Abstract
Background
Studies have reported differing frequencies of detection of polyomavirus simian virus 40 (SV40) in association with human lymphomas.
Objective
We addressed the hypothesis that SV40 positivity in lymphomas can vary among sampled populations.
Study design
Archival paraffin-embedded lymphoma specimens (n=171) from patients at two urban hospitals in Houston, Texas, USA, were analyzed following a cross-sectional study design. Extracted DNAs were characterized by quantitative polymerase chain reaction for the cellular RNase P gene and for SV40 and herpesvirus Epstein-Barr virus (EBV) sequences.
Results
Patient characteristics of the two study populations differed significantly whereas the classification of tumor types studied did not. SV40 DNA was detected more frequently in lymphomas from the public hospital population (10/44, 23%) than in lymphomas from the veterans’ hospital (VAMC) (4/127, 3%; P < 0.0001). EBV detection in lymphomas also differed between the two groups (17/44, 39% vs. 23/127, 18%; P = 0.01). SV40 positivity was associated with a younger age category of VAMC lymphoma patients (P = 0.02). Expression of T-antigen was detected by immunohistochemistry in half of lymphomas that contained SV40 DNA. Variation was observed in the quality and quantity of DNA recovered from paraffin-embedded specimens, but there was no difference in recoveries of DNA from samples from the two hospitals.
Conclusions
This study demonstrated that, in a direct comparison, the prevalence of SV40 DNA in lymphomas can differ significantly between groups with different demographic distributions.
Keywords: SV40, EBV, lymphoma, polyomavirus, T-antigen, EBER, real-time quantitative PCR, human infections
1. Background
Viral infections by polyomavirus simian virus 40 (SV40) and herpesvirus Epstein-Barr virus (EBV) have been implicated in lymphomagenesis.1 EBV is a ubiquitous human herpesvirus that produces a long-lasting infection in B cells in more than 90% of the population. It has frequently been associated with the pathogenesis of several neoplasms, especially lymphoid tumors and nasopharyngeal carcinoma.2 EBV has been classified as a group 1 carcinogen by the World Health Organization International Agency for Research on Cancer.3 More recently, it has been suggested that SV40 may be associated with some examples of lymphomagenesis, although causality has not been established.4–8
SV40 is a well-characterized tumor virus that encodes a powerful oncoprotein, the large tumor antigen (T-ag).4,9–12 The widespread use of early polio vaccines contaminated with SV40 due to production in primary rhesus monkey kidneys exposed millions of people worldwide to live SV40.4,11,13,14 There is strong evidence that SV40 can be found in humans today, both in those with and without malignancies. SV40 markers have been detected in tumor tissue from lymphoma patients in some studies15–23 but not in others.24–31 These discrepancies may reflect variations among sources of specimens, sample processing, or assays. In addition, the prevalence of SV40 infection in humans might vary among different populations and geographic regions.
2. Objectives
This study tested the hypothesis that SV40 positivity in lymphomas varies among population groups. Using the same analytical methodologies in the same laboratory, we determined the presence of SV40 and EBV in lymphoma samples from two patient groups in Houston, Texas with different demographics. We found striking differences in virus positivity of lymphomas between the two groups.
3. Study Design
3.1. Patients and tissue samples
A series of 171 archival, fixed, paraffin-embedded lymphoma specimens obtained between 1994 and 2006 in Houston, TX, were analyzed in separate cross-sectional studies. The series included 44 cases from the public hospital [Ben Taub General Hospital (BTGH) of the Harris County Hospital District] and 127 cases from the military veterans’ hospital [Michael E. DeBakey Veterans Affairs Medical Center (VAMC)]. The tumor samples included 167 non-Hodgkin lymphomas (NHL) and 4 Hodgkin lymphomas classified by the World Health Organization system.32 Lymphomas of the central nervous system were not included. In addition, 38 tissues were analyzed as controls (12 nonneoplastic lymphoid tissues, classified as benign lymphadenopathies, and 10 normal tissues from healthy individuals from BTGH, and 1 skin cancer and 15 normal tissues from VAMC). Institutional Review Board approval was obtained.
Patient demographics at the two hospitals in year 2006–2007 were as follows. For BTGH: Asian, 5%; Black, 34%; Hispanic, 46%; White, 13%; and Unknown/Other, 2%. For VAMC: Black, 28%; Hispanic, 4%; White, 55%; and Unknown/Other, 13%.
3.2. DNA extraction from paraffin sections
Two 20-µm sections were cut from each tissue sample block by the same person. To avoid cross-contamination, the microtome block was cleaned and the blade was replaced between specimens. Samples were deparaffinized and DNAs were extracted by a modification of a published procedure33 (see Supplemental Material). The method will recover low-molecular-weight DNA, an important consideration in case episomal viral DNA is present.
To avoid contamination and false-positive results, all DNA isolation procedures were performed under a biosafety hood in a dedicated room free from plasmids and viruses, physically separated from rooms used to prepare amplification reaction mixes and to carry out polymerase chain reaction (PCR) assays. In addition, filter tips, powder-free gloves, new disposable blades, and a separate set of pipettors were used. A negative extraction control lacking paraffin-embedded tissue was processed in parallel.
3.3. Real-time quantitative PCR
Real-time quantitative PCR (RQ-PCR) reaction mixes were prepared using a biosafety hood in a second dedicated room free from plasmids and viruses. The tubes were then taken to the DNA extraction room where sample DNAs were added; negative extraction controls and no-template controls were included. Finally, the tubes were taken to a third room where the positive control DNA was added and PCR amplification was carried out.
Samples were first screened for the single-copy human RNase P gene by RQ-PCR to evaluate the quality and concentration of DNA recovered. The possible presence of PCR inhibitors in the DNA preparations was examined using 10-fold sample dilutions. The number of human cell equivalents, measured by RNase P genome copy numbers, was used to determine variability in DNA yields and to normalize SV40 and EBV viral loads to human cell numbers in a sample.
Control plasmids were pSV40-B2E, SV40 strain Baylor,34 pBluescript II SK (+/−) containing the EBV-encoded RNA (EBER) gene, and RNase P.35 Standard curves were run with each viral assay. One set of SV40-specific large T-ag gene primers and probe was published previously,35 whereas the second one was designed as follows: SVT36-fwd primer 5′-CAACAAACAGTGTAGCCAAGCAA-3′ corresponding to nt 3624–3646 of SV40-776, SVT36-rev primer 5′-GGTTCTACAGGCTCTGCTGACATA-3′ corresponding to nt 3689–3666, and a fluorogenic probe 5′-FAM-CCAGCCATCCATTC-MGB-3′ corresponding to nt 3649–3662. EBV DNA reactions were performed using primers and probe from the EBER gene.36 The RQ-PCR assays were performed using an ABI Prism 7700 Sequence Detection System (Applied Biosystems, Foster City, CA). All PCR reactions were performed in duplicate.
RQ-PCR reactions were considered positive if ≥10 viral genome copies/reaction were detected. A sample was classified as SV40 positive if two independent tests were positive [i.e., two RQ-PCRs using different sets of primers or one RQ-PCR plus immunohistochemistry (IHC)]. Samples were considered positive for EBV on the basis of the EBER RQ-PCR assay.
3.4. Immunohistochemistry
The expression of SV40 T-ag was detected by IHC as described.19 After tissue sections were deparaffinized, antigen retrieval was performed in a steamer with citrate buffer (pH 6). A monoclonal antibody against the C-terminus of SV40 T-ag (PAb101, Santa Cruz Biotechnology Inc., Santa Cruz, CA) was used at a dilution of 1:5000. Sections were incubated with antibody at 4 °C overnight, then detected with the Mach3 Mouse probe-polymer kit (Biocare Medical, Walnut Creek, CA). The reaction was visualized with liquid diaminobenzidine (DAB+) Substrate Pack (DakoCytomation, Carpenteria, CA) and counterstained with Mayer’s modified hematoxylin. SV40-induced hamster tumors served as positive controls; normal mouse serum and diluent alone were included as negative controls.
3.5. Statistical analysis
Univariate analyses were conducted using descriptive statistics, including Chi-square analysis and Fisher’s Exact test for the evaluation of associations and means, standard deviations, medians, and ranges for dispersion. The binomial exact method was used to calculate 95% confidence intervals for proportions. The Z test was used to evaluate differences between proportions. An alpha level of 0.05 was used to determine statistical significance. Statistical analyses were conducted using the SAS statistical software, version 9.1.
4. Results
4.1. Study population
Two-hundred and nine paraffin-embedded tissue specimens were analyzed, including 171 cases of lymphoma and 38 control samples. Of the lymphomas, 44 cases were from the public hospital (BTGH) and 127 were from the military veterans’ hospital (VAMC). Identifying characteristics of the lymphoma patients differed significantly between the two hospitals, establishing that two different population groups were studied. However, the types of lymphoma studied from the two populations were similar (Table 1).
Table 1.
Demographic characteristics of lymphoma patients from two different population groups in Houston, Texas
| Factor | No. (%) of patients | P valueb | |
|---|---|---|---|
| BTGHa | VAMCa | ||
| All | 44 | 127 | |
| Gender | <0.0001 | ||
| Male | 30 (68) | 125 (98) | |
| Female | 14 (32) | 2 (2) | |
| Age (years) | <0.0001 | ||
| ≤50 | 21 (48) | 22 (17) | |
| >50 | 23 (52) | 105 (83) | |
| Race/ethnicityc | <0.0001 | ||
| Asian | 1 (2) | 0 (0) | |
| Black | 15 (34) | 32 (27) | |
| Hispanic | 23 (52) | 14 (12) | |
| White | 5 (11) | 71 (61) | |
| HIV statusd | 0.001 | ||
| Positive | 9 (47) | 10 (12) | |
| Negative | 10 (53) | 71 (88) | |
| Pathologye | NSf | ||
| Non-Hodgkin lymphoma | 41 (93) | 126 (99) | |
| B-cell lymphoma | 41 (93) | 117 (92) | |
| Diffuse large B-cell lymphoma | 27 (61) | 58 (46) | |
| Diffuse large B-cell lymphoma/T-cell/histiocyte-rich type | 0 (0) | 1 (1) | |
| Follicular lymphoma | 2 (4) | 24 (19) | |
| Plasmablastic lymphoma | 4 (9) | 0 (0) | |
| Mantle cell lymphoma | 3 (7) | 8 (6) | |
| Marginal zone B-cell lymphoma | 3 (7) | 16 (13) | |
| Nodal marginal zone B-cell lymphoma | 0 (0) | 2 (2) | |
| Extranodal marginal zone B-cell lymphoma | 0 (0) | 4 (3) | |
| Extranodal marginal zone B-cell lymphoma (of MALT type) | 3 (7) | 10 (8) | |
| Burkitt lymphoma | 1 (2) | 1 (1) | |
| Chronic lymphocytic leukemia/small lymphocytic lymphoma | 0 (0) | 8 (6) | |
| Lymphoplasmacytic lymphoma | 0 (0) | 1 (1) | |
| Unclassified | 1 (2) | 0 (0) | |
| NK/T-cell lymphoma | 0 (0) | 8 (6) | |
| Extranodal NK/T-cell lymphoma, nasal type | 0 (0) | 1 (1) | |
| Peripheral T-cell lymphoma, unspecified | 0 (0) | 2 (2) | |
| Anaplastic large cell lymphoma | 0 (0) | 2 (2) | |
| Mycosis fungoides | 0 (0) | 3 (2) | |
| Unclassified | 0 (0) | 1 (1) | |
| Hodgkin lymphoma | 3 (7) | 1 (1) | |
| Nodular sclerosis | 2 (4) | 0 (0) | |
| Mixed cellularity | 0 (0) | 1 (1) | |
| Unclassified | 1 (2) | 0 (0) | |
Abbreviations: BTGH = Ben Taub General Hospital; VAMC = Michael E. DeBakey Veterans Affairs Medical Center; NS = not significant.
Estimated between BTGH and VAMC patients with Z test for proportions.
Information on ethnicity was missing in 10 cases from VAMC.
Information on HIV status was missing in 25 cases from BTGH and in 46 cases from VAMC.
World Health Organization classification.
B-cell lymphoma vs. NK/T-cell lymphoma, P = 0.215. Among B-cell lymphoma: diffuse large B-cell vs. all other, P = 0.105.
4.2. Amplification of RNase P sequences by RQ-PCR from DNA derived from paraffin-embedded tissues
DNA was extracted from fixed tissues as described in Study Design. Dilutions of the DNA samples were analyzed by RQ-PCR for the human cellular housekeeping gene, RNase P. No PCR inhibitors were apparent in the undiluted DNA solutions from 93 of 184 (50.5%) samples. An inhibitory effect was observed with the other samples in that the level of RNase P amplification was less in undiluted samples than in samples diluted 10-fold. With one exception, a 1:10 dilution was adequate to dilute the PCR amplification inhibitors present in those DNA preparations and that dilution was used for further analysis.
Broad variation was observed in the amount of human cellular DNA recovered from the panel of paraffin-embedded lymphoid samples. Total DNA yields from the BTGH lymphoma samples ranged from 1100 to 8,234,300 cell equivalents (median = 552,675), whereas DNA yields from the VAMC samples ranged from 3700 to 19,163,000 cell equivalents (median = 664,204). These total DNA recoveries were not significantly different between the BTGH and VAMC samples (P = 0.21 by Wilcoxon rank-sum test). Also, the cell-equivalents of DNA tested per PCR reaction was not significantly different between the BTGH and VAMC specimens (P = 0.61 by Wilcoxon rank-sum test). We note that because of low DNA recoveries from a few specimens, <1000 cell equivalents of DNA were tested per PCR reaction for 8 of the lymphoma samples, 6 (4.7%) from the VAMC and 2 (4.5%) from BTGH. These findings highlight the dual problems of variable recoveries of cellular DNA and of variable presence of PCR inhibitors in DNA extracts and illustrate the importance of confirming the quantity and quality of DNA from paraffin-embedded tissues for viral PCR analyses. However, the quantity of human cellular DNA recovered and tested was comparable for samples from the two hospitals, ruling out that technical factor as a significant variable in this study.
4.3. Detection of SV40 and EBV DNAs in lymphomas
We analyzed 171 lymphomas and 38 controls for the presence of viral sequences and for viral load quantitation. SV40 DNA was detected in 10 of 44 (23%, 95% CI: 11–38%) lymphoma specimens obtained from BTGH and in only 4 of 127 (3%, 95% CI: 1–8%) samples from the VAMC (Table 2). This difference in proportions was significant (P < 0.0001). All 14 lymphomas found to be SV40 positive were classified as NHL with 13 cases classified as B cell and 1 as NK/T-cell lymphomas. The SV40-positive lymphoma types from BTGH included diffuse large B-cell (7), plasmablastic (1) and marginal zone B-cell (2), whereas those from VAMC were diffuse large B-cell (2), marginal zone B-cell (1), and NK/T-cell (1). The SV40 virus-positive lymphomas were confirmed by PCR using an independent set of primers directed against the middle of the T-ag gene or by detection of T-ag expression by IHC staining. One of 38 (2.6%, 95% CI: 0.03–14%) controls, a case of lymphadenopathy, was also positive for SV40.
Table 2.
Presence of SV40 and EBV DNA sequences in lymphomas
| Hospitala | Total no. of samples |
No. (%) | ||
|---|---|---|---|---|
| SV40 DNA-positive | EBV DNA-positive | SV40+EBV dual positive |
||
| BTGH | 44 | 10 (23)b | 17 (39)c | 4 (9) |
| VAMC | 127 | 4 (3)b | 23 (18)c | 2 (2) |
BTGH = Ben Taub General Hospital; VAMC = Michael E. DeBakey Veterans Affairs Medical Center.
BTGH vs. VAMC, P < 0.0001.
BTGH vs. VAMC, P < 0.01.
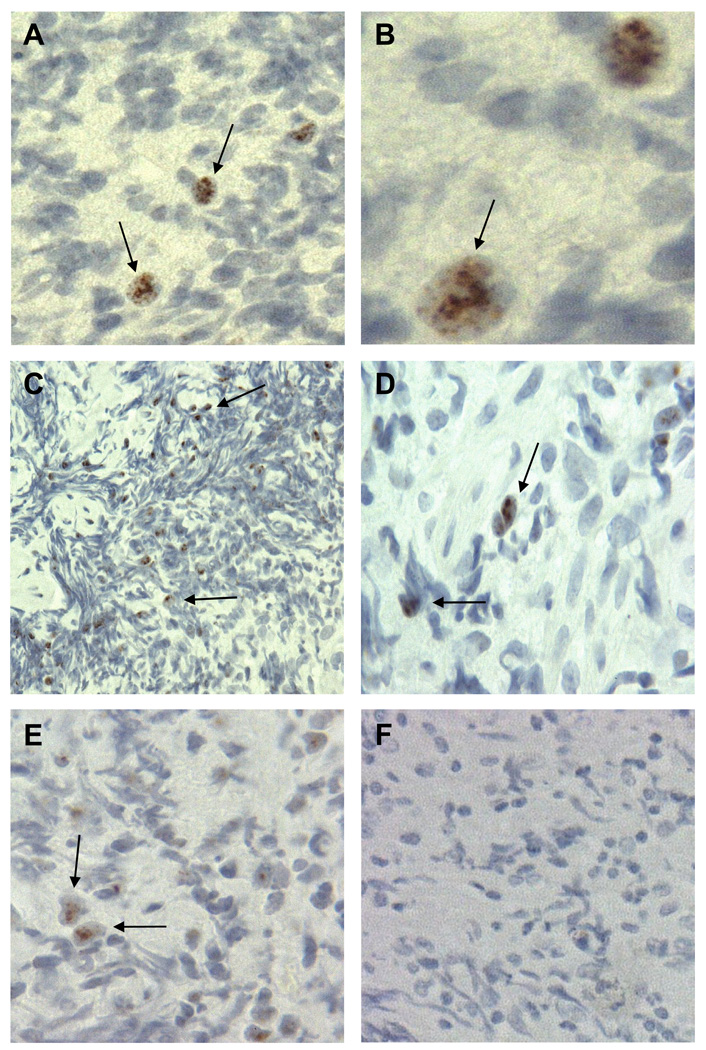
We examined 61 coded RQ-PCR SV40-positive and -negative specimens for expression of SV40 T-ag by IHC. When the code was broken, SV40 T-ag staining was detected in 8 of 14 (57%) specimens that were positive for SV40 DNA by PCR and in none of 47 SV40 DNA-negative samples (Fig. 1). The expression of the viral oncoprotein was observed in the nuclei of tumor cells. The intensity of the reaction varied among tumors and <25% of the cells in individual tumors appeared to be reactive.
Fig. 1.
SV40 T-ag expression in NHL. (A, B) Expression of SV40 T-ag in an SV40 DNA-positive diffuse large B-cell lymphoma from a 48-year-old white male. (C, D) Expression of SV40 T-ag in an SV40 DNA-positive B-cell lymphoma from a 47-year-old Hispanic male. (E) Expression of SV40 T-ag in an SV40 DNA-positive diffuse large B-cell lymphoma from a 23-year-old Hispanic male. (F) No T-ag expression in an SV40 DNA-negative B-cell lymphoma from a 39-year-old Hispanic male. Samples were stained with antibody PAb101. Arrows point to representative SV40 T-ag-positive cells in panels A–E. Original magnification for panels A, C, E, and F, 40×, and for panels B and D, 100×.
EBV genome sequences were detected in 17 of 44 (39%, 95% CI: 24–54%) lymphoma cases from BTGH and in 23 of 127 (18%, 95% CI: 12–26%) from the VAMC (Table 2). This difference in proportions was significant (P=0.01). EBV-positive lymphoma types from BTGH included diffuse large B-cell (12), plasmablastic (4), and Hodgkin (1), whereas EBV-positive lymphomas from the VAMC were diffuse large B-cell (9), diffuse large B-cell/T-cell/histiocyte-rich (1), follicular (2), mantle cell (3), marginal zone B-cell (3), NK/T-cell (4), and Hodgkin (1). These findings confirm other reports that EBV can be detected in some cases of follicular and mantle cell lymphomas.37–39 Two of 38 (5.3%, 95% CI: 0.6–18%) nonneoplastic lymphoid tissues harbored EBV sequences. A small number of lymphoma specimens (n=6) were positive for both viruses (Table 2). PCR negative controls included in all PCR assays excluded laboratory contamination.
Viral loads in virus-positive lymphomas were calculated from the RQ-PCR data. The content of SV40 DNA ranged from 1 to 90 genome copies per 1000 cells. For EBV, the amount of viral DNA detected ranged from 1 to >25,000 genome copies per 1000 cells, with 15 (38%) EBV-positive samples containing ≥1 genome copy per cell.
4.4. Patient characteristics and viral positivity of lymphomas
To attempt to identify risk factors for the presence of SV40 and EBV in lymphoid malignancies, we examined the associations between several patient characteristics and detection of SV40 and EBV DNAs (Table 3,Table 4). Most of the variables were not associated with the detection of either virus. However, an influence of patient age on SV40 positivity was found. As the BTGH and VAMC study populations differed significantly in age distribution (Table 1), frequency-driven cut-points were needed to classify the data by younger and older age within each sample: ≤40 and >40 years old for the BTGH group and ≤50 and >50 years old for the VAMC group. We found that SV40-positive lymphomas were more common in lymphoma patients ≤50 than in patients >50 from the VAMC (P=0.02) (Table 3). No significant associations of age category with EBV positivity of lymphomas were observed. However, considering those patients for whom HIV status was known, HIV-positive patients at the VAMC were more likely to have EBV-positive lymphomas than HIV-negative patients (Table 4). For the NHL-type subgroup of VAMC samples, those from patients who were HIV seropositive were more likely to be EBV-positive than those from patients who were HIV seronegative (5/9, 56% vs. 13/70, 19%; P=0.04). These data support previous observations of an association of EBV with systemic NHL in some HIV-positive subjects.40,41 In the BTGH group sample, none of the factors was significantly associated with risk of SV40 or EBV positivity in the subgroup analysis of NHL. However, it should be noted that the HIV status was unknown for many of the BTGH patients.
Table 3.
Patient demographics and SV40 positivity in lymphomas
| Factor | BTGH | VAMC | ||
|---|---|---|---|---|
| No. SV40 positive/Total no. of samples (%) |
P valuea | No. SV40 positive/Total no. of samples (%) |
P valuea | |
| Gender | NS | NS | ||
| Male | 6/30 (20) | 4/125 (3) | ||
| Female | 4/14 (29) | 0/2 (0) | ||
| Age categoryb | NS | 0.02 | ||
| Younger | 6/17 (35) | 3/22 (14) | ||
| Older | 4/27 (15) | 1/105 (1) | ||
| Race/ethnicityc | NS | NS | ||
| Asian | 0/1 (0) | 0/0 (0) | ||
| Black | 4/15 (27) | 0/32 (0) | ||
| Hispanic | 5/23 (22) | 1/14 (7) | ||
| White | 1/5 (20) | 3/71 (4) | ||
| HIV statusc | NS | NS | ||
| Positive | 2/9 (22) | 1/10 (10) | ||
| Negative | 1/10 (10) | 3/71 (4) | ||
| Pathology | NS | NS | ||
| Non-Hodgkin lymphoma | 10/41 (24) | 4/126 (3) | ||
| Hodgkin lymphoma | 0/3 (0) | 0/1 (0) | ||
Abbreviations: BTGH = Ben Taub General Hospital; VAMC = Michael E. DeBakey Veterans Affairs Medical Center; NS = not significant.
P values were evaluated between subgroups within a given hospital population.
Cut points for younger and older age variable: BTGH = ≤40 and >40 years, VAMC = ≤50 and >50 years.
For BTGH, 25 patients were of unknown HIV status; for VAMC, 10 patients were of unknown race/ethnicity and 46 were of unknown HIV status.
Table 4.
Patient demographics and EBV positivity in lymphomas
| Factor | BTGH | VAMC | ||
|---|---|---|---|---|
| No. EBV positive/Total no. of samples (%) |
P valuea | No. EBV positive/Total no. of samples (%) |
P valuea | |
| Gender | NS | NS | ||
| Male | 14/30 (47) | 23/125 (18) | ||
| Female | 3/14 (21) | 0/2 (0) | ||
| Age categoryb | NS | NS | ||
| Younger | 8/17 (47) | 5/22 (23) | ||
| Older | 9/27 (33) | 18/105 (17) | ||
| Race/ethnicity | NS | NS | ||
| Asian | 0/1 (0) | 0/0 (0) | ||
| Black | 6/15 (40) | 7/32 (22) | ||
| Hispanic | 9/23 (39) | 2/14 (14) | ||
| White | 2/5 (40) | 13/71 (18) | ||
| HIV status | NS | 0.03 | ||
| Positive | 6/9 (67) | 5/10 (50) | ||
| Negative | 5/10 (50) | 14/71 (20) | ||
| Pathology | NS | NS | ||
| Non-Hodgkin lymphoma | 16/41 (39) | 22/126 (17) | ||
| Hodgkin lymphoma | 1/3 (33) | 1/1 (100) | ||
Abbreviations: BTGH = Ben Taub General Hospital; VAMC = Michael E. DeBakey Veterans Affairs Medical Center; NS = not significant.
P values were evaluated between subgroups within a given hospital population.
Cut points for younger and older age variable: BTGH = ≤40 and >40 years, VAMC = ≤50 and >50 years.
5. Discussion
This report describes the detection of SV40 and EBV in lymphomas from two different sites in Houston, Texas. To our knowledge this is the first comparison in a single study of virus findings in lymphoma samples from sets of patients with different demographic characteristics. These findings support the hypothesis that the frequency of SV40 positivity in cancer specimens can vary among samples from different locations. The histologic classification of lymphomas and total DNA recoveries from archival specimens were similar between the two groups and technical procedures were not a variable as the same sectioning, processing, and analytic methodologies were applied to all samples by the same personnel.
Using an optimized DNA extraction protocol, we recovered DNA adequate to serve as a suitable RQ-PCR template from the majority of specimens. There was no significant difference in total DNA recoveries or in DNA tested per PCR reaction between samples from the two hospitals. Yields of DNA tended to be more variable from older archival samples. Also, PCR inhibitory activity was present in almost half of the DNA samples (which was circumvented by sample dilution). Such problems, if unrecognized and not addressed, lessen the chances of detecting viral sequences that might be present.
Efforts to determine the significance of SV40 association with lymphomas have been hampered by disparate findings reported in the literature. Sixteen controlled studies involving PCR assays for SV40 DNA performed in parallel in human lymphomas and control samples were reviewed recently.6 Overall, 19% of 2033 lymphoma samples were SV40 DNA-positive by PCR, as were 3.2% of 1207 control samples (P<0.001). The frequency of SV40 detection in lymphomas varied from 0% to 56% among the studies. The majority of control specimens that were SV40-positive were blood samples, and it is noteworthy that a number of reports have detected SV40 DNA in peripheral blood cells from healthy individuals.6 We have proposed that these discrepant findings could reflect technical matters pertaining to specimen analysis and/or population or geographic differences in the subjects studied.6,19 The study described here provides, for the first time, direct evidence from PCR analyses carried out in a single laboratory (thereby excluding technical differences) that SV40-positive lymphoma frequencies can vary significantly among groups.
A possible explanation for the observed differences between the two populations with respect to SV40 positivity of tumors is that their backgrounds may have reflected, in part, the localized use of SV40-contaminated oral (Sabin) poliovaccines almost 50 years ago. We hypothesize that exposure to prelicensure contaminated oral poliovaccine during field trials in 1959–1961 seeded SV40 into those populations and the virus established transmissible human infections that continue today. This hypothesis focuses on the oral poliovaccine because levels of contaminating SV40 were reportedly 100-fold or more higher than in contaminated killed (Salk) poliovaccines; we speculate that the successful establishment of human infections by SV40 was dose-dependent. Although the SV40-contaminated oral vaccine was used only sparingly in the US, it was used more widely in Mexico and several Central and South American countries. The relevance of the hypothesis to this study is that many immigrants in Houston from Mexico and Central America utilize the public hospital (BTGH) for their medical care. Patient country of origin information was not available for the samples analyzed, but future studies might be designed that could test this hypothesis.
Part of the explanation for the group-specific results might also relate to patient age. We observed that SV40 positivity was significantly associated with lymphoma patients ≤50 compared to those >50 from the VAMC, which in general has an elder population. This possible association of SV40-positive lymphomas with age has not been reported previously. Because it is known that the incidence of lymphoma increases with age,42 we speculate that SV40 infection could contribute to the early appearance of disease in some subjects.
SV40 viral loads in virus-positive lymphomas were low (1–90 genome copies per 1000 cells). The relevance of the viral sequences to the malignancies in which they were found was not addressed in this study. SV40 may have played a role in the induction and early promotion of lymphomagenesis, but as the lymphomas evolved and accumulated genetic alterations the viral growth-promoting effects may have become redundant and viral DNA was lost from some cells. Alternatively, SV40 may establish low-grade infections in lymphoid tissue and its detection in the tumors might be a sign of persistent infections. Finally, the DNAs recovered from paraffin-embedded tissues are highly fragmented and the efficiency of detection of viral sequences may not be optimal, resulting in an underestimation of viral content.
In summary, we have shown that the frequencies of SV40 and EBV associations with lymphomas vary between groups with different demographic characteristics. It appears that SV40 association with lymphomagenesis is affected by population characteristics. However, because of the numbers of samples tested, these observations should be confirmed in population-based studies involving larger numbers of samples.
Supplementary Material
Acknowledgements
We thank Dr. Paul Ling for providing the pBSK-EBER plasmid.
Conflicts of interest
Funding: This work was supported in part by training grant T32 AI007456 (ST) and research grants R01 CA104818 (JSB) and P30 CA125123 from the National Institutes of Health. The content is solely the responsibility of the authors and does not necessarily represent the official view of the National Cancer Institute, the National Institute of Allergy and Infectious Diseases, or the National Institutes of Health.
Abbreviations
- SV40
simian virus 40
- EBV
Epstein-Barr virus
- NHL
non-Hodgkin lymphoma
- T-ag
large tumor antigen
- BTGH
Ben Taub General Hospital
- VAMC
Michael E. DeBakey Veterans Affairs Medical Center
- PCR
polymerase chain reaction
- RQ-PCR
real-time quantitative PCR
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Competing interests: None.
Ethical approval: Approval for this study was received from the Institutional Review Board for Studies of Human Subject Research at Baylor College of Medicine.
References
- 1.Fisher SG, Fisher RI. The epidemiology of non-Hodgkin’s lymphoma. Oncogene. 2004;23:6524–6534. doi: 10.1038/sj.onc.1207843. [DOI] [PubMed] [Google Scholar]
- 2.Klein E, Kis LL, Klein G. Epstein-Barr virus infection in humans: from harmless to life endangering virus–lymphocyte interactions. Oncogene. 2007;26:1297–1305. doi: 10.1038/sj.onc.1210240. [DOI] [PubMed] [Google Scholar]
- 3.International Agency for Research on Cancer. Epstein-Barr virus and Kaposi’s sarcoma herpesvirus/human herpesvirus 8. vol. 70. Lyon: World Health Organization; 1997. IARC Monographs on the Evaluation of Carcinogenic Risks to Humans. [PMC free article] [PubMed] [Google Scholar]
- 4.Vilchez RA, Butel JS. Emergent human pathogen simian virus 40 and its role in cancer. Clin Microbiol Rev. 2004;17:495–508. doi: 10.1128/CMR.17.3.495-508.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gazdar AF, Butel JS, Carbone M. SV40 and human tumours: Myth, association or causality? Nat Rev Cancer. 2002;2:957–964. doi: 10.1038/nrc947. [DOI] [PubMed] [Google Scholar]
- 6.Butel JS. SV40, human infections, and cancer: emerging concepts and causality considerations. In: Khalili K, Jeang KT, editors. Viral Oncology: Basic Science and Clinical Applications. Wiley-Blackwell; 2009. in press. [Google Scholar]
- 7.Pagano JS, Blaser M, Buendia MA, Damania B, Khalili K, Raab-Traub N, et al. Infectious agents and cancer: criteria for a causal relation. Semin Cancer Biol. 2004;14:453–471. doi: 10.1016/j.semcancer.2004.06.009. [DOI] [PubMed] [Google Scholar]
- 8.Barbanti-Brodano G, Sabbioni S, Martini F, Negrini M, Corallini A, Tognon M. Simian virus 40 infection in humans and association with human diseases: Results and hypotheses. Virology. 2004;318:1–9. doi: 10.1016/j.virol.2003.09.004. [DOI] [PubMed] [Google Scholar]
- 9.Butel JS. Viral carcinogenesis: revelation of molecular mechanisms and etiology of human disease. Carcinogenesis. 2000;21:405–426. doi: 10.1093/carcin/21.3.405. [DOI] [PubMed] [Google Scholar]
- 10.Ahuja D, Sáenz-Robles MT, Pipas JM. SV40 large T antigen targets multiple cellular pathways to elicit cellular transformation. Oncogene. 2005;24:7729–7745. doi: 10.1038/sj.onc.1209046. [DOI] [PubMed] [Google Scholar]
- 11.Butel JS, Lednicky JA. Cell and molecular biology of simian virus 40: implications for human infections and disease. J Natl Cancer Inst. 1999;91:119–134. doi: 10.1093/jnci/91.2.119. [DOI] [PubMed] [Google Scholar]
- 12.Imperiale MJ, Major EO. Polyomaviruses. In: Knipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA, Roizman B, Straus SE, editors. Fields Virology. 5th ed. Philadelphia: Lippincott Williams & Wilkins; 2007. pp. 2263–2298. [Google Scholar]
- 13.Stratton K, Almario DA, McCormick MC. Immunization Safety Review: SV40 Contamination of Polio Vaccine and Cancer. Washington, DC: The National Academies Press; 2003. [PubMed] [Google Scholar]
- 14.Rollison DEM. Epidemiologic studies of polyomaviruses and cancer: Previous findings, methodologic challenges and future directions. In: Ahsan N, editor. Polyomaviruses and Human Diseases. Georgetown, TX: Landes Biosciences; 2006. pp. 342–356. [DOI] [PubMed] [Google Scholar]
- 15.Vilchez RA, Madden CR, Kozinetz CA, Halvorson SJ, White ZS, Jorgensen JL, et al. Association between simian virus 40 and non-Hodgkin lymphoma. Lancet. 2002;359:817–823. doi: 10.1016/S0140-6736(02)07950-3. [DOI] [PubMed] [Google Scholar]
- 16.Shivapurkar N, Harada K, Reddy J, Scheuermann RH, Xu Y, McKenna RW, et al. Presence of simian virus 40 DNA sequences in human lymphomas. Lancet. 2002;359:851–852. doi: 10.1016/S0140-6736(02)07921-7. [DOI] [PubMed] [Google Scholar]
- 17.Nakatsuka S-I, Liu A, Dong Z, Nomura S, Takakuwa T, Miyazato H, et al. Simian virus 40 sequences in malignant lymphomas in Japan. Cancer Res. 2003;63:7606–7608. [PubMed] [Google Scholar]
- 18.Shivapurkar N, Takahashi T, Reddy J, Zheng Y, Stastny V, Collins R, et al. Presence of simian virus 40 DNA sequences in human lymphoid and hematopoietic malignancies and their relationship to aberrant promoter methylation of multiple genes. Cancer Res. 2004;64:3757–3760. doi: 10.1158/0008-5472.CAN-03-3307. [DOI] [PubMed] [Google Scholar]
- 19.Meneses A, Lopez-Terrada D, Zanwar P, Killen DE, Monterroso V, Butel JS, et al. Lymphoproliferative disorders in Costa Rica and simian virus 40. Haematologica. 2005;90:1635–1642. [PubMed] [Google Scholar]
- 20.Amara K, Trimeche M, Ziadi S, Laatiri A, Hachana M, Sriha B, et al. Presence of simian virus 40 DNA sequences in diffuse large B-cell lymphomas in Tunisia correlates with aberrant promoter hypermethylation of multiple tumor suppressor genes. Int J Cancer. 2007;121:2693–2702. doi: 10.1002/ijc.23038. [DOI] [PubMed] [Google Scholar]
- 21.Zekri AR, Mohamed W, Bahnassy A, Refat L, Khaled M, Shalaby S, et al. Detection of simian virus 40 DNA sequences in Egyptian patients with different hematological malignancies. Leuk Lymphoma. 2007;48:1828–1834. doi: 10.1080/10428190701534408. [DOI] [PubMed] [Google Scholar]
- 22.Vilchez RA, Lopez-Terrada D, Middleton JR, Finch CJ, Killen DE, Zanwar P, et al. Simian virus 40 tumor antigen expression and immunophenotypic profile of AIDS-related non-Hodgkin’s lymphoma. Virology. 2005;342:38–46. doi: 10.1016/j.virol.2005.06.053. [DOI] [PubMed] [Google Scholar]
- 23.Daibata M, Nemoto Y, Kamioka M, Imai S, Taguchi H. Simian virus 40 in Japanese patients with lymphoproliferative disorders. Br J Haematol. 2003;121:190–191. doi: 10.1046/j.1365-2141.2003.04245_1.x. [DOI] [PubMed] [Google Scholar]
- 24.Shah KV. SV40 and human cancer: A review of recent data. Int J Cancer. 2006;120:215–223. doi: 10.1002/ijc.22425. [DOI] [PubMed] [Google Scholar]
- 25.Hernández-Losa J, Fedele CG, Pozo F, Tenorio A, Fernández V, Castellvi J, et al. Lack of association of polyomavirus and herpesvirus types 6 and 7 in human lymphomas. Cancer. 2005;103:293–298. doi: 10.1002/cncr.20801. [DOI] [PubMed] [Google Scholar]
- 26.Montesinos-Rongen M, Besleaga R, Heinsohn S, Siebert R, Kabisch H, Wiestler OD, et al. Absence of simian virus 40 DNA sequences in primary central nervous system lymphoma in HIV-negative patients. Virchows Archiv. 2004;444:436–438. doi: 10.1007/s00428-004-1001-9. [DOI] [PubMed] [Google Scholar]
- 27.Sui L-F, Williamson J, Lowenthal RM, Parker AJC. Absence of simian virus 40 (SV40) DNA in lymphoma samples from Tasmania, Australia. Pathology. 2005;37:157–159. doi: 10.1080/00313020500058474. [DOI] [PubMed] [Google Scholar]
- 28.Capello D, Rossi D, Gaudino G, Carbone A, Gaidano G. Simian virus 40 infection in lymphoproliferative disorders. Lancet. 2003;361:88–89. doi: 10.1016/S0140-6736(03)12157-5. [DOI] [PubMed] [Google Scholar]
- 29.MacKenzie J, Wilson KS, Perry J, Gallagher A, Jarrett RF. Association between simian virus 40 DNA and lymphoma in the United Kingdom. J Natl Cancer Inst. 2003;95:1001–1003. doi: 10.1093/jnci/95.13.1001. [DOI] [PubMed] [Google Scholar]
- 30.Brousset P, de Araujo V, Gascoyne RD. Immunohistochemical investigation of SV40 large T antigen in Hodgkin and non-Hodgkin’s lymphoma. Int J Cancer. 2004;112:533–535. doi: 10.1002/ijc.20397. [DOI] [PubMed] [Google Scholar]
- 31.Schüler F, Dölken SC, Hirt C, Dölken MT, Mentel R, Gürtler LG, et al. No evidence for simian virus 40 DNA sequences in malignant non-Hodgkin lymphomas. Int J Cancer. 2006;118:498–504. doi: 10.1002/ijc.21346. [DOI] [PubMed] [Google Scholar]
- 32.Jaffe ES, Harris NL, Stein H, Vardiman JW. Pathology & genetics: tumours of hematopoietic and lymphoid tissues. vol. III. Lyon, France: IARC Press; 2001. World Health Organization Classification of Tumours. [Google Scholar]
- 33.Lednicky JA, Butel JS. Consideration of PCR methods for the detection of SV40 in tissue and DNA specimens. Dev Biol Stand. 1998;94:155–164. [PubMed] [Google Scholar]
- 34.Lednicky JA, Butel JS. Tissue culture adaptation of natural isolates of simian virus 40: changes occur in viral regulatory region but not in carboxy-terminal domain of large T-antigen. J Gen Virol. 1997;78:1697–1705. doi: 10.1099/0022-1317-78-7-1697. [DOI] [PubMed] [Google Scholar]
- 35.McNees AL, White ZS, Zanwar P, Vilchez RA, Butel JS. Specific and quantitative detection of human polyomaviruses BKV, JCV, and SV40 by real time PCR. J Clin Virol. 2005;34:52–62. doi: 10.1016/j.jcv.2004.12.018. [DOI] [PubMed] [Google Scholar]
- 36.Ling PD, Vilchez RA, Keitel WA, Poston DG, Peng RS, White ZS, et al. Epstein-Barr virus DNA loads in adult human immunodeficiency virus type 1-infected patients receiving highly active antiretroviral therapy. Clin Infect Dis. 2003;37:1244–1249. doi: 10.1086/378808. [DOI] [PubMed] [Google Scholar]
- 37.Garbuglia AR, Iezzi T, Capobianchi MR, Pignoloni P, Pulsoni A, Sourdis J, et al. Detection of TT virus in lymph node biopsies of B-cell lymphoma and Hodgkin's disease, and its association with EBV infection. Int J Immunopathol Pharmacol. 2003;16:109–118. doi: 10.1177/039463200301600204. [DOI] [PubMed] [Google Scholar]
- 38.Sasikala PS, Nirmala K, Sundersingh S, Mahji U, Rajkumar T. Frequency and distribution of Epstein–Barr virus infection and its association with p53 expression in a series of primary nodal non-Hodgkin lymphoma patients from South India. Int J Lab Hematol. 2009 doi: 10.1111/j.1751-553X.2008.01125.x. in press. [DOI] [PubMed] [Google Scholar]
- 39.Wang J, Young L, Win W, Taylor CR. Distribution and ZAP-70 expression of WHO lymphoma categories in Shanxi, China: a review of 447 cases using a tissue microarray technique. Appl Immunohistochem Mol Morphol. 2005;13:323–332. doi: 10.1097/01.pai.0000176161.38402.b2. [DOI] [PubMed] [Google Scholar]
- 40.Levine AM. Acquired immunodeficiency syndrome-related lymphoma. Blood. 1992;80:8–20. [PubMed] [Google Scholar]
- 41.Ballerini P, Gaidano G, Gong JZ, Tassi V, Saglio G, Knowles DM, et al. Multiple genetic lesions in acquired immunodeficiency syndrome-related non-Hodgkin’s lymphoma. Blood. 1993;81:166–176. [PubMed] [Google Scholar]
- 42.Morton LM, Wang SS, Devesa SS, Hartge P, Weisenburger DD, Linet MS. Lymphoma incidence patterns by WHO subtype in the United States, 1992–2001. Blood. 2006;107:265–276. doi: 10.1182/blood-2005-06-2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.