Abstract
Dynamic interplay between the plasma membrane and underlying cytoskeleton is essential for cellular shape change. Spatial organization of actin filaments, whose growth generates membrane deformations during motility 1, phagocytosis 2, endocytosis 3, and cytokinesis 4, is mediated by specific protein-protein interactions that branch, crosslink, and bundle filaments into networks that interact with the membrane. Although membrane curvature has been found to influence binding of proteins with curvature-sensitive domains 5, the direct effect of membrane elasticity on cytoskeletal network organization is not clear. Here we show through in vitro reconstitution and elastic modeling that a lipid bilayer can drive the emergence of bundled actin filament protrusions from branched actin filament networks, thus playing a role normally attributed to actin-binding proteins. Formation of these filopodium-like protrusions with only a minimal set of purified proteins points to an active participation of the membrane in organizing actin filaments at the plasma membrane. In this way, elastic interactions between the membrane and cytoskeleton can cooperate with accessory proteins to drive cellular shape change.
Changes in cell shape require reorganization of both the cytoskeleton and the plasma membrane. The cytoskeleton provides cells with internal organization and mechanical rigidity and is composed of filamentous protein polymers assembled by accessory proteins into networks. In contrast, the membrane is a comparatively soft barrier that serves as a substrate for enzymatic reactions and compartmentalizes cells by restricting diffusion. In many modes of cellular shape change, the actin cytoskeleton adjacent to the membrane dynamically reorganizes from one specific arrangement of filaments to another. The growth of dendritic actin networks against the membrane at the leading edge of crawling cells is a well-studied example., A model that details the interactions of actin and actin-binding proteins in protruding lamellipodia, known as the dendritic nucleation model, has been supported by reconstitution of dendritic actin networks that propel solid objects 6-8. However, the influence of the cell membrane, which is both deformable and fluid, on actin network assembly is unclear. Cell-free reconstitution of vesicle motility in cytoplasmic extracts demonstrated that growth of a dendritic actin network can deform spherical vesicles to teardrop shapes in cell extracts 9,10, but it is difficult to attribute the shape change to specific molecular interactions due to the complexity of the extracts.
Here we used giant unilamellar vesicles (GUVs) that nucleated assembly of a dendritic actin network to study the interactions between actin network growth and deformable membranes. Reconstituted actin networks containing actin, N-WASP and Arp2/3 complex were assembled onto synthetic lipid bilayers as previously described 11. When the vesicles were incubated with the purified proteins, local activation of the nucleation promotion factor N-WASP led to Arp2/3-branched dendritic actin network formation on the external leaflet of the GUVs. Surprisingly, we observed thin protrusions emanating from the Arp2/3-branched network (Fig. 1) in nearly 95% of the vesicles over 10 μm in diameter. We observed similar phenotypes for NWASP activation by the synergistic activity of PIP2 and Cdc42 12, by TMR-PIP2 alone 11, and by DOGS-Ni-NTA, which binds and activates His-tagged N-WASP, in the absence of both PIP2 and Cdc42.
Figure 1.
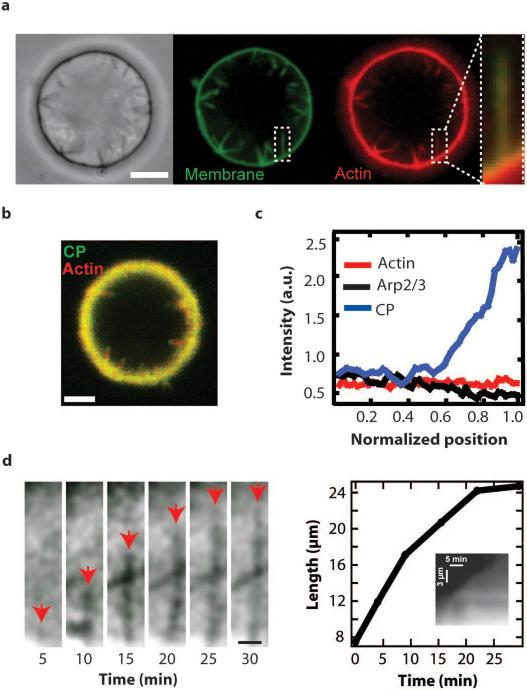
Thin actin protrusions emerge from dendritic actin networks. a, Phase contrast and spinning disk confocal images of membrane (green) and actin (red) show multiple protrusions in the lumen of a GUV. Overlay of the fluorescence images confirms that the membrane protrusions are supported by actin filaments. Scale bar, 5 μm. b, Localization of AF555 actin and AF488 CP fluorescence in thin actin filament protrusions shows that filament barbed ends are concentrated at the tip of the protrusions. AF488 CP was added to the reaction mixtures after > 30 minutes of actin polymerization to label filament barbed ends. Scale bar, 10 μm. c, Localization of actin, Arp2/3 complex, and CP along thin actin filament protrusions. The normalized Arp2/3 complex (n = 4) and CP (n = 3) traces were divided by the normalized actin (n = 7) linescans. d, Elongation of a thin protrusion visualized by phase contrast microscopy. Red arrows highlight the tip of the thin protrusion as it extends. A second, independent protrusion enters the field at 10 minutes and crosses the path of the protrusion that is tracked. Scale bar, 3 μm. The length of the protrusion was tracked through time showing that growth initially occurs quickly but slows down with decreasing monomer concentration. Kymograph analysis of the fluorescently labeled membrane confirms the elongation dynamics of the protrusion.
The membrane deformations generated by actin network growth and protruding into the vesicles can be easily identified with phase contrast microscopy. Using confocal microscopy, we confirmed that the thin protrusions consisted of actin filaments encapsulated by a membrane tube (Fig. 1a). To determine the arrangement of actin filaments within the membrane protrusions, we used fluorescently-labeled capping protein (CP) as a marker for F-actin barbed ends after elongation halted (Fig. 1b). Line scan analysis of the fluorescently-labeled protein demonstrated that F-actin barbed ends were concentrated at the tip of the thin actin filament protrusions (Fig. 1c), consistent with parallel filaments in the protrusion. As expected, the presence of CP during assembly of the actin network inhibited formation of the protrusions. Additional experiments with fluorescently-labeled N-WASP and Arp2/3 complex also suggested that the protrusions lacked dendritic architecture along their length (see Supplementary Information).
Typically, the thin actin filament protrusions formed from the dense membrane-associated dendritic actin networks within the first few minutes after introduction of purified proteins and grew into the lumen of the GUVs. Elongation was tracked over time using epi-fluorescence microscopy of fluorescently labeled lipids and confirmed with phase contrast microscopy (Fig. 1d). The thin actin filament protrusion in Figure 1d initially grew at a rate of ∼ 1 μm/min, gradually slowed, then halted at a terminal length of ∼ 24 μm. We found that the stationary length of thin actin filament protrusions ranged from 1−25 μm (n > 1000). During elongation, the protrusions remained straight with no visible lateral fluctuations. Using a kinetic model based on Mogilner and Rubinstein 13, we found that the observed elongation dynamics were consistent with growth of ∼ 10 filaments within a single protrusion (see Supplementary Information).
To further examine the growth of the thin actin filament protrusions, we conducted a photobleaching experiment combined with confocal imaging to determine where free actin monomers were added (Fig. 2a). Photobleached spots within protrusions showed little recovery (see Supplementary Information) as filament tips elongated (Fig. 2b), indicating monomer addition does not occur along the thin actin filament protrusions. These observations confirmed that protrusions only incorporate new fluorescently-labeled monomers proximal to the membrane at their tips.
Figure 2.
Elongation of a thin actin filament protrusion by polymerization proximal to the membrane. a, Laser scanning confocal images of fluorescence recovery of a photo-bleached region along a protrusion. The red arrow denotes the tip of the protrusion. Scale bar, 1 μm. b, Trajectories of positions along the photo-bleached protrusion. Blue circles mark the tip of the proto-filopodium. Black circles (white on the image) mark the edge of the photo-bleached spot that is proximal to the tip. Red circles mark the edge of the photo-bleached spot which is distal to the tip.
The observed actin-based protrusions show a striking resemblance to cellular filopodia in two respects: (i) elongation of the actin filaments occurs at the tip of the protrusion, and (ii) the protrusions lack the dendritic architecture of the networks from which they emerge 14 15. However, unlike in vivo filopodia, the formation and stability of the thin actin filament protrusions from dendritic actin networks did not require bundling proteins or tip complex proteins. In fact, the spontaneous initiation and growth of the protrusions in our experiments would be considered unlikely due to dendritic actin network geometry and resistance of the membrane to deformation 14. We propose that our observation of thin protrusion growth and stability in the absence of accessory proteins can be explained by considering the interplay between actin network and membrane mechanics.
Using a mechanical model of actin-membrane configurations, we found that a deformable membrane can overcome the bending rigidity of actin filaments to both gather and bundle nearby filaments into a single tubular protrusion, even in the absence of tip complex or bundling proteins. To illustrate how a deformable membrane can gather actin filaments, we computed the minimum energy configuration of an elastic sheet (representing the lipid bilayer) enveloping two semi-flexible protrusions (Fig. 3a, see Supplementary Information for calculation details). In our calculations, the protruding filaments are anchored 100 nm (L0) below and are orthogonal to the unperturbed membrane. For this geometry, we determine the energetically optimal separation of the tips of filaments with contour length L and separation D at their bases. We find that, indeed, membrane elasticity is sufficient to bring together the tips of two nearby nascent protrusions. For a membrane tension of 0.005 kB T/nm2 (ref. 16), the effective range of this membrane-mediated attraction extends beyond the filament thickness (∼ 10 nm) by only a few nanometers when the protrusion length is ∼ 80 nm, but the range of attraction grows rapidly with increasing protrusion length. Thus, if multiple filaments are gathered together by membrane-mediated attraction, their collective polymerization force can overcome the membrane's resistance, resulting in formation of long tubular deformations.
Figure 3.
Role of membrane in formation of thin actin filament protrusions. a, The total energy associated with membrane deformation and filament bending is calculated for two actin filaments anchored 100 nm below the membrane with protrusion length Lmem and separation D. The maximal distance at which it is energetically favorable for two filaments to merge into a single protrusion, or range of attraction, is shown as a function of protrusion length. The lightly shaded region under the curve represents the set of thermodynamically accessible states that will lead to filament bundling by the membrane. The darkly shaded region represents a subset of these states which are kinetically accessible, determined by the branching distance of ∼100 nm 1. Inset cartoons represent the merged and unmerged states (not to scale). b, Stability of thin actin filament protrusions against Euler buckling. (i) A schematic cartoon illustrates a straight protrusion whose total energy is due to the membrane tube. (ii) A hypothetical situation in which the filaments buckle under the load of the membrane. The membrane tube maintains a constant contour length and energy while the energetic cost of filament bending increases the total energy of the system, making buckling energetically unfavorable. c, Model of membrane-induced formation of a thin actin filament protrusion. (i) Small amplitude local deformations of the membrane arise as actin filaments polymerize against the membrane (denoted by solid arrows). (ii) Deformations that are within the range of attraction are able to merge to create a larger deformation that gathers additional filaments (denoted by the curly brackets). Deformations which fail to gather additional filaments are stalled and diminish (denoted by transparent arrows). After bundling enough filaments to overcome the membrane resistance to tube formation, a ‘proto-filopodium’ can elongate without further physical constraint until the free actin concentration is depleted.
The force required to initiate a tubular membrane protrusion exceeds the force resisting elongation, which is independent of tube length 17. Once formed, the thin actin filament protrusion should therefore grow in contour length under a constant force from the membrane. However, the load at which a semi-flexible filament (or a collection of parallel filaments) undergoes Euler buckling decreases steadily with increasing contour length. It is therefore not immediately clear how the long protrusion shown in the overlay of Figure 1a, with a buckling threshold on the order of femtonewtons 13, withstands a membrane load of tens of piconewtons.
The balance of forces that suggests buckling of long protrusions neglects the fact that a membrane tube must bend with the deflections of the filaments it encloses. While the additional bending energy from the curvature of the membrane tube is relatively small17, the steric constraint that it must follow the curvature of the enclosed actin filaments has important repercussions. For the case of a straight protrusion, the membrane energy is Emem = fmemLmem where fmem is the membrane resistance force and Lmem is the length of the membrane tube, and the bending energy of the filaments, Ebundle, is zero (Fig. 3b i). For the case of a buckled protrusion, the initial stages of buckling would involve membrane configurations conformal to the enclosed filaments. Because the membrane bends and does not shorten in these configurations, Emem exceeds fmemLmem while Ebundle is also positive (Fig. 3b ii). Thus, the total energy of the deformation is lower for a straight protrusion than for a buckled protrusion (see Supplementary Information).
The resulting stability against lateral deflections is consistent with our observation of long, straight actin filament protrusions as well as other long filopodia observed in vivo 18,19. This stabilization of membrane-enclosed filaments against buckling has in fact been observed for microtubules protruding from the interior of a liposome 20. Strikingly, these microtubules bend substantially inside the liposome body, but remain straight where surrounded by membrane tubes.
Our elastic model for membrane-induced bundling points to the protrusion length of individual actin filaments as a key parameter regulating the incidence of the filopodia-like protrusions. According to our calculations, increasing the protrusion length while holding membrane tension fixed should increase both the range and strength of lateral attraction between filaments. The maximum protrusion length is typically thought to be determined by a balance between polymerization force and membrane resistance (Fig. 3a). The force-limited length resulting from this balance, while limiting on average, could be transiently exceeded due to fluctuations in the network's internal stress. But it is also possible that single-filament protrusions rarely reach this limit before branching disrupts membrane configurations that favor bundling. In this case the branch-limited maximum protrusion length of a single filament is determined instead by the relative kinetics of branching and polymerization.
If protrusion lengths are branch limited, our model would predict that shorter protrusion lengths created by increased branching would reduce the likelihood of the thin actin filament protrusions. Indeed, when Arp2/3 complex concentration was increased by three fold while actin concentration was held constant, we observed a ∼ 60% reduction in the frequency of filopodia formation, consistent with expectations from the model and confirming that filament protrusions lengths are limited by kinetics rather than polymerization force. Conversely, the frequency of filopodium-like structures did not decrease when Arp2/3 complex concentration was decreased to as low as 10 nM (data not shown).
A second key parameter in the elastic model is membrane tension σ. Our calculations indicate that decreasing σ, while keeping fixed, should reduce the membrane's ability to gather filaments. Consistent with this prediction, we found that decreasing membrane tension by increasing solution osmolarity, while keeping concentrations of Arp2/3 complex and free actin monomer fixed, indeed lowered the frequency of forming thin actin filament protrusions by ∼ 15% (see Supplementary Information).
Based on in vitro experiments and electron microscopy studies, Svitkina et al. proposed the convergent elongation model to describe the formation of filopodia from lamellipodia 14. This model describes the transition from branched to parallel filament architecture in two stages: (i) elongation and lateral association of filaments by tip complex proteins with anti-capping and barbed-end clustering activities, followed by (ii) stabilization of the thin actin array of actin filaments by bundling proteins. Recent studies have identified additional filopodium regulators 21,22, and other studies have challenged whether an underlying dendritic network is necessary for filopodium formation 23,24. This debate has focused solely on the role of actin-binding proteins 25, overlooking the question of whether and how the membrane could play a role in filament organization during filopodium formation. The unexpected formation and stability of the filopodium-like protrusions in our in vitro system suggests that parallel actin-based structures are a natural result of the interaction between growing actin networks and an elastic membrane. Our results demonstrate that the lipid bilayer alone can facilitate free barbed-end clustering and alignment of filaments that can continue to elongate without buckling under the membrane load.
Formation of filopodium-like structures without bundling proteins or tip-complex proteins suggests that membrane-induced alignment of actin filaments could facilitate formation of filopodia in cells (Fig. 3c). We propose a model in which actin filaments are routinely brought together by elastic interactions with the membrane, forming a ‘protofilopodium’, which sets the stage for the formation of a mature filopodium through the participation of actin-binding proteins. Specifically, actin filaments initially gathered by the membrane would be stabilized by tip-complex proteins that associate with the filament ends to prevent capping and by bundling proteins such as fascin that reinforce parallel filament organization during membrane protrusion. In this way, accessory proteins exploit membrane-induced organization of actin filaments to provide regulatory control of protrusions but are not required for their initial formation. The minimal system for generating thin actin filament protrusions presented here would serve as an excellent platform for investigating and evaluating the role of various proteins required for filopodia formation in vivo; however, electron microscopy images of the actin filaments in these protrusions will be required to definitively confirm their organization. In a broader context, this work suggests that the mechanical properties of the plasma membrane may play a critical role in organizing the cytoskeletal filaments that drive many forms of cellular shape change.
METHODS
Protein expression, purification, and fluorescent labeling
Actin was purified from rabbit muscle acetone powder as described by Spudich and Watt 26. Fluorescently-labeled actin was prepared by reacting actin with AF350, AF488, or AF555 maleimide (Molecular Probes). His6-HA-tagged ΔEVH1 N-WASP was over-expressed in Escherichia coli and purified using Nickel affinity chromatography and HiTRAP Q (Amersham) 27. Arp2/3 complex was purified from bovine brains 28. AF488 N-WASP and AF488 Arp2/3 complex were labeled with AF488 maleimide. Protein purities were checked by sodium dodecyl sulfate polyacrylamide gel electrophoresis. Protein concentrations were determined by protein absorbance and BCA assays. N-WASP and Arp2/3 complex activities were confirmed by using in vitro bead motility assay with N-WASP-coated microspheres 29. AF488 capping protein, Cdc42/RhoGDI, and intersectin DH-PH were expressed in E. coli and were gifts from Jack Taunton.
GUV formation
GUVs were prepared in sucrose solution from synthetic and natural lipids as described previously 11. All lipids used were purchased from Avanti Polar Lipids unless otherwise stated. Three compositions were used primarily and all nucleated actin equally well. For canonical N-WASP activation with Cdc42 and PIP2, GUVs containing 75% egg PC, 20% DOPS, and 5% brain PIP2 were used 12. For some experiments, phase separating GUVs containing 24.3% DOPC, 35% DPPC, 40% cholesterol, and 0.7% BODIPY TMR PIP2 were used where actin nucleation occurs robustly with BODIPY TMR PIP2 alone (in the absence of Cdc42). DOGS-NiNTA vesicles, which binds His-tagged N-WASP, containing vesicles have 95% egg PC and 5% DOGS-NiNTA. Refer to Supplementary Information for specific conditions used in each experiment. BODIPY TMR PIP2 was purchased from Echelon Biosciences (Salt Lake City, Utah). For experiments where visualization of membrane was desired, either fluorescent lipid fluorescein-DOPE or C5 C8 BODIPY was used.
In vitro reconstitution of thin actin filament protrusions
GUVs were pre-incubated with 4 μM of N-WASP for a minimum of 10 minutes prior to initiating the reaction. For experiments requiring Cdc42, 0.75 μM of Cdc42/RhoGDI complex, 0.18 μM DH-PH, and 0.5 μM GTPγS were included during the pre-incubation step. Assembly of actin on GUVs was initiated by diluting the vesicle solution 10-fold into actin and Arp2/3 complex. The final mixture contains ∼8.5 μM G-actin, ∼160 nM Arp2/3 complex, and μ400 nM N-WASP in actin polymerization buffer. Osmolarity of the final protein mixtures was matched to the vesicles to within 5%. Large osmolarity differences are otherwise adjusted by varying the final salt concentration of the polymerization buffer (between 1× to 2 ×), which has no effect on the extent of actin polymerization. The protein vesicle mixture was added to a homemade chamber (0.1mm × 5mm × 20mm) consisting of a glass coverslip adhered to a glass slide using double-sided tape, then sealed with VALAP (1:1:1 of vaseline, lanoline, and paraffin) before imaging.
Microscopy
Actin associated vesicles were imaged by phase contrast and epifluorescence microscopy using a 100× 1.25 NA objective for most studies on an inverted microscope (Zeiss Axiovert 200) equipped with motorized stage control and a back-illuminated cooled CCD camera. All light microscopy operations were driven by Metamorph imaging software. Laser scanning confocal microscopy was used for protein localization and photo-bleaching experiments (Zeiss LSM 510). A small region along a single proto-filopodium was bleached using a 63× objective 1.25 NA with 18 mW argon laser at 488 nm and 458 nm. Fluorescence recovery was monitored over 10 min. Spinning disk confocal microscopy was also used to image actin networks on GUV.
Data analysis and modeling
Linescans of AF555 actin, AF488 Arp2/3 complex, and AF488 capping proteins along proto-filopodia were performed in Metamorph. Raw fluorescence intensity was corrected for background fluorescence and normalized with respect to the base of proto-filopodia (dendritic networks) for actin. For Arp2/3 complex and capping proteins, the linescans were further divided by the intensity traces for actin to represent the relative localization of actin binding proteins to actin.
For analysis of photobleaching experiment, fluorescence intensity across the length of the proto-filopodium was tracked over time. Regions of the fluorescence linescan were fitted to a sigmoidal function. The inflection point is identified and used as a metric to track movements of the marked positions.
Growth rate of filopodium-like protrusions was measured from phase contrast images. Integrated fluorescence actin intensity associated with the GUV was collected simultaneously with proto-filopodium elongation. After correcting for photo-bleaching and background fluorescence, the actin “polymerization curve” was fit to a sigmoid and time-dependent actin concentrations were interpolated (described in detail in Supplementary text).
Image processing
All images were processed using Adobe Photoshop. Any brightness or contrast adjustments were uniformly applied to the entire image field and stack.
Supplementary Material
ACKNOWLEDGEMENTS
We thank David Drubin and Eva Nogales, as well as Martijn van Duijn, Ross Rounsvell, Sapun Parekh and other members of the Fletcher lab for helpful discussions. We thank Viviana Risca for her help with confocal microscopy, and Jack Taunton, Derek Wong and Carl Co for providing various proteins. We are grateful for Chris Fromme for assisting us with FPLC. This work is supported in part by a fellowship from Natural Sciences and Engineering Research Council of Canada (A.P.L. and D.L.R.) and by funding from the NIH for the Cell Propulsion Lab as part of the Nanomedicine Roadmap Initiative (D.A.F).
Footnotes
AUTHOR CONTRIBUTIONS A.P. Liu and D.L. Richmond contributed equally to this work.
COMPETING FINANCIAL INTERESTS STATEMENT
The authors declare no competing financial interests.
REFERENCES
- 1.Pollard TD, Borisy GG. Cellular motility driven by assembly and disassembly of actin filaments. Cell. 2003;112:453–65. doi: 10.1016/s0092-8674(03)00120-x. [DOI] [PubMed] [Google Scholar]
- 2.Yeung T, Ozdamar B, Paroutis P, Grinstein S. Lipid metabolism and dynamics during phagocytosis. Curr Opin Cell Biol. 2006;18:429–37. doi: 10.1016/j.ceb.2006.06.006. [DOI] [PubMed] [Google Scholar]
- 3.Smythe E, Ayscough KR. Actin regulation in endocytosis. J Cell Sci. 2006;119:4589–98. doi: 10.1242/jcs.03247. [DOI] [PubMed] [Google Scholar]
- 4.Eggert US, Mitchison TJ, Field CM. Animal cytokinesis: from parts list to mechanisms. Annu Rev Biochem. 2006;75:543–66. doi: 10.1146/annurev.biochem.74.082803.133425. [DOI] [PubMed] [Google Scholar]
- 5.McMahon HT, Gallop JL. Membrane curvature and mechanisms of dynamic cell membrane remodelling. Nature. 2005;438:590–6. doi: 10.1038/nature04396. [DOI] [PubMed] [Google Scholar]
- 6.Mullins RD, Heuser JA, Pollard TD. The interaction of Arp2/3 complex with actin: nucleation, high affinity pointed end capping, and formation of branching networks of filaments. Proc Natl Acad Sci U S A. 1998;95:6181–6. doi: 10.1073/pnas.95.11.6181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Loisel TP, Boujemaa R, Pantaloni D, Carlier MF. Reconstitution of actin-based motility of Listeria and Shigella using pure proteins. Nature. 1999;401:613–6. doi: 10.1038/44183. [DOI] [PubMed] [Google Scholar]
- 8.Bernheim-Groswasser A, Wiesner S, Golsteyn RM, Carlier MF, Sykes C. The dynamics of actin-based motility depend on surface parameters. Nature. 2002;417:308–11. doi: 10.1038/417308a. [DOI] [PubMed] [Google Scholar]
- 9.Giardini PA, Fletcher DA, Theriot JA. Compression forces generated by actin comet tails on lipid vesicles. Proc Natl Acad Sci U S A. 2003;100:6493–8. doi: 10.1073/pnas.1031670100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Upadhyaya A, Chabot JR, Andreeva A, Samadani A, van Oudenaarden A. Probing polymerization forces by using actin-propelled lipid vesicles. Proc Natl Acad Sci U S A. 2003;100:4521–6. doi: 10.1073/pnas.0837027100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu AP, Fletcher DA. Actin polymerization serves as a membrane domain switch in model lipid bilayers. Biophys J. 2006;91:4064–70. doi: 10.1529/biophysj.106.090852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Co C, Wong DT, Gierke S, Chang V, Taunton J. Mechanism of actin network attachment to moving membranes: barbed end capture by N-WASP WH2 domains. Cell. 2007;128:901–13. doi: 10.1016/j.cell.2006.12.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mogilner A, Rubinstein B. The physics of filopodial protrusion. Biophys J. 2005;89:782–95. doi: 10.1529/biophysj.104.056515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Svitkina TM, et al. Mechanism of filopodia initiation by reorganization of a dendritic network. J Cell Biol. 2003;160:409–21. doi: 10.1083/jcb.200210174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Faix J, Rottner K. The making of filopodia. Curr Opin Cell Biol. 2006;18:18–25. doi: 10.1016/j.ceb.2005.11.002. [DOI] [PubMed] [Google Scholar]
- 16.Atilgan E, Wirtz D, Sun SX. Mechanics and dynamics of actin-driven thin membrane protrusions. Biophys J. 2006;90:65–76. doi: 10.1529/biophysj.105.071480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Derenyi I, Julicher F, Prost J. Formation and interaction of membrane tubes. Phys Rev Lett. 2002;88:238101. doi: 10.1103/PhysRevLett.88.238101. [DOI] [PubMed] [Google Scholar]
- 18.Wood W, et al. Wound healing recapitulates morphogenesis in Drosophila embryos. Nat Cell Biol. 2002;4:907–12. doi: 10.1038/ncb875. [DOI] [PubMed] [Google Scholar]
- 19.Onfelt B, et al. Structurally distinct membrane nanotubes between human macrophages support long-distance vesicular traffic or surfing of bacteria. J Immunol. 2006;177:8476–83. doi: 10.4049/jimmunol.177.12.8476. [DOI] [PubMed] [Google Scholar]
- 20.Fygenson DK, Elbaum M, Shraiman B, Libchaber A. Microtubues and vesicles under controlled tension. Physical Reivew E. 1997;55:850–859. [Google Scholar]
- 21.Sigal YJ, Quintero OA, Cheney RE, Morris AJ. Cdc42 and ARP2/3-independent regulation of filopodia by an integral membrane lipid-phosphatase-related protein. J Cell Sci. 2007;120:340–52. doi: 10.1242/jcs.03335. [DOI] [PubMed] [Google Scholar]
- 22.Mattila PK, et al. Missing-in-metastasis and IRSp53 deform PI(4,5)P2-rich membranes by an inverse BAR domain-like mechanism. J Cell Biol. 2007;176:953–64. doi: 10.1083/jcb.200609176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Snapper SB, et al. N-WASP deficiency reveals distinct pathways for cell surface projections and microbial actin-based motility. Nat Cell Biol. 2001;3:897–904. doi: 10.1038/ncb1001-897. [DOI] [PubMed] [Google Scholar]
- 24.Steffen A, et al. Filopodia formation in the absence of functional WAVE- and Arp2/3-complexes. Mol Biol Cell. 2006;17:2581–91. doi: 10.1091/mbc.E05-11-1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gupton SL, Gertler FB. Filopodia: the fingers that do the walking. Sci STKE. 2007;2007:re5. doi: 10.1126/stke.4002007re5. [DOI] [PubMed] [Google Scholar]
- 26.Spudich JA, Watt S. The regulation of rabbit skeletal muscle contraction. I. Biochemical studies of the interaction of the tropomyosin-troponin complex with actin and the proteolytic fragments of myosin. J Biol Chem. 1971;246:4866–71. [PubMed] [Google Scholar]
- 27.Papayannopoulos V, et al. A polybasic motif allows N-WASP to act as a sensor of PIP(2) density. Mol Cell. 2005;17:181–91. doi: 10.1016/j.molcel.2004.11.054. [DOI] [PubMed] [Google Scholar]
- 28.Egile C, et al. Activation of the CDC42 effector N-WASP by the Shigella flexneri IcsA protein promotes actin nucleation by Arp2/3 complex and bacterial actin-based motility. J Cell Biol. 1999;146:1319–32. doi: 10.1083/jcb.146.6.1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wiesner S, et al. A biomimetic motility assay provides insight into the mechanism of actin-based motility. J Cell Biol. 2003;160:387–98. doi: 10.1083/jcb.200207148. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.