Abstract
Macroautophagy (autophagy) is a bulk cytoplasmic degradation process that is conserved from yeast to mammals. Autophagy is an important cellular response to starvation and stress, and plays important roles in development, cell death, aging, immunity, and cancer. The fruit fly Drosophila melanogaster provides an excellent model system to study autophagy in vivo, in the context of a developing organism. Autophagy (atg) genes and their regulators are conserved in Drosophila, and autophagy is induced in response to nutrient starvation and hormones during development. In this review we provide an overview of how Drosophila research has contributed to our understanding of the role and regulation of autophagy in cell survival, growth, nutrient utilization, and cell death. Recent Drosophila research has also provided important mechanistic information about the role of autophagy in protein aggregation disorders, neurodegeneration, aging, and innate immunity. Differences in the role of autophagy in specific contexts and/or cell types suggest that there may be cell-context-specific regulators of autophagy, and studies in Drosophila are well-suited to yield discoveries about this specificity.
Introduction
Autophagy is a catabolic process that is ubiquitously implemented by eukaryotic cells. Three general types of autophagy have been described, macroautophagy, microautophagy, and chaperone-mediated autophagy [1, 2]. Of the three types, macroautophagy, a largely non-selective degradation process to eliminate macromolecules and organelles, has been best characterized and will hereby be referred to as autophagy. Whereas the Ubiquitin Proteasome System (UPS) is used to degrade short-lived proteins specifically targeted for degradation, autophagy is generally thought to be used for the bulk degradation of long-lived proteins. During autophagy, cytosolic components are sequestered by a double-membrane vesicle, the autophagosome, which delivers cargo to the lysosome for recycling.
Autophagy is an important cellular response to stress and a survival mechanism during starvation that is conserved in yeast, worms, flies, and mammals [3, 4]. In response to starvation, autophagy functions in the production of amino acids, providing the building blocks for new protein synthesis. Autophagy is also a mechanism for the production of mitochondrial substrates to produce the energy required to survive starvation [5]. In addition, autophagy is important for the elimination of damaged/unwanted organelles and protein aggregates [2, 6]. In animal cells, autophagy also plays a role in cellular remodeling during development and differentiation, and in the elimination of invasive microorganisms. Alterations and deficiencies in autophagy (atg) genes are associated with developmental and cell death defects, sensitivity to starvation and stress, decreased longevity, neurodegeneration, and tumor progression [2, 6–8].
The pathways that regulate autophagy are evolutionarily conserved [9, 10]. The protein kinase Target of Rapamycin (TOR) plays a central role in nutrient sensing and autophagy regulation. When nutrients are readily available, TOR is activated through the Class I phosphatidylinositol-3-kinase (PI3K) signaling pathway, and TOR inhibits autophagy through direct phosphorylation and repression of Atg1 [11, 12]. When nutrients are scarce, TOR becomes inactivated, its repression of Atg1 is relieved, and autophagy is induced. In yeast, autophagy induction requires Atg1, Atg13, and Atg17, and TOR has been shown to regulate the association of Atg1 with Atg13 via Atg13 phosphorylation [12, 13]. Nucleation of the autophagic membrane requires phosphatidylinositol phosphorylation, which in yeast requires the formation of a complex that includes Atg6/Beclin1, Vps34/Class III PI3K, Atg14 and Vps15 [14–16]. In mammalian cells, additional regulators of the Beclin1/Vps34 complex have been identified, including Ambra1, UVRAG and Bif-1, and the mammalian Atg14 homolog, Barkor, [17–20]. In addition, 2 distinct Beclin-1 complexes have been described in yeast and mammalian cells that may play distinct roles in membrane trafficking pathways [15, 16, 21].
Little is known about the origin of the autophagic membrane, and this is a subject of debate [22]. In yeast, autophagy proteins gather at a punctate structure, the Pre-Autophagosomal Structure (PAS), near the vacuole (the yeast lysosome) [4]. A recent study suggests that in mammalian cells, nucleation of the autophagic membrane begins in a punctate compartment that is in dynamic equilibrium with the Endoplasmic Reticulum (ER) [23]. This work is consistent with a previous study suggesting that ER membranes are used to form autophagosomes based on the localization of ER proteins in autophagosomal membranes [24].
Two ubiquitin-like conjugation systems, conserved from yeast to mammals, function in autophagosome formation [25, 26]. The formation of an Atg12-5-16 complex on the isolation membrane requires the E1-like activating enzyme Atg7 and the E2-like conjugating enzyme Atg10 [27–31]. This complex disassociates upon formation of the autophagosome. The second conjugation system requires the activity of Atg7 and the E2-like conjugating enzyme Atg3 [29, 32–34]. In this step, cytosolic Atg8 (LC3 in mammals) is modified by the attachment of the phospholipid anchor phosphatidyl-ethanolamine (PE) following its cleavage by the cysteine protease Atg4 [35–37]. This step results in the localization of Atg8-PE to the isolation membrane, and has been proposed to contribute to elongation of the autophagic membrane [38]. Atg8 remains associated with the autophagosome until it is trafficked to the lysosome, when the outer membrane of the autophagosome fuses with the lysosome to form the autolysosome, and Atg4 subsequently releases Atg8 from PE [35]. For these reasons, Atg8/LC3 is a widely used marker of autophagosomes [39]. Upon autophagosome formation, the inner membrane of the autophagosome and its contents are degraded by lysosomal hydrolases, and nutrients are subsequently released into the cytosol for recycling [3].
Autophagy in Drosophila Development
Pioneering genetic screens in the yeast Saccharomyces cerevisiae advanced our understanding of autophagy by identifying the genes that are required for this catabolic process [40–43]. The complexity of multicellular animals presents several interesting questions about autophagy and its relationship to nutrient utilization, cell growth, cell survival and cell death. For example, the ability of animals to respond to nutrient deprivation and adjust metabolic and catabolic processes to maintain homeostasis suggests that the mechanisms that regulate autophagy may differ under specific cellular contexts.
Drosophila melanogaster is an excellent genetic model for higher animals. Drosophila has a short life cycle, a wide variety of genetic tools available, and mutants and RNAi lines have been systematically generated (http://flybase.org). atg genes and their regulators are highly conserved in Drosophila[9, 44], and in many cases, in contrast to mammalian systems, single orthologs to atg genes exist in Drosophila, allowing for non-redundant genetic studies. Autophagy is induced in many Drosophila tissues in response to nutrient restriction; for example, in the fat body of starving larvae, and during the pupal stage upon the cessation of feeding. In addition, autophagy is induced in Drosophila in response to the steroid hormone 20-hydroxyecdysone (ecdysone) in both the fat body [45] and in dying larval structures such as the intestine and salivary glands [46, 47]. Thus, Drosophila serves as an excellent model to study autophagy in vivo, in response to nutrient restriction and also in the context of programmed cell death that occurs during development.
Drosophilaatg gene mutant phenotypes suggest a role for autophagy in development [9, 48]. Null mutations in atg1 are pupal lethal [49]. Surprisingly, however, mutations in some atg genes essential for autophagy are not lethal, even though these flies appear to possess greatly attenuated autophagy. Null atg7 mutants develop normally whereas strong atg8 hypomorphic mutations are semi-lethal [11, 50–52]. Both atg7 and atg8 mutants are hypersensitive to starvation and oxidative stress, exhibit degenerative neuronal defects, accumulate ubiquitin-positive aggregates in neurons, and have a shortened lifespan [11, 50–52]. This range in phenotypes suggests that some autophagy genes could play specialized roles, while others may be more pleiotropic and should be investigated for phenotypes that are not related to autophagy. The fact that atg7 null mutations are not lethal suggests a few different possibilities. Some atg genes may function redundantly, or it could be that other mechanisms may compensate for macroautophagy deficiencies during Drosophila development, such as chaperone-mediated autophagy or micro-autophagy [2]. Alternatively, given that certain autophagy gene mutations have cell-context-specific effects, there could be factors that determine specificity. Finding such factors will require studies to be carried out in nutritional contexts that are relevant to physiology and development, rather than in cell lines, and Drosophila is well suited for this type of study.
Autophagy in Growth and Nutrient Utilization
To develop to the proper size, animals require the coordination of cell growth, division and death within individual tissues, and this is influenced by environmental factors including nutrient availability [53]. Drosophila development provides a useful system to investigate the relationship between nutrients, autophagy, growth and development. Fly development lasts approximately 8 days in the laboratory; embryogenesis is 1 day long, the 3 larval feeding stages combine to last 3.5 days, and the transformation to an adult, known as metamorphosis, is 3.5 days long. Like vertebrate animals, fly development is controlled by growth factors and hormones. Insulin and Insulin-like growth factors are secreted when animals are feeding, stimulating protein synthesis, cell growth, and the processes that are required for animal growth [54]. In addition, the steroid ecdysone defines the length of the developmental period in Drosophila, as peaks in this steroid regulate larval molting and metamorphosis [55]. Several recent studies have investigated the relationship between growth factor and ecdysone signaling in flies [56, 57]. Surprisingly, few studies have investigated the relationship between these signaling pathways, autophagy and animal development.
Autophagy plays an important role in nutrient utilization during Drosophila larval development, and studies in the Drosophila fat body under conditions of starvation have provided important insights into the mechanisms that regulate autophagy. The fly fat body is a nutrient storage and mobilization organ akin to the mammalian liver and autophagy is induced in the fat body in response to amino acid starvation [49]. Either null mutations in TOR, or repression of TOR by modulation of components of the class I PI3K pathway, lead to the induction of autophagy in the fat body of feeding larvae, and result in the inhibition of growth, a reduction in cell size, and decreased viability [49]. Conversely, expression of either activated TOR, or activation of components of the PI3K signaling pathway, suppresses starvation-induced autophagy [49]. However, unlike in mammalian cells, the suppression of starvation-induced autophagy in this context does not require S6K phosphorylation [49].
Autophagy is also induced in several Drosophila tissues as a normal physiological response to the rise in hormones during metamorphosis. Autophagy and multiple atg gene RNA levels are induced following rises in ecdysone that trigger cell death of the larval midgut and salivary glands during metamorphosis [46, 47, 58], suggesting that autophagy, and possibly cell death, play important roles in the maintenance of nutrient homeostasis when developing animals are not feeding. In the fat body, Class I PI3K pathway signaling is down-regulated, and autophagy is induced, following the rise in ecdysone at the end of the third larval stage that triggers metamorphosis [45], indicating that regulation of the PI3K pathway is involved in the induction of autophagy in response to ecdysone as well as starvation. A recent screen for mutants that fail to induce autophagy in the fat body in response to ecdysone identified the Drosophila homologue of the AMP-activated protein kinase (AMPK) γ subunit, SNF4A γ [59]. AMPK has been implicated in the regulation of autophagy and cell survival following growth factor withdrawal in mammalian cells [60]. In addition, a recent study indicates that AMPK represses TOR and induces autophagy under nutrient-rich conditions in response to calcium signaling in mammals [61]. Therefore, these studies suggest a conserved role for AMPK in the regulation of TOR and autophagy.
Studies in the Drosophila fat body have also shown that mutations in genes required for endosomal biogenesis are required for autophagy [62]. Mutations in escrt genes encoding the ESCRT proteins I, II, and III, as well as mutations in vps4, the ESCRT complex regulatory ATPase, all lead to an accumulation of autophagosomes in the fat bodies of fed larvae [62, 63]. Furthermore, in escrt-II mutant cells, lysosomal associated membrane protein-1 (LAMP-1)-positive structures are distinct from Atg8-positive structures, suggesting a failure in the fusion of autophagosomes and lysosomes. This study also showed that mutations in fab1, an endosomal PI3 5-kinase, lead to the accumulation of autolysosomes that fail to degrade their contents, suggesting that fab1 is required for the maturation, and possibly the acidification, of autophagosomes [62].
The Vps34/Class III PI3K complex is a critical and conserved regulator of autophagy. This complex has been implicated in the regulation of endosomal maturation/lysosomal biogenesis as well as autophagy. It is possible that endosomal maturation/lysosomal biogenesis and autophagy may be co-regulated, although it is also possible that distinct Vps34 complexes regulate these vesicular trafficking pathways. Studies of mammalian cells suggest that amino acids stimulate TOR by activating Vps34/Class III PI3K [64, 65]. However, some differences in the regulation of autophagy may exist between insect and mammalian systems. In Drosophila, whereas vps34 is required for starvation-induced autophagy in the larval fat body, null mutations in vps34 do not affect TOR signaling [66]. It is not clear whether the differences between these results in flies and mammalian cells reflect evolutionary differences, or possibly cell context-specific differences between in vitro and in vivo studies.
Studies in Drosophila have also provided new insights into the relationship between autophagy and growth. Rapamycin treatment induces autophagy [67], and this revealed the critical regulatory role of TOR on autophagy that has been noted in many studies [10, 54, 68]. TOR represses autophagy through phosphorylation of Atg1 [11, 12]. Scott et al (2007) demonstrated that in the Drosophila larval fat body, over-expression of Atg1 inhibits cell growth, and this involves a negative feedback mechanism on TOR. In addition, atg1 mutant cells have a growth advantage when TOR signaling is reduced [11]. Thus, there is a reciprocal relationship between autophagy and cell growth. Interestingly, atg gene disruption in Drosophilator mutant fat body cells does not restore growth, but enhances the cell size reduction in tor mutants, suggesting that in this context, autophagy is required to supply nutrients in order for cells to grow [49]. Similarly, null atg5 or atg7 mutant mice lack the energy required to survive post-natal nutrient depletion [69, 70].
A recent study in mammalian cells, as well as in Drosophila, provided additional mechanistic information regarding the regulation of cell growth and autophagy. Kim et al (2008) found that Rag GTPases activate TOR in response to amino acid signals, and this activation is in parallel to Rheb signaling. Further, expression of constitutively active Rag GTPases in the absence of amino acids activates TOR, suppresses autophagy, and leads to an increase in organ size, whereas expression of dominant negative Rag GTPases leads to a decrease in organ size. Therefore, the reciprocal relationship between autophagy and cell growth involves the activation of Rag GTPases, and this mechanism is conserved [71, 72].
Autophagy and growth also exhibit a reciprocal relationship in the context of autophagy that occurs during cell death in Drosophila larval salivary glands. Growth arrest via regulation of the PI3K pathway is required for the induction of autophagy that occurs during salivary gland degradation [73]. Activation of positive regulators of growth, including Ras, Akt, or the PI3K catalytic subunit Dp110, inhibits autophagy and degradation of salivary glands [73]. Coexpression of the caspase inhibitor p35 with Dp110 enhances the Dp110-induced partial salivary gland degradation phenotype, suggesting that caspases function in parallel to growth arrest in this context. Importantly, expression of Atg1 and activation of autophagy suppresses the persistent phenotype in Dp110-expressing salivary glands, and atg loss of function mutations prevent the destruction of salivary glands [73]. Thus, growth arrest via down-regulation of Class I PI3K is required for the induction of autophagy and salivary gland degradation. Further, the inhibition of salivary gland degradation by positive regulators of cell growth requires the activity of TOR, suggesting that the inhibition of autophagy by cell growth regulators takes place through TOR signaling [73].
Recent work has elucidated how cell growth arrest is regulated in dying salivary glands. Loss-of-function mutations in the Warts/Hippo pathway, or over-expression of Wts downstream target Yorkie (Yki), the ortholog of mammalian Yes-associated protein (Yap), lead to tissue overgrowth [74]. However, it is not known how these genes influence cell growth and whether this occurs through signaling to the class I PI3K pathway. Significantly, warts (wts) is required for growth arrest and the induction of autophagy in dying salivary glands [75]. Mutations in wts, as well as knockdown of wts pathway components hpo, sav and mats, prevent salivary gland degradation. Surprisingly, however, over-expression of Yki in salivary glands does not phenocopy wts mutations, suggesting that in salivary glands, Wts signals independently of Yki. Significantly, wts mutants have altered class I PI3K markers and require the function of TOR and the insulin receptor substrate chico to inhibit salivary gland degradation [75]. Therefore, Wts influences PI3K signaling in salivary glands. These data provide mechanistic information about how cell growth is regulated during salivary gland cell death. It will be interesting to see whether Wts regulates cell growth in a PI3K-dependent manner in other systems.
Autophagy and Cell Death
Programmed cell death is a conserved and genetically regulated process that plays important roles throughout the lives of metazoans[76]. Cell death functions in the formation and shaping of structures during development, and in tissue homeostasis during adulthood. In 1973, Schweichel & Merker described three prominent types of programmed cell death that occur in developing mammalian cells and are distinguished by dying cell morphology and use of the lysosome [77, 78]. Type I cell death, or apoptosis, occurs in cells that die in isolation from one another, and is characterized by the condensation of nuclei and cytoplasm, DNA fragmentation, and phagocytosis by a secondary cell where the lysosome of the engulfing cell degrades the dead cell [79]. In type II, or autophagic cell death, cells die in groups, and cell destruction occurs with little or no phagocytosis. Type II cell death is characterized by the presence of autophagosomes that degrade cytoplasmic contents of the dying cell. The third and least common type, non-lysosomal cell death, is characterized by the lack of known lysosomal activity in the dying cell.
Type II cell death is observed at several stages during mammalian development, including regression of the corpeus luteum, involution of mammary and prostate glands, and regression of Mullerian duct structures during male genital development [78]. Type II cell death has also been observed during insect development, for example in dying flight muscle cells of the Hawkmoth Manduca sexta after which the term programmed cell death was originally coined [76]. Drosophila larval salivary glands that die during metamorphosis also exhibit Type II morphology; autophagosomes are present in the cytoplasm, and phagocytosis is not observed [46, 80]. While the presence of autophagosomes in cells that die by type II morphology has been well documented, the role of autophagy in cell death is controversial [81, 82].
Several studies demonstrate a role for autophagy in mammalian development [83]. atg5, as well as atg7, null mutant mice exhibit few developmental defects at birth, but die within 1 day of delivery, and when un-suckled, die earlier than wild-type mice [69, 70]. Therefore in this context, autophagy is required to generate the energy necessary to survive nutrient deficiency. Autophagy may also play a role in the context of cell death that occurs during mammalian development. A study utilizing an in vitro mouse stem cell model of embryogenesis demonstrated that null atg mutant embryonic stem cells are not deficient in the activation of programmed cell death, but fail to generate and display signals that are required for phagocytosis of dying cells [84]. Significantly, addition of methylpyruvate was sufficient to rescue atg mutant engulfment and clearance defects, indicating that the role of autophagy is to provide the metabolic substrates needed to complete this process. A subsequent study reported similar findings in the context of cell death that occurs in developing chick retina [85]. When autophagy was inhibited during retinal development, apoptotic cells accumulated, and this accumulation correlated with a deficiency in phosphatidylserine exposure [85]. Thus, these studies suggest that in the context of cell death that occurs during mammalian development, autophagy deficiency is associated with a failure in phagocytic clearance of apoptotic cells. Beyond these studies, little is known about the role of autophagy in developmental cell death in mammalian systems, and the cumbersome nature of mammalian genetics makes such in vivo studies difficult.
Schweichel & Merker’s observations of dying cell morphologies suggested that autophagy is induced in and promotes the death of certain types of dying cells [77]. We noted that autophagy and autophagy genes are induced in dying larval salivary gland cells [46, 58], suggesting that this process promotes cell death, but it was not until recently that this has been rigorously tested using genetic approaches. The availability of several in vivo modelsfor cell death in Drosophila has made it possible to directly test the role of autophagy in cell death. Several Drosophila studies demonstrate a positive role for autophagy in cell death. Atg1 over-expression in the Drosophila larval fat body is sufficient to induce autophagy and cell death in a caspase-dependent manner [11]. Mutations in atg8 or atg18, or decreased function of atg1, in addition to a number of other atg genes, all lead to the incomplete destruction of larval salivary glands, structures that die with Type II morphology [73]. Furthermore, knockdown of atg genes specifically in salivary gland cells leads to incomplete gland destruction, indicating that the role of autophagy occurring within these dying cells is tissue-autonomous [73]. However, in contrast to the results in the fat body [11], Atg1-overexpression in salivary glands induces cell death in a caspase-independent manner.
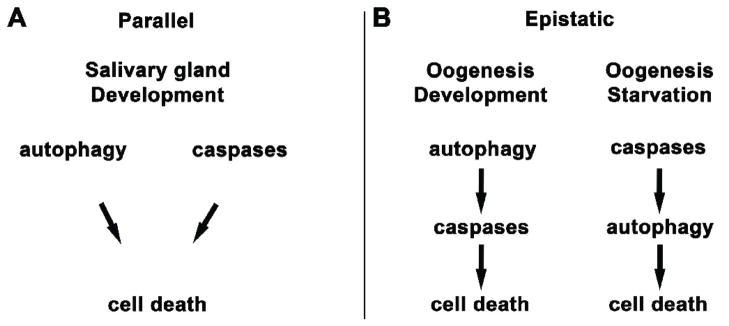
The relationship between autophagy and caspases may be context-specific. Several studies have suggested a synergistic role for autophagy and caspases in cell death that occurs during oogenesis in several Drosophila and higher Dipteran species [86–91]. Two recent studies suggest that there is an epistatic relationship between caspases and autophagy during oogenesis, however their findings differ in important ways (Figure 1) [92, 93]. Hou et al recently demonstrated that in germaria and midstage egg chambers, starvation-induced autophagy leads to degeneration [92]. The effector caspase DCP-1 and IAP protein Bruce are required for autophagy induction in these degenerating egg chambers in response to starvation [92]. Therefore this study suggests that caspases function upstream of starvation-induced autophagy and cell death in the ovary [92]. In contrast, Nezis et al recently demonstrated that during developmentally-induced cell death that occurs during germarium development and mid-oogenesis, both processed caspase3-like expression and autophagy induction occur, however mutations in autophagy genes result in reduced caspase activation [93]. Therefore in this case, developmentally-induced autophagy is required for caspase activation in dying female germline cells [93]. Thus the mechanisms underlying the hierarchical relationship between autophagy and caspases in cells that die during oogenesis remain to be resolved.
Fig. 1.
The relationship between autophagy and caspases during cell death may be context specific. A: During metamorphosis, Drosophila larval salivary glands undergo developmental programmed cell death. During destruction of salivary glands, both autophagy genes and caspases are required for complete degradation, and the combined inhibition of autophagy and caspases enhances this incomplete destruction [73, 94]. In addition, caspases are activated in autophagy mutants and autophagy occurs when caspases are inhibited, indicating that autophagy and caspases function in parallel pathways. B: During Drosophila oogenesis, starvation-induced autophagy leads to degeneration of egg chambers, and effector caspase DCP-1 and IAP protein Bruce are required for autophagy induction [92]. During developmentally-induced cell death that occurs during oogenesis, however, mutations in autophagy genes result in reduced caspase activation [93]. These studies indicate that autophagy and caspases function in an epistatic regulatory hierarchy.
Several studies also indicate that caspases and autophagy function in parallel in the destruction of certain tissues during Drosophila development (Figure 1). During the destruction of salivary glands, both autophagy and caspase activation occur, however, expression of the caspase inhibitor p35 only partially prevents salivary gland degradation [46]. In Drosophiladark (Apaf-1) mutants, salivary gland degradation is incomplete, but autophagy occurs normally, suggesting a role for this caspase regulator in type II cell death downstream or parallel to autophagy [94]. Significantly, Berry & Baehrecke demonstrate that the combined inhibition of autophagy and caspases enhances the incomplete destruction of salivary glands [73]. A recent study by Mohseni et al find that the destruction of the Drosophila amnioserosa, an extra-embryonic membrane that is eliminated late in embryogenesis, requires both autophagy and caspase activation [95]. It is not clear in this study whether the relationship between caspases and autophagy is either parallel or epistatic, but this finding provides additional support for the hypothesis that caspases and autophagy function cooperatively in the destruction of tissues during cell death. A similar process may occur in some types of mammalian cells, given that mammalian 3-dimensional cultures of MCF-10A mammary cells die with similar characteristics [96, 97]. Studies of MCF-10A mammary epithelial cell lines, a model for mammary lumen formation, implicate both autophagy and caspases in the elimination of cells during lumen formation [96, 97]. Suppression of either caspase activity or autophagy does not prevent lumen formation, whereas inhibition of both prevents cell elimination [97].
Although genetic evidence now indicates that autophagy contributes to cell death in some in vivo contexts [73, 92, 93, 95], how autophagy may function in cell death is not completely clear. One possibility is that autophagy is required to generate the metabolic substrates that are required for cell autonomous destruction. It is also possible that autophagy functions to specifically deplete a cell survival factor as has been shown in mammalian cells [98], and with accumulating models in which autophagy functions in physiological cell death, Drosophila may provide the best system to discover such a factor. Alternatively, autophagy may deplete critical resources, such as substrates for metabolism and mitochondria, which are required for cell survival, and this may lead to cell death.
Autophagy and Disease Models in Drosophila
Drosophila provides an ideal model system for studies of genes that have been implicated in human disease. More than 60% of the genes implicated in human disease have orthologs in flies, and studies in fly disease models have identified novel factors that regulate disease [99, 100]. Whereas knowledge of a gene associated with a human disorder may initially be limited to a genetic locus, the ease of fly genetics, the ability to generate mutants, knockdown or over-express genes of interest make it possible to identify the function of such genes in flies. In addition, many signaling pathways are highly conserved between flies and humans. Therefore, the use of genetic modifier screens in flies, to look for mutations that enhance or suppress a mutant phenotype, makes it possible to quickly place a gene in the context of a genetic pathway and identify novel factors not previously associated with a disease [99, 100].
Autophagy has been implicated in aging, protein aggregate disorders, neurodegeneration, immunity, and cancer [6–8]. While there are no cancer models in Drosophila, fly atg7 and atg8 mutants, like atg mutant mice, have a decreased life span and exhibit neurodegenerative defects [11, 50, 52, 101, 102]. Autophagy is also required for Drosophila innate immune responses [103]. Fly disease models have proven useful in identifying new factors that may regulate disease, and have also provided mechanistic evidence for how autophagy is regulated in the context of neurodegenerative disorders, immune responses, and aging.
Protein Aggregate Disorders
The accumulation of protein aggregates in degenerating neurons is associated with many human neurodegenerative diseases [6]. These protein aggregates are often ubiquitin-positive, suggesting defects in protein degradation by the UPS, and a number of studies have shown that the UPS is impaired in neurodegenerative disorders. However, an increase in the number of autophagosomes in degenerating neurons has also been noted in patients with many polyglutamine diseases, as well as Alzheimer’s and Parkinson’s. Autophagy gene mutant phenotypes also suggest a role for autophagy in aggregate protein disorders and neurodegeneration. atg7 conditional knockout mice are impaired in autophagosome formation, and accumulate ubiquitin-positive aggregates [70]. Loss of either atg5 or atg7 specifically in neurons leads to the accumulation of ubiquitin-positive aggregates and neurodegeneration in mice [101, 102]. Drosophilaatg7 and atg8 null mutants also exhibit the accumulation of ubiquitin-positive aggregates in degenerating neurons [50, 51, 104].
Although an increase in the number of autophagosomes is associated with neurodegeneration, whether autophagy functions as a cytoprotective or as a cell death mechanism in this context has been a subject of debate. In both Drosophila and mouse models of Huntington disease, TOR inhibition induced autophagy and led to a decrease in huntingtin protein accumulation and cell death, whereas autophagy inhibition enhanced polyglutamine toxicity [105]. In a Drosophila model of the neurodegenerative disease spinobulbar muscular atrophy (SBMA), polyglutamine expansion led to cell degeneration and atg gene-knockdown enhanced this degeneration [106]. Conversely, induction of autophagy via rapamycin treatment in this study suppressed cell degeneration [106]. Thus, these studies suggest that autophagy plays a cytoprotective role in the context of aggregate protein disorders.
While protein aggregate disorders are characterized by defects in the UPS and autophagy, these two catabolic systems have typically been thought to act independently of one another. However recent studies suggest that there may be a relationship between the UPS and autophagy, and several proteins have been implicated in a possible mechanism linking these processes. In the Drosophila model for SBMA mentioned above, genetic impairment of the proteasome led to the induction of autophagy [106]. Furthermore, this study demonstrated that the microtubule-associated protein Histone Deacetylase 6 (HDAC6), previously shown to interact with polyubiquitinated proteins [107], is required for autophagy induction upon proteasome impairment [106]. In addition, expression of HDAC6 during proteasome impairment was sufficient to rescue cell degeneration in an autophagy-dependent manner.
Other proteins have also been implicated in providing a link between autophagy and the UPS. p62/Sequestosome 1 (SQSTM1) binds both poly-ubiquitin and atg8/LC3, and loss of p62 leads to an increase in huntingtin-induced cell death in cell lines [108]. In autophagy-deficient mice, p62 is required for the formation of ubiquitin-positive protein aggregates [109]. Similarly, the Drosophila p62 ortholog Ref(2)P is required for ubiquitin-positive aggregate formation in atg8 mutants [51]. In addition, autophagy-linked FYVE protein (Alfy) localizes to ubiquitinated protein aggregates and autophagosomes, and mutations in its Drosophila homolog, blue cheese, lead to an accumulation of ubiquitin-positive aggregates in neurons [110, 111]. In line with these findings, Drosophila null mutations in vps15, the Vps34/Class III PI3K regulatory kinase required for autophagy, exhibit ubiquitin- and Ref(2)P-positive aggregates [112].
Studies in Drosophila also suggest a role for endosomal biogenesis genes in aggregate protein disorders. Mutations in escrt genes are linked to several human neurodegenerative disorders, including frontotemporal dementia and amyotrophic lateral sclerosis [113, 114]. In a Drosophila model for Huntington disease, reducing the genetic dosage of ESCRT proteins, or expressing a dominant-negative form of the ESCRT regulatory ATPase Vps4, led to the accumulation of autophagosomes and ubiquitin-positive aggregates [62]. In ESCRT-depleted HeLa cells, ubiquitin-positive aggregates also contained p62 and Alfy [115]. Rab5, a gene known to be involved in endocytosis, was also shown to be required for autophagosome formation, and inhibition of Rab5 enhanced polyglutamine toxicity in a Drosophila model of Huntington disease [116].
Autophagy and Aging
Drosophilaatg7 and atg8 mutants have a decreased lifespan and are sensitive to oxidative stress, demonstrating a role for autophagy in aging [50, 52]. In Drosophila, autophagy gene expression is reduced with age [52]. Conversely, Atg8 over-expression in adult neurons results in an increase in resistance to oxidative stress, a process that has been linked to aging [117], and the extension of lifespan [52]. Further, atg7 conditional knockout mice accumulate peroxisomes in their livers and exhibit oxidative stress, demonstrating a direct role for autophagy in peroxisomal degradation in mammals [118].
Evidence in mice suggests that the relationship between autophagy and aging involves chaperone-mediated autophagy (CMA) [119]. CMA is a selective degradation process in which soluble proteins are delivered directly to the lysosome [2]. Like macroautophagy, CMA is activated in response to oxidative stress [120, 121]. In addition, LAMP2A, a receptor required for CMA, decreases with age due to a loss in LAMP2A protein stability [122, 123]. Furthermore, inducing the expression of LAMP2A in the livers of transgenic mice increases the levels of CMA, and these mice accumulate fewer damaged proteins in their livers [119]. CMA has not been investigated in Drosophila, and it would be useful to apply the strength of fly genetics to this important catabolic process.
Autophagy and Immunity
Evidence suggests that autophagy plays important roles in mammalian innate and adaptive immunity in the elimination of pathogens, and antigen processing and presentation [8, 124]. Autophagy is important during T-cell selection to generate self-tolerance, and autophagy deficiency leads to multi-organ inflammation [125]. Furthermore, mutations in atg16 have been linked to inflammatory bowel disease in humans and mice [126, 127]. Autophagy is also important as a host-defense mechanism to clear intracellular pathogens [128–132], and some invading pathogens and viruses utilize or inhibit the autophagic machinery to evade host-defense mechanisms [133–135]. During M. tuberculosis infection, autophagy is induced, and this induction requires the immunity-related GTPase family member IRGM [129, 136]. However, mechanistic evidence demonstrating how autophagy is regulated in the context of immune signaling has been lacking.
Drosophila lack adaptive immunity, but can mount innate immune responses. Recent in vivo studies in Drosophila have helped to elucidate the mechanism of autophagy induction during pathogenic infection. Yano et al found that Drosophila resistance to the gram-positive bacteria L. monocytogenes requires the pattern-recognition receptor PGRP-LE, a member of a family of receptors that recognize bacterial peptidoglycans, induce antimicrobial peptide (AMP) genes, and activate the innate immune signaling IMD and Toll pathways [137]. Significantly, Yano et al showed that autophagy in blood cells is necessary to inhibit the intracellular growth of L. monocytogenes, and to survive infection. Further, the PGRP-LE receptor is required for the induction of autophagy following L. monocytogenes infection [137]. However, autophagy induction in this case is independent of the Toll and IMD immune signaling pathways, suggesting the possibility that the PGRP-LE receptor may signal through an unknown factor to induce autophagy [137].
Another recent Drosophila study has provided new information about the crosstalk between autophagy and innate immune signaling pathways. A screen for genes required for Drosophila innate immune responses to bacterial infection identified the gene immune response-deficient 1 (ird1), the Drosophila homolog of the Vps34/Class III PI3K regulator, vps15 [138]. AMP genes are induced during starvation [139], but ird1 mutants fail to express AMP genes upon infection [138]. Furthermore, ird1 mutants cannot activate the IMD pathway, demonstrating that ird1/vps15 is an important regulator of the innate immune response [138]. In addition, this study showed that if wild-type flies are starved during bacterial infection, the expression of AMP genes is suppressed, and these flies exhibit decreased survival upon infection, suggesting that autophagy plays an important role in modulating nutrient sensing and immune signaling [138]. This result provides insight into studies suggesting that in human and mouse populations, malnutrition leads to an increased susceptibility to infections [140–142].
Conclusions
Drosophila is well-suited to in vivo studies of autophagy and its relationship to cell survival, growth, and death in the context of a developing multi-cellular organism. Drosophila research has produced many new discoveries about the role and regulation of autophagy during development, aging, innate immunity, and in the context of diseases such as protein aggregate disorders and neurodegeneration. Genetic screens for genes that are required for autophagy in Drosophila have yielded new discoveries [143, 144], but additional screens are needed to identify the genes that regulate autophagy in the context of nutrient deprivation, pathogenic invasion, and other processes. Importantly, these screens should provide key mechanistic information about the regulation of autophagy, and it is likely that such genes will have conserved functions in mammals.
Autophagy is induced in Drosophila upon starvation in tissues such as the fat body, muscle, and ovaries. Autophagy is also induced in response to the steroid hormone ecdysone during metamorphosis in tissues such as the fat body, intestine, and salivary glands. This allows studies to be carried out in different cell types and in response to different stimuli. Autophagy regulation may differ in the contexts of nutrient deprivation compared to ecdysone induction, in the context of cell survival versus cell death, or in response to different pathogens. Thus, there may be a cell-context specific component to the proper regulation of autophagy, and it is possible that as-yet undiscovered regulators that influence the induction of autophagy in response to different stimuli, and/or in specific cell-types, may exist, and flies are well-suited to such discoveries.
Acknowledgments
We thank the NIH (GM59136 and GM079431) for support.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Klionsky DJ. The molecular machinery of autophagy: unanswered questions. J Cell Sci. 2005;118:7–18. doi: 10.1242/jcs.01620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cuervo AM. Autophagy and aging: keeping that old broom working. Trends Genet. 2008;24:604–612. doi: 10.1016/j.tig.2008.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xie Z, Klionsky DJ. Autophagosome formation: core machinery and adaptations. Nat Cell Biol. 2007;9:1102–1109. doi: 10.1038/ncb1007-1102. [DOI] [PubMed] [Google Scholar]
- 4.Mizushima N. Autophagy: process and function. Genes Dev. 2007;21:2861–2873. doi: 10.1101/gad.1599207. [DOI] [PubMed] [Google Scholar]
- 5.Lum JJ, DeBerardinis RJ, Thompson CB. Autophagy in metazoans: cell survival in the land of plenty. Nat Rev Mol Cell Biol. 2005;6:439–448. doi: 10.1038/nrm1660. [DOI] [PubMed] [Google Scholar]
- 6.Nedelsky NB, Todd PK, Taylor JP. Autophagy and the ubiquitin-proteasome system: Collaborators in neuroprotection. Biochim Biophys Acta. 2008;12:691–699. doi: 10.1016/j.bbadis.2008.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mathew R, Karantza-Wadsworth V, White E. Role of autophagy in cancer. Nat Rev Cancer. 2007;7:961–967. doi: 10.1038/nrc2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–1075. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Meléndez A, Neufeld TP. The cell biology of autophagy in metazoans: a developing story. Development. 2008;135:2347–2360. doi: 10.1242/dev.016105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pattingre S, Espert L, Biard-Piechaczyk M, Codogno P. Regulation of macroautophagy by mTOR and Beclin 1 complexes. Biochimie. 2007;2:313–323. doi: 10.1016/j.biochi.2007.08.014. [DOI] [PubMed] [Google Scholar]
- 11.Scott RC, Juhász G, Neufeld TP. Direct induction of autophagy by Atg1 inhibits cell growth and induces apoptotic cell death. Curr Biol. 2007;17:1–11. doi: 10.1016/j.cub.2006.10.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kamada Y, Funakoshi T, Shintani T, Nagano K, Ohsumi M, Ohsumi Y. Tor-mediated induction of autophagy via an Apg1 protein kinase complex. J Cell Biol. 2000;150:1507–1513. doi: 10.1083/jcb.150.6.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Scott SV, Nice DC, 3rd, Nau JJ, Weisman LS, Kamada Y, Keizer-Gunnink I, Funakoshi T, Veenhuis M, Ohsumi Y, Klionsky DJ. Apg13p and Vac8p are part of a complex of phosphoproteins that are required for cytoplasm to vacuole targeting. J Biol Chem. 2000;275:25840–25849. doi: 10.1074/jbc.M002813200. [DOI] [PubMed] [Google Scholar]
- 14.Suzuki K, Kirisako T, Kamada Y, Mizushima N, Noda TOY. The pre-autophagosomal structure organized by concerted functions of APG genes is essential for autophagosome formation. EMBO J. 2001;20:5971–5981. doi: 10.1093/emboj/20.21.5971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kihara A, Noda T, Ishihara N, Ohsumi Y. Two distinct Vps34 phosphatidylinositol 3-kinase complexes function in autophagy and carboxypeptidase Y sorting in Saccharomyces cerevisiae. J Cell Biol. 2001;152:519–530. doi: 10.1083/jcb.152.3.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kametaka S, Okano T, Ohsumi M, Ohsumi Y. Apg14p and Apg6/Vps30p form a protein complex essential for autophagy in the yeast, Saccharomyces cerevisiae. J Biol Chem. 1998;273:22284–22291. doi: 10.1074/jbc.273.35.22284. [DOI] [PubMed] [Google Scholar]
- 17.Liang C, Feng P, Ku B, Dotan I, Canaani D, Oh BH, Jung JU. Autophagic and tumour suppressor activity of a novel Beclin1-binding protein UVRAG. Nat Cell Biol. 2006;8:688–699. doi: 10.1038/ncb1426. [DOI] [PubMed] [Google Scholar]
- 18.Fimia GM, Stoykova A, Romagnoli A, Giunta L, Di Bartolomeo S, Nardacci R, Corazzari M, Fuoco C, Ucar A, Schwartz P, Gruss P, Piacentini M, Chowdhury K, Cecconi F. Ambra1 regulates autophagy and development of the nervous system. Nature. 2007;447:1121–1125. doi: 10.1038/nature05925. [DOI] [PubMed] [Google Scholar]
- 19.Takahashi Y, Coppola D, Matsushita N, Cualing HD, Sun M, Sato Y, Liang C, Jung JU, Cheng JQ, Mul JJ, Pledger WJ, Wang HG. Bif-1 interacts with Beclin 1 through UVRAG and regulates autophagy and tumorigenesis. Nat Cell Biol. 2007;9:1142–1151. doi: 10.1038/ncb1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sun Q, Fan W, Chen K, Ding X, Chen S, Zhong Q. Identification of Barkor as a mammalian autophagy-specific factor for Beclin 1 and class III phosphatidylinositol 3-kinase. Proc Natl Acad Sci U S A. 2008;105:19211–19216. doi: 10.1073/pnas.0810452105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Itakura E, Kishi C, Inoue K, Mizushima N. Beclin 1 Forms Two Distinct Phosphatidylinositol 3-Kinase Complexes with Mammalian Atg14 and UVRAG. Mol Biol Cell. 2008;12:5360–5372. doi: 10.1091/mbc.E08-01-0080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Juhasz G, Neufeld TP. Autophagy: a forty-year search for a missing membrane source. PLoS Biol. 2006;4:e36. doi: 10.1371/journal.pbio.0040036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Axe EL, Walker SA, Manifava M, Chandra P, Roderick HL, Habermann A, Griffiths G, Ktistakis NT. Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. J Cell Biol. 2008;182:685–701. doi: 10.1083/jcb.200803137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dunn WAJ. Studies on the mechanisms of autophagy: formation of the autophagic vacuole. J Cell Biol. 1990;110:1923–1933. doi: 10.1083/jcb.110.6.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ohsumi Y. Molecular dissection of autophagy: two ubiquitin-like systems. Nature Reviews Mol Cell Biol. 2001;2:211–216. doi: 10.1038/35056522. [DOI] [PubMed] [Google Scholar]
- 26.Reggiori F, Klionsky DJ. Autophagy in the eukaryotic cell. Eukaryot Cell. 2002;1:11–21. doi: 10.1128/EC.01.1.11-21.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mizushima N, Noda T, Yoshimori T, Tanaka Y, Ishii T, George MD, Klionsky DJ, Ohsumi M, Ohsumi Y. A protein conjugation system essential for autophagy. Nature. 1998;395:395–398. doi: 10.1038/26506. [DOI] [PubMed] [Google Scholar]
- 28.Mizushima N, Noda T, Ohsumi Y. Apg16p is required for the function of the Apg12p-Apg5p conjugate in the yeast autophagy pathway. Embo J. 1999;18:3888–3896. doi: 10.1093/emboj/18.14.3888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tanida I, Mizushima N, Kiyooka M, Ohsumi M, Ueno T, Ohsumi Y, Kominami E. Apg7p/Cvt2p: A novel protein-activating enzyme essential for autophagy. Mol Biol Cell. 1999;10:1367–1379. doi: 10.1091/mbc.10.5.1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shintani T, Mizushima N, Ogawa Y, Matsuura A, Noda T, Ohsumi Y. Apg10p, a novel protein-conjugating enzyme essential for autophagy in yeast. EMBO J. 1999;18:5234–5241. doi: 10.1093/emboj/18.19.5234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kuma A, Mizushima N, Ishihara N, Ohsumi Y. Formation of the approximately 350-kDa Apg12-Apg5.Apg16 multimeric complex, mediated by Apg16 oligomerization, is essential for autophagy in yeast. J Biol Chem. 2002;277:18619–18625. doi: 10.1074/jbc.M111889200. [DOI] [PubMed] [Google Scholar]
- 32.Kirisako T, Baba M, Ishihara N, Miyazawa K, Ohsumi M, Yoshimori T, Noda T, Ohsumi Y. Formation process of autophagosome is traced with Apg8/Aut7p in yeast. J Cell Biol. 1999;147:435–446. doi: 10.1083/jcb.147.2.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huang WP, Scott SV, Kim J, Klionsky DJ. The itinerary of a vesicle component, Aut7p/Cvt5p, terminates in the yeast vacuole via the autophagy/Cvt pathways. J Biol Chem. 2000;275:5845–5851. doi: 10.1074/jbc.275.8.5845. [DOI] [PubMed] [Google Scholar]
- 34.Lang T, Schaeffeler E, Bernreuther D, Bredschneider M, Wolf DH, Thumm M. Aut2p and Aut7p, two novel microtubule-associated proteins are essential for delivery of autophagic vesicles to the vacuole. Embo J. 1998;17:3597–3607. doi: 10.1093/emboj/17.13.3597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ichimura Y, Kirisako T, Takao T, Satomi Y, Shimonishi Y, Ishihara N, Mizushima N, Tanida I, Kominami E, Ohsumi M, Noda T, Ohsumi Y. A ubiquitin-like system mediates protein lipidation. Nature. 2000;408:488–492. doi: 10.1038/35044114. [DOI] [PubMed] [Google Scholar]
- 36.Kabeya Y, Mizushima N, Ueno T, Yamamoto A, Kirisako T, Noda T, Kominami E, Ohsumi Y, Yoshimori T. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 2000;19:5720–5728. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kirisako T, Ichimura Y, Okada H, Kabeya Y, Mizushima N, Yoshimori T, Ohsumi M, Takao T, Noda T, Ohsumi Y. The reversible modification regulates the membrane-binding state of Apg8/Aut7 essential for autophagy and the cytoplasm to vacuole targeting pathway. J Cell Biol. 2000;151:263–276. doi: 10.1083/jcb.151.2.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nakatogawa H, Ichimura Y, Ohsumi Y. Atg8, a ubiquitin-like protein required for autophagosome formation, mediates membrane tethering and hemifusion. Cell. 2007;130:165–178. doi: 10.1016/j.cell.2007.05.021. [DOI] [PubMed] [Google Scholar]
- 39.Klionsky DJ, Abeliovich H, Agostinis P, Agrawal DK, Aliev G, Askew DS, Baba M, Baehrecke EH, Bahr BA, Ballabio A, Bamber BA, Bassham DC, Bergamini E, Bi X, Biard-Piechaczyk M, Blum JS, Bredesen DE, Brodsky JL, Brumell JH, Brunk UT, Bursch W, Camougrand N, Cebollero E, Cecconi F, Chen Y, Chin LS, Choi A, Chu CT, Chung J, Clarke PG, Clark RS, Clarke SG, Clavé C, Cleveland JL, Codogno P, Colombo MI, Coto-Montes A, Cregg JM, Cuervo AM, Debnath J, Demarchi F, Dennis PB, Dennis PA, Deretic V, Devenish RJ, Di Sano F, Dice JF, Difiglia M, Dinesh-Kumar S, Distelhorst CW, Djavaheri-Mergny M, Dorsey FC, Dröge W, Dron M, Dunn WAJ, Duszenko M, Eissa NT, Elazar Z, Esclatine A, Eskelinen EL, Fésüs L, Finley KD, Fuentes JM, Fueyo J, Fujisaki K, Galliot B, Gao FB, Gewirtz DA, Gibson SB, Gohla A, Goldberg AL, Gonzalez R, González-Estévez C, Gorski S, Gottlieb RA, Häussinger D, He YW, Heidenreich K, Hill JA, Høyer-Hansen M, Hu X, Huang WP, Iwasaki A, Jäättelä M, Jackson WT, Jiang X, Jin S, Johansen T, Jung JU, Kadowaki M, Kang C, Kelekar A, Kessel DH, Kiel JA, Kim HP, Kimchi A, Kinsella TJ, Kiselyov K, Kitamoto K, Knecht E, Komatsu M, Kominami E, Kondo S, Kovács AL, Kroemer G, Kuan CY, Kumar R, Kundu M, Landry J, Laporte M, Le W, Lei HY, Lenardo MJ, Levine B, Lieberman A, Lim KL, Lin FC, Liou W, Liu LF, Lopez-Berestein G, López-Otín C, Lu B, Macleod KF, Malorni W, Martinet W, Matsuoka K, Mautner J, Meijer AJ, Meléndez A, Michels P, Miotto G, Mistiaen WP, Mizushima N, Mograbi B, Monastyrska I, Moore MN, Moreira PI, Moriyasu Y, Motyl T, Münz C, Murphy LO, Naqvi NI, Neufeld TP, Nishino I, Nixon RA, Noda T, Nürnberg B, Ogawa M, Oleinick NL, Olsen LJ, Ozpolat B, Paglin S, Palmer GE, Papassideri I, Parkes M, Perlmutter DH, Perry G, Piacentini M, Pinkas-Kramarski R, Prescott M, Proikas-Cezanne T, Raben N, Rami A, Reggiori F, Rohrer B, Rubinsztein DC, Ryan KM, Sadoshima J, Sakagami H, Sakai Y, Sandri M, Sasakawa C, Sass M, Schneider C, Seglen PO, Seleverstov O, Settleman J, Shacka JJ, Shapiro IM, Sibirny A, Silva-Zacarin EC, Simon HU, Simone C, Simonsen A, Smith MA, Spanel-Borowski K, Srinivas V, Steeves M, Stenmark H, Stromhaug PE, Subauste CS, Sugimoto S, Sulzer D, Suzuki T, Swanson MS, Tabas I, Takeshita F, Talbot NJ, Tallóczy Z, Tanaka K, Tanaka K, Tanida I, Taylor GS, Taylor JP, Terman A, Tettamanti G, Thompson CB, Thumm M, Tolkovsky AM, Tooze SA, Truant R, Tumanovska LV, Uchiyama Y, Ueno T, Uzcátegui NL, van der Klei I, Vaquero EC, Vellai T, Vogel MW, Wang HG, Webster P, Wiley JW, Xi Z, Xiao G, Yahalom J, Yang JM, Yap G, Yin XM, Yoshimori T, Yu L, Yue Z, Yuzaki M, Zabirnyk O, Zheng X, Zhu X, Deter RL. Guidelines for the use and interpretation of assays for monitoring autophagy in higher eukaryotes. Autophagy. 2008;4:151–175. doi: 10.4161/auto.5338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Harding TM, Hefner-Gravink A, Thumm M, Klionsky DJ. Genetic and phenotypic overlap between autophagy and the cytoplasm to vacuole protein. J Biol Chem. 1996;271:17621–17624. doi: 10.1074/jbc.271.30.17621. [DOI] [PubMed] [Google Scholar]
- 41.Thumm M, Egner R, Koch B, Schlumpberger M, Straub M, Veenhuis M, Wolf DH. Isolation of autophagocytosis mutants of Saccharomyces cerevisiae. FEBS Lett. 1994;349:275–280. doi: 10.1016/0014-5793(94)00672-5. [DOI] [PubMed] [Google Scholar]
- 42.Harding TM, Morano KA, Scott SV, Klionsky DJ. Isolation and characterization of yeast mutants in the cytoplasm to vacuole protein targeting pathway. J Cell Bio. 1995;131:591–602. doi: 10.1083/jcb.131.3.591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tsukada M, Ohsumi Y. Isolation and characterization of autophagy-defective mutants of Saccharomyces cerevisiae. FEBS Lett. 1993;1–2:169–174. doi: 10.1016/0014-5793(93)80398-e. [DOI] [PubMed] [Google Scholar]
- 44.Baehrecke EH. Autophagic programmed cell death in Drosophila. Cell Death & Differ. 2003;10:940–945. doi: 10.1038/sj.cdd.4401280. [DOI] [PubMed] [Google Scholar]
- 45.Rusten TE, Lindmo K, Juhasz G, Sass M, Seglen PO, Brech A, Stenmark H. Programmed autophagy in the Drosophila fat body is induced by ecdysone through regulation of the PI3K pathway. Dev Cell. 2004;7:179–192. doi: 10.1016/j.devcel.2004.07.005. [DOI] [PubMed] [Google Scholar]
- 46.Lee CY, Baehrecke EH. Steroid regulation of autophagic programmed cell death during development. Development. 2001;128:1443–1455. doi: 10.1242/dev.128.8.1443. [DOI] [PubMed] [Google Scholar]
- 47.Lee CY, Cooksey BAK, Baehrecke EH. Steroid regulation of midgut cell death during Drosophila development. Dev Biol. 2002;250:101–111. doi: 10.1006/dbio.2002.0784. [DOI] [PubMed] [Google Scholar]
- 48.Neufeld TP, Baehrecke EH. Eating on the fly: function and regulation of autophagy during cell growth, survival and death in Drosophila. Autophagy. 2008;4:557–562. doi: 10.4161/auto.5782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Scott RC, Schuldiner ONTP. Role and regulation of starvation-induced autophagy in the Drosophila fat body. Dev Cell. 2004;7:167–178. doi: 10.1016/j.devcel.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 50.Juhász G, Erdi B, Sass M, Neufeld TP. Atg7-dependent autophagy promotes neuronal health, stress tolerance, and longevity but is dispensable for metamorphosis in Drosophila. Genes Dev. 2007;21:3061–3066. doi: 10.1101/gad.1600707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nezis IP, Simonsen A, Sagona AP, Finley K, Gaumer S, Contamine D, Rusten TE, Stenmark H, Brech A. Ref(2)P, the Drosophila melanogaster homologue of mammalian p62, is required for the formation of protein aggregates in adult brain. J Cell Biol. 2008;180:1065–1071. doi: 10.1083/jcb.200711108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Simonsen A, Cumming RC, Brech A, Isakson P, Schubert DR, Finley KD. Promoting basal levels of autophagy in the nervous system enhances longevity and oxidant resistance in adult Drosophila. Autophagy. 2008;4:176–184. doi: 10.4161/auto.5269. [DOI] [PubMed] [Google Scholar]
- 53.Conlon I, Raff M. Size control in animal development. Cell. 1999;96:235–244. doi: 10.1016/s0092-8674(00)80563-2. [DOI] [PubMed] [Google Scholar]
- 54.Arsham AM, Neufeld TP. Thinking globally and acting locally with TOR. Curr Opin Cell Biol. 2006;18:589–597. doi: 10.1016/j.ceb.2006.09.005. [DOI] [PubMed] [Google Scholar]
- 55.King-Jones K, Thummel CS. Developmental biology. Less steroids make bigger flies. Science. 2005;310:630–631. doi: 10.1126/science.1120410. [DOI] [PubMed] [Google Scholar]
- 56.Colombani J, Bianchini L, Layalle S, Pondeville E, Dauphin-Villemant C, Antoniewski C, Carre C, Noselli S, Leopold P. Antagonistic actions of ecdysone and insulins determine final size in Drosophila. Science. 2005;310:667–670. doi: 10.1126/science.1119432. [DOI] [PubMed] [Google Scholar]
- 57.Layalle S, Arquier N, Leopold P. The TOR pathway couples nutrition and developmental timing in Drosophila. Dev Cell. 2008;15:568–577. doi: 10.1016/j.devcel.2008.08.003. [DOI] [PubMed] [Google Scholar]
- 58.Lee CY, Clough EA, Yellon P, Teslovich TM, Stephan DA, Baehrecke EH. Genome-wide analyses of steroid- and radiation-triggered programmed cell death in Drosophila. Curr Biol. 2003;13:350–357. doi: 10.1016/s0960-9822(03)00085-x. [DOI] [PubMed] [Google Scholar]
- 59.Lippai M, Csikos G, Maroy P, Lukacsovich T, Juhasz G, Sass M. SNF4Agamma, the Drosophila AMPK gamma subunit is required for regulation of developmental and stress-induced autophagy. Autophagy. 2008;4:476–486. doi: 10.4161/auto.5719. [DOI] [PubMed] [Google Scholar]
- 60.Liang J, Shao SH, Xu ZX, Hennessy B, Ding Z, Larrea M, Kondo S, Dumont DJ, Gutterman JU, Walker CL, Slingerland JM, Mills GB. The energy sensing LKB1-AMPK pathway regulates p27(kip1) phosphorylation mediating the decision to enter autophagy or apoptosis. Nat Cell Biol. 2007;9:218–224. doi: 10.1038/ncb1537. [DOI] [PubMed] [Google Scholar]
- 61.Hoyer-Hansen M, Bastholm L, Szyniarowski P, Campanella M, Szabadkai G, Farkas T, Bianchi K, Fehrenbacher N, Elling F, Rizzuto R, Mathiasen IS, Jaattela M. Control of macroautophagy by calcium, calmodulin-dependent kinase kinase-beta, and Bcl-2. Mol Cell. 2007;25:193–205. doi: 10.1016/j.molcel.2006.12.009. [DOI] [PubMed] [Google Scholar]
- 62.Rusten TE, Vaccari T, Lindmo K, Rodahl LM, Nezis IP, Sem-Jacobsen C, Wendler F, Vincent JP, Brech A, Bilder D, Stenmark H. ESCRTs and Fab1 regulate distinct steps of autophagy. Curr Biol. 2007;17:1817–1825. doi: 10.1016/j.cub.2007.09.032. [DOI] [PubMed] [Google Scholar]
- 63.Lee JA, Beigneux A, Ahmad ST, Young SG, Gao FB. ESCRT-III dysfunction causes autophagosome accumulation and neurodegeneration. Curr Biol. 2007;17:1561–1567. doi: 10.1016/j.cub.2007.07.029. [DOI] [PubMed] [Google Scholar]
- 64.Byfield MP, Murray JT, Backer JM. hVps34 is a nutrient-regulated lipid kinase required for activation of p70 S6 kinase. J Biol Chem. 2005;280:33076–33082. doi: 10.1074/jbc.M507201200. [DOI] [PubMed] [Google Scholar]
- 65.Nobukuni T, Joaquin M, Roccio M, Dann SG, Kim SY, Gulati P, Byfield MP, Backer JM, Natt F, Bos JL, Zwartkruis FJ, Thomas G. Amino acids mediate mTOR/raptor signaling through activation of class 3 phosphatidylinositol 3OH-kinase. Proc Natl Acad Sci U S A. 2005;102:14238–14243. doi: 10.1073/pnas.0506925102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Juhász G, Hill JH, Yan Y, Sass M, Baehrecke EH, Backer JM, Neufeld TP. The class III PI(3)K Vps34 promotes autophagy and endocytosis but not TOR signaling in Drosophila. J Cell Biol. 2008;181:655–666. doi: 10.1083/jcb.200712051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Blommaart EF, Luiken JJ, Blommaart PJ, van Woerkom GM, Meijer AJ. Phosphorylation of ribosomal protein S6 is inhibitory for autophagy in isolated rat hepatocytes. J Biol Chem. 1995;270:2320–2326. doi: 10.1074/jbc.270.5.2320. [DOI] [PubMed] [Google Scholar]
- 68.Wullschleger S, Loewith R, Hall MN. TOR signaling in growth and metabolism. Cell. 2006;124:471–484. doi: 10.1016/j.cell.2006.01.016. [DOI] [PubMed] [Google Scholar]
- 69.Kuma A, Hatano M, Matsui M, Yamamoto A, Nakaya H, Yoshimori T, Ohsumi Y, Tokuhisa T, Mizushima N. The role of autophagy during the early neonatal starvation period. Nature. 2004;432:1032–1036. doi: 10.1038/nature03029. [DOI] [PubMed] [Google Scholar]
- 70.Komatsu M, Waguri S, Ueno T, Iwata J, Murata S, Tanida I, Ezaki J, Mizushima N, Ohsumi Y, Uchiyama Y, Kominami E, Tanaka K, Chiba T. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J Cell Biol. 2005;169:425–434. doi: 10.1083/jcb.200412022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kim E, Goraksha-Hicks P, Li L, Neufeld TP, Guan KL. Regulation of TORC1 by Rag GTPases in nutrient response. Nat Cell Biol. 2008;10:935–945. doi: 10.1038/ncb1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sancak Y, Peterson TR, Shaul YD, Lindquist RA, Thoreen CC, Bar-Peled L, Sabatini DM. The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science. 2008;320:1496–1501. doi: 10.1126/science.1157535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Berry DL, Baehrecke EH. Growth arrest and autophagy are required for salivary gland cell degradation in Drosophila. Cell. 2007;131:1137–1148. doi: 10.1016/j.cell.2007.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Edgar BA. From cell structure to transcription: Hippo forges a new path. Cell. 2006;124:267–273. doi: 10.1016/j.cell.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 75.Dutta S, Baehrecke EH. Warts Is Required for PI3K-Regulated Growth Arrest, Autophagy, and Autophagic Cell Death in Drosophila. Curr Biol. 2008;18:1466–1475. doi: 10.1016/j.cub.2008.08.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lockshin RA, Williams CM. Programmed cell death-I. Cytology of degeneration in the intersegmental muscles of the pernyi silkmoth. J Insect Physiol. 1965;11:123–133. doi: 10.1016/0022-1910(65)90099-5. [DOI] [PubMed] [Google Scholar]
- 77.Schweichel JU, Merker HJ. The morphology of various types of cell death in prenatal tissues. Teratology. 1973;7:253–266. doi: 10.1002/tera.1420070306. [DOI] [PubMed] [Google Scholar]
- 78.Clarke PGH. Developmental cell death: morphological diversity and multiple mechanisms. Anat Embryol. 1990;181:195–213. doi: 10.1007/BF00174615. [DOI] [PubMed] [Google Scholar]
- 79.Kerr JF, Wyllie AH, Currie AR. Apoptosis: a basic biological phenomenon with wide-ranging implications in tissue kinetics. Br J Cancer. 1972;26:239–257. doi: 10.1038/bjc.1972.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Martin DN, Baehrecke EH. Caspases function in autophagic cell death in Drosophila. Development. 2004;131:275–284. doi: 10.1242/dev.00933. [DOI] [PubMed] [Google Scholar]
- 81.Levine B, Yuan J. Autophagy in cell death: an innocent convict? J Clin Invest. 2005;115:2679–2688. doi: 10.1172/JCI26390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Baehrecke EH. Autophagy: dual roles in life and death? Nature Reviews Mol Cell Biol. 2005;6:505–510. doi: 10.1038/nrm1666. [DOI] [PubMed] [Google Scholar]
- 83.Cecconi F, Levine B. The role of autophagy in mammalian development: cell makeover rather than cell death. Dev Cell. 2008;15:344–357. doi: 10.1016/j.devcel.2008.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Qu X, Zou Z, Sun Q, Luby-Phelps K, Cheng P, Hogan RN, Gilpin C, Levine B. Autophagy gene-dependent clearance of apoptotic cells during embryonic development. Cell. 2007;128:931–946. doi: 10.1016/j.cell.2006.12.044. [DOI] [PubMed] [Google Scholar]
- 85.Mellen MA, de la Rosa EJ, Boya P. The autophagic machinery is necessary for removal of cell corpses from the developing retinal neuroepithelium. Cell Death Differ. 2008;15:1279–1290. doi: 10.1038/cdd.2008.40. [DOI] [PubMed] [Google Scholar]
- 86.Nezis IP, Stravopodis DJ, Margaritis LH, Papassideri IS. Programmed cell death of follicular epithelium during the late developmental stages of oogenesis in the fruit flies Bactrocera oleae and Ceratitis capitata (Diptera, Tephritidae) is mediated by autophagy. Dev Growth Differ. 2006;48:189–198. doi: 10.1111/j.1440-169X.2006.00856.x. [DOI] [PubMed] [Google Scholar]
- 87.Nezis IP, Stravopodis DJ, Margaritis LH, Papassideri IS. Autophagy is required for the degeneration of the ovarian follicular epithelium in higher Diptera. Autophagy. 2006;2:297–298. doi: 10.4161/auto.2858. [DOI] [PubMed] [Google Scholar]
- 88.Mpakou VE, Nezis IP, Stravopodis DJ, Margaritis LH, Papassideri IS. Programmed cell death of the ovarian nurse cells during oogenesis of the silkmoth Bombyx mori. Dev Growth Differ. 2006;48:419–428. doi: 10.1111/j.1440-169X.2006.00878.x. [DOI] [PubMed] [Google Scholar]
- 89.Velentzas AD, Nezis IP, Stravopodis DJ, Papassideri IS, Margaritis LH. Stage-specific regulation of programmed cell death during oogenesis of the medfly Ceratitis capitata (Diptera, Tephritidae) Int J Dev Biol. 2007;51:57–66. doi: 10.1387/ijdb.062164av. [DOI] [PubMed] [Google Scholar]
- 90.Velentzas AD, Nezis IP, Stravopodis DJ, Papassideri IS, Margaritis LH. Mechanisms of programmed cell death during oogenesis in Drosophila virilis. Cell Tissue Res. 2007;327:399–414. doi: 10.1007/s00441-006-0298-x. [DOI] [PubMed] [Google Scholar]
- 91.Velentzas AD, Nezis IP, Stravopodis DJ, Papassideri IS, Margaritis LH. Apoptosis and autophagy function cooperatively for the efficacious execution of programmed nurse cell death during Drosophila virilis oogenesis. Autophagy. 2007;3:130–132. doi: 10.4161/auto.3582. [DOI] [PubMed] [Google Scholar]
- 92.Hou YC, Chittaranjan S, Barbosa SG, McCall K, Gorski SM. Effector caspase Dcp-1 and IAP protein Bruce regulate starvation-induced autophagy during Drosophila melanogaster oogenesis. J Cell Biol. 2008;182:1127–1139. doi: 10.1083/jcb.200712091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Nezis IP, Lamark T, Velentzas AD, Rusten TE, Bjorkoy G, Johansen T, Papassideri IS, Stravopodis DJ, Margaritis LH, Stenmark H, Brech A. Cell death during Drosophila melanogaster early oogenesis is mediated through autophagy. Autophagy. 2009;5 doi: 10.4161/auto.5.3.7454. [DOI] [PubMed] [Google Scholar]
- 94.Akdemir F, Farkas R, Chen P, Juhasz G, Medved’ova L, Sass M, Wang L, Wang X, Chittaranjan S, Gorski SM, Rodriguez A, Abrams JM. Autophagy occurs upstream or parallel to the apoptosome during histolytic cell death. Development. 2006;133:1457–1465. doi: 10.1242/dev.02332. [DOI] [PubMed] [Google Scholar]
- 95.Mohseni N, McMillan SC, Chaudhary R, Mok J, Reed BH. Autophagy promotes caspase-dependent cell death during Drosophila development. Autophagy. 2009;5 doi: 10.4161/auto.5.3.7444. [DOI] [PubMed] [Google Scholar]
- 96.Debnath J, Mills KR, Collins NL, Reginato MJ, Muthuswamy SK, Brugge JS. The role of apoptosis in creating and maintaining luminal space within normal and oncogene-expressing mammary acini. Cell. 2002;111:29–40. doi: 10.1016/s0092-8674(02)01001-2. [DOI] [PubMed] [Google Scholar]
- 97.Mills KR, Reginato M, Debnath J, Queenan B, Brugge JS. Tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) is required for induction of autophagy during lumen formation in vitro. Proc Natl Acad Sci USA. 2004;101:3438–3443. doi: 10.1073/pnas.0400443101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Yu L, Wan F, Dutta S, Welsh S, Liu Z, Freundt E, Baehrecke EH, Lenardo MJ. Autophagic programmed cell death by selective catalase degradation. Proc Natl Acad Sci USA. 2006;103:4952–4957. doi: 10.1073/pnas.0511288103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Bernards A, Hariharan IK. Of flies and men--studying human disease in Drosophila. Curr Opin Genet Dev. 2001;11:274–278. doi: 10.1016/s0959-437x(00)00190-8. [DOI] [PubMed] [Google Scholar]
- 100.Hariharan IK, Haber DA. Yeast, flies, worms, and fish in the study of human disease. N Engl J Med. 2003;348:2457–2463. doi: 10.1056/NEJMon023158. [DOI] [PubMed] [Google Scholar]
- 101.Komatsu M, Waguri S, Chiba T, Murata S, Iwata J, Tanida I, Ueno T, Koike M, Uchiyama Y, Kominami E, Tanaka K. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature. 2006;441:880–884. doi: 10.1038/nature04723. [DOI] [PubMed] [Google Scholar]
- 102.Hara T, Nakamura K, Matsui M, Yamamoto A, Nakahara Y, Suzuki-Migishima R, Yokoyama M, Mishima K, Saito I, Okano H, Mizushima N. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 2006;441:885–889. doi: 10.1038/nature04724. [DOI] [PubMed] [Google Scholar]
- 103.Ferrandon D, Imler JL, Hetru C, Hoffmann JA. The Drosophila systemic immune response: sensing and signalling during bacterial and fungal infections. Nat Rev Immunol. 2007;7:862–874. doi: 10.1038/nri2194. [DOI] [PubMed] [Google Scholar]
- 104.Simonsen A, Cumming RC, Finley KD. Linking lysosomal trafficking defects with changes in aging and stress response in Drosophila. Autophagy. 2007;3:499–501. doi: 10.4161/auto.4604. [DOI] [PubMed] [Google Scholar]
- 105.Ravikumar B, Vacher C, Berger Z, Davies JE, Luo S, Oroz LG, Scaravilli F, Easton DF, Duden R, O’Kane CJ, Rubinsztein DC. Inhibition of mTOR induces autophagy and reduces toxicity of polyglutamine expansions in fly and mouse models of Huntington disease. Nat Genetics. 2004;36:585–595. doi: 10.1038/ng1362. [DOI] [PubMed] [Google Scholar]
- 106.Pandey UB, Nie Z, Batlevi Y, McCray BA, Ritson GP, Nedelsky NB, Schwartz SL, DiProspero NA, Knight MA, Schuldiner O, Padmanabhan R, Hild M, Berry DL, Garza D, Hubbert CC, Yao TP, Baehrecke EH, Taylor JP. HDAC6 rescues neurodegeneration and provides an essential link between autophagy and the UPS. Nature. 2007;447:859–863. doi: 10.1038/nature05853. [DOI] [PubMed] [Google Scholar]
- 107.Kawaguchi Y, Kovacs JJ, McLaurin A, Vance JM, Ito A, Yao TP. The deacetylase HDAC6 regulates aggresome formation and cell viability in response to misfolded protein stress. Cell. 2003;115:727–738. doi: 10.1016/s0092-8674(03)00939-5. [DOI] [PubMed] [Google Scholar]
- 108.Bjorkoy G, Lamark T, Brech A, Outzen H, Perander M, Overvatn A, Stenmark H, Johansen T. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J Cell Biol. 2005;171:603–614. doi: 10.1083/jcb.200507002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Komatsu M, Waguri S, Koike M, Sou YS, Ueno T, Hara T, Mizushima N, Iwata J, Ezaki J, Murata S, Hamazaki J, Nishito Y, Iemura S, Natsume T, Yanagawa T, Uwayama J, Warabi E, Yoshida H, Ishii T, Kobayashi A, Yamamoto M, Yue Z, Uchiyama Y, Kominami E, Tanaka K. Homeostatic levels of p62 control cytoplasmic inclusion body formation in autophagy-deficient mice. Cell. 2007;131:1149–1163. doi: 10.1016/j.cell.2007.10.035. [DOI] [PubMed] [Google Scholar]
- 110.Finley KD, Edeen PT, Cumming RC, Mardahl-Dumesnil MD, Taylor BJ, Rodriguez MH, Hwang CE, Benedetti M, McKeown M. blue cheese mutations define a novel, conserved gene involved in progressive neural degeneration. J Neurosci. 2003;23:1254–1264. doi: 10.1523/JNEUROSCI.23-04-01254.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Simonsen A, Birkeland HC, Gillooly DJ, Mizushima N, Kuma A, Yoshimori T, Slagsvold T, Brech A, Stenmark H. Alfy, a novel FYVE-domain-containing protein associated with protein granules and autophagic membranes. J Cell Sci. 2004;117:4239–4251. doi: 10.1242/jcs.01287. [DOI] [PubMed] [Google Scholar]
- 112.Lindmo K, Brech A, Finley KD, Gaumer S, Contamine D, Rusten TE, Stenmark H. The PI 3-kinase regulator Vps15 is required for autophagic clearance of protein aggregates. Autophagy. 2008;4:500–506. doi: 10.4161/auto.5829. [DOI] [PubMed] [Google Scholar]
- 113.Skibinski G, Parkinson NJ, Brown JM, Chakrabarti L, Lloyd SL, Hummerich H, Nielsen JE, Hodges JR, Spillantini MG, Thusgaard T, Brandner S, Brun A, Rossor MN, Gade A, Johannsen P, Sorensen SA, Gydesen S, Fisher EM, Collinge J. Mutations in the endosomal ESCRTIII-complex subunit CHMP2B in frontotemporal dementia. Nat Genet. 2005;37:806–808. doi: 10.1038/ng1609. [DOI] [PubMed] [Google Scholar]
- 114.Parkinson N, Ince PG, Smith MO, Highley R, Skibinski G, Andersen PM, Morrison KE, Pall HS, Hardiman O, Collinge J, Shaw PJ, Fisher EM. ALS phenotypes with mutations in CHMP2B (charged multivesicular body protein 2B) Neurology. 2006;67:1074–1077. doi: 10.1212/01.wnl.0000231510.89311.8b. [DOI] [PubMed] [Google Scholar]
- 115.Filimonenko M, Stuffers S, Raiborg C, Yamamoto A, Malerod L, Fisher EM, Isaacs A, Brech A, Stenmark H, Simonsen A. Functional multivesicular bodies are required for autophagic clearance of protein aggregates associated with neurodegenerative disease. J Cell Biol. 2007;179:485–500. doi: 10.1083/jcb.200702115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Ravikumar B, Imarisio S, Sarkar S, O’Kane CJ, Rubinsztein DC. Rab5 modulates aggregation and toxicity of mutant huntingtin through macroautophagy in cell and fly models of Huntington disease. J Cell Sci. 2008;121:1649–1660. doi: 10.1242/jcs.025726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Terlecky SR, Koepke JI, Walton PA. Peroxisomes and aging. Biochim Biophys Acta. 2006;1763:1749–1754. doi: 10.1016/j.bbamcr.2006.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Iwata J, Ezaki J, Komatsu M, Yokota S, Ueno T, Tanida I, Chiba T, Tanaka K, Kominami E. Excess peroxisomes are degraded by autophagic machinery in mammals. J Biol Chem. 2006;281:4035–4041. doi: 10.1074/jbc.M512283200. [DOI] [PubMed] [Google Scholar]
- 119.Zhang C, Cuervo AM. Restoration of chaperone-mediated autophagy in aging liver improves cellular maintenance and hepatic function. Nat Med. 2008;14:959–965. doi: 10.1038/nm.1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Kiffin R, Christian C, Knecht E, Cuervo AM. Activation of chaperone-mediated autophagy during oxidative stress. Mol Biol Cell. 2004;15:4829–4840. doi: 10.1091/mbc.E04-06-0477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Massey AC, Kaushik S, Sovak G, Kiffin R, Cuervo AM. Consequences of the selective blockage of chaperone-mediated autophagy. Proc Natl Acad Sci U S A. 2006;103:5805–5810. doi: 10.1073/pnas.0507436103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Cuervo AM, Dice JF. Age-related decline in chaperone-mediated autophagy. J Biol Chem. 2000;275:31505–31513. doi: 10.1074/jbc.M002102200. [DOI] [PubMed] [Google Scholar]
- 123.Kiffin R, Kaushik S, Zeng M, Bandyopadhyay U, Zhang C, Massey AC, Martinez-Vicente M, Cuervo AM. Altered dynamics of the lysosomal receptor for chaperone-mediated autophagy with age. J Cell Sci. 2007;120:782–791. doi: 10.1242/jcs.001073. [DOI] [PubMed] [Google Scholar]
- 124.Vyas JM, Van der Veen AG, Ploegh HL. The known unknowns of antigen processing and presentation. Nat Rev Immunol. 2008;8:607–618. doi: 10.1038/nri2368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Nedjic J, Aichinger M, Emmerich J, Mizushima N, Klein L. Autophagy in thymic epithelium shapes the T-cell repertoire and is essential for tolerance. Nature. 2008;455:396–400. doi: 10.1038/nature07208. [DOI] [PubMed] [Google Scholar]
- 126.Cadwell K, Liu JY, Brown SL, Miyoshi H, Loh J, Lennerz JK, Kishi C, Kc W, Carrero JA, Hunt S, Stone CD, Brunt EM, Xavier RJ, Sleckman BP, Li E, Mizushima N, Stappenbeck TS, Virgin HWt. A key role for autophagy and the autophagy gene Atg16l1 in mouse and human intestinal Paneth cells. Nature. 2008;456:259–263. doi: 10.1038/nature07416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Saitoh T, Fujita N, Jang MH, Uematsu S, Yang BG, Satoh T, Omori H, Noda T, Yamamoto N, Komatsu M, Tanaka K, Kawai T, Tsujimura T, Takeuchi O, Yoshimori T, Akira S. Loss of the autophagy protein Atg16L1 enhances endotoxin-induced IL-1beta production. Nature. 2008;456:264–268. doi: 10.1038/nature07383. [DOI] [PubMed] [Google Scholar]
- 128.Nakagawa I, Amano A, Mizushima N, Yamamoto A, Yamaguchi H, Kamimoto T, Nara A, Funao J, Nakata M, Tsuda K, Hamada S, Yoshimori T. Autophagy defends cells against invading group A Streptococcus. Science. 2004;306:1037–1040. doi: 10.1126/science.1103966. [DOI] [PubMed] [Google Scholar]
- 129.Gutierrez MG, Master SS, Singh SB, Taylor GA, Colombo MI, Deretic V. Autophagy is a defense mechanism inhibiting BCG and Mycobacterium tuberculosis survival in infected macrophages. Cell. 2004;119:753–766. doi: 10.1016/j.cell.2004.11.038. [DOI] [PubMed] [Google Scholar]
- 130.Ogawa M, Yoshimori T, Suzuki T, Sagara H, Mizushima N, Sasakawa C. Escape of intracellular Shigella from autophagy. Science. 2005;307:727–731. doi: 10.1126/science.1106036. [DOI] [PubMed] [Google Scholar]
- 131.Ling YM, Shaw MH, Ayala C, Coppens I, Taylor GA, Ferguson DJ, Yap GS. Vacuolar and plasma membrane stripping and autophagic elimination of Toxoplasma gondii in primed effector macrophages. J Exp Med. 2006;203:2063–2071. doi: 10.1084/jem.20061318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Andrade RM, Wessendarp M, Gubbels MJ, Striepen B, Subauste CS. CD40 induces macrophage anti-Toxoplasma gondii activity by triggering autophagy-dependent fusion of pathogen-containing vacuoles and lysosomes. J Clin Invest. 2006;116:2366–2377. doi: 10.1172/JCI28796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Jounai N, Takeshita F, Kobiyama K, Sawano A, Miyawaki A, Xin KQ, Ishii KJ, Kawai T, Akira S, Suzuki K, Okuda K. The Atg5 Atg12 conjugate associates with innate antiviral immune responses. Proc Natl Acad Sci U S A. 2007;104:14050–14055. doi: 10.1073/pnas.0704014104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Huang J, Klionsky DJ. Autophagy and human disease. Cell Cycle. 2007;6:1837–1849. doi: 10.4161/cc.6.15.4511. [DOI] [PubMed] [Google Scholar]
- 135.Kirkegaard K, Taylor MP, Jackson WT. Cellular autophagy: surrender, avoidance and subversion by microorganisms. Nat Rev Microbiol. 2004;2:301–314. doi: 10.1038/nrmicro865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Singh SB, Davis AS, Taylor GA, Deretic V. Human IRGM induces autophagy to eliminate intracellular mycobacteria. Science. 2006;313:1438–1441. doi: 10.1126/science.1129577. [DOI] [PubMed] [Google Scholar]
- 137.Yano T, Mita S, Ohmori H, Oshima Y, Fujimoto Y, Ueda R, Takada H, Goldman WE, Fukase K, Silverman N, Yoshimori T, Kurata S. Autophagic control of listeria through intracellular innate immune recognition in drosophila. Nat Immunol. 2008;9:908–916. doi: 10.1038/ni.1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Wu J, Randle KE, Wu LP. ird1 is a Vps15 homologue important for antibacterial immune responses in Drosophila. Cell Microbiol. 2007;9:1073–1085. doi: 10.1111/j.1462-5822.2006.00853.x. [DOI] [PubMed] [Google Scholar]
- 139.Zinke I, Schutz CS, Katzenberger JD, Bauer M, Pankratz MJ. Nutrient control of gene expression in Drosophila: microarray analysis of starvation and sugar-dependent response. Embo J. 2002;21:6162–6173. doi: 10.1093/emboj/cdf600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Chandra RK. Nutrition, immunity and infection: from basic knowledge of dietary manipulation of immune responses to practical application of ameliorating suffering and improving survival. Proc Natl Acad Sci U S A. 1996;93:14304–14307. doi: 10.1073/pnas.93.25.14304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Faggioni R, Moser A, Feingold KR, Grunfeld C. Reduced leptin levels in starvation increase susceptibility to endotoxic shock. Am J Pathol. 2000;156:1781–1787. doi: 10.1016/S0002-9440(10)65049-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Chan J, Tian Y, Tanaka KE, Tsang MS, Yu K, Salgame P, Carroll D, Kress Y, Teitelbaum R, Bloom BR. Effects of protein calorie malnutrition on tuberculosis in mice. Proc Natl Acad Sci U S A. 1996;93:14857–14861. doi: 10.1073/pnas.93.25.14857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Simonsen A, Cumming RC, Lindmo K, Galaviz V, Cheng S, Rusten TE, Finley KD. Genetic modifiers of the Drosophila blue cheese gene link defects in lysosomal transport with decreased life span and altered ubiquitinated-protein profiles. Genetics. 2007;176:1283–1297. doi: 10.1534/genetics.106.065011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Juhasz G, Puskas LG, Komonyi O, Erdi B, Maroy P, Neufeld TP, Sass M. Gene expression profiling identifies FKBP39 as an inhibitor of autophagy in larval Drosophila fat body. Cell Death Differ. 2007;14:1181–1190. doi: 10.1038/sj.cdd.4402123. [DOI] [PMC free article] [PubMed] [Google Scholar]