monkey kidney COS7 cells [224]. The mutation spectrum revealed that preferential incorporation opposite the AP sites is dA (48%) > dC (39%) > dG (13%) ≫ dT (none). Thus, if either dA or dC is incorporated opposite the AP sites derived from depurination of N7-guanine adducts, GC->TA transversions are expected as the primary mutation induced by AP sites. In addition, a small proportion (16%) of deletions was also observed [224]. During BER, the AP sites are converted into single strand breaks (SSB) by AP endonuclease 1, which leads to the formation of 5′dRp and 3′ hydroxyl termini. In the same vector system, the mutation analysis showed that preferential incorporation opposite the 5′-dRp was dA (46%) > dG (41%) ≫ dC (13%) ≫ dT (none). As with AP sites, a small proportion (9%) of deletions was observed. Therefore, N7-guanine adducts are expected to cause G->T transversions, G->C transversions, and deletions if they result from products of BER intermediates 5′dRp and AP sites. In contrast, the nitrosamine DMN and ENU induce mainly G->A transition mutations in the BigBlue transgenenic mice model [225-228]. In CHO cells, MMS primarily increases GC to AT transitions in the HPRT gene [207]. In splenic T-lymphocytes of mice, EO induced four A:T transversions, three A:T transitions, two G:C transversions, and two G:C transitions [229]. In in vitro and in vivo test systems, BD and its metabolites, EB, DEB, and EB-diol induce predominantly deletions and base substitutions at A:T [230-232]. Consequently, the mutation spectra are quite different for compounds known to form N7-guanine adducts that can convert to AP sites, than the mutation spectrum produced by AP sites.
4.5 Ring-opened formamidopyrimidine adducts (FAPy adducts)
N7-guanine adducts are susceptible to attack by hydroxide on the C8 carbon and subsequent ring opening to form 5-N-alkyl-2,6,-diamino-4-hydroxyformamidopyrimidine (Alkyl-FAPy) [233-241]. Early studies [233, 239] proposed placement of a formyl group (-CHO) on the N7-position and this has been confirmed by NMR examination of 5-N-methyl-2,6,-diamino-4-hydroxyformamidopyrimidine (Me-FAPy) and 5-N-(9-hydroxyaflatoxin)-2,6,-diamino-4-hydroxyformamidopyrimidine (AFB1-FAPy) [242, 243]. During the ring opening, the negative charge on the N9-position is delocalized to an α-β-unsaturated carbonyl by the pyrimidine ring, subsequently stabilizing the glycosidic bond. This has important biological implications since, unlike the N7-guanine adduct, the Alkyl-FAPy will not spontaneously depurinate and be lost from DNA. In fact it has been shown that the Me-FAPy was more persistent in rat bladder epithelium than O6-Me-Gua after 21 days and accounted for approximately 72% of the total MNU-derived adducts [244]. In rat liver Me-FAPy was the main persistent adduct after DMN and SDMH treatment [245]. There is an urgent need for more data on the amount of Me-FAPy and other FAPy adducts in cells and tissues in exposed and control animals.
For example, aflatoxin, one of the most potent liver carcinogens, is metabolically activated to the 8,9-epoxide which forms primarily N7-AFB1-Gua [246-252]. The N7-AFB1-Gua converts to secondary lesions including AP sites and AFB1-FAPy. Compared to N7-AFB1-Gua, AFB1-FAPy is highly persistent in rat liver DNA, reaching maximum amounts 2 weeks after exposure [249]. While the structures of N7-AFB1-Gua and AFB1-FAPy are similar, they alter the secondary DNA structure differently [242, 253]. The increased chemical stability (persistence) and altered secondary DNA structure are believed to be responsible for the much greater mutagenicity of AFB1-FAPy compared to N7-AFB1-Gua. In fact, N7-AFB1-Gua and/or AFB1-FAPy cause primarily G to T mutations, consistent with the observed G to T mutations in codon 249 of the p53 tumor suppressor gene in 50% of hepatocellular carcinomas and in aflatoxin-treated human hepatocytes cultures [254, 255]. Additionally, aflatoxin-induced G to T mutations in the ras oncogene are assumed to be important in tumor progression [256-258]. Although both N7-AFB1-Gua and AFB1-FAPy cause the types of mutation consistent with the ones observed in aflatoxin-exposed biological systems, the MF is 6-fold lower (4% vs 32%) for N7-AFB1-Gua compared to AFB1-Fapy, respectively [259, 260]. Whether the N7-AFB1-Gua converted to AFB1-FAPy during the mutation assay was not determined, however, if this were the case, the mutagenic potency for the N7-AFB1-Gua would be even less, making the AFB1-FAPy even more important. This suggests that the AFB1-FAPy, and potentially other ring-opened N7-guanine adducts, may be the ultimate lesions responsible for mutagenesis and genotoxicity of aflatoxin and other carcinogens. Unfortunately, there are no data on the formation and persistence of other FAPy adducts and future investigations of N7-guanine adducts should include studies on the ring-opened FAPy derivates.
5 Conclusions
After decades of research on N7-guanine adducts in animals and humans, it has become clear that specific N7-guanine adducts are excellent biomarkers for internal exposure when they are determined in tissue DNA. In contrast, N7-guanine adducts that can be formed from endogenous or background sources are less reliable for estimating low external exposures. While the presence of N7-guanine adducts clearly demonstrate exposure to the tissues or cells, subsequent interpretations and conclusions need to consider that the value of this endpoint is complicated by endogenous and background formation, and can vary according to species, age, duration of exposure, and tissue. From our review of the literature, we did not find any evidence that N7-guanine adducts can be used beyond confirmation of exposure to the target tissue and demonstration of the molecular dose. This is especially important for translation to human studies, since most studies consist of a single time point analysis of an individual adduct. If the goal is to confirm exposure or identify factors that increase endogenous formation, analysis of N7-guanine adducts in DNA may be sufficient. Consequently, values of N7-guanine adducts in human urine (not reviewed herein) may be sufficient to identify exposures or factors increasing endogenous formation between population groups, even if no information about the site of formation will be available [246, 261, 262]. Importantly, a correlation with other endpoints like chromosome aberrations or mutation frequency cannot be expected to be definitive, since such biomarkers of effect are not chemical-specific and are products of complex biological processes that may only partly (or even not at all) involve N7-guanine adducts.
The main challenge in studying the mutagenic potency of N7-guanine adducts is that their chemical instability has prevented systematic investigation of the lesions in site-directed mutagenesis studies. In fact, oligodeoxynucleotides containing N7-Me-Gua have been prepared, using DNA polymerase and structural analysis by NMR revealed no disturbance of the b-DNA helix [263]. However, since N7-guanine adducts are not considered stable enough to undergo site-directed mutagenesis studies, an alternative would be to stabilize the glycosidic bond by either substitution of guanine with 2-amino-8,9-dihydro-1H-purin-6(7H)-one or by incorporation of a fluorine at the 2′-position of the 2′deoxyribose moiety as recently reported by Lee et al. [264]. The ring opened N7-Me-FAPy adduct of N7-Me-Gua have been shown to block DNA polymerase in vitro [265, 266] but its relevance in vivo needs to be determined. Finally, the formation of N7-guanine adducts is often accompanied by formation of other adducts (e.g., O6-Me-Gua) that are known to be highly mutagenic lesions. Mechanistically, the lack of mutagenicity is linked to the fact that the N7-position does not participate in hydrogen bonding in the DNA double helix, unlike the N1, N2, or O6-positions of Gua [2].
It is noteworthy that covalent binding to DNA and the ability to form N7-guanine adducts alone has long been considered evidence of genotoxicity for several drugs and chemicals [267]. Conversely, N7-guanine adducts easily depurinate to produce AP sites, however, these lesions are the most common endogenous DNA damage, and their number does not appear to increase even with extremely high exposures to alkylating agents. Therefore, one needs to be cautious in classifying compounds and drugs based on formation of N7-guanine adducts alone. This becomes very important for risk assessment of low chronic exposures, when data are limited to high and single dose exposures. The default is to assume a linear dose-response to zero and subsequent extrapolation of the DNA damage data from high to low dose. However, examples have been reported that show that at low exposures, biological processes like mutagenesis reach a background that is much different in slope than N7-guanine adducts [27, 200]. It was suggested that these background mutations are driven by endogenous processes rather than by external exposure. For example, using [13C]MMS for cell treatment and mass spectrometry quantition of [13C]N7-Me-Gua and N7-Me-Gua, Swenberg et al. demonstrated that the adduct formation by MMS from exogenous sources is linear towards zero, while the dose-response for mutation frequency follows a “hockeystick” threshold dose-response (Figure 6)[27]. Similar data have also been presented for EO, EMS, and acrylamide [27, 200, 268]. Based on the information presented in Section 4, it is very unlikely that exposures resulting in elevated amounts of N7-guanine adducts are sufficient to induce mutations, since pro-mutagenic lesions, such as O6-guanine adducts, will also increase and are expected to drive mutagenesis.
Figure 6.
Formation of endogenous and exogenous N7-Me-Gua in AHH-1 cells exposed to [13C2] MMS for 24 h (Modified from Swenberg et al [27]). Exogenous N7-Me-Gua adducts were distinguished form endogenous N7-Me-Gua base on mass differences due to stable isotope labeled [13C2] MMS used for cell treatment. Adduct amounts were compared to mutation frequency in HPRT gene as reported by Doak et al. [283].
A careful review of the literature related to N7-guanine adducts revealed that there is little evidence that N7-guanine adducts cause mutations. In addition, there is mounting evidence that they do not cause mutations, since they do not participate in hydrogen bonding and easily depurinate. In addition, formation of N7-guanine adducts is often accompanied by the formation of other adducts, known to be mutagenic, and therefore it is difficult to credit mutation events to N7-guanine adducts. As discussed in detail before, the relative ratio between N7-guanine adducts and other adducts (e.g., O6-alkylguanine) is dependent upon the chemical and modified by species, strain, tissue, or cell types, differences in activation, and differences in DNA-repair. Consequently, the formation of N7-guanine adducts cannot be used in isolation as a quantitative biomarker for promutagenic DNA lesions, mutagenic response or as a surrogate for other biological processes.
Supplementary Material
Table S1: Amounts of N7-guanine adducts in mice.
Table S2: Amounts of N7-guanine adducts in rats.
Acknowledgments
The authors are thankful to Lynn Pottenger for constructive and productive discussion and editorical assistance. This work was supported in part by NIH grants P42-ES05948, P30-ES10126, R01-ES012689 and the American Chemistry Council.
Abbreviations
- AFB1-FAPy
5-N-(9-hydroxyaflatoxin)-2,6,-diamino-4-hydroxyformamidopyrimidine
- Alkyl-FAPy
5-N-alkyl-2,6,-diamino-4-hydroxyformamidopyrimidine
- AGE
allyl glycidyl ether
- AP-sites
apurinic sites
- BER
base excision repair
- bisN7-Gua-BD-diol
1,4-bis(guan-7-yl)-2,3-butanediol
- DEB
1,2;3,4-diepoxybutane
- DEN
diethylnitrosamine
- DES
diethylsulfate
- EB
1,2-epoxy-3-butene
- EB-diol
1,2-epoxy-3,4-butanediol
- ECH
epichlorohydrin
- EMS
ethylmethanesulfonate
- ENU
ethylnitrosourea
- EO
ethylene oxide
- ET
ethylene
- HENU
1-(2-hydroxyethyl)-1-nitrosourea
- HPLC-ECD
High pressure liquid chromatography with electro chemical detection
- ISB
Immunoslot blot
- KC
Kupffer cells
- LC-MS/MS
Liquid chromatography tandem mass spectrometry
- Me-FAPy
5-N-methyl-2,6,-diamino-4-hydroxyformamidopyrimidine
- MF
mutation frequency
- MMS
methylmethanesulfonate
- MN
micronucleus
- MNU
N-methyl-N-nitrosourea
- N7-AFB1-Gua
N7-(9-hydroxyaflatoxin)-guanine
- N7-Et-Gua
N7-ethyl-guanine
- N7-Gua-N1-Ade-BD-diol
1-(guan-7-yl)-4-(aden-1-yl)-2,3-butanediol
- N7-HB-Gua
N7-hydroxybutenly-guanine
- N7-HE-Gua
N7-hydroxyethyl-guanine
- N7-HMGSH-Gua
S-[1-(hydroxymethyl)-2-(N7-guanyl)ethyl]-glutathione
- N7-HP-Gua
N7-hydroxypropyl-guanine
- N7-Me-Gua
N7-methyl-guanine
- N7-THB-Gua
N7-trihydroxybutanly-guanine
- nnt
normal nucleotides
- NPC
non-parenchymal cells
- NS
non smokers
- O2-Et-Thy
O2-ethyl-thymidine
- O4-Et-Thy
O4-ethyl-thymidine
- O6-Et-Gua
O6-ethyl-guanine
- O6-Me-Gua
O6-methyl-guanine
- O6MT
O6-methylguanine DNA-methyltransferase
- 32P
32P-postlabeling
- 32P-AEC
32P-postlabeling anion exchange chromatography
- PO
propylene oxide
- S
smokers
- SDMH
1,2-dimethylhydrazine
- SEC
sinusoidal endothelia cells
- SO
styrene oxide
- SSB
single strand breaks
- TCP
1,2,3-Trichloropropane
- THB-Val
trihydroxybutanly–valine
Footnotes
The abbreviation N7-HB-Gua will be used for both the N7-(2-hydroxy-3-butenyl)-guanine and N7-(1-hydroxy-3-butenyl)-guanine (N7-HB-Gua).
- pmol adduct/ μmol Gua = 1.95 adducts/107 nnt
- fmol/ μg DNA = 3.25 adducts/ 107 nnt
If data were reported graphically, values were estimated from figures. These values need to be used with some caution, as they do not represent exact values.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Reiner B, Zamenhof S. Studies on the chemically reactive groups of deoxyribonucleic acids. J Biol Chem. 1957;228:475–486. [PubMed] [Google Scholar]
- 2.Watson JD, Crick FH. Molecular structure of nucleic acids; a structure for deoxyribose nucleic acid. Nature. 1953;171:737–738. doi: 10.1038/171737a0. [DOI] [PubMed] [Google Scholar]
- 3.Boysen G, Hecht SS. Analysis of DNA and protein adducts of benzo[a]pyrene in human tissues using structure-specific methods. Mutat Res. 2003;543:17–30. doi: 10.1016/s1383-5742(02)00068-6. [DOI] [PubMed] [Google Scholar]
- 4.van Zeeland AA. Molecular dosimetry of chemical mutagens. Relationship between DNA adduct formation and genetic changes analyzed at the molecular level. Mutat Res. 1996;353:123–150. doi: 10.1016/0027-5107(95)00245-6. [DOI] [PubMed] [Google Scholar]
- 5.De Bont R, Van Larebeke N. Endogenous DNA damage in humans: a review of quantitative data. Mutagenesis. 2004;19:169–185. doi: 10.1093/mutage/geh025. [DOI] [PubMed] [Google Scholar]
- 6.Rundle A. Carcinogen-DNA adducts as a biomarker for cancer risk. Mutat Res. 2006;600:23–36. doi: 10.1016/j.mrfmmm.2006.05.031. [DOI] [PubMed] [Google Scholar]
- 7.Bartsch H. DNA adducts in human carcinogenesis: etiological relevance and structure-activity relationship. Mutat Res. 1996;340:67–79. doi: 10.1016/s0165-1110(96)90040-8. [DOI] [PubMed] [Google Scholar]
- 8.Beranek DT. Distribution of methyl and ethyl adducts following alkylation with monofunctional alkylating agents. Mutat Res. 1990;231:11–30. doi: 10.1016/0027-5107(90)90173-2. [DOI] [PubMed] [Google Scholar]
- 9.Pauwels W, Veulemans H. Comparison of ethylene, propylene and styrene 7,8-oxide in vitro adduct formation on N-terminal valine in human haemoglobin and on N-7-guanine in human DNA. Mutat Res. 1998;418:21–33. doi: 10.1016/s1383-5718(98)00106-5. [DOI] [PubMed] [Google Scholar]
- 10.Hemminki K. DNA adducts, mutations and cancer. Carcinogenesis. 1993;14:2007–2012. doi: 10.1093/carcin/14.10.2007. [DOI] [PubMed] [Google Scholar]
- 11.Guengerich FP. Mechanisms of formation of DNA adducts from ethylene dihalides, vinyl halides, and arylamines. Drug Metab Rev. 1994;26:47–66. doi: 10.3109/03602539409029784. [DOI] [PubMed] [Google Scholar]
- 12.La DK, Swenberg JA. DNA adducts: biological markers of exposure and potential applications to risk assessment. Mutat Res. 1996;365:129–146. doi: 10.1016/s0165-1110(96)90017-2. [DOI] [PubMed] [Google Scholar]
- 13.Swenberg JA, Ham A, Koc H, Morinello E, Ranasinghe A, Tretyakova N, Upton PB, Wu KY. DNA adducts: effects of low exposure to ethylene oxide, vinyl chloride and butadiene. Mutat Res. 2000;464:77–86. doi: 10.1016/s1383-5718(99)00168-0. [DOI] [PubMed] [Google Scholar]
- 14.Chang LW, Hsia SM, Chan PC, Hsieh LL. Macromolecular adducts: biomarkers for toxicity and carcinogenesis. Annu Rev Pharmacol Toxicol. 1994;34:41–67. doi: 10.1146/annurev.pa.34.040194.000353. [DOI] [PubMed] [Google Scholar]
- 15.Doerge DR, da Costa GG, McDaniel LP, Churchwell MI, Twaddle NC, Beland FA. DNA adducts derived from administration of acrylamide and glycidamide to mice and rats. Mutat Res. 2005;580:131–141. doi: 10.1016/j.mrgentox.2004.10.013. [DOI] [PubMed] [Google Scholar]
- 16.Doerge DR, Churchwell MI, Beland FA. Analysis of DNA adducts from chemical carcinogens and lipid peroxidation using liquid chromatography and electrospray mass spectrometry. J Environ Sci Health C Environ Carcinog Ecotoxicol Rev. 2002;20:1–20. doi: 10.1081/GNC-120003925. [DOI] [PubMed] [Google Scholar]
- 17.Miller EC, Miller JA. The presence and significance of bound aminoazo dyes in liver of rats fed p-dimethylaminoazobenzene. Cancer Res. 1947;7:468–480. [Google Scholar]
- 18.Miller EC, Miller JA. In vivo combinations between carcinogens and tissue constituents and their possible role in carcinogenesis. Cancer Res. 1952;12:547–556. [PubMed] [Google Scholar]
- 19.Wheeler GP, Skipper HE. Studies with mustards. III. In vivo fixation of C14 from nitrogen mustard-C14H3 in nucleic acid fractions of animal tissues. Arch Biochem Biophys. 1957;72:465–475. doi: 10.1016/0003-9861(57)90222-9. [DOI] [PubMed] [Google Scholar]
- 20.Brookes P, Lawley PD. The reaction of mono- and di-functional alkylating agents with nucleic acids. Biochem J. 1961;80:496–503. doi: 10.1042/bj0800496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Singer B, Grunberger D. Molecular Biology of Mutagens and Carcinogens, Plenum Press, New York. 1983 [Google Scholar]
- 22.Swenson DH. Significance of electrophilic reactivity and especially DNA alkylation in carcinogenesis and mutagenesis. Dev Toxicol Environ Sci. 1983;11:247–254. [PubMed] [Google Scholar]
- 23.O'Connor PJ. Interaction of chemical carcinogens with macromolecules. J Cancer Res Clin Oncol. 1981;99:167–186. doi: 10.1007/BF00412452. [DOI] [PubMed] [Google Scholar]
- 24.Lawley PD. Some chemical aspects of dose-response relationships in alkylation mutagenesis. Mutat Res. 1974;23:283–295. doi: 10.1016/0027-5107(74)90102-x. [DOI] [PubMed] [Google Scholar]
- 25.Singer B, Bodell WJ, Cleaver JE, Thomas GH, Rajewsky MF, Thon W. Oxygens in DNA are main targets for ethylnitrosourea in normal and xeroderma pigmentosum fibroblasts and fetal rat brain cells. Nature. 1978;276:85–88. doi: 10.1038/276085a0. [DOI] [PubMed] [Google Scholar]
- 26.Singer B. Reaction of guanosine with ethylating agents. Biochemistry. 1972;11:3939–3947. doi: 10.1021/bi00771a017. [DOI] [PubMed] [Google Scholar]
- 27.Swenberg JA, Fryar-Tita E, Jeong YC, Boysen G, Starr T, Walker VE, Albertini RJ. Biomarkers in Toxicology and Risk Assessment: Informing Critical Dose-Response Relationships. Chem Res Toxicol. 2008;21:253–265. doi: 10.1021/tx700408t. [DOI] [PubMed] [Google Scholar]
- 28.Citti L, Gervasi PG, Turchi G, Bellucci G, Bianchini R. The reaction of 3,4-epoxy-1-butene with deoxyguanosine and DNA in vitro: synthesis and characterization of the main adducts. Carcinogenesis. 1984;5:47–52. doi: 10.1093/carcin/5.1.47. [DOI] [PubMed] [Google Scholar]
- 29.Margison GP, Margison JM, Montesano R. Methylated purines in the deoxyribonucleic acid of various Syrian-golden-hamster tissues after administration of a hepatocarcinogenic dose of dimethylnitrosamine. Biochem J. 1976;157:627–634. doi: 10.1042/bj1570627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.King HW, Osborne MR, Brookes P. The in vitro and in vivo reaction at the N7-position of guanine of the ultimate carcinogen derived from benzo(a)pyrene. Chem Biol Interact. 1979;24:345–353. doi: 10.1016/0009-2797(79)90082-6. [DOI] [PubMed] [Google Scholar]
- 31.Osborne M, Merrifield K. Depurination of benzo[a]pyrene-diolepoxide treated DNA. Chem Biol Interact. 1985;53:183–195. doi: 10.1016/s0009-2797(85)80095-8. [DOI] [PubMed] [Google Scholar]
- 32.Lewis JG, Swenberg JA. Differential repair of O(6)-methylguanine in DNA of rat hepatocytes and nonparenchymal cells. Nature. 1980;288:185–41. doi: 10.1038/288185a0. [DOI] [PubMed] [Google Scholar]
- 33.Walker VE, Wu KY, Upton PB, Ranasinghe A, Scheller N, Cho MH, Vergnes JS, Skopek TR, Swenberg JA. Biomarkers of exposure and effect as indicators of potential carcinogenic risk arising from in vivo metabolism of ethylene to ethylene oxide. Carcinogenesis. 2000;21:1661–1669. doi: 10.1093/carcin/21.9.1661. [DOI] [PubMed] [Google Scholar]
- 34.Young JF, Luecke RH, Doerge DR. Physiologically based pharmacokinetic/pharmacodynamic model for acrylamide and its metabolites in mice, rats, and humans. Chem Res Toxicol. 2007;20:388–399. doi: 10.1021/tx600287w. [DOI] [PubMed] [Google Scholar]
- 35.Boucheron JA, Richardson FC, Morgan PH, Swenberg JA. Molecular dosimetry of O4-ethyldeoxythymidine in rats continuously exposed to diethylnitrosamine. Cancer Res. 1987;47:1577–1581. [PubMed] [Google Scholar]
- 36.Kleihues P, Bucheler J. Long-term persistence of O6-methylguanine in rat brain DNA. Nature. 1977;269:625–626. doi: 10.1038/269625a0. [DOI] [PubMed] [Google Scholar]
- 37.Gates KS, Nooner T, Dutta S. Biologically Relevant Chemical Reactions of N7-Alkylguanine Residues in DNA. Chem Res Tox. 2004;17:839–856. doi: 10.1021/tx049965c. [DOI] [PubMed] [Google Scholar]
- 38.Miller JA. Carcinogenesis by Chemicals: An Overview--G. H. A. Clowes Memorial Lecture. Cancer Res. 1970;30:559–576. [PubMed] [Google Scholar]
- 39.Schwarz M, Wiesbeck G, Hummel J, Kunz W. Effect of ethanol on dimethylnitrosamine activation and DNA synthesis in rat liver. Carcinogenesis. 1982;3:1071–1075. doi: 10.1093/carcin/3.9.1071. [DOI] [PubMed] [Google Scholar]
- 40.Goth R, Rajewsky MF. Persistence of O6-ethylguanine in rat-brain DNA: correlation with nervous system-specific carcinogenesis by ethylnitrosourea. Proc Natl Acad Sci U S A. 1974;71:639–643. doi: 10.1073/pnas.71.3.639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kleihues P, Margison GP. Carcinogenicity of N-methyl-N-nitrosourea: possible role of excision repair of O6-methylguanine from DNA. J Natl Cancer Inst. 1974;53:1839–1841. [PubMed] [Google Scholar]
- 42.Swenberg JA, Dyroff MC, Bedell MA, Popp JA, Huh N, Kirstein U, Rajewsky MF. O4-Ethyldeoxythymidine, but not O6-ethyldeoxyguanosine, accumulate in hepatocyte DNA of rats exposed continuously to diethylnitrosamine. Proc Natl Acad Sci USA. 1984;81:1692–1695. doi: 10.1073/pnas.81.6.1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gupta RC, Reddy MV, Randerath K. 32P-postlabeling analysis of non-radioactive aromatic carcinogen--DNA adducts. Carcinogenesis. 1982;3:1081–1092. doi: 10.1093/carcin/3.9.1081. [DOI] [PubMed] [Google Scholar]
- 44.Randerath K, Randerath E, Agrawal HP, Gupta RC, Schurdak ME, Reddy MV. Postlabeling methods for carcinogen-DNA adduct analysis. Environ Health Perspect. 1985;62:57–65. doi: 10.1289/ehp.856257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Phillips DH. Detection of DNA modifications by the 32P-postlabelling assay. Mutat Res. 1997;378:1–12. doi: 10.1016/s0027-5107(97)00092-4. [DOI] [PubMed] [Google Scholar]
- 46.Savela K, Hemminki K, Hewer A, Phillips DH, Putman KL, Randerath K. Interlaboratory comparison of the 32P-postlabelling assay for aromatic DNA adducts in white blood cells of iron foundry workers. Mutat Res. 1989;224:485–492. doi: 10.1016/0165-1218(89)90074-8. [DOI] [PubMed] [Google Scholar]
- 47.Arab K, Pedersen M, Nair J, Meerang M, Knudsen LE, Bartsch H. Typical signature of DNA damage in white blood cells: A pilot study on etheno adducts in Danish mother-newborn child pairs. Carcinogenesis. 2008 doi: 10.1093/carcin/bgn264. [DOI] [PubMed] [Google Scholar]
- 48.Bartsch H, Nair J, Velic I. Etheno-DNA base adducts as tools in human cancer aetiology and chemoprevention. Eur J Cancer Prev. 1997;6:529–534. doi: 10.1097/00008469-199712000-00007. [DOI] [PubMed] [Google Scholar]
- 49.Nair J, Barbin A, Guichard Y, Bartsch H. 1,N6-ethenodeoxyadenosine and 3,N4-ethenodeoxycytine in liver DNA from humans and untreated rodents detected by immunoaffinity/32P-postlabeling. Carcinogenesis. 1995;16:613–617. doi: 10.1093/carcin/16.3.613. [DOI] [PubMed] [Google Scholar]
- 50.Phillips DH, Arlt VM. The 32P-postlabeling assay for DNA adducts. Nat Protoc. 2007;2:2772–2781. doi: 10.1038/nprot.2007.394. [DOI] [PubMed] [Google Scholar]
- 51.Boysen G, Kenney PM, Upadhyaya P, Wang M, Hecht SS. Effects of benzyl isothiocyanate and 2-phenethyl isothiocyanate on benzo[a]pyrene and 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone metabolism in F-344 rats. Carcinogenesis. 2003;24:517–525. doi: 10.1093/carcin/24.3.517. [DOI] [PubMed] [Google Scholar]
- 52.Foiles PG, Akerkar SA, Carmella SG, Kagan M, Stoner GD, Resau JH, Hecht SS. Mass spectrometric analysis of tobacco-specific nitrosamine-DNA adducts in smokers and nonsmokers. Chem Res Tox. 1991;4:364–368. doi: 10.1021/tx00021a017. [DOI] [PubMed] [Google Scholar]
- 53.Lin D, Lay JO, Jr, Bryant MS, Malaveille C, Friesen M, Bartsch H, Lang NP, Kadlubar FF. Analysis of 4-aminobiphenyl-DNA adducts in human urinary bladder and lung by alkaline hydrolysis and negative ion gas chromatography-mass spectrometry. Environ Health Perspect. 1994;102 Suppl 6:11–16. doi: 10.1289/ehp.94102s611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Melikian AA, Sun P, Coleman S, Amin S, Hecht SS. Detection of DNA and globin adducts of polynuclear aromatic hydrocarbon diol epoxides by gas chromatography-mass spectrometry and 3H-CH3I postlabeling of released tetraols. Chem Res Tox. 1996;9:508–516. doi: 10.1021/tx950165z. [DOI] [PubMed] [Google Scholar]
- 55.Fedtke N, Boucheron JA, Turner MJ, Swenberg JA. Vinyl chloride-induced DNA adducts. I: Quantitative determination of N2,3-ethenoguanine based on electrophore labeling. Carcinogenesis. 1990;11:1279–1285. doi: 10.1093/carcin/11.8.1279. [DOI] [PubMed] [Google Scholar]
- 56.Pastorelli R, Cerri A, Mezzetti M, Consonni E, Airoldi L. Effect of DNA repair gene polymorphisms on BPDE-DNA Adducts in human lymphocytes. Int J Cancer. 2002;100:9–13. doi: 10.1002/ijc.10463. [DOI] [PubMed] [Google Scholar]
- 57.Singh R, Farmer PB. Liquid chromatography-electrospray ionization-mass spectrometry: the future of DNA adduct detection. Carcinogenesis. 2006;27:178–196. doi: 10.1093/carcin/bgi260. [DOI] [PubMed] [Google Scholar]
- 58.Esmans EL, Broes D, Hoes I, Lemiere F, Vanhoutte K. Liquid chromatography-mass spectrometry in nucleosid, nucleotide and modified nucleotide characterization. J Chromatogr A. 1998;794:109–127. [Google Scholar]
- 59.Apruzzese WA, Vouros P. Analysis of DNA adducts by capillary methods coupled to mass spectrometry: a perspective. J Chromatogr A. 1998;794:97–108. doi: 10.1016/s0021-9673(97)00820-0. [DOI] [PubMed] [Google Scholar]
- 60.Andrews CL, Vouros P, Harsch A. Analysis of DNA adducts using high-performance separation techniques coupled to electrospray ionization mass spectrometry. J Chromatogr A. 1999;856:515–526. doi: 10.1016/s0021-9673(99)00779-7. [DOI] [PubMed] [Google Scholar]
- 61.Tarun M, Rusling JF. Quantitative Measurement of DNA Adducts Using Neutral Hydrolysis and LC-MS. Validation of Genotoxicity Sensors. Analytical Chemistry. 2005;77:2056–2062. doi: 10.1021/ac048283r. [DOI] [PubMed] [Google Scholar]
- 62.Koc H, Swenberg JA. Applications of mass spectrometry for quantitation of DNA adducts. J Chromatogr B. 2002;778:323–343. doi: 10.1016/s1570-0232(02)00135-6. [DOI] [PubMed] [Google Scholar]
- 63.Farmer PB, Brown K, Tompkins E, Emms VL, Jones DJ, Singh R, Phillips DH. DNA adducts: Mass spectrometry methods and future prospects. Toxicol Appl Pharmacol. 2005;207:293–301. doi: 10.1016/j.taap.2004.12.020. [DOI] [PubMed] [Google Scholar]
- 64.Hah SS, Sumbad RA, Vere White RW, Turteltaub KW, Henderson PT. Characterization of oxaliplatin-DNA adduct formation in DNA and differentiation of cancer cell drug sensitivity at microdose concentrations. Chem Res Toxicol. 2007;20:1745–1751. doi: 10.1021/tx700376a. [DOI] [PubMed] [Google Scholar]
- 65.Brown K, Tompkins EM, Boocock DJ, Martin EA, Farmer PB, Turteltaub KW, Ubick E, Hemingway D, Horner-Glister E, White IN. Tamoxifen forms DNA adducts in human colon after administration of a single [14C]-labeled therapeutic dose. Cancer Res. 2007;67:6995–7002. doi: 10.1158/0008-5472.CAN-07-0913. [DOI] [PubMed] [Google Scholar]
- 66.Vodicka P, Koskinen M, Vodickova L, Stetina R, Smerak P, Barta I, Hemminki K. DNA adducts, strand breaks and micronuclei in mice exposed to styrene by inhalation. Chem Biol Interact. 2001;137:213–227. doi: 10.1016/s0009-2797(01)00253-8. [DOI] [PubMed] [Google Scholar]
- 67.Belinsky SA, White CM, Devereux TR, Swenberg JA, Anderson MW. Cell selective alkylation of DNA in rat lung following low dose exposure to the tobacco specific carcinogen 4-(N-methyl-N-nitrosamino)-1-(3-pyridyl)-1-butanone. Cancer Res. 1987;47:1143–1148. [PubMed] [Google Scholar]
- 68.Koc H, Tretyakova NY, Walker VE, Henderson RF, Swenberg JA. Molecular dosimetry of N-7 guanine adduct formation in mice and rats exposed to 1,3-butadiene. Chem Res Tox. 1999;12:566–574. doi: 10.1021/tx980265f. [DOI] [PubMed] [Google Scholar]
- 69.Belinsky SA, Dolan ME, White CM, Maronpot RR, Pegg AE, Anderson MW. Cell specific differences in O6-methylguanine-DNA methyltransferase activity and removal of O6-methylguanine in rat pulmonary cells. Carcinogenesis. 1988;9:2053–2058. doi: 10.1093/carcin/9.11.2053. [DOI] [PubMed] [Google Scholar]
- 70.Gaubatz JW, Tan BH. Introduction, distribution, and removal of 7-methylguanine in different liver chromatin fractions of young and old mice. Mutat Res. 1997;375:25–35. doi: 10.1016/s0027-5107(96)00246-1. [DOI] [PubMed] [Google Scholar]
- 71.Park JW, Ames BN. 7-Methylguanine adducts in DNA are normally present at high levels and increase on aging: analysis by HPLC with electrochemical detection. Proc Natl Acad Sci U S A. 1988;85:7467–7470. doi: 10.1073/pnas.85.20.7467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Druckrey H, Preussmann R. N-Nitroso-N-methylurethane: a potent carcinogen. Nature. 1962;195:1111. doi: 10.1038/1951111a0. [DOI] [PubMed] [Google Scholar]
- 73.Kleihues P, Lantos PL, Magee PN. Chemical carcinogenesis in the nervous system. Int Rev Exp Pathol. 1976;15:153–232. [PubMed] [Google Scholar]
- 74.IARC. IARC monographs on the evaluation of the carcinogenic risk of chemicals to humans: some N-nitroso compounds. IARC Monogr Eval Carcinog Risk Chem Man. 1978;17:1–349. [PubMed] [Google Scholar]
- 75.Buecheler J, Kleihues P. Excision of O6-methylguanine from DNA of various mouse tissues following a single injection of N-methyl-N-nitrosourea. Chem Biol Interact. 1977;16:325–333. doi: 10.1016/0009-2797(77)90112-0. [DOI] [PubMed] [Google Scholar]
- 76.Margison GP, Kleihues P. Chemical carcinogenesis in the nervous system. Preferential accumulation of O6-methylguanine in rat brain deoxyribonucleic acid during repetitive administration of N-methyl-N-nitrosourea. Biochem J. 1975;148:521–525. doi: 10.1042/bj1480521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wechsler W, Rice JM, Vesselinovitch SD. Transplacental and neonatal induction of neurogenic tumors in mice: comparison with related species and with human pediatric neoplasms. Natl Cancer Inst Monogr. 1979:219–226. [PubMed] [Google Scholar]
- 78.Swenberg JA. Current approaches to the experimental investigation of chemicals in relation to cancer of the brain. Ann N Y Acad Sci. 1982;381:43–53. doi: 10.1111/j.1749-6632.1982.tb50365.x. [DOI] [PubMed] [Google Scholar]
- 79.Singer B. Alkylation of the O6 of guanine is only one of many chemical events that may initiate carcinogenesis. Cancer Invest. 1984;2:233–238. doi: 10.3109/07357908409104377. [DOI] [PubMed] [Google Scholar]
- 80.Engelbergs J, Thomale J, Rajewsky MF. Role of DNA repair in carcinogen-induced ras mutation. Mutat Res. 2000;450:139–153. doi: 10.1016/s0027-5107(00)00021-x. [DOI] [PubMed] [Google Scholar]
- 81.Magee PN, Barnes JM. The production of malignant primary hepatic tumours in the rat by feeding dimethylnitrosamine. Br J Cancer. 1956;10:114–122. doi: 10.1038/bjc.1956.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hecht SS. Biochemistry, biology, and carcinogenicity of tobacco-specific N-nitrosamines. Chem Res Tox. 1998;11:559–603. doi: 10.1021/tx980005y. [DOI] [PubMed] [Google Scholar]
- 83.Hecht SS. Progress and challenges in selected areas of tobacco carcinogenesis. Chem Res Tox. 2008;21:160–171. doi: 10.1021/tx7002068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Shuker DE, Bartsch H. DNA adducts of nitrosamines. IARC Sci Publ. 1994:73–89. [PubMed] [Google Scholar]
- 85.Morse MA, Eklind KI, Hecht SS, Chung FL. Inhibition of tobacco-specific nitrosamine 4-(N-nitrosomethylamino)-1-(3-pyridyl)-1-butanone (NNK) tumorigenesis with aromatic isothiocyanates. IARC Sci Publ. 1991:529–534. [PubMed] [Google Scholar]
- 86.Olajos EJ. Biological interactions of N-nitroso compounds: A review. Ecotoxicology and Environmental Safety. 1977;1:175–196. doi: 10.1016/0147-6513(77)90034-3. [DOI] [PubMed] [Google Scholar]
- 87.Preussmann R. Carcinogenic N-nitroso compounds and their environmental significance. Naturwissenschaften. 1984;71:25–30. doi: 10.1007/BF00365976. [DOI] [PubMed] [Google Scholar]
- 88.Lindamood C, III, Bedell MA, Billings KC, Dyroff MC, Swenberg JA. Dose response for DNA alkylation, [3H]thymidine uptake into DNA, and O6-methylguanine-DNA methyltransferase activity in hepatocytes of rats and mice continuously exposed to dimethylnitrosamine. Cancer Res. 1984;44:196–200. [PubMed] [Google Scholar]
- 89.Pegg AE, Hui G. Formation and subsequent removal of O6-methylguanine from deoxyribonucleic acid in rat liver and kidney after small doses of dimethylnitrosamine. Biochem J. 1978;173:739–748. doi: 10.1042/bj1730739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zang H, Fang Q, Pegg AE, Guengerich FP. Kinetic Analysis of Steps in the Repair of Damaged DNA by Human O6-Alkylguanine-DNA Alkyltransferase. J Biol Chem. 2005;280:30873–30881. doi: 10.1074/jbc.M505283200. [DOI] [PubMed] [Google Scholar]
- 91.Jalas JR, Hecht SS, Murphy SE. Cytochrome P450 enzymes as catalysts of metabolism of 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone, a tobacco specific carcinogen. Chem Res Toxicol. 2005;18:95–110. doi: 10.1021/tx049847p. [DOI] [PubMed] [Google Scholar]
- 92.Upadhyaya P, Kalscheuer S, Hochalter JB, Villalta PW, Hecht SS. Quantitation of pyridylhydroxybutyl-DNA adducts in liver and lung of F-344 rats treated with 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone and enantiomers of its metabolite 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanol. Chem Res Toxicol. 2008;21:1468–1476. doi: 10.1021/tx8001109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Upadhyaya P, Sturla SJ, Tretyakova N, Ziegel R, Villalta PW, Wang M, Hecht SS. Identification of Adducts Produced by the Reaction of 4-(Acetoxymethylnitrosamino)-1-(3-pyridyl)-1-butanol with Deoxyguanosine and DNA. Chem Res Tox. 2003;16:180–190. doi: 10.1021/tx0256376. [DOI] [PubMed] [Google Scholar]
- 94.Castonguay A, Foiles PG, Trushin N, Hecht SS. Study of DNA methylation by tobacco-specific N-nitrosamines. Environ Health Perspect. 1985;62:197–202. doi: 10.1289/ehp.8562197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Devereux TR, Anderson MW, Belinsky SA. Factors regulating activation and DNA alkylation by 4-(N-methyl-N-nitrosamino)-1-(3-pyridyl)-1-butanone and nitrosodimethylamine in rat lung and isolated lung cells, and the relationship to carcinogenicity. Cancer Res. 1988;48:4215–4221. [PubMed] [Google Scholar]
- 96.Foiles PG, Trushin N, Castonguay A. Measurement of O6-methyldeoxyguanosine in DNA methylated by the tobacco- specific carcinogen 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone using a biotin-avidin enzyme-linked immunosorbent assay. Carcinogenesis. 1985;6:989–993. doi: 10.1093/carcin/6.7.989. [DOI] [PubMed] [Google Scholar]
- 97.Guo Z, Smith TJ, Thomas PE, Yang CS. Metabolic activation of 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone as measured by DNA alkylation in vitro and its inhibition by isothiocyanates. Cancer Res. 1991;51:4798–4803. [PubMed] [Google Scholar]
- 98.Murphy SE, Heiblum R, Trushin N. Comparative metabolism of N′-nitrosonornicotine and 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone by cultured F344 rat oral tissue and esophagus. Cancer Res. 1990;50:4685–4691. [PubMed] [Google Scholar]
- 99.Sticha KR, Staretz ME, Wang M, Liang H, Kenney PM, Hecht SS. Effects of benzyl isothiocyanate and phenethyl isothiocyanate on benzo[a]pyrene metabolism and DNA adduct formation in the A/J mouse. Carcinogenesis. 2000;21:1711–1719. doi: 10.1093/carcin/21.9.1711. [DOI] [PubMed] [Google Scholar]
- 100.Sticha KR, Kenney PM, Boysen G, Liang H, Su X, Wang M, Upadhyaya P, Hecht SS. Effects of benzyl isothiocyanate and phenethyl isothiocyanate on DNA adduct formation by a mixture of benzo[a]pyrene and 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone in A/J mouse lung. Carcinogenesis. 2002;23:1433–1439. doi: 10.1093/carcin/23.9.1433. [DOI] [PubMed] [Google Scholar]
- 101.Hecht SS. DNA adduct formation from tobacco-specific N-nitrosamines. Mutat Res. 1999;424:127–142. doi: 10.1016/s0027-5107(99)00014-7. [DOI] [PubMed] [Google Scholar]
- 102.Murphy SE, Palomino A, Hecht SS, Hoffmann D. Dose-response study of DNA and hemoglobin adduct formation by 4- (methylnitrosamino)-1-(3-pyridyl)-1-butanone in F344 rats. Cancer Res. 1990;50:5446–5452. [PubMed] [Google Scholar]
- 103.Belinsky SA, White CM, Trushin N, Hecht SS. Cell specificity for the pulmonary metabolism of tobacco-specific nitrosamines in the Fischer rat. Carcinogenesis. 1989;10:2269–2274. doi: 10.1093/carcin/10.12.2269. [DOI] [PubMed] [Google Scholar]
- 104.Belinsky SA, Foley JF, White CM, Anderson MW, Maronpot RR. Dose-response relationship between O6-methylguanine formation in Clara cells and induction of pulmonary neoplasia in the rat by 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone. Cancer Res. 1990;50:3772–3780. [PubMed] [Google Scholar]
- 105.IARC. IARC monographs on the evaluation of carcinogenic risks to humans. Solar and ultraviolet radiation. IARC Monogr Eval Carcinog Risks Hum. 1992;55:1–316. [PMC free article] [PubMed] [Google Scholar]
- 106.Toth B. Synthetic and naturally occurring hydrazines as possible cancer causative agents. Cancer Res. 1975;35:3693–3697. [PubMed] [Google Scholar]
- 107.NTP. Hydrazine and hydrazine sulfate. Rep Carcinog. 2002;10:138–139. [PubMed] [Google Scholar]
- 108.Jenner AM, Timbrell JA. In vitro microsomal metabolism of hydrazine. Xenobiotica. 1995;25:599–609. doi: 10.3109/00498259509061878. [DOI] [PubMed] [Google Scholar]
- 109.Herron DC, Shank RC. DNA methylation during chronic administration of 1,2-dimethylhydrazine in a carcinogenic regimen. Carcinogenesis. 1982;3:857–860. doi: 10.1093/carcin/3.8.857. [DOI] [PubMed] [Google Scholar]
- 110.Harbach PR, Swenberg JA. Effects of selenium on 1,2-dimethylhydrazine metabolism and DNA alkylation. Carcinogenesis. 1981;2:575–580. doi: 10.1093/carcin/2.7.575. [DOI] [PubMed] [Google Scholar]
- 111.Swenberg JA, Cooper HK, Bucheler J, Kleihues P. 1,2-Dimethylhydrazine-induced methylation of DNA bases in various rat organs and the effect of pretreatment with disulfiram. Cancer Res. 1979;39:465–467. [PubMed] [Google Scholar]
- 112.Swenberg JA, Richardson FC, Boucheron JA, Dyroff MC. Relationships between DNA adduct formation and carcinogenesis. Environ Health Perspect. 1985;62:177–183. doi: 10.1289/ehp.8562177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Herron DC, Shank RC. In vivo kinetics of O6-methylguanine and 7-methylguanine formation and persistence in DNA of rats treated with symmetrical dimethylhydrazine. Cancer Res. 1981;41:3967–3972. [PubMed] [Google Scholar]
- 114.Pegg AE. Repair of alkylated DNA by cell extracts from various organs and species. Basic Life Sci. 1983;24:545–563. doi: 10.1007/978-1-4684-4400-1_29. [DOI] [PubMed] [Google Scholar]
- 115.Bedell MA, Lewis JG, Billings KC, Swenberg JA. Cell specificity in hepatocarcinogenesis: preferential accumulation of O6-methylguanine in target cell DNA during continuous exposure to rats to 1,2-dimethylhydrazine. Cancer Res. 1982;42:3079–3083. [PubMed] [Google Scholar]
- 116.Druckrey H, Preussmann R, Matzkies F, Ivankovic S. [Selective production of intestinal cancer in rats by 1,2-dimethylhydrazine] Naturwissenschaften. 1967;54:285–286. doi: 10.1007/BF00620890. [DOI] [PubMed] [Google Scholar]
- 117.Delaney V, Mullaney J, Bourke E. Juvenile nephronophthisis, congenital hepatic fibrosis and retinal hypoplasia in twins. Q J Med. 1978;47:281–290. [PubMed] [Google Scholar]
- 118.Weisburger JH, Madison RM, Ward JM, Viguera C, Weisburger EK. Modification of diethylnitrosamine liver carcinogenesis with phenobarbital but not with immunosuppression. J Natl Cancer Inst. 1975;54:1185–1188. doi: 10.1093/jnci/54.5.1185. [DOI] [PubMed] [Google Scholar]
- 119.Swenberg JA, Bedell MA, Billings KC, Umbenhauer DR, Pegg AE. Cell-specific differences in O6-alkylguanine DNA repair activity during continuous exposure to carcinogen. Proc Natl Acad Sci USA. 1982;79:5499–5502. doi: 10.1073/pnas.79.18.5499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.IARC. IARC monographs on the evaluation of the carcinogenic risk of chemicals to humans: 1,3-Butadiene (in preparation) IARC Monogr Eval Carcinog Risk Chem Man. 2007;97 [PubMed] [Google Scholar]
- 121.Cheng H, Sathiakumar N, Graff J, Matthews R, Delzell E. 1,3-Butadiene and leukemia among synthetic rubber industry workers: exposure-response relationships. Chem Biol Interact. 2007;166:15–24. doi: 10.1016/j.cbi.2006.10.004. [DOI] [PubMed] [Google Scholar]
- 122.Delzell E, Sathiakumar N, Graff J, Macaluso M, Maldonado G, Matthews R. An updated study of mortality among North American synthetic rubber industry workers. HEI Report. 2006:1–63. [PubMed] [Google Scholar]
- 123.Sathiakumar N, Delzell E, Cheng H, Lynch J, Sparks W, Macaluso M. Validation of 1,3-butadiene exposure estimates for workers at a synthetic rubber plant. Chem Biol Interact. 2007;166:29–43. doi: 10.1016/j.cbi.2006.08.016. [DOI] [PubMed] [Google Scholar]
- 124.Divine BJ, Hartman CM. A cohort mortality study among workers at a 1,3 butadiene facility. Chem Biol Interact. 2001;135-136:535–553. doi: 10.1016/s0009-2797(01)00212-5. [DOI] [PubMed] [Google Scholar]
- 125.Koskinen M, Plna K. Specific DNA adducts induced by some mono-substituted epoxides in vitro and in vivo. Chem Biol Interact. 2000;129:209–229. doi: 10.1016/s0009-2797(00)00206-4. [DOI] [PubMed] [Google Scholar]
- 126.Rios-Blanco MN, Ranasinghe A, Upton P, Lee MS, Filser JG, Swenberg JA. Exposure-dependent accumulation of N-(2-hydroxypropyl)valine in hemoglobin of F344 rats exposed to propylene oxide by the inhalation route. Journal of Chromatography B. 2002;778:383–391. doi: 10.1016/s1570-0232(02)00115-0. [DOI] [PubMed] [Google Scholar]
- 127.Rios-Blanco MN, Ranasinghe A, Lee MS, Faller T, Filser JG, Swenberg JA. Molecular dosimetry of N7-(2-hydroxypropyl)guanine in tissues of F344 rats after inhalation exposure to propylene oxide. Carcinogenesis. 2003;24:1233–1238. doi: 10.1093/carcin/bgg087. [DOI] [PubMed] [Google Scholar]
- 128.Wu KY, Scheller N, Ranasinghe A, Yen TY, Sangaiah R, Giese R, Swenberg JA. A gas chromatography/electron capture/negative chemical ionization high-resolution mass spectrometry method for analysis of endogenous and exogenous N7-(2-hydroxyethyl)guanine in rodents and its potential for human biological monitoring. Chem Res Toxicol. 1999;12:722–729. doi: 10.1021/tx990059n. [DOI] [PubMed] [Google Scholar]
- 129.Rusyn I, Asakura S, Li Y, Kosyk O, Koc H, Nakamura J, Upton PB, Swenberg JA. Effects of ethylene oxide and ethylene inhalation on DNA adducts, apurinic/apyrimidinic sites and expression of base excision DNA repair genes in rat brain, spleen, and liver. DNA Repair. 2005;4:1099–1110. doi: 10.1016/j.dnarep.2005.05.009. [DOI] [PubMed] [Google Scholar]
- 130.Bolt HM, Filser JG. Kinetics and disposition in toxicology. Example: Carcinogenic risk estimate for ethylene. Arch Toxicol. 1987;60:73–76. doi: 10.1007/BF00296951. [DOI] [PubMed] [Google Scholar]
- 131.Bolt HM, Filser JG, Stormer F. Inhalation pharmacokinetics based on gas uptake studies. V. Comparative pharmacokinetics of ethylene and 1,3-butadiene in rats. Arch Toxicol. 1984;55:213–218. doi: 10.1007/BF00341013. [DOI] [PubMed] [Google Scholar]
- 132.Föst U, Marczynski B, Kasemann R, Peter H. Determination of 7-(2-hydroxyethyl)guanine with gas chromatography/mass spectrometry as a parameter for genotoxicity of ethylene oxide. Arch Toxicol. 1989 Suppl 13:250–253. doi: 10.1007/978-3-642-74117-3_43. [DOI] [PubMed] [Google Scholar]
- 133.Walker VE, Fennell TR, Upton PB, Skopek TR, Prevost V, Shuker DE, Swenberg JA. Molecular dosimetry of ethylene oxide: formation and persistence of 7-(2-hydroxyethyl)guanine in DNA following repeated exposures of rats and mice. Cancer Res. 1992;52:4328–4334. [PubMed] [Google Scholar]
- 134.Walker VE, Fennell TR, Upton PB, MacNeela JP, Swenberg JA. Molecular dosimetry of DNA and hemoglobin adducts in mice and rats exposed to ethylene oxide. Environ Health Perspect. 1993;99:11–17. doi: 10.1289/ehp.939911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Filser JG, Denk B, Törnqvist M, Kessler W, Ehrenberg L. Pharmacokinetics of ethylene in man: body burden with ethylene oxide and hydroxyethylation of hemoglobin due to endogenous and environmental ethylene. Arch Toxicol. 1992;66:157–163. doi: 10.1007/BF01974008. [DOI] [PubMed] [Google Scholar]
- 136.Filser JG, Bolt HM. Exhalation of ethylene oxide by rats on exposure to ethylene. Mutat Res. 1983;120:57–60. doi: 10.1016/0165-7992(83)90074-x. [DOI] [PubMed] [Google Scholar]
- 137.Jensen KG. Determination of ethylene oxide residues in processed food products by gas-liquid chromatography after derivatization. Z Lebensm Unters Forsch. 1988;187:535–540. doi: 10.1007/BF01042385. [DOI] [PubMed] [Google Scholar]
- 138.Tompkins EM, Jones DJ, Lamb JH, Marsden DA, Farmer PB, Brown K. Simultaneous detection of five different 2-hydroxyethyl-DNA adducts formed by ethylene oxide exposure, using a high-performance liquid chromatography/electrospray ionisation tandem mass spectrometry assay. Rapid Commun Mass Spectrom. 2008;22:19–28. doi: 10.1002/rcm.3328. [DOI] [PubMed] [Google Scholar]
- 139.Marsden DA, Jones DJ, Lamb JH, Tompkins EM, Farmer PB, Brown K. Determination of endogenous and exogenously derived N7-(2-hydroxyethyl)guanine adducts in ethylene oxide-treated rats. Chem Res Toxicol. 2007;20:290–299. doi: 10.1021/tx600264t. [DOI] [PubMed] [Google Scholar]
- 140.van Sittert NJ, Boogaard PJ, Natarajan AT, Tates AD, Ehrenberg LG, Tornqvist MA. Formation of DNA adducts and induction of mutagenic effects in rats following 4 weeks inhalation exposure to ethylene oxide as a basis for cancer risk assessment. Mutat Res. 2000;447:27–48. doi: 10.1016/s0027-5107(99)00208-0. [DOI] [PubMed] [Google Scholar]
- 141.Bolt HM, Leutbecher M, Golka K. A note on the physiological background of the ethylene oxide adduct 7-(2-hydroxyethyl)guanine in DNA from human blood. Arch Toxicol. 1997;71:719–721. doi: 10.1007/s002040050451. [DOI] [PubMed] [Google Scholar]
- 142.Kato S, Petruzzelli S, Bowman ED, Turteltaub KW, Blomeke B, Weston A, Shields PG. 7-Alkyldeoxyguanosine adduct detection by two-step HPLC and the 32P-postlabeling assay. Carcinogenesis. 1993;14:545–550. doi: 10.1093/carcin/14.4.545. [DOI] [PubMed] [Google Scholar]
- 143.Yong LC, Schulte PA, Kao CY, Giese RW, Boeniger MF, Strauss GH, Petersen MR, Wiencke JK. DNA adducts in granulocytes of hospital workers exposed to ethylene oxide. Am J Ind Med. 2007;50:293–302. doi: 10.1002/ajim.20443. [DOI] [PubMed] [Google Scholar]
- 144.Törnqvist M, Magnusson AL, Farmer PB, Tang YS, Jeffrey AM, Wazneh L, Beulink GD, van der WH, van Sittert NJ. Ring test for low levels of N-(2-hydroxyethyl)valine in human hemoglobin. Anal Biochem. 1992;203:357–360. doi: 10.1016/0003-2697(92)90325-2. [DOI] [PubMed] [Google Scholar]
- 145.Wu KY, Chiang SY, Huang TH, Tseng YS, Chen YL, Kuo HW, Hsieh CL. Formation of N-(2-hydroxyethyl)valine in human hemoglobin-effect of lifestyle factors. Mutat Res. 2004;559:73–82. doi: 10.1016/j.mrgentox.2003.12.011. [DOI] [PubMed] [Google Scholar]
- 146.Zhao C, Tyndyk M, Eide I, Hemminki K. Endogenous and background DNA adducts by methylating and 2-hydroxyethylating agents. Mutat Res. 1999;424:117–125. doi: 10.1016/s0027-5107(99)00013-5. [DOI] [PubMed] [Google Scholar]
- 147.Zhao CY, Kumar R, Zahlsen K, Sundmark HB, Hemminki K, Eide I. Persistence of 7-(2-hydroxyethyl)guanine-DNA adducts in rats exposed to ethene by inhalation. Biomarkers. 1997;2:355–359. doi: 10.1080/135475097231445. [DOI] [PubMed] [Google Scholar]
- 148.Widlak P, Zheng X, Österdahl BG, Drettner B, Christensson B, Kumar R, Hemminki K. N-nitrosodimethylamine and 7-methylguanine DNA adducts in tissues of rats fed Chinese salted fish. Cancer Lett. 1995;94:85–90. doi: 10.1016/0304-3835(95)03828-k. [DOI] [PubMed] [Google Scholar]
- 149.Pottenger LH, Malley LA, Bogdanffy MS, Donner EM, Upton PB, Li Y, Walker VE, Harkema JR, Banton MI, Swenberg JA. Evaluation of effects from repeated inhalation exposure of F344 rats to high concentrations of propylene. Toxicol Sci. 2007;97:336–347. doi: 10.1093/toxsci/kfm038. [DOI] [PubMed] [Google Scholar]
- 150.Himmelstein MW, Acquavella JF, Recio L, Medinsky MA, Bond JA. Toxicology and epidemiology of 1,3-butadiene. Crit Rev Toxicol. 1997;27:1–108. doi: 10.3109/10408449709037482. [DOI] [PubMed] [Google Scholar]
- 151.Csanady GA, Guengerich FP, Bond JA. Comparison of the biotransformation of 1,3-butadiene and its metabolite, butadiene monoepoxide, by hepatic and pulmonary tissues from humans, rats and mice. Carcinogenesis. 1992;13:1143–1153. doi: 10.1093/carcin/13.7.1143. [DOI] [PubMed] [Google Scholar]
- 152.Duescher RJ, Elfarra AA. Human liver microsomes are efficient catalysts of 1,3-butadiene oxidation: evidence for major roles by cytochromes P450 2A6 and 2E1. Arch Biochem Biophys. 1994;311:342–349. doi: 10.1006/abbi.1994.1246. [DOI] [PubMed] [Google Scholar]
- 153.Zhao CY, Koskinen M, Hemminki K. 32P-postlabelling analysis of 1,3-butadiene-induced DNA adducts in vivo and in vitro. Biomarkers. 2000;5:168–181. doi: 10.1080/135475000230334. [DOI] [PubMed] [Google Scholar]
- 154.Tretyakova N, Sangaiah R, Yen TY, Gold A, Swenberg JA. Adenine adducts with diepoxybutane: isolation and analysis in exposed calf thymus DNA. Chem Res Tox. 1997;10:1171–1179. doi: 10.1021/tx9700681. [DOI] [PubMed] [Google Scholar]
- 155.Tretyakova N, Lin Y, Sangaiah R, Upton PB, Swenberg JA. Identification and quantitation of DNA adducts from calf thymus DNA exposed to 3,4-epoxy-1-butene. Carcinogenesis. 1997;18:137–147. doi: 10.1093/carcin/18.1.137. [DOI] [PubMed] [Google Scholar]
- 156.Kanuri M, Nechev LV, Tamura PJ, Harris CM, Harris TM, Lloyd RS. Mutagenic spectrum of butadiene-derived N1-deoxyinosine adducts and N6,N6-deoxyadenosine intrastrand cross-links in Mammalian cells. Chem Res Tox. 2002;15:1572–1580. doi: 10.1021/tx025591g. [DOI] [PubMed] [Google Scholar]
- 157.Rodriguez DA, Kowalczyk A, Ward JB, Jr, Harris CM, Harris TM, Lloyd RS. Point mutations induced by 1,2-epoxy-3-butene N1 deoxyinosine adducts. Environ Mol Mutagen. 2001;38:292–296. doi: 10.1002/em.10026. [DOI] [PubMed] [Google Scholar]
- 158.Carmical JR, Nechev LV, Harris CM, Harris TM, Lloyd RS. Mutagenic potential of adenine N6 adducts of monoepoxide and diolepoxide derivatives of butadiene. Environ Mol Mutagen. 2000;35:48–56. [PubMed] [Google Scholar]
- 159.Krause RJ, Elfarra AA. Oxidation of butadiene monoxide to meso- and (+/-)-diepoxybutane by cDNA-expressed human cytochrome P450s and by mouse, rat, and human liver microsomes: evidence for preferential hydration of meso-diepoxybutane in rat and human liver microsomes. Arch Biochem Biophys. 1997;337:176–184. doi: 10.1006/abbi.1996.9781. [DOI] [PubMed] [Google Scholar]
- 160.Goggin M, Anderson C, Park S, Swenberg J, Walker V, Tretyakova N. Quantitative high-performance liquid chromatography-electrospray ionization-tandem mass spectrometry analysis of the adenine-guanine cross-links of 1,2,3,4-diepoxybutane in tissues of butadiene-exposed B6C3F1 mice. Chem Res Toxicol. 2008;21:1163–1170. doi: 10.1021/tx800051y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Goggin M, Loeber R, Park S, Walker V, Wickliffe J, Tretyakova N. HPLC-ESI+-MS/MS analysis of N7-guanine-N7-guanine DNA cross-links in tissues of mice exposed to 1,3-butadiene. Chem Res Toxicol. 2007;20:839–847. doi: 10.1021/tx700020q. [DOI] [PubMed] [Google Scholar]
- 162.Tretyakova N, Livshits A, Park S, Bisht B, Goggin M. Structural elucidation of a novel DNA-DNA cross-link of 1,2,3,4-diepoxybutane. Chem Res Toxicol. 2007;20:284–289. doi: 10.1021/tx060204e. [DOI] [PubMed] [Google Scholar]
- 163.Loeber R, Michaelson E, Fang Q, Campbell C, Pegg AE, Tretyakova N. Cross-linking of the DNA repair protein O(6)-alkylguanine DNA alkyltransferase to DNA in the presence of antitumor nitrogen mustards. Chem Res Toxicol. 2008;21:787–795. doi: 10.1021/tx7004508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Loeber R, Rajesh M, Fang Q, Pegg AE, Tretyakova N. Cross-linking of the human DNA repair protein O6-alkylguanine DNA alkyltransferase to DNA in the presence of 1,2,3,4-diepoxybutane. Chem Res Toxicol. 2006;19:645–654. doi: 10.1021/tx0600088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Goggin M, Swenberg JA, Walker VE, Tretyakova N. Molecular dosimetry of 1,2,3,4-diepoxybutane-induced DNA-DNA cross-links in B6C3F1 mice and F344 rats exposed to 1,3-butadiene by inhalation. Cancer Res. 2009;69:2479–2486. doi: 10.1158/0008-5472.CAN-08-4152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Bechtold WE, Strunk MR, Chang IY, Ward JB, Henderson RF. Species-Differences in Urinary Butadiene Metabolites - Comparisons of Metabolite Ratios Between Mice, Rats, and Humans. Toxicology and Applied Pharmacology. 1994;127:44–49. doi: 10.1006/taap.1994.1137. [DOI] [PubMed] [Google Scholar]
- 167.Cheng X, Ruth JA. A simplified methodology for quantitation of butadiene metabolites. Application to the study of 1,3-butadiene metabolism by rat liver microsomes. Drug Metab Dispos. 1993;21:121–124. [PubMed] [Google Scholar]
- 168.Kreuzer PE, Kessler W, Welter HF, Baur C, Filser JG. Enzyme specific kinetics of 1,2-epoxybutene-3 in microsomes and cytosol from livers of mouse, rat, and man. Arch Toxicol. 1991;65:59–67. doi: 10.1007/BF01973504. [DOI] [PubMed] [Google Scholar]
- 169.Nauhaus SK, Fennell TR, Asgharian B, Bond JA, Sumner SC. Characterization of urinary metabolites from Sprague-Dawley rats and B6C3F1 mice exposed to [1,2,3,4-13C]butadiene. Chem Res Toxicol. 1996;9:764–773. doi: 10.1021/tx950196u. [DOI] [PubMed] [Google Scholar]
- 170.Sabourin PJ, Burka LT, Bechtold WE, Dahl AR, Hoover MD, Chang LY, Henderson RF. Species differences in urinary butadiene metabolites; identification of 1,2-dihydroxy-4-(N-acetylcysteinyl)butane, a novel metabolite of butadiene. Carcinogenesis. 1992;13:1633–1638. doi: 10.1093/carcin/13.9.1633. [DOI] [PubMed] [Google Scholar]
- 171.Malvoisin E, Roberfroid M. Hepatic microsomal metabolism of 1,3-butadiene. Xenobiotica. 1982;12:137–144. doi: 10.3109/00498258209046787. [DOI] [PubMed] [Google Scholar]
- 172.Lewis DF, Bird MG, Parke DV. Molecular modelling of CYP2E1 enzymes from rat, mouse and man: an explanation for species differences in butadiene metabolism and potential carcinogenicity, and rationalization of CYP2E substrate specificity. Toxicology. 1997;118:93–113. doi: 10.1016/s0300-483x(96)03583-4. [DOI] [PubMed] [Google Scholar]
- 173.Boysen G, Georgieva NI, Upton PB, Jayaraj K, Li Y, Walker VE, Swenberg JA. Analysis of diepoxide-specific cyclic N-terminal globin adducts in mice and rats after inhalation exposure to 1,3-butadiene. Cancer Res. 2004;64:8517–8520. doi: 10.1158/0008-5472.CAN-04-3184. [DOI] [PubMed] [Google Scholar]
- 174.Boysen G, Georgieva NI, Upton PB, Walker VE, Swenberg JA. N-terminal globin adducts as biomarkers for formation of butadiene derived epoxides. Chem Biol Interact. 2007;166:84–92. doi: 10.1016/j.cbi.2006.10.005. [DOI] [PubMed] [Google Scholar]
- 175.Boysen G, Scarlett CO, Temple B, Combs TP, Brooks NL, Borchers CH, Swenberg JA. Identification of covalent modifications in P450 2E1 by 1,2-epoxy-3-butene in vitro. Chem Biol Interact. 2007;166:170–175. doi: 10.1016/j.cbi.2007.01.007. [DOI] [PubMed] [Google Scholar]
- 176.Blobaum AL, Kent UM, Alworth WL, Hollenberg PF. Mechanism-based inactivation of cytochromes P450 2E1 and 2E1 T303A by tert-butyl acetylenes: characterization of reactive intermediate adducts to the heme and apoprotein. Chem Res Toxicol. 2002;15:1561–1571. doi: 10.1021/tx020052x. [DOI] [PubMed] [Google Scholar]
- 177.von Weymarn LB, Sridar C, Hollenberg PF. Identification of amino acid residues involved in the inactivation of cytochrome P450 2B1 by two acetylenic compounds: the role of three residues in nonsubstrate recognition Sites. J Pharmacol Exp Ther. 2004;311:71–79. doi: 10.1124/jpet.104.069757. [DOI] [PubMed] [Google Scholar]
- 178.von Weymarn LB, Blobaum AL, Hollenberg PF. The mechanism-based inactivation of P450 2B4 by tert-butyl 1-methyl-2-propynyl ether: structural determination of the adducts to the P450 heme. Archives of Biochemistry and Biophysics. 2004;425:95–105. doi: 10.1016/j.abb.2004.03.014. [DOI] [PubMed] [Google Scholar]
- 179.Yang J, Liao M, Shou M, Jamei M, Yeo KR, Tucker GT, Rostami-Hodjegan A. Cytochrome p450 turnover: regulation of synthesis and degradation, methods for determining rates, and implications for the prediction of drug interactions. Curr Drug Metab. 2008;9:384–394. doi: 10.2174/138920008784746382. [DOI] [PubMed] [Google Scholar]
- 180.Correia MA. Cytochrome P450 turnover. Methods Enzymol. 1991;206:315–325. doi: 10.1016/0076-6879(91)06101-8. [DOI] [PubMed] [Google Scholar]
- 181.Kartha JS, Yost GS. Mechanism-based inactivation of lung-selective cytochrome P450 CYP2F enzymes. Drug Metab Dispos. 2008;36:155–162. doi: 10.1124/dmd.107.017897. [DOI] [PubMed] [Google Scholar]
- 182.Moreno RL, Goosen T, Kent UM, Chung FL, Hollenberg PF. Differential effects of naturally occurring isothiocyanates on the activities of cytochrome P450 2E1 and the mutant P450 2E1 T303A. Arch Biochem Biophys. 2001;391:99–110. doi: 10.1006/abbi.2001.2390. [DOI] [PubMed] [Google Scholar]
- 183.Ortiz de Montellano PR, Mico BA. Destruction of cytochrome P-450 by ethylene and other olefins. Mol Pharmacol. 1980;18:128–135. [PubMed] [Google Scholar]
- 184.Ortiz de Montellano PR, Kunze KL, Mico BA. Destruction of cytochrome P-450 by olefins: N-alkylation of prosthetic heme. Mol Pharmacol. 1980;18:602–605. [PubMed] [Google Scholar]
- 185.Ortiz de Montellano PR, Beilan HS, Kunze KL, Mico BA. Destruction of cytochrome P-450 by ethylene. Structure of the resulting prosthetic heme adduct. J Biol Chem. 1981;256:4395–4399. [PubMed] [Google Scholar]
- 186.Masubuchi Y, Horie T. Toxicological significance of mechanism-based inactivation of cytochrome p450 enzymes by drugs. Crit Rev Toxicol. 2007;37:389–412. doi: 10.1080/10408440701215233. [DOI] [PubMed] [Google Scholar]
- 187.Harrison KL, Khan NS, Dey P, Povey AC. N7-methyldeoxyguanosine levels in DNA isolated from cervical cytology samples are associated with smoking. Int J Cancer. 2006;119:961–963. doi: 10.1002/ijc.21900. [DOI] [PubMed] [Google Scholar]
- 188.Zhao C, Hemminki K. The in vivo levels of DNA alkylation products in human lymphocytes are not age dependent: an assay of 7-methyl- and 7-(2-hydroxyethyl)-guanine DNA adducts. Carcinogenesis. 2002;23:307–310. doi: 10.1093/carcin/23.2.307. [DOI] [PubMed] [Google Scholar]
- 189.Mustonen R, Hemminki K. 7-Methylguanine levels in DNA of smokers' and non-smokers' total white blood cells, granulocytes and lymphocytes. Carcinogenesis. 1992;13:1951–1955. doi: 10.1093/carcin/13.11.1951. [DOI] [PubMed] [Google Scholar]
- 190.Mustonen R, Schoket B, Hemminki K. Smoking-related DNA adducts: 32P-postlabeling analysis of 7-methylguanine in human bronchial and lymphocyte DNA. Carcinogenesis. 1993;14:151–154. doi: 10.1093/carcin/14.1.151. [DOI] [PubMed] [Google Scholar]
- 191.Mustonen R, Forsti A, Hietanen P, Hemminki K. Measurement by 32P-postlabelling of 7-methylguanine levels in white blood cell DNA of healthy individuals and cancer patients treated with dacarbazine and procarbazine. Human data and method development for 7-alkylguanines. Carcinogenesis. 1991;12:1423–1431. doi: 10.1093/carcin/12.8.1423. [DOI] [PubMed] [Google Scholar]
- 192.Harrison KL, Wood M, Lees NP, Hall CN, Margison GP, Povey AC. Development and application of a sensitive and rapid immunoassay for the quantitation of N7-methyldeoxyguanosine in DNA samples. Chem Res Toxicol. 2001;14:295–301. doi: 10.1021/tx000071b. [DOI] [PubMed] [Google Scholar]
- 193.Harrison KL, Crosbie PA, Agius RM, Barber PV, Carus M, Margison GP, Povey AC. No association between N7-methyldeoxyguanosine and 8-oxodeoxyguanosine levels in human lymphocyte DNA. Mutat Res. 2006;600:125–130. doi: 10.1016/j.mrfmmm.2006.03.005. [DOI] [PubMed] [Google Scholar]
- 194.Saad AA, O'Connor PJ, Mostafa MH, Metwalli NE, Cooper DP, Margison GP, Povey AC. Bladder tumor contains higher N7-methylguanine levels in DNA than adjacent normal bladder epithelium. Cancer Epidemiol Biomarkers Prev. 2006;15:740–743. doi: 10.1158/1055-9965.EPI-05-0813. [DOI] [PubMed] [Google Scholar]
- 195.Lewis SJ, Cherry NM, Niven RM, Barber PV, Povey AC. Associations between smoking, GST genotypes and N7-methylguanine levels in DNA extracted from bronchial lavage cells. Mutat Res. 2004;559:11–18. doi: 10.1016/j.mrgentox.2003.11.011. [DOI] [PubMed] [Google Scholar]
- 196.Szyfter K, Hemminki K, Szyfter W, Szmeja Z, Banaszewski J, Pabiszczak M. Tobacco smoke-associated N7-alkylguanine in DNA of larynx tissue and leucocytes. Carcinogenesis. 1996;17:501–506. doi: 10.1093/carcin/17.3.501. [DOI] [PubMed] [Google Scholar]
- 197.Filser JG, Denk B, Tornqvist M, Kessler W, Ehrenberg L. Pharmacokinetics of ethylene in man; body burden with ethylene oxide and hydroxyethylation of hemoglobin due to endogenous and environmental ethylene. Arch Toxicol. 1992;66:157–163. doi: 10.1007/BF01974008. [DOI] [PubMed] [Google Scholar]
- 198.Törnqvist M, Mowrer J, Jensen S, Ehrenberg L. Monitoring of environmental cancer initiators through hemoglobin adducts by a modified Edman degradation method. Analytical Biochemistry. 1986;154:255–266. doi: 10.1016/0003-2697(86)90524-5. [DOI] [PubMed] [Google Scholar]
- 199.Törnqvist M, Almberg JG, Bergmark E, Nilsson S, Osterman-Golkar SM. Ethylene oxide doses in ethene-exposed fruit store workers. Scandanavian Journal of Work & Environmental Health. 1989;15:436–438. doi: 10.5271/sjweh.1829. [DOI] [PubMed] [Google Scholar]
- 200.Tompkins EM, McLukie KIE, Jones DJL, Farmer PB, Brown K. Mutagenicity of DNA adducts derived from ethylene oxide exposure in the pSP189 shuttle vector replicated in human Ad293 cells. Mutat Res. 2009 doi: 10.1016/j.mrgentox.2009.05.011. [DOI] [PubMed] [Google Scholar]
- 201.Lieberman M, Kunishi A, Mapson LW, Wardale DA. Stimulation of Ethylene Production in Apple Tissue Slices by Methionine. Plant Physiol. 1966;41:376–382. doi: 10.1104/pp.41.3.376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Baker JE, Anderson JD, Adams DO, Apelbaum A, Lieberman M. Biosynthesis of ethylene from methionine in aminoethoxyvinylglycine-resistant avocado tissue. Plant Physiol. 1982;69:93–97. doi: 10.1104/pp.69.1.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Swenberg JA, Koc H, Upton PB, Georgieva N, Ranasinghe A, Walker VE, Henderson R. Using DNA and hemoglobin adducts to improve the risk assessment of butadiene. Chem Biol Interact. 2001;135-136:387–403. doi: 10.1016/s0009-2797(01)00221-6. [DOI] [PubMed] [Google Scholar]
- 204.Tornqvist M, Kautiainen A. Adducted proteins for identification of endogenous electrophiles. Environ Health Perspect. 1993;99:39–44. doi: 10.1289/ehp.99-1567046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Beranek DT, Heflich RH, Kodell RL, Morris SM, Casciano DA. Correlation between specific DNA-methylation products and mutation induction at the HGPRT locus in Chinese hamster ovary cells. Mutat Res. 1983;110:171–180. doi: 10.1016/0027-5107(83)90026-x. [DOI] [PubMed] [Google Scholar]
- 206.Fortini P, Calcagnile A, Di Muccio A, Bignami M, Dogliotti E. Quantitative relationship between ethylated DNA bases and gene mutation at two loci in CHO cells. Environ Mol Mutagen. 1993;21:154–159. doi: 10.1002/em.2850210209. [DOI] [PubMed] [Google Scholar]
- 207.Calleja F, Jansen JG, Vrieling H, Laval F, van Zeeland AA. Modulation of the toxic and mutagenic effects induced by methyl methanesulfonate in Chinese hamster ovary cells by overexpression of the rat N-alkylpurine-DNA glycosylase. Mutat Res. 1999;425:185–194. doi: 10.1016/s0027-5107(99)00034-2. [DOI] [PubMed] [Google Scholar]
- 208.Op het Veld CW, Jansen J, Zdzienicka MZ, Vrieling H, van Zeeland AA. Methyl methanesulfonate-induced hprt mutation spectra in the Chinese hamster cell line CHO9 and its xrcc1-deficient derivative EM-C11. Mutat Res. 1998;398:83–92. doi: 10.1016/s0027-5107(97)00243-1. [DOI] [PubMed] [Google Scholar]
- 209.Klungland A, Laake K, Hoff E, Seeberg E. Spectrum of mutations induced by methyl and ethyl methanesulfonate at the hprt locus of normal and tag expressing Chinese hamster fibroblasts. Carcinogenesis. 1995;16:1281–1285. doi: 10.1093/carcin/16.6.1281. [DOI] [PubMed] [Google Scholar]
- 210.Loechler EL, Green CL, Essigmann JM. In vivo mutagenesis by O6-methylguanine built into a unique site in a viral genome. Proc Natl Acad Sci U S A. 1984;81:6271–6275. doi: 10.1073/pnas.81.20.6271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Ellison KS, Dogliotti E, Connors TD, Basu AK, Essigmann JM. Site-specific mutagenesis by O6-alkylguanines located in the chromosomes of mammalian cells: influence of the mammalian O6- alkylguanine-DNA alkyltransferase. Proc Natl Acad Sci U S A. 1989;86:8620–8624. doi: 10.1073/pnas.86.22.8620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Nivard MJM, Czene K, Segerback D, Vogel EW. Mutagenic activity of ethylene oxide and propylene oxide under XPG proficient and deficient conditions in relation to N-7-(2-hydroxyalkyl)guanine levels in Drosophila. Mutat Res. 2003;529:95–107. doi: 10.1016/s0027-5107(03)00111-8. [DOI] [PubMed] [Google Scholar]
- 213.Zhang S, Glickman BW, de Boer JG. Spontaneous mutation of the lacI transgene in rodents: absence of species, strain, and insertion-site influence. Environ Mol Mutagen. 2001;37:141–146. doi: 10.1002/em.1021. [DOI] [PubMed] [Google Scholar]
- 214.Vergnes JS, Pritts IM. Effects of ethylene on micronucleus formation in the bone marrow of rats and mice following four weeks of inhalation exposure. Mutat Res. 1994;324:87–91. doi: 10.1016/0165-7992(94)90051-5. [DOI] [PubMed] [Google Scholar]
- 215.Rios-Blanco MN, Faller TH, Nakamura J, Kessler W, Kreuzer PE, Ranasinghe A, Filser JG, Swenberg JA. Quantitation of DNA and hemoglobin adducts and apurinic/apyrimidinic sites in tissues of F344 rats exposed to propylene oxide by inhalation. Carcinogenesis. 2000;21:2011–2018. doi: 10.1093/carcin/21.11.2011. [DOI] [PubMed] [Google Scholar]
- 216.O'Brien PJ, Ellenberger T. Dissecting the broad substrate specificity of human 3-methyladenine-DNA glycosylase. J Biol Chem. 2004;279:9750–9757. doi: 10.1074/jbc.M312232200. [DOI] [PubMed] [Google Scholar]
- 217.O'Brien PJ, Ellenberger T. The Escherichia coli 3-methyladenine DNA glycosylase AlkA has a remarkably versatile active site. J Biol Chem. 2004;279:26876–26884. doi: 10.1074/jbc.M403860200. [DOI] [PubMed] [Google Scholar]
- 218.Wyatt MD, Pittman DL. Methylating agents and DNA repair responses: Methylated bases and sources of strand breaks. Chem Res Toxicol. 2006;19:1580–1594. doi: 10.1021/tx060164e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.McCarthy TV, Karran P, Lindahl T. Inducible repair of O-alkylated DNA pyrimidines in Escherichia coli. EMBO J. 1984;3:545–550. doi: 10.1002/j.1460-2075.1984.tb01844.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Bjelland S, Bjoras M, Seeberg E. Excision of 3-methylguanine from alkylated DNA by 3-methyladenine DNA glycosylase I of Escherichia coli. Nucleic Acids Res. 1993;21:2045–2049. doi: 10.1093/nar/21.9.2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Bjelland S, Birkeland NK, Benneche T, Volden G, Seeberg E. DNA glycosylase activities for thymine residues oxidized in the methyl group are functions of the AlkA enzyme in Escherichia coli. J Biol Chem. 1994;269:30489–30495. [PubMed] [Google Scholar]
- 222.Wood RD, Mitchell M, Sgouros J, Lindahl T. Human DNA repair genes. Science. 2001;291:1284–1289. doi: 10.1126/science.1056154. [DOI] [PubMed] [Google Scholar]
- 223.Wyatt MD, Allan JM, Lau AY, Ellenberger TE, Samson LD. 3-methyladenine DNA glycosylases: structure, function, and biological importance. Bioessays. 1999;21:668–676. doi: 10.1002/(SICI)1521-1878(199908)21:8<668::AID-BIES6>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 224.Simonelli V, Narciso L, Dogliotti E, Fortini P. Base excision repair intermediates are mutagenic in mammalian cells. Nucleic Acids Res. 2005;33:4404–4411. doi: 10.1093/nar/gki749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Shane BS, Smith-Dunn DL, de Boer JG, Glickman BW, Cunningham ML. Mutant frequencies and mutation spectra of dimethylnitrosamine (DMN) at the lacI and cII loci in the livers of Big Blue transgenic mice. Mutat Res. 2000;452:197–210. doi: 10.1016/s0027-5107(00)00081-6. [DOI] [PubMed] [Google Scholar]
- 226.Wang X, Suzuki T, Itoh T, Honma M, Nishikawa A, Furukawa F, Takahashi M, Hayashi M, Kato T, Sofuni T. Specific mutational spectrum of dimethylnitrosamine in the lacI transgene of Big Blue C57BL/6 mice. Mutagenesis. 1998;13:625–630. doi: 10.1093/mutage/13.6.625. [DOI] [PubMed] [Google Scholar]
- 227.Souliotis VL, van Delft JH, Steenwinkel MJ, Baan RA, Kyrtopoulos SA. DNA adducts, mutant frequencies and mutation spectra in lambda lacZ transgenic mice treated with N-nitrosodimethylamine. Carcinogenesis. 1998;19:731–739. doi: 10.1093/carcin/19.5.731. [DOI] [PubMed] [Google Scholar]
- 228.Delker DA, Geter DR, Kleinert KM, Gollapudi BB. Frequency and spectrum of lacI mutations in the liver of Big Blue mice following the administration of genotoxic carcinogens singly and in series. Int J Toxicol. 2008;27:35–42. doi: 10.1080/10915810701876620. [DOI] [PubMed] [Google Scholar]
- 229.Walker VE, Skopek TR. A mouse model for the study of in vivo mutational spectra: sequence specificity of ethylene oxide at the hprt locus. Mutat Res. 1993;288:151–162. doi: 10.1016/0027-5107(93)90216-3. [DOI] [PubMed] [Google Scholar]
- 230.Recio L, Steen AM, Pluta LJ, Meyer KG, Saranko CJ. Mutational spectrum of 1,3-butadiene and metabolites 1,2-epoxybutene and 1,2,3,4-diepoxybutane to assess mutagenic mechanisms. Chem Biol Interact. 2001;135-136:325–341. doi: 10.1016/s0009-2797(01)00220-4. [DOI] [PubMed] [Google Scholar]
- 231.Lee DH, Kin TH, Lee SY, Kim HJ, Rhee SK, Yoon B, Pfeifer GP, Lee CS. Mutations induced by 1,3-butadiene metabolites, butadiene diolepoxide, and 1,2,3,4-diepoxybutane at the Hprt locus in CHO-K1 cells. Mol Cells. 2002;14:411–419. [PubMed] [Google Scholar]
- 232.Recio L, Pluta LJ, Meyer KG. The in vivo mutagenicity and mutational spectrum at the lacI transgene recovered from the spleens of B6C3F1 lacI transgenic mice following a 4-week inhalation exposure to 1,3-butadiene. Mutat Res. 1998;401:99–110. doi: 10.1016/s0027-5107(97)00319-9. [DOI] [PubMed] [Google Scholar]
- 233.Townsend L, Robins RK. Ring cleavage of purine nucleosides to yield possible biogenetic precursors of pteridines and riboflavin. J Am Chem Soc. 1963;85:242–243. [Google Scholar]
- 234.Chetsanga CJ, Bearie B, Makaroff C. Alkaline opening of imidazole ring of 7-methylguanosine. 1. Analysis of the resulting pyrimidine derivatives. Chem Biol Interact. 1982;41:217–233. doi: 10.1016/0009-2797(82)90091-6. [DOI] [PubMed] [Google Scholar]
- 235.Chetsanga CJ, Makaroff C. Alkaline opening of imidazole ring of 7-methylguanosine. 2. Further studies on reaction mechanisms and products. Chem Biol Interact. 1982;41:235–249. doi: 10.1016/0009-2797(82)90092-8. [DOI] [PubMed] [Google Scholar]
- 236.Chetsanga CJ, Polidori G, Mainwaring M. Analysis and excision of ring-opened phosphoramide mustard-deoxyguanine adducts in DNA. Cancer Res. 1982;42:2616–2621. [PubMed] [Google Scholar]
- 237.Box HC, Lilga KT, Potienko G. 13C nuclear magnetic resonance studies of radiation damage: radiation-induced degradation of glycine. Proc Natl Acad Sci U S A. 1977;74:2394–2396. doi: 10.1073/pnas.74.6.2394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Darzynkiewicz E, Labadi I, Haber D, Burger K, Lonnberg H. 7-Methylguanine nucleotides and their structural analogues; protolytic equilibria, complexing with magnesium (II) ion and kinetics for alkaline opening of the imidazole ring. Acta Chem Scand B. 1988;42:86–92. doi: 10.3891/acta.chem.scand.42b-0086. [DOI] [PubMed] [Google Scholar]
- 239.Griffin BE, Haines JA, Reese CB. The methylation of adenylyl-(3′-5′)-uridine and uridylyl-(3′-5′)-adenosine with diazomethane. Biochim Biophys Acta. 1967;142:536–538. doi: 10.1016/0005-2787(67)90634-x. [DOI] [PubMed] [Google Scholar]
- 240.Wang JS, Groopman JD. DNA damage by mycotoxins. Mutat Res. 1999;424:167–181. doi: 10.1016/s0027-5107(99)00017-2. [DOI] [PubMed] [Google Scholar]
- 241.Tudek B. Imidazole ring-opened DNA purines and their biological significance. J Biochem Mol Biol. 2003;36:12–19. doi: 10.5483/bmbrep.2003.36.1.012. [DOI] [PubMed] [Google Scholar]
- 242.Mao H, Deng Z, Wang F, Harris TM, Stone MP. An intercalated and thermally stable FAPY adduct of aflatoxin B1 in a DNA duplex: structural refinement from 1H NMR. Biochemistry. 1998;37:4374–4387. doi: 10.1021/bi9718292. [DOI] [PubMed] [Google Scholar]
- 243.Humphreys WG, Guengerich FP. Structure of formamidopyrimidine adducts as determined by NMR using specifically 15N-labeled guanosine. Chem Res Toxicol. 1991;4:632–636. doi: 10.1021/tx00024a005. [DOI] [PubMed] [Google Scholar]
- 244.Kadlubar FF, Beranek DT, Weis CC, Evans FE, Cox R, Irving CC. Characterization of the purine ring-opened 7-methylguanine and its persistence in rat bladder epithelial DNA after treatment with the carcinogen N-methylnitrosourea. Carcinogenesis. 1984;5:587–592. doi: 10.1093/carcin/5.5.587. [DOI] [PubMed] [Google Scholar]
- 245.Beranek DT, Weis CC, Evans FE, Chetsanga CJ, Kadlubar FF. Identification of N5-methyl-N5-formyl-2,5,6-triamino-4-hydroxypyrimidine as a major adduct in rat liver DNA after treatment with the carcinogens, N,N-dimethylnitrosamine or 1,2-dimethylhydrazine. Biochem Biophys Res Commun. 1983;110:625–631. doi: 10.1016/0006-291x(83)91195-6. [DOI] [PubMed] [Google Scholar]
- 246.Busby WF, Wogan GN. Aflatoxins. In: Searle C, editor. Chemical Carcinogenesis. Am Chem Soc; Washington, DC: 1984. pp. 945–1136. [Google Scholar]
- 247.Croy RG, Essigmann JM, Reinhold VN, Wogan GN. Identification of the principal aflatoxin B1-DNA adduct formed in vivo in rat liver. Proc Natl Acad Sci U S A. 1978;75:1745–1749. doi: 10.1073/pnas.75.4.1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Lin JK, Miller JA, Miller EC. 2,3-Dihydro-2-(guan-7-yl)-3-hydroxy-aflatoxin B1, a major acid hydrolysis product of aflatoxin B1-DNA or -ribosomal RNA adducts formed in hepatic microsome-mediated reactions and in rat liver in vivo. Cancer Res. 1977;37:4430–4438. [PubMed] [Google Scholar]
- 249.Croy RG, Wogan GN. Quantitative comparison of covalent aflatoxin-DNA adducts formed in rat and mouse livers and kidneys. J Natl Cancer Inst. 1981;66:761–768. [PubMed] [Google Scholar]
- 250.Croy RG, Wogan GN. Temporal patterns of covalent DNA adducts in rat liver after single and multiple doses of aflatoxin B1. Cancer Res. 1981;41:197–203. [PubMed] [Google Scholar]
- 251.Groopman JD, Croy RG, Wogan GN. In vitro reactions of aflatoxin B1-adducted DNA. Proc Natl Acad Sci U S A. 1981;78:5445–5449. doi: 10.1073/pnas.78.9.5445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Martin CN, Garner RC. Aflatoxin B -oxide generated by chemical or enzymic oxidation of aflatoxin B1 causes guanine substitution in nucleic acids. Nature. 1977;267:863–865. doi: 10.1038/267863a0. [DOI] [PubMed] [Google Scholar]
- 253.Gopalakrishnan S, Harris TM, Stone MP. Intercalation of aflatoxin B1 in two oligodeoxynucleotide adducts: comparative 1H NMR analysis of d(ATCAFBGAT).d(ATCGAT) and d(ATAFBGCAT)2. Biochemistry. 1990;29:10438–10448. doi: 10.1021/bi00498a002. [DOI] [PubMed] [Google Scholar]
- 254.Hsu IC, Metcalf RA, Sun T, Welsh JA, Wang NJ, Harris CC. Mutational hotspot in the p53 gene in human hepatocellular carcinomas. Nature. 1991;350:427–428. doi: 10.1038/350427a0. [DOI] [PubMed] [Google Scholar]
- 255.Bressac B, Kew M, Wands J, Ozturk M. Selective G to T mutations of p53 gene in hepatocellular carcinoma from southern Africa. Nature. 1991;350:429–431. doi: 10.1038/350429a0. [DOI] [PubMed] [Google Scholar]
- 256.Vogelstein B, Fearon ER, Hamilton SR, Kern SE, Preisinger AC, Leppert M, Nakamura Y, White R, Smits AM, Bos JL. Genetic alterations during colorectal-tumor development. N Engl J Med. 1988;319:525–532. doi: 10.1056/NEJM198809013190901. [DOI] [PubMed] [Google Scholar]
- 257.Bos JL, Fearon ER, Hamilton SR, Verlaan-de VM, van Boom JH, van der Eb AJ, Vogelstein B. Prevalence of ras gene mutations in human colorectal cancers. Nature. 1987;327:293–297. doi: 10.1038/327293a0. [DOI] [PubMed] [Google Scholar]
- 258.McMahon G, Hanson L, Lee JJ, Wogan GN. Identification of an activated c-Ki-ras oncogene in rat liver tumors induced by aflatoxin B1. PNAS. 1986;83:9418–9422. doi: 10.1073/pnas.83.24.9418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Smela ME, Hamm ML, Henderson PT, Harris CM, Harris TM, Essigmann JM. The aflatoxin B(1) formamidopyrimidine adduct plays a major role in causing the types of mutations observed in human hepatocellular carcinoma. Proc Natl Acad Sci U S A. 2002;99:6655–6660. doi: 10.1073/pnas.102167699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Bailey EA, Iyer RS, Stone MP, Harris TM, Essigmann JM. Mutational properties of the primary aflatoxin B1-DNA adduct. PNAS. 1996;93:1535–1539. doi: 10.1073/pnas.93.4.1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Groopman JD. Do aflatoxin-DNA adduct measurements in humans provide accurate data for cancer risk assessment? IARC Sci Publ. 1988:55–62. [PubMed] [Google Scholar]
- 262.Groopman JD, Cain LG, Kensler TW. Aflatoxin exposure in human populations: measurements and relationship to cancer. Crit Rev Toxicol. 1988;19:113–145. doi: 10.3109/10408448809014902. [DOI] [PubMed] [Google Scholar]
- 263.Ezaz-Nikpay K, Verdine GL. The effects of N7-methylguanine on duplex DNA structure. Chem Biol. 1994;1:235–240. doi: 10.1016/1074-5521(94)90016-7. [DOI] [PubMed] [Google Scholar]
- 264.Lee S, Bowman BR, Ueno Y, Wang S, Verdine GL. Synthesis and structure of duplex DNA containing the genotoxic nucleobase lesion N7-methylguanine. J Am Chem Soc. 2008;130:11570–11571. doi: 10.1021/ja8025328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.O'Connor TR, Boiteux S, Laval J. Ring-opened 7-methylguanine residues in DNA are a block to in vitro DNA synthesis. Nucl Acids Res. 1988;16:5879–5894. doi: 10.1093/nar/16.13.5879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Boiteux S, Laval J. Imidazole open ring 7-methylguanine: an inhibitor of DNA synthesis. Biochem Biophys Res Commun. 1983;110:552–558. doi: 10.1016/0006-291x(83)91185-3. [DOI] [PubMed] [Google Scholar]
- 267.Bolt HM, Foth H, Hengstler JG, Degen GH. Carcinogenicity categorization of chemicals-new aspects to be considered in a European perspective. Toxicol Lett. 2004;151:29–41. doi: 10.1016/j.toxlet.2004.04.004. [DOI] [PubMed] [Google Scholar]
- 268.Gocke E, Müller L. In vivo studies in the mouse to define a threshold for genotoxicity of EMS. Muta Res. 2009 doi: 10.1016/j.mrgentox.2009.04.005. [DOI] [PubMed] [Google Scholar]
- 269.Segerback D, Calleman CJ, Schroeder JL, Costa LG, Faustman EM. Formation of N-7-(2-carbamoyl-2-hydroxyethyl)guanine in DNA of the mouse and the rat following intraperitoneal administration of [14C]acrylamide. Carcinogenesis. 1995;16:1161–1165. doi: 10.1093/carcin/16.5.1161. [DOI] [PubMed] [Google Scholar]
- 270.Plna K, Nilsson R, Koskinen M, Segerback D. 32P-postlabelling of propylene oxide 1-and N-6-substituted adenine and 3-substituted cytosine/uracil: formation and persistence in vitro and in vivo. Carcinogenesis. 1999;20:2025–2032. doi: 10.1093/carcin/20.10.2025. [DOI] [PubMed] [Google Scholar]
- 271.Plna K, Segerback D, Schweda EK. DNA adduct formation by allyl glycidyl ether. Carcinogenesis. 1996;17:1465–1471. doi: 10.1093/carcin/17.7.1465. [DOI] [PubMed] [Google Scholar]
- 272.Selzer RR, Elfarra AA. In vitro reactions of butadiene monoxide with single- and double-stranded DNA: characterization and quantitation of several purine and pyrimidine adducts. Carcinogenesis. 1999;20:285–292. doi: 10.1093/carcin/20.2.285. [DOI] [PubMed] [Google Scholar]
- 273.Tretyakova NY, Chiang SY, Walker VE, Swenberg JA. Quantitative analysis of 1,3-butadiene-induced DNA adducts in vivo and in vitro using liquid chromatography electrospray ionization tandem mass spectrometry. J Mass Spectrom. 1998;33:363–376. doi: 10.1002/(SICI)1096-9888(199804)33:4<363::AID-JMS643>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 274.Montesano R. Alkylation of DNA and tissue specificity in nitrosamine carcinogenesis. J Supramol Struct Cell Biochem. 1981;17:259–273. doi: 10.1002/jsscb.380170307. [DOI] [PubMed] [Google Scholar]
- 275.Beranek DT, Weis CC, Swenson DH. A comprehensive quantitative analysis of methylated and ethylated DNA using high pressure liquid chromatography. Carcinogenesis. 1980;1:595–606. doi: 10.1093/carcin/1.7.595. [DOI] [PubMed] [Google Scholar]
- 276.Shields PG, Povey AC, Wilson VL, Weston A, Harris CC. Combined high-performance liquid chromatography/32P-postlabeling assay of N7-methyldeoxyguanosine. Cancer Res. 1990;50:6580–6584. [PubMed] [Google Scholar]
- 277.Petruzzelli S, Tavanti LM, Celi A, Giuntini C. Detection of N7-methyldeoxyguanosine adducts in human pulmonary alveolar cells. Am J Respir Cell Mol Biol. 1996;15:216–223. doi: 10.1165/ajrcmb.15.2.8703477. [DOI] [PubMed] [Google Scholar]
- 278.Blomeke B, Greenblatt MJ, Doan VD, Bowman ED, Murphy SE, Chen CC, Kato S, Shields PG. Distribution of 7-alkyl-2′-deoxyguanosine adduct levels in human lung. Carcinogenesis. 1996;17:741–748. doi: 10.1093/carcin/17.4.741. [DOI] [PubMed] [Google Scholar]
- 279.Chen L, Wang M, Villalta PW, Hecht SS. Liquid chromatography-electrospray ionization tandem mass spectrometry analysis of 7-ethylguanine in human liver DNA. Chem Res Toxicol. 2007 doi: 10.1021/tx700147f. [DOI] [PubMed] [Google Scholar]
- 280.Van Delft JHM, Van Winden MJM, Van Den Ende AMC, Baan RA. Determining N7-Alkylguanine Adducts by Immunochemical Methods and HPLC with Electrochemical Detection: Applications in Animal Studies and in Monitoring Human Exposure to Alkylating Agents. Environ Health Perspect. 1993;99:25–32. doi: 10.1289/ehp.939925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Bodell WJ, Giannini DD, Hassenbusch S, Levin VA. Levels of N7-(2-hydroxyethyl)guanine as a molecular dosimeter of drug delivery to human brain tumors. Neuro Oncol. 2001;3:241–245. doi: 10.1093/neuonc/3.4.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Khan SA, Carmichael PL, Taylor-Robinson SD, Habib N, Thomas HC. DNA adducts, detected by 32P postlabelling, in human cholangiocarcinoma. Gut. 2003;52:586–591. doi: 10.1136/gut.52.4.586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Doak SH, Jenkins GJS, Johnson GE, Quick E, Parry EM, Parry JM. Mechanistic Influences for Mutation Induction Curves after Exposure to DNA-Reactive Carcinogens. Cancer Res. 2007;67:3904–3911. doi: 10.1158/0008-5472.CAN-06-4061. [DOI] [PubMed] [Google Scholar]
- 284.Chiang SY, Huang TH, Uang SN, Wu HD, Wei YC, Lin HY, Swenberg JA, Wu KY. Analysis of 7-methylguanine using isotope dilution and gas chromatography/electron-capture negative chemical ionization mass spectrometry. Rapid Commun Mass Spectrom. 2005;19:1915–1920. doi: 10.1002/rcm.2003. [DOI] [PubMed] [Google Scholar]
- 285.Frei JV, Swenson DH, Warren W, Lawley PD. Alkylation of deoxyribonucleic acid in vivo in various organs of C57BL mice by the carcinogens N-methyl-N-nitrosourea, N-ethyl-N-nitrosourea and ethyl methanesulfonate in relation to induction of thymic lymphoma. Biochemical J. 1978;174:1031–1044. doi: 10.1042/bj1741031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.van Zeeland AA, Mohn GR, Neuhauser-Klaus A, Ehling UH. Quantitative comparison of genetic effects of ethylating agents on the basis of DNA adduct formation. Use of O6-ethylguanine as molecular dosimeter for extrapolation from cells in culture to the mouse. Environ Health Perspect. 1985;62:163–169. doi: 10.1289/ehp.8562163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Swenson DH, Petzold GL, Harbach PR. The binding of 1-(2-hydroxyethyl)-1-nitrosourea to DNA in vitro and to DNA of thymus and marrow in C57BL mice in vivo. Cancer Lett. 1986;33:75–81. doi: 10.1016/0304-3835(86)90103-5. [DOI] [PubMed] [Google Scholar]
- 288.Segerback D, Osterman-Golkar S, Molholt B, Nilsson R. In vivo tissue dosimetry as a basis for cross-species extrapolation in cancer risk assessment of propylene oxide. Regul Toxicol Pharmacol. 1994;20:1–14. doi: 10.1006/rtph.1994.1033. [DOI] [PubMed] [Google Scholar]
- 289.Vodicka PE, Linhart I, Novak J, Koskinen M, Vodickova L, Hemminki K. 7-Alkylguanine adduct levels in urine, lungs and liver of mice exposed to styrene by inhalation. Toxicol Appl Pharmacol. 2006;210:1–8. doi: 10.1016/j.taap.2005.04.020. [DOI] [PubMed] [Google Scholar]
- 290.Pauwels W, Vodiceka P, Severi M, Plna K, Veulemans H, Hemminki K. Adduct formation on DNA and haemoglobin in mice intraperitoneally administered with styrene. Carcinogenesis. 1996;17:2673–2680. doi: 10.1093/carcin/17.12.2673. [DOI] [PubMed] [Google Scholar]
- 291.La DK, Schoonhoven R, Ito N, Swenberg JA. The effects of exposure route on DNA adduct formation and cellular proliferation by 1,2,3-trichloropropane. Toxicol Appl Pharmacol. 1996;140:108–114. doi: 10.1006/taap.1996.0203. [DOI] [PubMed] [Google Scholar]
- 292.Zeiger E, Recio L, Fennell TR, Haseman JK, Snyder RW, Friedman M. Investigation of the low-dose response in the in vivo induction of micronuclei and adducts by acrylamide. Toxicol Sci. 2009;107:247–257. doi: 10.1093/toxsci/kfn214. [DOI] [PubMed] [Google Scholar]
- 293.Ghanayem BI, McDaniel LP, Churchwell MI, Twaddle NC, Snyder R, Fennell TR, Doerge DR. Role of CYP2E1 in the epoxidation of acrylamide to glycidamide and formation of DNA and hemoglobin adducts. Toxicol Sci. 2005;88:311–318. doi: 10.1093/toxsci/kfi307. [DOI] [PubMed] [Google Scholar]
- 294.Swenberg JA, Bedell MA. Banbury Report 113: Indicators of Genotoxic Exposure. Cold Spring Harbor Laboratory; Cold Spring Harbor: 1982. Cell-specific DNA alkylation and repair: Application of new fluorometric techniques to detect adducts; pp. 205–220. [Google Scholar]
- 295.Lewis JG, Swenberg JA. Effect of 1,2-dimethylhydrazine and diethylnitrosamine on cell replication and unscheduled DNA synthesis in target and nontarget cell populations in rat liver following chronic administration. Cancer Res. 1982;42:89–92. [PubMed] [Google Scholar]
- 296.Lewis JG, Swenberg JA. The kinetics of DNA alkylation, repair and replication in hepatocytes, Kupffer cells, and sinusoidal endothelial cells in rat liver during continuous exposure to 1,2-dimethylhydrazine. Carcinogenesis. 1983;4:529–536. doi: 10.1093/carcin/4.5.529. [DOI] [PubMed] [Google Scholar]
- 297.Brink A, Schulz B, Stopper H, Lutz WK. Biological significance of DNA adducts investigated by simultaneous analysis of different endpoints of genotoxicity in L5178Y mouse lymphoma cells treated with methyl methanesulfonate. Mutat Res. 2007;625:94–101. doi: 10.1016/j.mrfmmm.2007.05.007. [DOI] [PubMed] [Google Scholar]
- 298.Souliotis VL, Henneman JR, Reed CD, Chhabra SK, Diwan BA, Anderson LM, Kyrtopoulos SA. DNA adducts and liver DNA replication in rats during chronic exposure to N-nitrosodimethylamine (NDMA) and their relationships to the dose-dependence of NDMA hepatocarcinogenesis. Mutat Res. 2002;500:75–87. doi: 10.1016/s0027-5107(01)00301-3. [DOI] [PubMed] [Google Scholar]
- 299.Koepke SR, Kroeger-Koepke MB, Bosan W, Thomas BJ, Alvord WG, Michejda CJ. Alkylation of DNA in rats by N-nitrosomethyl-(2-hydroxyethyl)amine: dose response and persistence of the alkylated lesions in vivo. Cancer Res. 1988;48:1537–1542. [PubMed] [Google Scholar]
- 300.Swann PF, Pegg AE, Hawks A, Farber E, Magee PN. Evidence for ethylation of rat liver deoxyribonucleic acid after administration of ethionine. Biochem J. 1971;123:175–181. doi: 10.1042/bj1230175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301.Swann PF, Magee PN. Nitrosamine-induced carcinogenesis. The alkylation of N-7 of guanine of nucleic acids of the rat by diethylnitrosamine, N-ethyl-N-nitrosourea and ethyl methanesulphonate. Biochem J. 1971;125:841–847. doi: 10.1042/bj1250841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302.Netto LE, RamaKrishna NV, Kolar C, Cavalieri EL, Rogan EG, Lawson TA, Augusto O. Identification of C8-methylguanine in the hydrolysates of DNA from rats administered 1,2-dimethylhydrazine. Evidence for in vivo DNA alkylation by methyl radicals. J Biol Chem. 1992;267:21524–21527. [PubMed] [Google Scholar]
- 303.Papanikolaou A, Shank RC, Delker DA, Povey A, Cooper DP, Rosenberg DW. Initial levels of azoxymethane-induced DNA methyl adducts are not predictive of tumor susceptibility in inbred mice. Toxicol Appl Pharmacol. 1998;150:196–203. doi: 10.1006/taap.1998.8393. [DOI] [PubMed] [Google Scholar]
- 304.Fedtke N, Walker VE, Swenberg JA. Determination of 7-(2-oxoethyl)guanine and N2,3-ethenoguanine in DNA hydrolysates by HPLC. Arch Toxicol. 1989 Supp 13:214–218. doi: 10.1007/978-3-642-74117-3_33. [DOI] [PubMed] [Google Scholar]
- 305.Maniere I, Godard T, Doerge DR, Churchwell MI, Guffroy M, Laurentie M, Poul JM. DNA damage and DNA adduct formation in rat tissues following oral administration of acrylamide. Mutat Res. 2005;580:119–129. doi: 10.1016/j.mrgentox.2004.10.012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1: Amounts of N7-guanine adducts in mice.
Table S2: Amounts of N7-guanine adducts in rats.