Abstract
Background
The Drosophila gene erupted (ept) encodes the fly homolog of human Tumor Susceptibility Gene-101 (TSG101), which functions as part of the conserved ESCRT-1 complex to facilitate the movement of cargoes through the endolysosomal pathway. Loss of ept or other genes that encode components of the endocytic machinery (e.g. synatxin7/avalanche, rab5, and vps25) produces disorganized overgrowth of imaginal disc tissue. Excess cell division is postulated to be a primary cause of these ‘neoplastic’ phenotypes, but the autonomous effect of these mutations on cell cycle control has not been examined.
Principal Findings
Here we show that disc cells lacking ept function display an altered cell cycle profile indicative of deregulated progression through the G1-to-S phase transition and express reduced levels of the tumor suppressor ortholog and G1/S inhibitor Rbf1. Genetic reductions of the Drosophila aPKC kinase (DaPKC), which has been shown to promote tumor growth in other fly tumor models, prevent both the ept neoplastic phenotype and the reduction in Rbf1 levels that otherwise occurs in clones of ept mutant cells; this effect is coincident with changes in localization of Notch and Crumbs, two proteins whose sorting is altered in ept mutant cells. The effect on Rbf1 can also be blocked by removal of the γ-secretase component presenilin, suggesting that cleavage of a γ-secretase target influences Rbf1 levels in ept mutant cells. Expression of exogenous rbf1 completely ablates ept mutant eye tissues but only mildly affects the development of discs composed of cells with wild type ept.
Conclusions
Together, these data show that loss of ept alters nuclear cell cycle control in developing imaginal discs and identify the DaPKC, presenilin, and rbf1 genes as modifiers of molecular and cellular phenotypes that result from loss of ept.
Introduction
Genetic screens in Drosophila have identified a relatively small group of mutations that disrupt normal epithelial architecture and lead to neoplastic overgrowth of developing larval imaginal discs, a set of polarized epithelial tissues that grow during larval stages and develop into the majority of adult structures [reviewed in 1]. The genes affected by these mutations encode proteins with conserved human homologs and fall generally into two functional classes: those involved in the establishment and maintenance of apicobasal polarity [reviewed in 2], and those involved in vesicular trafficking of transmembrane proteins [3]–[8]. Genes in this latter group have been termed ‘endocytic tumor suppressor genes’ and include rab5, syntaxin-7/avalanche (syx7/avl), erupted/tumor susceptibility gene-101 (ept/tsg101 and referred to hereafter as ept), and vps25. Each of these genes is required at distinct steps in the trafficking proteins from the apical membrane to the lysosome. The latter two genes, ept and vps25, respectively encode components of the ESCRT (endosomal sorting complex required for transport)-I and ESCRT-II complexes which promote maturation of late-endosomes into multi-vesicular bodies (MVBs) prior to subsequent fusion with the lysosome [9], [10].
Though it is assumed that mutations in these vesicular trafficking factors promote tissue growth in part by removing developmental blocks to excess cell division, there is little direct evidence that links ept, vps25, or syx7/avl mutations to specific cell cycle transitions or to core components of the nuclear cell cycle machinery. Mutations in ept are known to block the trafficking and degradation of certain apically localized trans-membrane proteins, including the apical membrane determinant Crumbs and the transmembrane receptor Notch [3], [5], [6], [8], but the effects these molecules have on the cell division process in ept mutant cells is not known. Notch has many context-specific links to the cell cycle including controlling levels of the mitotic regulator Cyclin A [11], activity of the dE2f1 transcription factor [11], [12], and expression of the dacapo, string, and fizzy related genes in ovarian follicle cells [13], [14]. Notch has also been reported to collaborate with chromatin modifying factors to silence expression of the rbf1 gene in eye imaginal disc tumors [15]. Thus, there are many pathways through which Notch could potentially affect either the G2/M or G1/S cell cycle transitions in ept mutant cells. The ability of crb overexpression to drive imaginal disc neoplasia [4] argues that Crb can also directly or indirectly affect the cell division process. Yet the potential links between Crb–an integral membrane scaffolding molecule with no known intrinsic signaling activity – and the cell division process are not well understood. crb indirectly regulates Notch in the larval wing by modulating activity of the γ-secretase complex [16]. However, since mechanisms that deregulate cell division in endocytic tumor suppressor mutants are poorly understood, it is difficult to discern specific pathways through which Notch, Crb, and the myriad of other receptors that are candidate targets of the ESCRT pathway (for example those shown to be affected by loss of the hrs gene [17]) might exert pro-proliferative effects in these mutant backgrounds.
We have taken a dual approach to examine cell division control in ept mutant eye-antennal tumors: we have sought to identify genetic manipulations that suppress ept tumor growth, and in parallel we have characterized the effect of ept loss on cell cycle phasing and expression of core cell cycle regulatory factors. We have found that genetic reduction of the DaPKC apical-membrane kinase effectively suppress the growth of ept mutant eye-antennal tumors. In parallel, we have found that ept mutant eye and wing imaginal discs are enriched for cells in the G2/M phase and depleted for those G1 phase, and this correlates with reduced expression of the nuclear S-phase inhibitor and tumor suppressor homolog Rbf1. These two phenotypes are linked by the observation that expression of a dominant-negative DaPKC transgene (DN-DaPKC) in ept eye cells is sufficient to prevent the reduction in Rbf1 levels. It is also sufficient to prevent high-level expression of the Upd protein, which is induced in a Notch-dependent manner in vps25 mutant cells [3]. A similar rescue of Rbf1 levels is observed following removal of the presenilin gene (psn) from ept mutant clones, indicating that cleavage of a Psn substrate(s) contributes to the effect of the ept genotype on Rbf1 levels. To test the physiologic significance of the Rbf1 reduction in ept cells, we have re-expressed exogenous rbf1 in either wild type or ept mutant eye-antennal tissue and found that excess rbf1 is able to completely ablate mutant tissue while having little effect on normal tissue. These data indicate that DaPKC- and psn-dependent loss of Rbf1 from ept cells may be a significant factor in their overgrowth.
Results
DaPKC is required for ept tumor growth
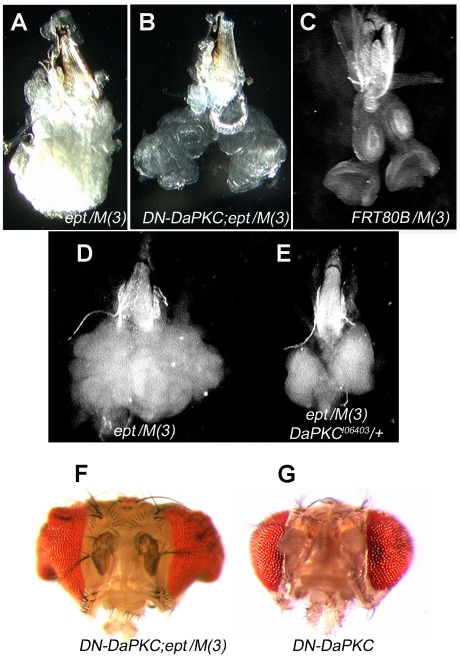
Under normal circumstances, ept mutant eye-antennal tumors created using the cell-lethal Minute (M) technique (the genotype eyFLP;;ept2,FRT80B/P[m-w+]RpL141,FRT80B is hereafter referred to as ept/M(3)) grow into large unstructured masses (Fig. 1A) that fail to differentiate into recognizable eye tissue and kill the animal bearing them during the late larval and pupal phases [6]. To test the genetic requirements of this tumor-like phenotype, we screened a small collection of alleles of signaling, polarity, and growth regulatory genes (stat92E, crb, lgl, Drosophila aPKC, yki, cyclinD, dMyc, s6k and others) for their ability to suppress size and/or architectural phenotypes associated with loss of ept/tsg101. Alleles of two of these genes had significant effects on the morphology of ept/M(3) tumors: the DaPKC gene, which encodes an apical-membrane kinase that controls epithelial polarity and endocytosis [18]–[21], and the stat92E gene, which encodes the sole fly homolog of the Stat family of mammalian transcription factors [22], [23]. Analysis of the effect of DaPKC on ept tumor growth is presented here. Expression of a transgene encoding a dominant-negative version of the DaPKC kinase (DN-DaPKC; [24]) in ept/M(3) mutant discs caused these tissues to develop as enlarged eye/antennal structures (Fig. 1B) that are morphologically similar to normal eye discs (Fig. 1C) and contain differentiated photoreceptor neurons (data not shown). Most animals bearing these DN-DaPKC;ept/M(3) discs survive to late-pupal and pharate adult stages well beyond the point where animals bearing ept/M(3) tumors normally die; a few survive to eclosion and emerge with irregular heads and eyes (Fig. 1F) that are enlarged relative to those expressing the DN-DaPKC transgene alone (Fig. 1G). To confirm that this genetic interaction is not an artifact of the DN-DaPKC transgene, ept/M(3) tumors were also generated in a background heterozygous for the genomic DaPKCk06403 loss-of-function allele [25]. This DaPKCk06403/+ genotype also shrank the size of ept/M(3) tumors (Figs. 1D,E), confirming that DaPKC is required for the ept tumor phenotype.
Figure 1. DaPKC is required for ept tumor growth.
Bright-field images of ept/M(3) mutant (A), ept/M(3) mutant expressing DN-DaPKC (B), or FRT80B/M(3) (C) eye-antennal discs dissected from wandering 3rd instar larvae. (D,E) Side-by-side image of an ept/M(3) mutant eye antennal disc and an ept/M(3) disc that is heterozygous for the DaPKCk06403 allele. (F,G) Heads from surviving ept/M(3)+DN-DaPKC adults or DN-aPKC control animals. Grouped images are to scale.
ept mutant cells exhibit G1/S cell cycle deregulation
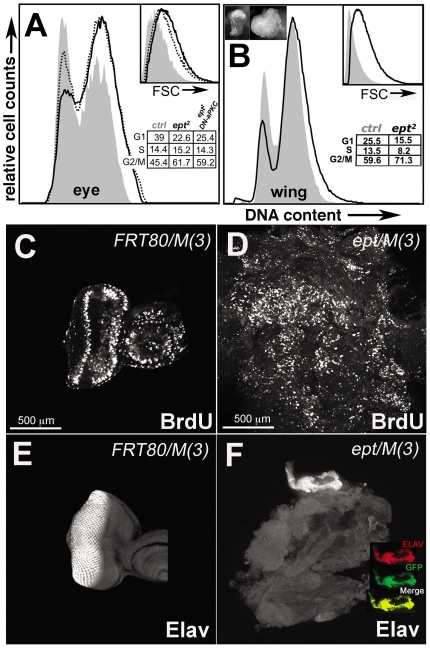
Because excess cell proliferation is a key factor in tissue hypertrophy, we examined the effect of ept loss on the cell cycling properties of cells in eye-antennal discs and whether DaPKC might influence this effect. Fluorescence-activated cell sorting (FACS) analysis of ept/M(3) eye-antennal discs shows that the population of cells within them is under-represented for 2N G1-phase cells and enriched for S- and G2/M-phase cells relative to control FRT/M(3) discs (Fig. 2A). Cell size is also increased in ept/M(3) tumors relative to control cells (Fig. 2A, see inset). ept/M(3) eye-antennal discs show widespread BrdU incorporation relative to control discs (Figs. 2C–D) and lack cells that express the neuronal marker Elav (Figs. 2E–F). FACS analysis of ept mutant wing disc tumors [genotype UbxFlp;;M(3),FRT80B/ept2,FRT80B] show similar cell cycle and cell size shifts as ept/M(3) eye-antennal disc cells (Fig. 2B), although the G2/M-shift is less pronounced in the wing. Thus cell cycling changes associated with loss of ept function are not solely due to a block in progression of the eye-specific morphogenetic furrow [26], indicating that loss of ept affects G1/S progression in multiple larval discs.
Figure 2. Cell cycle deregulation in ept mutant imaginal discs.
(A–B) Flow cytometric analysis of cells in control FRT80B/M(3) (grey fill), ept mutant (black line), or DN-DaPKC,ept/M(3) (dotted line) wing or eye imaginal discs. Percent of cells in each cell cycle phase is indicated. Inset images in (B) show a normal wing disc and one composed of cells homozygous for the ept2 allele. Forward scatter (FSC) plot of cell size is also included for each sample. Patterns of BrdU incorporation (C–D) and Elav expression (E–F) in control FRT80B/M(3) (C,E) and ept mutant (D,F) eye-antennal disc tumors dissected from wandering-stage larvae. Inset in (F) shows expression of Elav (red) only in surviving GFP-positive Min/+ cells (green).
Expression of DN-DaPKC in the background of ept/M(3) eye-antennal discs led to a reproducible shift of a fraction of cells back into the G1-phase (dotted line, Fig. 2C). The cell cycle shift induced by the DN-DaPKC transgene only partially restored cell cycle phasing and had no discernable effect on the enlarged size of ept cells, suggesting that additional factors contribute to each of these phenotypes. Consistent with this hypothesis, a reduction in the dose of the stat92E gene affects both cell cycle phasing and cell size in ept/M(3) tumors (see accompanying paper by Gilbert et al.).
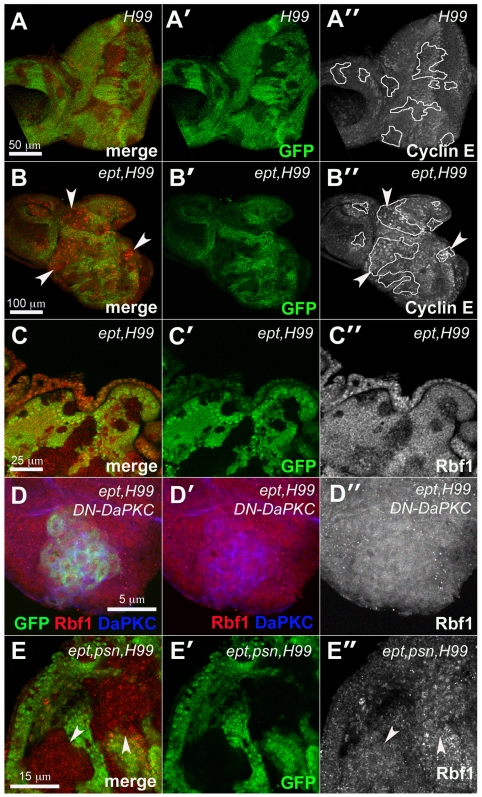
We next examined the levels of G1/S regulatory proteins in clones of eye disc cells doubly mutant for ept and the H99 chromosomal deletion, which removes the genes rpr, grim, and hid [27] and prevents activation of the pro-death caspase enzymes [28]–[33] that are otherwise detected at high levels in ept mutant clones of cells (Figure S1); their removal thus allows recovery of larger ept mutant clones and permits molecular analysis of protein epitopes that might otherwise be degraded. The expression of two key regulators of G1/S, the pro-division protein Cyclin E (CycE) and Rbf1, the Drosophila homolog of the retinoblastoma (Rb) tumor suppressor protein [34], were found to be affected by the ept genotype. Compared to H99 control clones (Fig. 3A–A″), some ept,H99 mutant clones express elevated levels of CycE protein (Fig. 3B–B″, see arrows). CycE is also slightly elevated in some normal cells that surround mutant clones, which is likely a reflection of the previously described non-autonomous mitogenic effect of ept mutant cells [6]. Neither of these effects on CycE are not apparent in all clones, and are thus not a strongly penetrant part of the ept,H99 phenotype. By contrast, immunostaining with a monoclonal antibody specific to Rbf1 [34] shows that levels of this protein are clearly reduced in ept,H99 disc cells compared to surrounding control cells (Fig. 3C–C″). Clones of H99 mutant cells do not show the same effect (Figure S2), indicating that ept is required to maintain normal levels of Rbf1 protein in eye-antennal disc cells.
Figure 3. ept mutations reduce levels of the Drosophila Rb ortholog Rbf1.
Confocal images of larval eye discs containing clones of H99 mutant cells (A–A″), ept,H99 double-mutant cells (B–C″) or ept,H99,psn triple mutant cells (E–E″) marked by the absence of GFP (green) co-stained for CycE (red in A–B″), or Rbf1 (red in C–C″, and E–E″). Tracing in A″ and B″ outlines H99 and eptH99 mutant clones respectively. The anti-CycE signal was recorded at the same optical settings in panels A″ and B″. Arrowheads in panels B and B″ denote ept,H99 double mutant clones that express CycE. (D–D″) MARCM-mediated expression of DN-DaPKC (blue) in ept,H99 double-mutant cells marked by GFP (green) restores levels of Rbf1 (red). Arrows in panel E denote clones of ept,H99,psn cells that express normal levels of Rbf1 relative to adjacent control cells.
DaPKC and psn are required for the effect on Rbf1 levels in ept mutant cells
Given the effect of the DaPKC alleles on ept/M(3) tumor growth, we next examined whether DaPKC activity might affect Rbf1 levels in ept mutant cells. To do this, the DN-DaPKC transgene was expressed specifically in ept,H99 mutant cells using the MARCM technique [35]. Although this led to a significant reduction in the size of ept,H99 clones (paralleling the effect of the DN-DaPKC transgene on ept tumor size), close examination of DN-DaPKC+ept,H99 clones stained with the anti-Rbf1 antibody revealed no obvious difference in Rbf1 levels relative to surrounding normal cells (Fig 3D–D″). By this measure, DaPKC activity is required for the effect of the ept,H99 genotype on Rbf1 levels. Because of the somewhat variable effect of the ept,H99 genotype on CycE, CycE levels were not examined in this MARCM DN-DaPKC background. DaPKC is known to regulate a number of cellular processes and pathways ([18] and reviewed in [19]), including the Notch pathway in larval brain neuroblasts [36]. To test whether an allele of a Notch pathway component might also alter the effect of the ept,H99 genotype on Rbf1, a strong loss-of-function allele of the presenilin gene (psn227;[37]) was recombined onto the ept,H99 chromosome. psn encodes a required component of the γ-secretase that cleaves and releases the Notch intracellular domain and it's activity is needed for Notch activation in vivo [38], [39], Because all three loci are on the left arm of chromosome 3, this ept,H99,psn mutant chromosome allows for the production of somatic clones of triple mutant cells. These cells are deficient in ept function and Psn activity, but give rise to easily detectable clones due to their inability to die. Immunostaining with the anti-Rbf1 antibody indicates that the level of Rbf1 in these ept,H99,psn cells (arrows in Fig. 3E–E″) is similar to that in surrounding normal cells. Thus, loss of psn has a similar effect on levels of the anti-Rbf1 epitope in the ept,H99 genotype as does expression of the DN-DaPKC allele.
Effect of DN-DaPKC on Notch and Upd in ept cells
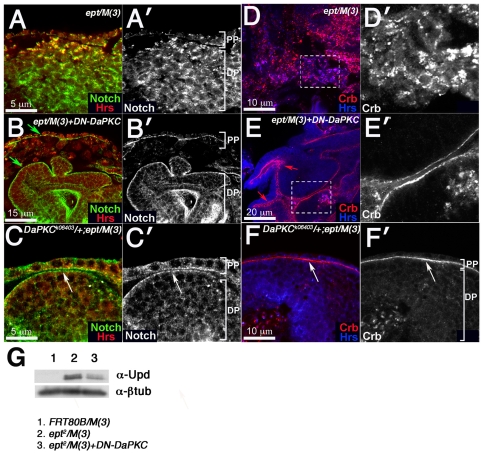
The similar effect of the psn227 allele and the DN-DaPKC transgene on the Rbf1 epitope in ept mutant cells led us to examine Notch protein in ept mutant cells in backgrounds in which DaPKC activity is reduced. ept/M(3) tumors grow as disorganized masses that lack ‘landmarks’ normally associated with the eye disc (e.g. morphogenetic furrow, optic stalk, eye-antennal boundary, etc) thus depriving the tissue of any A/P or D/V reference points. We have therefore used the sole remaining disc feature, the overlying peripodial membrane, to orient each image. As shown in a prior study [6], loss of ept causes the eye disc proper (DP) to grow as disorganized groups of cells surrounded by a layer of peripodial cells (PP) in which the Notch protein shows increased co-staining with the endosomal protein Hrs (Fig. 4A–A′). This co-localization of Notch and Hrs is most apparent in PP cells and in more cortical regions of the DP (upper portion of image in Fig. 4A), suggesting that Notch/Hrs-positive endosomes may accumulate in more apical regions of DP cells. Notch-Hrs co-localization has also been observed in cells lacking the ESCRT-II subunit gene vps25 [3], [5] and has been interpreted as an indication that Notch normally traffics through ESCRT-I and -II vesicular compartments on its way to the lysosome. Interestingly reducing DaPKC activity, either by expression of the DN-DaPKC transgene (Fig. 4B–B′) or heterozygosity for the DaPKCk06403 loss-of-function allele (Fig. 4C–C′), alters the pattern of Notch protein localization in ept/M(3): a larger proportion of the anti-Notch signal is detected on outer surface of the DP and on the apical face of PP cells (arrows in Fig. 4B), with a corresponding drop in the proportion detected in Hrs-positive structures in cytoplasm of cells in the DP. DaPKC alleles have a similar effect DP and PP populations of Crumbs (Fig. 4D–F), a protein that is normally trapped in cytoplasmic puncta in ept mutant cells [6] and that is known to be regulated by DaPKC [24]. As recent studies have suggested that Notch cleavage and activation requires internalization of Notch protein into specific endosomal compartments [40], we sought to determine if changes in Notch localization following expression DN-DaPKC might correlate with changes in expression of a validated Notch target. It has previously been shown that expression of a Notch RNA interference ‘knock-down’ construct in vps25 mutant eye disc cells is sufficient to block overproduction of the Unpaired protein (Upd), which is otherwise expressed at very high levels in vps25 mutant cells [3]. The overexpression of Upd in ept mutant cells has also been shown to occur by a Notch-dependent mechanism [6]. We find that expression of the DN-DaPKC is sufficient to substantially blunt the elevation in Upd levels as measured by immunoblotting for total Upd protein present in ept/M(3) eye-antennal discs (Fig. 4G). Thus, in addition to its effect on ept/M(3) tumor growth, Rbf1 levels, and Notch localization, DaPKC is also required for Notch-dependent hyper-accumulation of Upd in ept/M(3) tumors.
Figure 4. DaPKC is required for cytoplasmic accumulation of Crb and Notch in ept mutant tissues.
Confocal images of ept/M(3) (A–A′ and D–D′), ept/M(3)+DN-DaPKC (B–B′ and E–E′), or ept/M(3),DaPKCk06403/+ (C–C′ and F–F′) tumors stained for Notch and Hrs (green and red respectively in A–C) or Crb and Hrs (red and blue respectively in D–F). Dashed boxes in panels D and E are enlarged in panels D′ and E′. Arrows in panels B and C mark membrane-associated Notch in disc and peripodial cells. Arrows in panels E and F mark membrane-associated and apical junctional Crb staining. Peripodial cell layer (PP) and disc proper (DP) are labeled in panels A′, B′, C′, and F′ so as to orient the image relative to overall disc structure. Image in panel D′ is of ept mutant DP cells; image in E′ is structured lobes of ept mutant DP cells that are able to form the presence of the DN-DaPKC transgene. (G) Western blot analysis of Upd levels in FRT80B/M(3) (lane 1), ept/M(3) (lane 2), and ept/M(3)+DN-DaPKC (lane 3) larval eye-antennal discs. The blot was stripped and re-probed with an anti-β−gal antibody as a loading control.
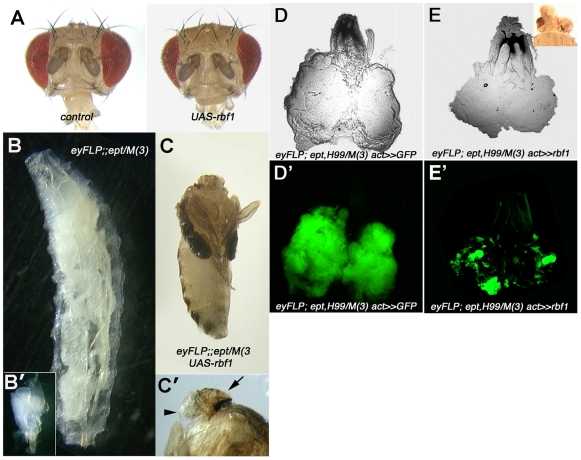
Transgenic expression of rbf1 in ept/M(3) tumors
In light of the well-known role Rb family proteins play in regulating the G1-to-S phase cell cycle transition [reviewed in 41], reduced Rbf1 expression in ept mutant cells might be predicted to impair the Rbf1-mediated block to unregulated S-phase entry. To test what effect re-expression of rbf1 might then have on the ept tumor phenotype, a UAS-rbf1 transgene was driven in the background of either normal eye-antennal discs or ept mutant eye/antennal tumors. rbf1 expression in control FRT80B/M(3) eyes/heads leads to a moderate reduction in eye and head size (Fig. 5A). In contrast, re-expression of rbf1 in ept eye/antennal tumors completely blocks the growth of the mutant tissue and results in headless pharate adults (Fig. 5B–C). A residual lump of cuticle (arrow in Fig. 5C′) is all that remains of tumors that would otherwise overgrow and kill the animal as an enlarged larva (e.g. Fig. 5B–B″). Expression of rbf1 thus has a significant and specific effect on the terminal organismal phenotype resulting from ept eye-antennal tumors. To test if the effect of rbf1 on ept/M(3) tumor growth is due to enhanced rates of apoptosis, the previous experiment was repeated in ept,H99/M(3) tumors that express UAS-GFP under control of the eyFLP and Actin5c>CD2>Gal4 transgenes [42], such that each ‘flip-out’ event creates a clone of GFP-positive mutant cells. In the absence of the UAS-rbf1 transgene, these ept,H99 mutant tumors are uniformly green (Fig. 5D–D″), indicating that clones of GFP-expressing ept,H99 mutant cells take over the majority of the organ. However, in the presence of the UAS-rbf1 transgene, GFP-positive clones of ept,H99 mutant cells remain quite small (Fig. 5E′), indicating that exogenously produced Rbf1 retains the ability to retard the overgrowth of ept,H99 mutant cells. At an organ level, re-expression of rbf1 in the apoptosis-compromised background of eptH99/M(3) tumors results in pharate adults with small heads and eyes (inset, Fig. 5E). Thus, the ability of rbf1 to block ept tumor growth is only partly reduced in a background in which cell death is compromised. These observations demonstrate that an eye-antennal disc composed of ept mutant cells responds differently to over-expression of rbf1 than a disc composed of normal cells eye antennal cells, and suggests that re-introducing rbf1 either slows proliferation of ept,H99 mutant cells or kills them by an H99-independent mechanism; alternatively, specification of early progenitors of the head/eye fate may be defective in cells that simultaneously overexpress rbf1 and lack ept.
Figure 5. Effect of re-expressing rbf1 in ept mutant eye-antennal discs.
(A) Overexpression of rbf1 in the developing eye and head of control animals results in a mild reduction in adult organ size. Overexpression of rbf1 in the developing eye and head tissue of animals bearing ept eye-antennal tumors blocks the ept/M(3) giant larval phenotype (B) and results in headless adults (C) with small maxillary palps and a remnant of head cuticle (indicated by arrowhead and arrow respectively in C′). Inset in B′ shows an ept mutant eye-antennal tumor to scale with respect to B and C. Actin ‘Flp-out’ mediated overexpression of GFP (D–D′) or GFP and rbf1 (E–E′) in apoptosis-compromised ept,H99 mutant eye antennal discs reduces clone size (D′ vs E′).
Discussion
The data presented here indicate that the DaPKC kinase plays a significant role in the growth of ept mutant imaginal disc tumors. DaPKC is also required for tumor growth in other Drosophila mutants [36], [43]–[45], suggesting that DaPKC may be generally required for excess proliferation in many different backgrounds. The molecular mechanism through which reduced DaPKC activity exerts these effects in ept/M(3) cells is not known, although it correlates with increased apical membrane localization of two membrane-bound proteins, Crumbs and Notch, that otherwise aggregate in vesicle-associated structures in the cytoplasm of ept mutant cells [6]. DaPKC directly phosphorylates Crumbs and the endocytic adaptor Numb [36], [44], [46], which can in turn inhibit Notch [reviewed in 47]. Thus the effect of reduced DaPKC activity on ept tumor growth could be mediated exclusively through effects on Notch and Crb. However two considerations suggest this is unlikely: first, mutations in the hrs gene affect the vesicular trafficking of many different receptors [17], suggesting that signals transduced through many receptors may contribute to the overall ept tumor phenotype; second, aPKC kinases appear to play a general role in promoting endocytic internalization [18], again suggesting that reduced DaPKC function may affect many different pathways. In light of these considerations, it seems more likely that DaPKC alleles partially rescue ept phenotypes not because they specifically affect the endocytic uptake of one or two apical-membrane proteins (e.g. Crb and Notch), but because they generally lessen the endocytic uptake of a spectrum of proteins that are otherwise trapped in the late-endosome of ept mutant cells. Thus DaPKC might act as a ‘permissive factor’ in ept tumor growth via a positive role in endosomal trafficking upstream of ESCRT-1; in its absence a set of proteins that normally enter the endolysosomal pathway and become trapped in ept mutant endosomes are shunted toward an alternate fate that precludes their accumulation in late endosomal compartments. Properly testing this model will require a comprehensive analysis of the requirement for DaPKC in sorting individual candidate proteins, and in controlling the activity of specific polarity, signaling, and proliferative pathways in which these proteins act.
The effect of ept loss on G1/S control in imaginal discs confirms that one element of the ept tumor phenotype is deregulated cell division. Additionally, the data show that this correlates with a requirement for ept to maintain normal levels of the key S-phase inhibitor Rbf1 in imaginal disc cells. Loss of Rb homlogs in multiple species is sufficient to accelerate tumor progression [reviewed in 48]. Thus the effect of ept on Rbf1 provides a link between a member of the ‘neoplastic’ tumor suppressor genes and an established cell cycle regulator and tumor suppressor protein. Our experiments argue that this effect on Rbf1 is significant to the physiology of ept mutant tissues and identify two factors, the DaPKC kinase and the γ-secretase subunit Psn, as required for the reduction of Rbf1 in ept mutant cells. ept alleles do not affect on Psn protein levels in the eye disc (data not shown), indicating that this requirement for Psn is probably indirect via cleavage of another factor whose activity controls Rbf1 levels. The Notch receptor is a candidate for this role: it is trapped and hyper-activated in ept mutant endosomes [6], and its activation mechanism requires a cleavage event that studies have found correlates with Notch endosomal localization [40]. Moreover, ectopic Notch activity has been reported to facilitate transcriptional silencing of the rbf1 gene in an eye disc tumor model [15]. Thus the effect of the psn allele on Rbf1 levels in ept cells could indicate a role for Notch upstream of Rbf1. Consistent with this, the ability of the dominant-negative DaPKC allele to restore Rbf1 levels in ept cells correlates with its ability to block overproduction of the Upd protein, a Notch-target in ESCRT mutant cells. The ability of DaPKC to act as a pro-growth ‘proliferation factor’ in larval brain neuroblasts (NBs) a cell type that uses asymmetric divisions to regulate the fate and proliferative potential of daughter cells, has also been genetically linked to Notch activity [36], [43], [44]. Though these observations suggest that the effect of DaPKC and psn on Rbf1 levels in ept mutant cells may reflect a Notch-regulatory role for both genes, it remains possible that psn and DaPKC are also involved in a Notch-independent signaling pathway that is responsible for the drop in Rbf1 levels in ept mutant disc cells. As certain endocytic tumor suppressors activate Notch (e.g. ept and vps25) and others do so to a much lesser extent (e.g. ayx7/avl), one way to begin to address this question may be to examine the pattern of Rbf1 expression in each of these mutant backgrounds.
As a component of the ESCRT-1 complex, ept developmental phenotypes are expected to reflect the combined deregulation of the myriad proteins that traffic through ESCRT-1 dependent compartments of the endolysosomal pathway. Indeed certain elements of the ept mutant phenotype (e.g. increased cell size) do not appear to respond to reduced DaPKC activity. Many other prominent signaling molecules are mislocalized in Drosophila wing disc cells mutant for the endocytic regulator hrs [17], which acts at a sorting step prior to the ESCRT-1 complex [49]. The pathways in which these molecules act should thus be considered as additional candidate effectors of ept mutations. This expanding array of potential ESCRT targets raises the possibility that inactivation of ept will have tissue- and stage-specific phenotypes that reflect the changing pattern of proteins targeted for endolysosomal degradation in various cell types. If targeted endocytosis regulates a similar array of proteins in certain mammalian epithelial cells as it does in Drosophila imaginal discs, it may be that defects in this process will act via an aPKC- and Psn-dependent pathway to produce neoplastic phenotypes similar to those observed with alleles of Drosophila ept.
Materials and Methods
Genetics
Crosses were carried out at 25°C unless otherwise indicated. DN-DaPKC-mediated rescue of ept tumor phenotypes was optimal at 20°C. ept mutant eye clones were generated as described previously [4], [6]. ept,H99 and ept,H99,psn mutant eye clones were generated by crossing either w;;eptX1,Df(3L)H99,FRT80B/TM6B or w;;eptX1,Df(3L)H99,psn227,FRT80B/TM6B males to yweyFLP;;P[m-w+;ubiGFP],FRT80B females. ept mutant eye-antennal tumors were generated by crossing w;;ept2,FRT80B males and yweyFLP;;P[m-w+]RpL141,FRT80B/TM6B females. ‘DN-DaPKC,ept/M(3)’ animals were generated by crossing the w;UAS-DN-aPKC;ept2,FRT80B/TM6B stock to the y,w,eyFLP; act>y+>Gal4/CyO:twi-GFP;P[m-w+]RpL141,FRT80B/TM6B stock. Expression of DN-DaPKC in ept,H99 clones was achieved by crossing w;UAS-DN-DaPKC;eptX1,Df(3L)H99,FRT80B/TM6B males to eyFLP;tub-Gal4;tub-Gal80,FRT80B females. ‘rbf1,ept/M(3)’ and ‘rbf1,eptH99/M(3)’ animals were generated by crossing the w;UAS-rbf1/CyO;ept2,FRT80B/TM6B or w;UAS-rbf1/CyO;eptX1,Df(3L)H99,FRT80B/TM6B stocks to the y,w,eyFLP; act>y+>Gal4/CyO:twi-GFP;P[m-w+]RpL141,FRT80B/TM6B stock. The DaPKCk06403 and psn227 were obtained from the Bloomington Drosophila Stock Center (BDSC). The Df(3L)H99 stock was a gift of K. White. The UAS-DN-DaPKC chromosome 2 stock was a gift of D. Bilder. DN-DaPKC-expressing ept mutant clones were generated by crossing the yweyFLP;tub-Gal4/CyO;tub-Gal80,FRT80B stock (gift of J. Treisman) to w;UAS-DN-DaPKC/CyO,twi-GFP;ept2,FRT80B/TM6B.
Flow Cytometry
Eye or wings discs were dissociated in PBS Trypsin-EDTA and stained with 20 µM DRAQ-5 (Biostatus Limited). Data were acquired on a Becton Dickinson LSR II flow cytometer via a 755 nM Red laser with a 780/60 nM BP collection filter and were analyzed with FACSDiva Software.
Microscopy & Immunohistochemistry
Immunostaining and confocal microscopy was performed as described previously [50]. Antibodies used were: rat anti-Crb-extra (gift of U. Tepass and E. Knust) 1∶500; guinea pig anti-Hrs (gift of H. Bellen) 1∶1000; mouse anti-BrdU (Becton Dickinson) 1∶100; mouse anti-CycE, 1∶5 (gift of H. Richardson); mouse 9C6 anti-Notch (DSHB) 1∶50; mouse anti-Rbf1 DX5 (gift of W. Du), 1∶50; rat anti-ELAV (DSHB) 1∶1000; goat anti-rabbit Cy5, goat anti-mouse Cy3, goat anti-guinea pig Cy3, and goat anti-rat Cy3 (Jackson Laboratories) each at 1∶50. BrdU incorporation assays were performed as described previously [51].
Supporting Information
Levels of cleaved Caspase-3 are elevated in ept/tsg101 mutant eye clones. Clones of ept/tsg101 mutant cells marked by the absence of GFP (green) contain many dying cells, as indicated by staining for cleaved Caspase-3 (blue).
(1.84 MB TIF)
Rbf1 levels are unaffected by the H99 deletion. Confocal image of H99 clones (lacking GFP) in a mosaic 3rd instar eye imaginal disc stained with the anti-Rbf1 antibody (red).
(0.73 MB TIF)
Acknowledgments
We apologize to those whose work could not be cited due to space constraints. We thank W. Du, D. Bilder, H. Richardson, S. Campuzano, J. Treisman, D. Ready, H. Bellen, U. Tepass, E. Knust, and K. White for gifts of fly stocks and reagents. Other reagents were obtained from the BDSC, DGRC, and DSHB.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: NIH F32-GM077898 to MMG http://www.nih.gov/ NIH CA123368-01 to KHM http://www.cancer.gov/ The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Hariharan IK, Bilder D. Regulation of imaginal disc growth by tumor-suppressor genes in Drosophila. Annu Rev Genet. 2006;40:335–361. doi: 10.1146/annurev.genet.39.073003.100738. [DOI] [PubMed] [Google Scholar]
- 2.Bilder D. Epithelial polarity and proliferation control: links from the Drosophila neoplastic tumor suppressors. Genes Dev. 2004;18:1909–1925. doi: 10.1101/gad.1211604. [DOI] [PubMed] [Google Scholar]
- 3.Vaccari T, Bilder D. The Drosophila tumor suppressor vps25 prevents nonautonomous overproliferation by regulating notch trafficking. Dev Cell. 2005;9:687–698. doi: 10.1016/j.devcel.2005.09.019. [DOI] [PubMed] [Google Scholar]
- 4.Lu H, Bilder D. Endocytic control of epithelial polarity and proliferation in Drosophila. Nat Cell Biol. 2005;7:1132–1139. doi: 10.1038/ncb1324. [DOI] [PubMed] [Google Scholar]
- 5.Thompson BJ, Mathieu J, Sung HH, Loeser E, Rorth P, et al. Tumor suppressor properties of the ESCRT-II complex component Vps25 in Drosophila. Dev Cell. 2005;9:711–720. doi: 10.1016/j.devcel.2005.09.020. [DOI] [PubMed] [Google Scholar]
- 6.Moberg KH, Schelble S, Burdick SK, Hariharan IK. Mutations in erupted, the Drosophila ortholog of mammalian tumor susceptibility gene 101, elicit non-cell-autonomous overgrowth. Dev Cell. 2005;9:699–710. doi: 10.1016/j.devcel.2005.09.018. [DOI] [PubMed] [Google Scholar]
- 7.Childress JL, Acar M, Tao C, Halder G. Lethal giant discs, a novel C2-domain protein, restricts notch activation during endocytosis. Curr Biol. 2006;16:2228–2233. doi: 10.1016/j.cub.2006.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Herz HM, Chen Z, Scherr H, Lackey M, Bolduc C, et al. vps25 mosaics display non-autonomous cell survival and overgrowth, and autonomous apoptosis. Development. 2006;133:1871–1880. doi: 10.1242/dev.02356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bishop N, Woodman P. TSG101/mammalian VPS23 and mammalian VPS28 interact directly and are recruited to VPS4-induced endosomes. J Biol Chem. 2001;276:11735–11742. doi: 10.1074/jbc.M009863200. [DOI] [PubMed] [Google Scholar]
- 10.Katzmann DJ, Babst M, Emr SD. Ubiquitin-dependent sorting into the multivesicular body pathway requires the function of a conserved endosomal protein sorting complex, ESCRT-I. Cell. 2001;106:145–155. doi: 10.1016/s0092-8674(01)00434-2. [DOI] [PubMed] [Google Scholar]
- 11.Baonza A, Freeman M. Control of cell proliferation in the Drosophila eye by Notch signaling. Dev Cell. 2005;8:529–539. doi: 10.1016/j.devcel.2005.01.019. [DOI] [PubMed] [Google Scholar]
- 12.Firth LC, Baker NE. Extracellular signals responsible for spatially regulated proliferation in the differentiating Drosophila eye. Dev Cell. 2005;8:541–551. doi: 10.1016/j.devcel.2005.01.017. [DOI] [PubMed] [Google Scholar]
- 13.Schaeffer V, Althauser C, Shcherbata HR, Deng WM, Ruohola-Baker H. Notch-dependent Fizzy-related/Hec1/Cdh1 expression is required for the mitotic-to-endocycle transition in Drosophila follicle cells. Curr Biol. 2004;14:630–636. doi: 10.1016/j.cub.2004.03.040. [DOI] [PubMed] [Google Scholar]
- 14.Shcherbata HR, Althauser C, Findley SD, Ruohola-Baker H. The mitotic-to-endocycle switch in Drosophila follicle cells is executed by Notch-dependent regulation of G1/S, G2/M and M/G1 cell-cycle transitions. Development. 2004;131:3169–3181. doi: 10.1242/dev.01172. [DOI] [PubMed] [Google Scholar]
- 15.Ferres-Marco D, Gutierrez-Garcia I, Vallejo DM, Bolivar J, Gutierrez-Avino FJ, et al. Epigenetic silencers and Notch collaborate to promote malignant tumours by Rb silencing. Nature. 2006;439:430–436. doi: 10.1038/nature04376. [DOI] [PubMed] [Google Scholar]
- 16.Herranz H, Stamataki E, Feiguin F, Milán M. Self-refinement of Notch activity through the transmembrane protein Crumbs: modulation of gamma-secretase activity. EMBO. 2006;7:297–302. doi: 10.1038/sj.embor.7400617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jekely G, Rorth P. Hrs mediates downregulation of multiple signalling receptors in Drosophila. EMBO Rep. 2003;4:1163–1168. doi: 10.1038/sj.embor.7400019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Balklava Z, Pant S, Fares H, Grant BD. Genome-wide analysis identifies a general requirement for polarity proteins in endocytic traffic. Nat Cell Biol. 2007;9:1066–1073. doi: 10.1038/ncb1627. [DOI] [PubMed] [Google Scholar]
- 19.Assemat E, Bazellieres E, Pallesi-Pocachard E, Le Bivic A, Massey-Harroche D. Polarity complex proteins. Biochim Biophys Acta. 2008;1778:614–630. doi: 10.1016/j.bbamem.2007.08.029. [DOI] [PubMed] [Google Scholar]
- 20.Suzuki A, Ohno S. The PAR-aPKC system: lessons in polarity. J Cell Sci. 2006;119:979–987. doi: 10.1242/jcs.02898. [DOI] [PubMed] [Google Scholar]
- 21.Henrique D, Schweisguth F. Cell polarity: the ups and downs of the Par6/aPKC complex. Curr Opin Genet Dev. 2003;13:341–350. doi: 10.1016/s0959-437x(03)00077-7. [DOI] [PubMed] [Google Scholar]
- 22.Yan R, Small S, Desplan C, Dearolf CR, Darnell JE., Jr Identification of a Stat gene that functions in Drosophila development. Cell. 1996;84:421–430. doi: 10.1016/s0092-8674(00)81287-8. [DOI] [PubMed] [Google Scholar]
- 23.Hou XS, Melnick MB, Perrimon N. Marelle acts downstream of the Drosophila HOP/JAK kinase and encodes a protein similar to the mammalian STATs. Cell. 1996;84:411–419. doi: 10.1016/s0092-8674(00)81286-6. [DOI] [PubMed] [Google Scholar]
- 24.Sotillos S, Diaz-Meco MT, Caminero E, Moscat J, Campuzano S. DaPKC-dependent phosphorylation of Crumbs is required for epithelial cell polarity in Drosophila. J Cell Biol. 2004;166:549–557. doi: 10.1083/jcb.200311031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Spradling AC, Stern D, Beaton A, Rhem EJ, Laverty T, et al. The Berkeley Drosophila Genome Project gene disruption project: Single P-element insertions mutating 25% of vital Drosophila genes. Genetics. 1999;153:135–177. doi: 10.1093/genetics/153.1.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wolff T, Ready DF. The beginning of pattern formation in the Drosophila compound eye: the morphogenetic furrow and the second mitotic wave. Development. 1991;113:841–850. doi: 10.1242/dev.113.3.841. [DOI] [PubMed] [Google Scholar]
- 27.White K, Grether ME, Abrams JM, Young L, Farrell K, et al. Genetic control of programmed cell death in Drosophila. Science. 1994;264:677–683. doi: 10.1126/science.8171319. [DOI] [PubMed] [Google Scholar]
- 28.Ryoo HD, Bergmann A, Gonen H, Ciechanover A, Steller H. Regulation of Drosophila IAP1 degradation and apoptosis by reaper and ubcD1. Nat Cell Biol. 2002;20:20. doi: 10.1038/ncb795. [DOI] [PubMed] [Google Scholar]
- 29.Holley CL, Olson MR, Colon-Ramos DA, Kornbluth S. Reaper eliminates IAP proteins through stimulated IAP degradation and generalized translational inhibition. Nat Cell Biol. 2002;14:14. doi: 10.1038/ncb798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yoo SJ, Huh JR, Muro I, Yu H, Wang L, et al. Hid, Rpr and Grim negatively regulate DIAP1 levels through distinct mechanisms. Nat Cell Biol. 2002;4:416–424. doi: 10.1038/ncb793. [DOI] [PubMed] [Google Scholar]
- 31.Lisi S, Mazzon I, White K. Diverse domains of THREAD/DIAP1 are required to inhibit apoptosis induced by REAPER and HID in Drosophila. Genetics. 2000;154:669–678. doi: 10.1093/genetics/154.2.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Goyal L, McCall K, Agapite J, Hartwieg E, Steller H. Induction of apoptosis by Drosophila reaper, hid and grim through inhibition of IAP function. Embo J. 2000;19:589–597. doi: 10.1093/emboj/19.4.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang SL, Hawkins CJ, Yoo SJ, Muller HA, Hay BA. The Drosophila caspase inhibitor DIAP1 is essential for cell survival and is negatively regulated by HID. Cell. 1999;98:453–463. doi: 10.1016/s0092-8674(00)81974-1. [DOI] [PubMed] [Google Scholar]
- 34.Du W, Vidal M, Xie JE, Dyson N. RBF, a novel RB-related gene that regulates E2F activity and interacts with cyclin E in Drosophila. Genes Dev. 1996;10:1206–1218. doi: 10.1101/gad.10.10.1206. [DOI] [PubMed] [Google Scholar]
- 35.Lee T, Luo L. Mosaic analysis with a repressible cell marker (MARCM) for Drosophila neural development. Trends Neurosci. 2001;24:251–254. doi: 10.1016/s0166-2236(00)01791-4. [DOI] [PubMed] [Google Scholar]
- 36.Wang H, Somers GW, Bashirullah A, Heberlein U, Yu F, et al. Aurora-A acts as a tumor suppressor and regulates self-renewal of Drosophila neuroblasts. Genes Dev. 2006;20:3453–3463. doi: 10.1101/gad.1487506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mahoney MB, Parks AL, Ruddy DA, Tiong SY, Esengil H, et al. Presenilin-based genetic screens in Drosophila melanogaster identify novel notch pathway modifiers. Genetics. 2006;172:2309–2324. doi: 10.1534/genetics.104.035170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Struhl G, Adachi A. Requirements for presenilin-dependent cleavage of notch and other transmembrane proteins. Mol Cell. 2000;6:625–636. doi: 10.1016/s1097-2765(00)00061-7. [DOI] [PubMed] [Google Scholar]
- 39.Struhl G, Greenwald I. Presenilin-mediated transmembrane cleavage is required for Notch signal transduction in Drosophila. Proc Natl Acad Sci U S A. 2001;98:229–234. doi: 10.1073/pnas.011530298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vaccari T, Lu H, Kanwar R, Fortini ME, Bilder D. Endosomal entry regulates Notch receptor activation in Drosophila melanogaster. J Cell Biol. 2008;180:755–762. doi: 10.1083/jcb.200708127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dyson N. The regulation of E2F by pRB-family proteins. Genes Dev. 1998;12:2245–2262. doi: 10.1101/gad.12.15.2245. [DOI] [PubMed] [Google Scholar]
- 42.Pignoni F, Zipursky SL. Induction of Drosophila eye development by decapentaplegic. Development. 1997;124:271–278. doi: 10.1242/dev.124.2.271. [DOI] [PubMed] [Google Scholar]
- 43.Lee CY, Robinson KJ, Doe CQ. Lgl, Pins and aPKC regulate neuroblast self-renewal versus differentiation. Nature. 2006;439:594–598. doi: 10.1038/nature04299. [DOI] [PubMed] [Google Scholar]
- 44.Lee CY, Andersen RO, Cabernard C, Manning L, Tran KD, et al. Drosophila Aurora-A kinase inhibits neuroblast self-renewal by regulating aPKC/Numb cortical polarity and spindle orientation. Genes Dev. 2006;20:3464–3474. doi: 10.1101/gad.1489406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rolls MM, Albertson R, Shih HP, Lee CY, Doe CQ. Drosophila aPKC regulates cell polarity and cell proliferation in neuroblasts and epithelia. J Cell Biol. 2003;163:1089–1098. doi: 10.1083/jcb.200306079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Smith CA, Lau KM, Rahmani Z, Dho SE, Brothers G, et al. aPKC-mediated phosphorylation regulates asymmetric membrane localization of the cell fate determinant Numb. Embo J. 2007;26:468–480. doi: 10.1038/sj.emboj.7601495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lai EC. Protein degradation: four E3s for the notch pathway. Curr Biol. 2002;12:R74–78. doi: 10.1016/s0960-9822(01)00679-0. [DOI] [PubMed] [Google Scholar]
- 48.van den Heuvel S, Dyson NJ. Conserved functions of the pRB and E2F families. Nat Rev Mol Cell Biol. 2008;9:713–724. doi: 10.1038/nrm2469. [DOI] [PubMed] [Google Scholar]
- 49.Lloyd TE, Atkinson R, Wu MN, Zhou Y, Pennetta G, et al. Hrs regulates endosome membrane invagination and tyrosine kinase receptor signaling in Drosophila. Cell. 2002;108:261–269. doi: 10.1016/s0092-8674(02)00611-6. [DOI] [PubMed] [Google Scholar]
- 50.Moberg KH, Bell DW, Wahrer DC, Haber DA, Hariharan IK. Archipelago regulates Cyclin E levels in Drosophila and is mutated in human cancer cell lines. Nature. 2001;413:311–316. doi: 10.1038/35095068. [DOI] [PubMed] [Google Scholar]
- 51.Pellock BJ, Buff E, White K, Hariharan IK. The Drosophila tumor suppressors Expanded and Merlin differentially regulate cell cycle exit, apoptosis, and Wingless signaling. Dev Biol. 2007;304:102–115. doi: 10.1016/j.ydbio.2006.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Levels of cleaved Caspase-3 are elevated in ept/tsg101 mutant eye clones. Clones of ept/tsg101 mutant cells marked by the absence of GFP (green) contain many dying cells, as indicated by staining for cleaved Caspase-3 (blue).
(1.84 MB TIF)
Rbf1 levels are unaffected by the H99 deletion. Confocal image of H99 clones (lacking GFP) in a mosaic 3rd instar eye imaginal disc stained with the anti-Rbf1 antibody (red).
(0.73 MB TIF)