Abstract
Caenorhabditis elegans F09E10.3 (dhs-25) was identified as encoding a 3-oxoacyl-thioester reductase, potentially of the mitochondrial type 2 fatty acid synthase (FASII) system. Mitochondrial FASII is a relatively recent discovery in metazoans, and the relevance of this process to animal physiology has not been elucidated. A good animal model to study the role of FASII is the nematode C. elegans. However, the components of nematode mitochondrial FASII have hitherto evaded positive identification. The nematode F09E10.3 protein was ectopically expressed without an additional mitochondrial targeting sequence in Saccharomyces cerevisiae mutant cells lacking the homologous mitochondrial FASII enzyme 3-oxoacyl-ACP reductase Oar1p. These yeast oar1Δ mutants are unable to respire, grow on nonfermentable carbon sources, or synthesize sufficient levels of lipoic acid. Mutant yeast cells producing a full-length mitochondrial F09E10.3 protein contained NAD+-dependent 3-oxoacyl-thioester reductase activity and resembled the corresponding mutant overexpressing native Oar1p for the above-mentioned phenotype characteristics. This is the first identification of a metazoan 3-oxoacyl-thioester reductase (see Note Added in Proof).
1. Introduction
Eukaryotes have long been thought to undertake de novo fatty acid biosynthesis exclusively in their cytosol. This acyl carrier protein-(ACP-) dependent process relies on a single associative type 1 fatty acid synthase (FASI), which accommodates all the necessary enzyme activities. However, this traditional view has since been updated following the discovery of an additional mitochondrial FASII in yeast and mammals (reviewed in [1, 2]). FASII represents a dissociated system in which each of the enzyme activities occurs within a discrete protein. Although all of the fungal—and most of the mammalian—FASII constituents have been identified, the metazoan 3-oxoacyl-ACP reductase component has hitherto not been reported.
The present work is concerned with identifying the nematode mitochondrial FASII enzyme corresponding to 3-oxoacyl-ACP reductase. The C. elegans genome has been fully sequenced [5], and a cursory glance at the databases (WormBase, http://www.wormbase.org/) reveals that the nematode contains numerous enzymes with amino acid similarities to the mitochondrial 3-oxoacyl-ACP reductase Oar1p of Saccharomyces cerevisiae FASII [6]. In S. cerevisiae, FASII is critical for mitochondrial function and morphology, and a lesion to any one of the genes encoding FASII proteins results in a respiratory growth phenotype, underdeveloped mitochondria lacking cytochromes and, in some cases, loss of mitochondrial DNA [1, 2]. Here, S. cerevisiae was employed as a heterologous system for determining whether any of the candidate nematode proteins might represent a mitochondrial 3-oxoacyl-ACP reductase, by monitoring their ability to replace fungal Oar1p. S. cerevisiae oar1Δ cells heterologously expressing an active nematode 3-oxoacyl-thioester reductase were compared to an otherwise isogenic strain relying on an ectopically expressed native reductase for growth on glycerol, respiration, and lipoic acid production. The results in yeast are discussed with reference to the potential for a dysfunctional FASII in extending nematode lifespan.
2. Materials and Methods
2.1. S. cerevisiae Strain, Plasmids, and Oligonucleotides
Plasmids and oligonucleotides used are outlined in Table 1. The Escherichia coli strain TOP10 F′ was used for all plasmid amplifications and isolations. The mutant yeast strain BY4741oar1Δ (MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 ykl055c::kanMX) was purchased from EUROSCARF (http://www.uni-frankfurt.de/). Expression plasmids were introduced into BY4741oar1Δ cells using a published method [7], and transformants were enriched on solid SD-Ura medium, described below.
Table 1.
Plasmids and oligonucleotides used.
| Plasmids or oligonucleotides | Description | Source/reference |
|---|---|---|
| Plasmida | ||
| (1) pBluescript KS II (+) | pKS cloning vector | Stratagene |
| pKS:F09E10.31 | F09E10.3 in pBluescript | This study |
| pKS:D1054.81 | D1054.8 in pBluescript | This study |
| (2) YEp352 | URA3-marked multicopy plasmid | [3] |
| (3) YEp:CTA12 | CTA1 behind its own promoter | [4] |
| YEp:F09E10.33 | F09E10.3 behind the CTA1 promoter | This study |
| YEp:D1054.83 | D1054.8 behind the CTA1 promoter | This study |
| YEp:OAR13 | OAR1 behind the CTA1 promoter | Z.-J. Chen |
| oligonucleotideb | ||
| Ce OAR1-1 5′ XbaI | TTATTCTAGATGAGTGCTTTGCTGGGAGG | This study |
| Ce OAR1-1 3′ XhoI | TATTCTCGAGCTACATACTAAATCCACCGG | This study |
| Ce OAR1-2 5′ XbaI | TTATTCTAGATGACTCGTTTTGCTGAAAAAGTC | This study |
| Ce OAR1-2 3′ XhoI | TATTCTCGAGTTAATGTAAAAGTTTTGCAAAG | This study |
aThe superscripts following the plasmid designations refer to their parental vectors, for example, pKS:F09E10.31 was derived from (1) pBluescript KS II (+).
bOligonucleotides Ce OAR1-1 refer to F09E10.3 and Ce OAR1-2 to D1054.8.
2.2. Plasmid Constructions
DNA work and plasmid assembly were undertaken according to standard methods [8]. The F09E10.3 and D1054.8 genes were isolated from cDNA by polymerase chain reaction using Phusion high fidelity DNA polymerase (Finnzymes Oy, Espoo, Finland) and the appropriate oligonucleotide pairs (Table 1). Subsequent processing of the resultant amplicons, including their electrophoresis, purification, and insertion into an EcoRV-digested pBluescript plasmid vector (Stratagene, La Jolla, CA) was performed as described [9]. The pBluescript inserts were isolated following digestion with XbaI and XhoI restriction enzymes, and ligated behind the CTA1 promoter to pYE352:CTA1 [4] from which the CTA1 open reading frame encoding yeast peroxisomal catalase A was removed using the same enzymes. Sequencing of insert nucleotides ensured that no mutations were introduced during the amplification process. Construction of plasmid YEp352:OAR1 expressing yeast OAR1 behind the CTA1 promoter will be described elsewhere. (See Note Added in Proof).
2.3. Media, Growth, Enzyme Assays, Respiration Competence and Lipoic Acid Measurements
Standard media were prepared for yeast [10] and E. coli [11]. Episomal plasmids based on the URA3-marked YEp352 plasmid vector [3] were retained in transformed S. cerevisiae strains by applying selective pressure using solid SD-Ura medium consisting of 0.67% (wt/vol) yeast nitrogen base without amino acids, 2% (wt/vol) D-glucose, 3% (wt/vol) agar, and with all necessary supplements added except for uracil (Sigma-Aldrich Inc, St. Louis, MO USA). Solid synthetic complete glycerol medium (SCglycerol) was prepared essentially as the aforementioned SD-Ura medium, with the exceptions that uracil was added and glucose was replaced with 3% (wt/vol) glycerol as the sole carbon source.
For high levels of protein expression prior to enzyme assays, yeast cells were grown on oleic acid medium as described [12]. Cells were broken with glass beads in a buffer consisting of 50 mM KPi (pH 7.0), 200 mM KCl, and 0.1% (wt/vol) Triton X-100. Protein concentrations [13] and reductase activities [14] were determined as described. The reductase activity assay mixture consisted of 50 mM KPi (pH 8.0) and 50 μg/mL bovine serum albumin, 2.0 μL purified protein representing the hydratase 2 domain from human MFE2 (1/10 dilution of a 3 mg/ml stock), 125 μM NAD+ or NADP+, and 60 μM 2-trans-hexenoyl-CoA [15]. In this assay, the reverse reaction was monitored, whereby the 2-trans-hexenoyl-CoA substrate was first hydrated by the aforementioned hydratase 2 domain of human MFE2 into the 3-hydroxyacyl-CoA, followed by the removal of two electrons from C3 by 3-oxoacyl-thioester reductase leaving behind a carbonyl group within the 3-oxoacyl-CoA product. To improve the sensitivity of the reductase assay, which was designed to be performed on crude extracts, the coincidental reduction of NAD+ or NADP+ into the corresponding NAD(H) and NADP(H) species was monitored spectrophotometrically at 340–385 nm, instead of just 340 nm, using a dual wavelength spectrophotometer (Shimadzu Corp., Kyoto, Japan) [16]. Respiration competence was monitored using an overlay consisting of 0.1% (wt/vol) 2,3,5-triphenyltetrazolium chloride (TTC) in phosphate-buffered saline and 1.5% (wt/vol) low-melting temperature agarose [17].
The content of lipoic acid in yeast strains was estimated using a biological assay relying on lipoic acid-deficient bacterial cells described previously [18, 19]. The assay determined the growth of these defective bacterial cells in liquid medium to which extracts were added that were generated from control and tester yeast strains, and the growth values were converted to the amount of lipoic acid present in the yeast cells based on a standard curve. Each yeast strain was grown in triplicates for 48 h in liquid SD-Ura medium selecting for plasmid presence. Acid hydrolysis was carried out in 0.5 ml 9 N H2SO4, and E. coli JRG33 (lipA9) cultures were inoculated to an initial OD600 of 0.015 in 2 ml of 1 × basal growth medium [20] containing 50 mM sodium succinate and grown for 36–48 h.
2.4. Mitochondrial Localisation Predictions and Sequence Comparisons
MitoProt (http://www.mips.biochem.mpg.de/cgi-bin/proj/medgen/mitofilter), PSORT II (http://psort.ims.u-tokyo.ac.jp/form2.html), and TargetP (http://www.cbs.dtu.dk/services/TargetP/) were used to calculate the N-terminal region of proteins that could support a mitochondrial targeting sequence [21, 22] and Multalin (http://npsa-pbil.ibcp.fr/cgi-bin/npsa_automat.pl?page=/NPSA/npsa_multalin.html) and Genedoc (http://www.nrbsc.org/gfx/genedoc/index.html) to generate the sequence comparison in Figure 1.
Figure 1.
Similarity of the C. elegans D1054.8 and F09E10.3 gene products to their bacterial or yeast homologues, FabG and YKL055c (Oar1p), respectively. Comparison of the deduced amino acid sequences was undertaken using Multalin and Genedoc. Dashes indicate the arrangement of the sequences for best fit. Black shadings refer to strictly conserved amino acid residues among all four sequences whereas the darker and lighter grey shadings denote regions with more relaxed residue similarities not necessarily shared by the full set of sequences. MitoProt predictions of cleaved sequences are indicated (D1054.8, *; F09E1.03, +). The catalytic triad of FabG consisting of the amino acid residues serine 148 (S), tyrosine 151 (Y), and lysine 155 (K) are shown in bold below the sequences. Accession numbers used were FabG; AP_001719; D1054.8, CAA98438; F09E10.3, AAB52488; YKL055c, P35731.
3. Results
3.1. Selection of Nematode F09E10.3 and D1054.8 for Analysis
The WormBase homology group identifier KOG1200, which refers to “mitochondrial/plastidial beta-ketoacyl-ACP reductases” (3-oxoacyl-ACP reductases), lists Y39A1A.11/dhs-11 and F09E1.03/dhs-25 as potentially encoding this activity in the nematode. However, there are approximately 28 additional genes annotated with the dhs designation (alcohol dehydrogenase/short chain). Moreover, the C. elegans genome contains a multitude of genes whose products are homologous to yeast mitochondrial Oar1p. To determine which of these genes might encode proteins residing in mitochondria, the MitoProt (MP), PSORT II (PS), and TargetP (TP) algorithms were used to analyze the deduced amino acid sequences of the candidate genes, resulting in a prediction that the sequence relating to F09E10.3 (MP, 0.9119; PS, 0.1; TP, 0.540) had a probability of representing a mitochondrial protein that was equivalent to or higher than that calculated for the known yeast mitochondrial protein Oar1p (0.9166, 0.1, 0.244). In addition to F09E10.3, D1054.8 (0.9232, 0.439, 0.505) was also selected for investigation as it was previously chronicled as the sole potential FASII-like 3-oxoacyl-ACP reductase gene whose RNAi inactivation affected worm longevity [23]. The conceptual translation products of both D1054.8 and F09E10.3 contained amino acid residues that appeared to correspond to the catalytic triad previously characterized in bacterial FabG [24], albeit only within the amino acid sequence encoded by the latter nematode gene was the distance across the triad region fully conserved (Figure 1).
3.2. Nematode F09E10.3 Restores Respiratory Growth to Yeast oar1Δ Cells
To determine whether F09E10.3 or D1054.8 might code for 3-oxoacyl-ACP reductases, the genes were expressed behind the promoter of yeast CTA1 encoding peroxisomal catalase A (Cta1p), which is only moderately derepressed on nonfermentable carbon sources such as glycerol, used here for complementation assays. On glycerol medium, yeast cells shift their metabolism to respiration, which requires an operational FASII within functional mitochondria [1, 2]. An extra fungal mitochondrial leader sequence was not fused to the nematode proteins. Yeast oar1Δ mutant cells were transformed with plasmids expressing fungal Oar1p (positive control), native Cta1p (negative control), or each of the nematode F09E10.3 or D1054.8 protein products and were applied to solid SCglycerol medium. This demonstrated that only cells expressing F09E10.3 or OAR1 could grow and divide (see below).
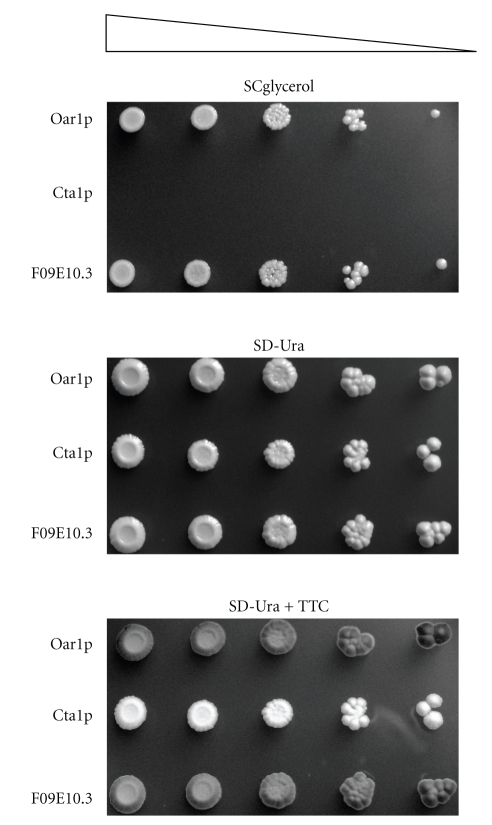
To assess the efficiency of the nematode F09E10.3 protein at replacing yeast Oar1p, the aforementioned transformants were propagated on SD-Ura medium, and following tenfold serial dilution, cultures were spotted onto SD-Ura or SCglycerol media, and the plates were incubated at 30°C until single colonies were detectable. The results in Figure 2 demonstrated that like the situation with the self-complemented strain, mutant cells expressing F09E10.3 could thrive on glycerol, whereas those mutant cells expressing CTA1 were not able to proliferate on this medium type. Hence, the respiratory-deficient phenotype of mutant oar1Δ cells was rescued following the expression of F09E10.3, indicating functional complementation.
Figure 2.
Phenotype rescue of a yeast oar1Δ strain expressing F09E10.3. Mutant S. cerevisiae BY4741oar1Δ cells producing native mitochondrial Oar1p (positive control), peroxisomal catalase A Cta1p (negative control), or the nematode mitochondrial protein F09E10.3 (tester strain), were grown on SD-Ura medium selecting for plasmid presence, and following tenfold serial dilution (triangle) were spotted onto the indicated solid media. Respiration competence was examined by applying 2,3,5-triphenyltetrazolium chloride (TTC) as an overlay to the SD-Ura medium.
Yeast cells lacking the native OAR1 gene have been shown before to lack assembled cytochrome complexes [6], and so their rehabilitation to normal respiratory growth was expected to occur contemporaneously with the assembly of cytochrome complexes and the regeneration of the electron transport chain. To examine whether mutant cells expressing Oar1p or the F09E10.3 protein contained a regenerated electron transfer chain, respiration competence was assayed by applying to the cells spotted onto solid SD-Ura medium an overlay consisting of 2,3,5-triphenyltetrazolium chloride (TTC). The results revealed that only mutant oar1Δ cells expressing Oar1p or the F09E10.3 protein could rapidly metabolize TTC to generate the red chromophore, whereas those expressing Cta1p were essentially white (lower panel; Figure 2).
3.3. Yeast oar1Δ Cells Expressing F09E10.3 Contain 3-Oxoacyl-Thioester Reductase Activity
FASII enzyme activities can often be measured in soluble protein extracts made from mutant yeast cells ectopically expressing heterologous proteins. For example, enzyme activities for 2-trans-enoyl-thioester reductase can be detected in protein extracts made from yeast etr1Δ mutant cells producing S. cerevisiae Etr1p [25], Candida tropicalis Etr1p or Etr2p [25, 26], and Mycobacterium tuberculosis InhA [9]. However, other FASII enzyme activities are less amenable to detection using this procedure. Although discernible levels of 3-hydroxyacyl-thioester dehydratase activity can be measured in extracts made from yeast htd2Δ mutants producing mycobacterial HadAB, HadBC, HtdX, HtdY, and HtdZ [27, 28], the sole documented measurement of dehydratase activity in mutant cells enriched for native Htd2p could only be undertaken following the laborious preparation of pure mitochondria [29].
To examine whether the F09E10.3 protein contained reductase activity, yeast cells were grown in triplicate cultures on oleic acid medium [12] under which conditions the CTA1 promoter is highly induced [30]. Oleic acid has not been shown previously to interfere with yeast FASII [9, 25, 27, 28]. The cells were broken with glass beads, and the ensuant soluble protein extracts were subjected to spectrophotometric analysis for reductase activity (in the reverse orientation), which ostensibly could depend on either NAD+ or NADP+ as co-factor. Enzyme assays performed on extracts from oar1Δ cells over-expressing native Oar1p, a presumed NADP+-dependent enzyme, did not yield appreciable levels of activity using the present assay. Likewise, mutant cells over-expressing Cta1p also failed to produce extracts with a specific reductase activity. On the other hand, soluble protein extracts obtained from mutant cells over-expressing the F09E10.3 protein gave rise to an NAD+-dependent oxidation of the 2-trans-hexenoyl-CoA substrate at the rate of 0.97 ± 0.18 (S.D., n = 3) μmol/mg protein x min−1. NADP+ did not act as co-factor when examining the nematode protein for reductase activity. Hence, ectopic expression of C. elegans F09E10.3 in yeast oar1Δ mutant cells resulted in clearly discernible levels of catalytic activity in an assay designed to expose 3-oxoacyl-thioester reductases.
3.4. Expression of F09E10.3 Restores Lipoic Acid Production in the oar1Δ Mutant
Lastly, to couple the expression of the F09E10.3 protein with fatty acid biosynthesis in yeast mitochondria, lipoic acid synthesis was measured in these strains. The results showed that the negative control strain (oar1Δ cells expressing peroxisomal catalase A) yielded extracts supporting only a minimal growth level of the mutant bacterial cells that was equivalent to 25 ± 3 ng lipoic acid per gram wet weight yeast cells, whereas levels of lipoic acid in mutant cells expressing fungal Oar1p or the nematode F09E10.3 protein gave rise to a bacterial growth that corresponded to 138 ± 22 or 80 ± 4 ng lipoic acid per or gram wet weight, respectively. Hence, the combined results demonstrated that mutant oar1Δ cells could grow on glycerol since their formerly defective mitochondrial functions were restored due to the expression of the full-length F09E10.3 protein, indicating that it represented a putative mitochondrial 3-oxoacyl-thioester reductase.
4. Discussion
The present study demonstrated a physiological function for the gene product of C. elegans F09E10.3/dhs-25 as a possible 3-oxoacyl-thioester reductase of mitochondrial FASII. The F09E10.3 protein could reach its designated subcellular compartment in fungi without resorting to a dedicated yeast mitochondrial targeting sequence, so as to replace the native mitochondrial 3-oxoacyl-ACP reductase Oar1p in S. cerevisiae oar1Δ cells. The efficiency of this interchange was underscored by monitoring F09E10.3-complemented yeast mutant cells for respiratory growth on glycerol medium, metabolism of TTC, and synthesis of lipoic acid. Although the corresponding FASII protein in protozoans has been exposed previously, for example, Plasmodium falciparum FabG was crystallized and characterized enzymatically [31] and potential Oar1p homologues in Trypanosome brucei were shown to be mitochondrial [32], F09E10.3 represents the first metazoan 3-oxoacyl-thioester reductase gene to be positively identified (see Note Added in Proof). WormBase reports no RNAi phenotypes for F09E10.3, but this could be due to redundancy as the result of additional mitochondrial reductases not yet disclosed.
Although all previously studied 3-oxoacyl-thioester reductases have been shown to depend on NADP(H) for catalysis, the finding that the F09E10.3 protein relied on NAD(H) was not altogether surprising. This is because other mitochondrial enzyme activities, such as isocitrate dehydrogenase, are also known to require each of these two cofactors. WormBase lists at least 10 potential NAD(H)-dependent and an additional 10 potential NADP(H)-dependent isocitrate dehydrogenases (gene onthologies GO:0004449 and GO:0004450, resp.). It is tempting to speculate that future studies will uncover additional 3-oxoacyl-thioester reductases in metazoan mitochondria that would similarly depend on NAD(H) rather than on NADP(H).
The role of mitochondrial processes in affecting nematode longevity has been studied extensively using RNAi technology [23, 33, 34]. Dysfunctional mitochondrial activities that have been previously linked to a prolonged nematode lifespan include the electron transport chain, RNA editing, and cytochrome complex formation. However, since mitochondrial FASII has hitherto not been identified in nematodes, its involvement in C. elegans lifespan extension could not be specifically addressed. If the lifespan extension observed with the RNAi intervention of C. elegans W09H1.5 [23], which encodes a potential homologue of the aforementioned yeast Etr1p, is in any way indicative, then a dysfunctional mitochondrial FASII might be conducive to longer nematode life. Worm longevity notwithstanding, the issue is that yeast FASII mutants will likely continue to play an important role in exposing FASII enzymes in metazoan mitochondria.
Note Added in Proof —
Identification of human NAD(H)-dependent 3-oxoacyl-ACP reductase, HsKAR, of mitochondrial FASII, and the description of pYE352:OAR1 have been published online [35].
Acknowledgments
J. Kalervo Hiltunen is kindly thanked for setting up the enzyme assays and providing additional assistance whenever required. The author also wishes to express his gratitude to Alexander J. Kastaniotis for his advice and help throughout this work and for undertaking the lipoic acid measurements together with Katri Näppä. The unpublished plasmid YEp:OAR1 was a generous gift by Zhi-Jun Chen. The author is grateful to Tatu Haataja for donating the hydratase 2 domain of human MFE2 (all from the University of Oulu, Finland). This work was supported in part by Grant P20764-B03 from the Austrian Science Fund (FWF) as well as Grant H 1668/2006 from the Hochschuljubiläumsstiftung der Stadt Wien.
References
- 1.Hiltunen JK, Okubo F, Kursu VAS, Autio KJ, Kastaniotis AJ. Mitochondrial fatty acid synthesis and maintenance of respiratory competent mitochondria in yeast. Biochemical Society Transactions. 2005;33(5):1162–1165. doi: 10.1042/BST20051162. [DOI] [PubMed] [Google Scholar]
- 2.Hiltunen JK, Schonauer MS, Autio KJ, Mittelmeier TM, Kastaniotis AJ, Dieckmann CL. Mitochondrial fatty acid synthesis type II: more than just fatty acids. The Journal of Biological Chemistry. 2009;284(14):9011–9015. doi: 10.1074/jbc.R800068200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hill JE, Myers AM, Koerner TJ, Tzagoloff A. Yeast/E. coli shuttle vectors with multiple unique restriction sites. Yeast. 1986;2(3):163–167. doi: 10.1002/yea.320020304. [DOI] [PubMed] [Google Scholar]
- 4.Flippula SA, Sormunen RT, Hartig A, Kunau W-H, Hiltunen JK. Changing stereochemistry for a metabolic pathway in vivo. Experiments with the peroxisomal β-oxidation in yeast. The Journal of Biological Chemistry. 1995;270(46):27453–27457. doi: 10.1074/jbc.270.46.27453. [DOI] [PubMed] [Google Scholar]
- 5.Consortium. Genome sequence of the nematode C. elegans: a platform for investigating biology. Science. 1998;282(5396):2012–2018. doi: 10.1126/science.282.5396.2012. [DOI] [PubMed] [Google Scholar]
- 6.Schneider R, Brors B, Burger F, Camrath S, Weiss H. Two genes of the putative mitochondrial fatty acid synthase in the genome of Saccharomyces cerevisiae. Current Genetics. 1997;32(6):384–388. doi: 10.1007/s002940050292. [DOI] [PubMed] [Google Scholar]
- 7.Chen D-C, Yang B-C, Kuo T-T. One-step transformation of yeast in stationary phase. Current Genetics. 1992;21(1):83–84. doi: 10.1007/BF00318659. [DOI] [PubMed] [Google Scholar]
- 8.Ausubel FM, Brent R, Kingston RE, et al. Current Protocols in Molecular Biology. New York, NY, USA: John Wiley & Sons; 1989. [Google Scholar]
- 9.Gurvitz A, Hiltunen JK, Kastaniotis AJ. Function of heterologous Mycobacterium tuberculosis InhA, a type 2 fatty acid synthase enzyme involved in extending C20 fatty acids to C60-to-C90 mycolic acids, during de novo lipoic acid synthesis in Saccharomyces cerevisiae. Applied and Environmental Microbiology. 2008;74(16):5078–5085. doi: 10.1128/AEM.00655-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rose MD, Winston F, Heiter P. Methods in Yeast Genetics: A Laboratory Manual. Cold Spring Harbor, NY, USA: Cold Spring Harbor Laboratory; 1990. [Google Scholar]
- 11.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY, USA: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 12.Gurvitz A, Rottensteiner H, Kilpeläinen SH, et al. The Saccharomyces cerevisiae peroxisomal 2,4-dienoyl-coA reductase is encoded by the oleate-inducible gene SPS19. The Journal of Biological Chemistry. 1997;272(35):22140–22147. doi: 10.1074/jbc.272.35.22140. [DOI] [PubMed] [Google Scholar]
- 13.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein dye binding. Analytical Biochemistry. 1976;72(1-2):248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- 14.Dommes V, Kunau WH. 2,4-dienoyl coenzyme A reductases from bovine liver and Escherichia coli. Comparison of properties. The Journal of Biological Chemistry. 1984;259(3):1781–1788. [PubMed] [Google Scholar]
- 15.Goldman P, Vagelos PR. The specificity of triglyceride synthesis from diglycerides in chicken adipose tissue. The Journal of Biological Chemistry. 1961;236:2620–2623. [PubMed] [Google Scholar]
- 16.Hiltunen JK. Metabolic effects of pent-4-enoate in isolated perfused rat heart. Biochemical Journal. 1978;170(2):241–247. doi: 10.1042/bj1700241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Böker-Schmitt E, Francisci S, Schweyen RJ. Mutations releasing mitochondrial biogenesis from glucose repression in Saccharomyces cerevisiae. Journal of Bacteriology. 1982;151(1):303–310. doi: 10.1128/jb.151.1.303-310.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hayden MA, Huang IY, Iliopoulos G, Orozco M, Ashley GW. Biosynthesis of lipoic acid: characterization of the lipoic acid auxotrophs Escherichia coli W1485-lip2 and JRG33-lip9. Biochemistry. 1993;32(14):3778–3782. doi: 10.1021/bi00065a033. [DOI] [PubMed] [Google Scholar]
- 19.Brody S, Oh C, Hoja U, Schweizer E. Mitochondrial acyl carrier protein is involved in lipoic acid synthesis in Saccharomyces cerevisiae. FEBS Letters. 1997;408(2):217–220. doi: 10.1016/s0014-5793(97)00428-6. [DOI] [PubMed] [Google Scholar]
- 20.Herbert AA, Guest JR. Turbidimetric and polarographic assays for lipoic acid using mutants of Escherichia coli. Methods in Enzymology. 1970;18:269–272. [Google Scholar]
- 21.Claros MG, Vincens P. Computational method to predict mitochondrially imported proteins and their targeting sequences. European Journal of Biochemistry. 1996;241(3):779–786. doi: 10.1111/j.1432-1033.1996.00779.x. [DOI] [PubMed] [Google Scholar]
- 22.Emanuelsson O, Nielsen H, Brunak S, von Heijne G. Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. Journal of Molecular Biology. 2000;300(4):1005–1016. doi: 10.1006/jmbi.2000.3903. [DOI] [PubMed] [Google Scholar]
- 23.Hamilton B, Dong Y, Shindo M, et al. A systematic RNAi screen for longevity genes in C. elegans. Genes and Development. 2005;19(13):1544–1555. doi: 10.1101/gad.1308205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Price AC, Zhang Y-M, Rock CO, White SW. Structure of β-ketoacyl-[acyl carrier protein] reductase from Escherichia coli: negative cooperativity and its structural basis. Biochemistry. 2001;40(43):12772–12781. doi: 10.1021/bi010737g. [DOI] [PubMed] [Google Scholar]
- 25.Torkko JM, Koivuranta KT, Miinalainen IJ, et al. Candida tropicalis Etr1p and Saccharomyces cerevisiae Ybr026p (Mrf1'p), 2-enoyl thioester reductases essential for mitochondrial respiratory competence. Molecular and Cellular Biology. 2001;21(18):6243–6253. doi: 10.1128/MCB.21.18.6243-6253.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Torkko JM, Koivuranta KT, Kastaniotis AJ, et al. Candida tropicalis expresses two mitochondrial 2-enoyl thioester reductases that are able to form both homodimers and heterodimers. The Journal of Biological Chemistry. 2003;278(42):41213–41220. doi: 10.1074/jbc.M307664200. [DOI] [PubMed] [Google Scholar]
- 27.Gurvitz A, Hiltunen JK, Kastaniotis AJ. Identification of a novel mycobacterial 3-hydroxyacyl-thioester dehydratase, HtdZ (Rv0130), by functional complementation in yeast. Journal of Bacteriology. 2008;190(11):4088–4090. doi: 10.1128/JB.00016-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gurvitz A, Hiltunen JK, Kastaniotis AJ. Heterologous expression of mycobacterial proteins in Saccharomyces cerevisiae reveals two physiologically functional 3-hydroxyacyl-thioester dehydratases, HtdX and HtdY, in addition to HadABC and HtdZ. Journal of Bacteriology. 2009;191(8):2683–2690. doi: 10.1128/JB.01046-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kastaniotis AJ, Autio KJ, Sormunen RT, Hiltunen JK. Htd2p/Yhr067p is a yeast 3-hydroxyacyl-ACP dehydratase essential for mitochondrial function and morphology. Molecular Microbiology. 2004;53(5):1407–1421. doi: 10.1111/j.1365-2958.2004.04191.x. [DOI] [PubMed] [Google Scholar]
- 30.Filipits M, Simon MM, Rapatz W, Hamilton B, Ruis H. A Saccharomyces cerevisiae upstream activating sequence mediates induction of peroxisome proliferation by fatty acids. Gene. 1993;132(1):49–55. doi: 10.1016/0378-1119(93)90513-3. [DOI] [PubMed] [Google Scholar]
- 31.Wickramasinghe SR, Inglis KA, Urch JE, Muller S, van Aalten DMF, Fairlamb AH. Kinetic, inhibition and structural studies on 3-oxoacyl-ACP reductase from Plasmodium falciparum, a key enzyme in fatty acid biosynthesis. Biochemical Journal. 2006;393(2):447–457. doi: 10.1042/BJ20050832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stephens JL, Lee SH, Paul KS, Englund PT. Mitochondrial fatty acid synthesis in Trypanosoma brucei. The Journal of Biological Chemistry. 2007;282(7):4427–4436. doi: 10.1074/jbc.M609037200. [DOI] [PubMed] [Google Scholar]
- 33.Lee SS, Lee RYN, Fraser AG, Kamath RS, Ahringer J, Ruvkun G. A systematic RNAi screen identifies a critical role for mitochondria in C. elegans longevity. Nature Genetics. 2003;33(1):40–48. doi: 10.1038/ng1056. [DOI] [PubMed] [Google Scholar]
- 34.Curran SP, Ruvkun G. Lifespan regulation by evolutionarily conserved genes essential for viability. PLoS Genetics. 2007;3(4, article e56) doi: 10.1371/journal.pgen.0030056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen Z, Kastaniotis AJ, Miinalainen IJ, Rajaram V, Wierenga RK, Hiltunen JK. 17β-Hydroxysteroid dehydrogenase type 8 and carbonyl reductase type 4 assemble as a ketoacyl reductase of human mitochondrial FAS. The FASEB Journal. 2009;2009(23) doi: 10.1096/fj.09-133587. [DOI] [PubMed] [Google Scholar]