Abstract
We have extended our previous studies of calculating acidity constants for the acidic residues found in the turkey ovomucoid third domain protein (OMTKY3) by determining the relative pKa values for the basic residues (Lys13, Arg21, Lys29, Lys34, His52, and Lys55). A polarizable force field (PFF) was employed. The values of the pKa were found by direct comparison of energies of solvated protonated and deprotonated forms of the protein. Poisson Boltzmann (PBF) and Generalized Born (SGB) continuum solvation models represent the hydration, and a non-polarizable fixed-charges OPLS-AA force field was used for comparison. Our results indicate that (i) the pKa values of the basic residues can be found in close agreement with the experimental values when a PFF is used in conjunction with the PBF solvation model, (ii) it is sufficient to take into the account only the residues which are in close proximity (hydrogen bonded) to the residue in question, and (iii) The PBF solvation model is superior to the SGB solvation model for these pKa calculations. The average error with the PBF/PFF model is only 0.7 pH units, compared with 2.2 and 6.1 units for the PBF/OPLS and SGB/OPLS, respectively. The maximum deviation of the PBF/PFF results from the experimental values is 1.7 pH units compared with 6.0 pH units for the PBF/OPLS. Moreover, the best results were obtained while using an advanced non-polar energy calculation scheme. The overall conclusion is that this methodology and force field are suitable for accurate assessment of pKa shifts for both acidic basic protein residues.
Keywords: Polarizable force fields, relative pKa values, continuum solvation models, turkey ovomucoid third domain, OMTKY3
I. Introduction
Acidity constants of protein residues play an important role in protein stability and function. Therefore, predicting protein pKa shifts is very beneficial because obtaining pKa values experimentally is not always feasible. Calculating these values, however, is a highly challenging task. When a chemical group (residue) is positioned in a protein, its electrostatic environment is very different from the same group being hydrated in bulk water. Moreover, the protein conformation affects the geometry of the residues. pKa shifts, therefore, depend on many-body effects by nature.
Several groups have attempted to calculate the relative pKa values including pKa shifts for proteins and for protein residues within different proteins1–25. Three general methods have been employed for this task: quantum mechanic (QM) methods, linear free energy relationships, and empirical force fields. Quantum mechanical or combined quantum mechanical/molecular mechanical calculations can produce pKa values in a good agreement with experimental data10,14–16. However, this approach is limited by the size of the system to be considered and the level of the quantum mechanical theory. Such calculations also require a dielectric continuum approximation for the solvation, and the solvation model has to be well parameterized and suitable for the task, which in itself is a very significant challenge. A linear free energy relationship can be used to fit pKa values to an extensive database20. However, transferability of the parameters to a system not present in the fitting set is always questionable.
Finally, simulations with empirical force fields can be used to assess the pKa shifts for protein residues. The hydration can be simulated with an explicit model9,12,23, or more commonly, a continuum dielectric model1–7,15–19,21,23,24. The most typical technique is to employ a fixed-charge description for the protein and to assign an effective dielectric constant to the protein (e.g., the Tanford-Kirkwood method8,26). However, assigning an effective dielectric constant to the protein interior has a vague physical meaning, and the optimal value of this constant also remains a controversy (values between 2 and ca 203,5,6,19,21), and the accuracy is not guaranteed. Additionally, these calculations seem to reqire inclusion of the electrostatic interactions of residues separated by large distances, but experimental and theoretical evidence supports the primary importance of the immediate environment of the ionizable residue16,27.
We have previously published results of successfully reproducing pKa shifts of carboxyl residues in the turkey ovomucoid third domain (OMTKY3) with a polarizable force field for proteins17. Briefly, the following calculations were carried out. Acidic residues together with the residues with hydrogen bonds with the former ones were simulated explicitly. These systems were surrounded by continuum solvent, in both Generalized Born and Poisson-Boltzmann formalisms. Geometry optimizations were carried out, with the backbone parts fixed, but with most of the side-chain atoms permitted to move. These calculations were carried out for both protonated and deprotonated forms of the acidic residues (Asp and Glu), as well as the reference systems (acids), the relative energies were used to produce the pKa shifts. This formalism is essentially the same as described in more detail below. The accuracy was uniformly good with the average error of 0.58 pH units and with the maximum deviation of only 0.8 units. This result was achieved without any specific fitting of the parameters to the acidity constant calculations. The polarizable model was parameterized to reproduce electrostatic properties of the systems in an arbitrary environment. We believe it was this physical robustness which led to the successful reproduction of the protein pKa shifts. The electrostatic part of the model employed point charges and point dipoles, and the dielectric constant was equal to 1.0 within the protein.
In this article we extend the range of the considered system to the basic residues of OMTKY3. The system was chosen because of the extensive amount of experimental and computational pKa data available. The relative pKa values of the acidic residues have been studied comprehensively compared with the basic ones.
OMTKY3 is a 56-residue protein solved by both x-ray crystallography and NMR28,29. The protein has five acidic residues (Asp7, Glu10, Glu19, Asp27, and Glu43) and six basic residues (Lys13, Arg21, Lys29, Lys34, His52, and Lys55). Of the six basic residues, Lys29 is in close proximity to Asp27 for a potential interaction of the side chains. Lys34 lies within the protein while the other basic residues are exposed to the solvent. Lys55 resides 3.7 Å from Tyr20, and Lys34 resides 3.8 Å from Tyr11. Nδ of His52 has a potential hydrogen bond interaction with the carboxyl group within its backbone.
We present calculations for the relative pKa values of the six basic residues in OMTKY3. We have employed both the polarizable force field (PFF)30 and the fixed-charges OPLS-AA31. Comparisons were also made between the Poisson Boltzmann (PBF) and Surface Generalized Born (SGB) continuum solvation models when using the OPLS-AA force field. Moreover, an advanced treatment of the non-polar contribution to the solvation energy32 available in the Impact software package33 was used and its superiority for the task was confirmed. The simulation details are presented in the next section followed by the results and discussion in section III and by the conclusions in section IV.
II. Method
Force Fields
Both a polarizable force field (PFF)30 and the fixed-charges OPLS-AA31 force field were employed. The details of building the PFF has been described elsewhere30,34. A brief outline is as follows.
The total energy Etotal is computed as a sum of five energy terms: the electrostatic term Eelectrostatic, the nonelectrostatic van der Waals (vdW) term EvdW, bond stretching Ebonds, angle bending Eangles, and torsional rotation Etorsion.
| (1) |
The total electrostatic energy Eelectrostatic of the model is defined by the interactions of fixed-magnitudes atomic charges, point dipoles, and inducible point dipoles.
| (2) |
where Jij,kl is a scalar coupling between bond-charge increments on sites i,j and k,l for qij and qkl. Sij,k is a vector coupling between a bond-charge increment on sites i,j and a dipole on site k µk; a rank-two tensor coupling, Ti,j, describes the interactions between dipoles on sites i and j. ai is the polarizability of site i. Parameter χi describes “dipole affinity” of site i.
In accordance with the Coulomb formalism, we assume:
| (3) |
| (4) |
| (5) |
where inducible dipoles are placed on all heavy atoms and on some polar hydrogens34. Fixed charges q and permanent dipoles defined by the “dipole affinities” χ are present on all atoms. Point charges representing electron lone pairs are placed on sp2 and sp3 oxygen atoms with the virtual site – oxygen distances set to 0.47 Å (“virtual sites”).
The electrostatic parameters were fitted to reproduce the quantum mechanically determined electrostatic potential and its reaction to perturbations; B3LYP density functional theory (DFT) with the cc-pVTZ(−f) basis set of Dunning35 was used for the QM calculations.
The overall non-electrostatic pair potential form is:
| (6) |
where the A parameter is set so that the 1/r12 term is close to zero in the hydrogen bonding region but is large enough to prevent atoms from being positioned too close, thus penetrating the nonphysical region of the phase space. After the electrostatic part of the force field is ready, fitting of the vdW parameters is completed. The repulsion parameters C and B are produced to ensure the correct gas-phase dimerization properties, as compared to the high-level ab initio results. The procedure is based on an extrapolation scheme utilizing LMP2/cc-pVTZ(−f) and LMP2/cc-pVQZ data34,36.
The bond stretching and angle bending terms have the standard harmonic functional form, and the torsional energy is described by a sum of Fourier series.
| (7) |
| (8) |
| (9) |
where Kr and KΘ represent the force constants; r, req, Θ, and Θeq are actual and equilibrium values of bond lengths and angles, respectively; and φ represents the dihedral angles. All the parameters in Equations are taken directly from the OPLS-AA force field31.
For the non-polarized calculations, the standard OPLS-AA functional form was used, with the Eelectrostatic and EvdW terms in eqn. ( 1 ) replaced by the sum of the Coulomb and Lennard-Jones contributions for pairwise intra- and intermolecular interactions:
| (10) |
The coefficient fij is equal to 0.0 for any i-j pairs connected by a valence bond (1–2 pairs) or a valence bond angle (1–3 pairs). fij = 0.5 for 1,4-interactions (atoms separated by exactly 3 bonds) and fij = 1.0 for all other cases. Standard OPLS-AA parameters are employed.
pKa Calculations
Previously, our group calculated the pKa shifts for the acidic amino acids in turkey ovomucoid third domain (OMTKY3)28. We are using the same formalism for the basic residues. The relative pKa values are calculated from the free energies of both the residue (A) and its acidic or basic reference system (acid, or side chain).
| (11) |
| (12) |
The relative pKa values can then be defined as
| (13) |
| (14) |
From there, the difference in pKa values between the residue and its corresponding acidic or basic side chain can then be calculated. When rearranged, the equation can yield the relative pKa value of the residue.
| (15) |
| (16) |
Values of the energies G(A−), G(A-H), G(acid−), and G(acid-H) are obtained via geometry optimizations with Poisson-Boltzmann (PBF) and Surface Generalized Born (SGB) continuum solvent models. All the geometry optimizations were carried out with the Impact software suite33.
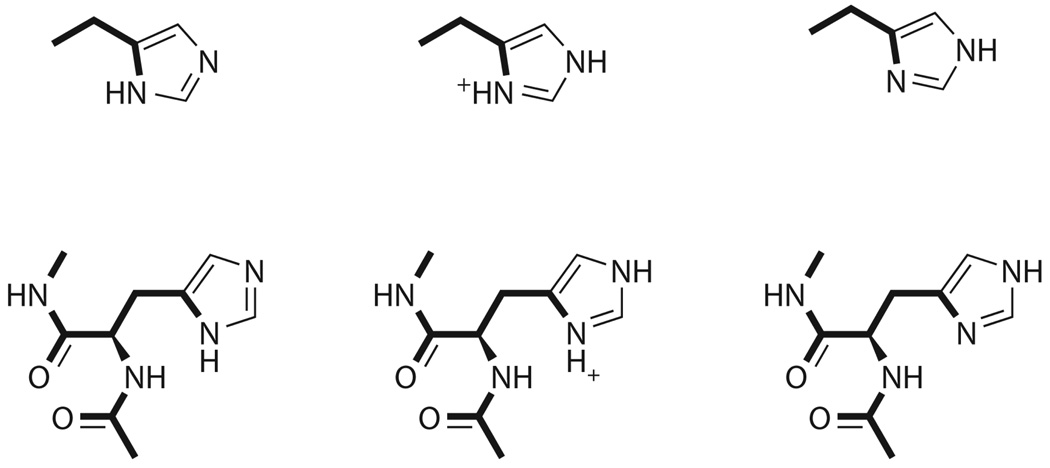
The initial conformations for the six basic residues (Lys13, Arg21, Lys29, Lys34, His52, and Lys55) were obtained from the experimental OMTKY3 geometry (pdb ID 1omu28). The structure of OMTKY3 was determined in an aqueous solution with a neutral pH, and therefore the basic residues were protonated. In this project, the residues were simulated in both the protonated and the deprotonated states. Each deprotonated state was represented by a different number of conformations depending upon the number of hydrogens bonded to the protonated residue (Figure 1 – Figure 4). For instance, histidine had two hydrogens covalently bonded to two different nitrogens (Nδ and Nε); and the deprotonated histidine had two structures simulated.
Figure 1.
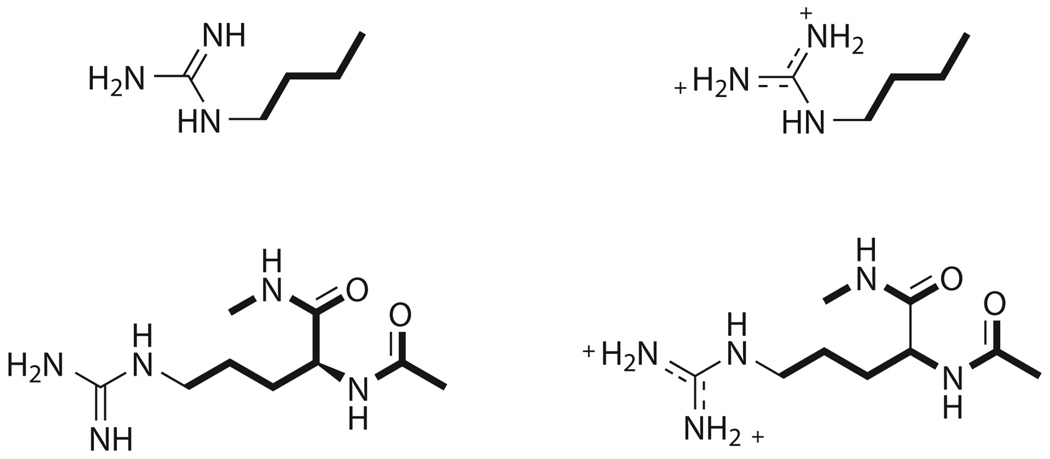
Arginine reference system (top) and capped residue (bottom) simulated for Arg21 calculations. The geometry of the residue was that found in the actual OMTKY structure, further optimized with IMPACT.33 The bold bonds were restrained throughout the simulations while the rest were permitted to move freely. The reference system (n-butylguanidine) pKa was 12.48 (n-butylguanidine).
Figure 4.
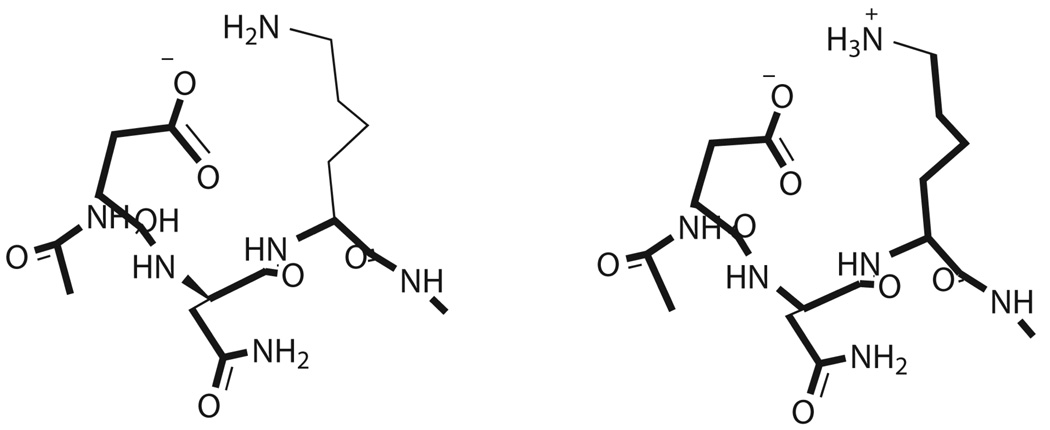
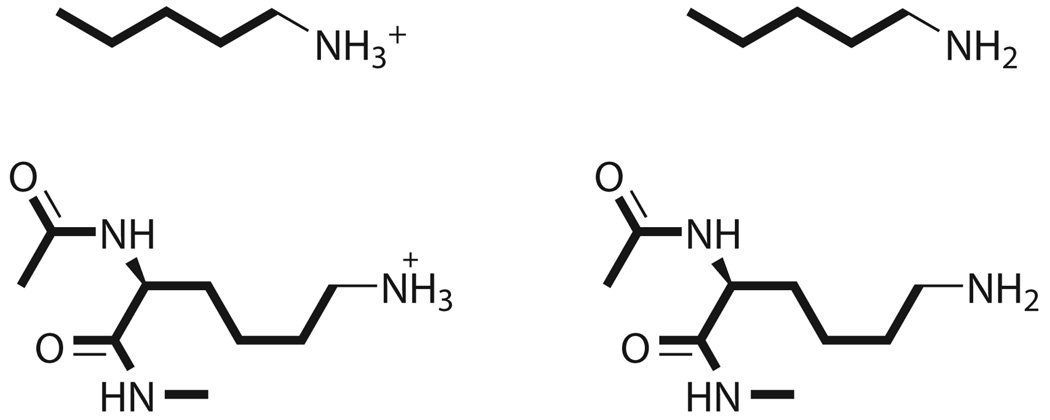
Lys29 was within proximity to Asp27 for potential hydrogen-bonded interactions. Ace-Asp-Asn-Lys-NMe was the sequence simulated for the complete residue calculations. The entire neutral side chain of Lys29 was unconstrained. The reference structure was similar to the other lysines (n-pentylamine, pKa 10.6338).
We follow the formalism introduced in the previous work17 for choosing only a part of the OMTKY3 protein to be represented explicitly. Results of both experimental and computational studies7,15,17 suggest that the immediate chemical environment of ionizable residues plays the most significant role in defining the acidity constant shifts for these residues. Therefore, we chose to work with a relatively small subset of the protein. The models for the acidic residues were originally suggested and used for quantum mechanical calculations15. In a nutshell, we included the residues in question and those residues with hydrogen bonds to them.
The residues were capped with an acetyl group on the N-terminus and an N-methylamine group on the C-terminus except for Lys29. Because Lys29 has a potential interaction with Asp27, the residue sequence was Ac-Asp-Asn-Lys-NMe. Additionally, coordinates for the reference systems were taken from the residues. Lysine was represented by n-pentylamine (pKa 10.638), histidine by 4-ethylimidazole (pKa 7.5538), and arginine by n-butylguanidine (pKa 12.48 from methylguanidine38).
All structures were minimized with Impact 3.033. Geometries were constrained, except as follows. In order to avoid errors in energy caused by using a completely rigid structure, some atoms in the residues at hand were allowed to move in the course of the geometry optimizations. The exact breakdown of fixed and movable atoms is shown on Figure 1–Figure 4. The parts of the molecules shown in bold were kept fixed, while the non-bold ones were permitted a complete flexibility. Therefore, we believe that the structures were relaxed enough to avoid any force-field artifacts which could result from employing the unmodified PDB structures and, more importantly, this flexibility permitted the differences in geometries resulting from the protonated-deprotonated transition to manifest and to be taken into the account. At the same time, geometry optimizations were used and therefore, no thermodynamic sampling which would be present in Monte Carlo or molecular dynamics simulations was present.
Lys29, however, had no constraints on the neutral side chain allowing the side chain to move freely. The reasons for making this exception are considered in more detail in the next section. The minimizations were carried out with either the Generalized Born or the Poisson Boltzmann continuum solvent model39,40. Both solvent models had an internal dielectric constant of 1.0 and a nonbonded cutoff of 100.0 Å. The nonbonded list was updated every 1000 steps. Two different force fields were employed for this work: OPLS-AA31 and the polarizable force field (PFF)30; the PFF was only used in conjunction with the Poisson Boltzmann continuum solvent model. Additionally, an advanced treatment of nonpolar solvent effects were included for some of the simulations32 to represent better the hydrophobic protein environment.
Briefly, the formalism was as follows (see Reference 32 for a complete description and discussion). The non-polar terms was generally in the following form:
| (17) |
The summation runs over all the atoms. γ(ti) and α(ti) denote parameters for the atom type ti of the atom i, and Ai stands for the solute-accessible area of the same atom. Therefore, the parameter . γ(ti) is in the essence a representation of the surface tension, while the presence of α(ti) means that even a buried atom contributes to the solute-solvent interaction. The expression was further modified to include a function of the Born radius Bi, and the final form of the latter was:32
| (18) |
where
| (19) |
a, b, and c were parameters of the theory.32 The nonpolar term was used as implemented in Impact 3.0.33
Simulations for each minimized structure consisted of ten cycles of conjugate gradient minimizations. Each minimization cycle ran for a maximum of 10,000 steps before the next cycle of minimizations began. The r.m.s.d. of the system had to converge within 0.05 Å, and the energy change had a convergence criterion of 1.0 × 10−6 kcal/mol. Minimizations began with an initial step size of 0.05 and ended with a maximum step size of 1.0. The conformation with the lowest free energy was then used as the initial guess for the next minimization. This procedure was repeated for a total of five runs per structure to ensure convergence.
III. Results and Discussion
A comparison was made between our results, the experimental pKa values, and relative pKa values calculated from NMR models. The NMR-calculated pKa values were determined using CHARMM with the Poisson Boltzmann continuum solvent6. All the acidity constant results are shown in Table 1 and Table 2. Overall, the PBF/PFF combination yields the best result, with the average error of only 0.7 pH units – if the advanced nonpolar energy treatment is employed in the solvation model (Table 2). If the non-polarizable OPLS force field is used, the PBF solvation model is clearly superior to the SGB model with ca. 2.1–2.2 pH units average error. No overall accuracy gain seems to be exhbited with the nonpolar term if the fixed-charges OPLS-AA force field was used; yet, it offers an advantage for the polarizable model as seen by comparing the results in Table 1 and Table 2. We believe that the lack of improvement which would be produced by the advanced nonpolar term in the OPLS-AA case is explained by the intrinsic inability of the fixed-charges model to accurately capture energetic effects introduced by such strong perturbations as the deprotonation considered in calculating the pKa shifts of the residues. It can be seen from the data in Table 1 and Table 2, that the errors in the OPLS-AA numbers are not systematically increasing or decreasing with the introduction of the new non-polar term, not do the number systematically shift up or down. Therefore, it appears that the errors in the energies introduced by the electrostatic rigidity of the fixed-charges model are simply too great to permit the pKa values to become more accurate by simply changing the solvation model. At the same time, the polarizable force field is by nature more accurate in its response to the changing electrostatic environment, and thus the improvement of the solvation model is removing a more significant part of the deviation from the experimental data. Moreover, since the intrinsic error of the polarizable force field is smaller than for the fixed-charges one, it can be expected that a higher level of accuracy could be achieved in creating a new solvent model for the former. The solvation parameters will be representing the solvent effect itself, and will not be used to compensate for the deficiencies of the non-adjusting fixed charges. Therefore, one would expect that such a solvation model will be more transferable.
Table 1.
pKa values for basic residues of OMTKY3 compared with the experimental and the NMR-calculated ones – no advanced nonpolar solvation energy term.
| without advanced nonpolar solution term | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| SGB/OPLS | PBF/OPLS | PBF/PFF | |||||||||
| Expt.a | NMRa | pKa | Errexp | Errnmr | pKa | Errexp | Errnmr | pKa | Errexp | Errnmr | |
| Lys13 | 9.9 | 11.2 | 17.0 | 7.1 | 5.8 | 10.1 | 0.3 | 1.1 | 10.7 | 0.8 | 0.5 |
| Arg21 | 12.8 | 11.4 | 1.4 | 11.9 | 0.9 | 26.7 | 13.9 | ||||
| Lys29 | 11.1 | 11.2 | 27.0 | 15.9 | 15.8 | 17.5 | 6.4 | 6.3 | 13.9 | 2.7 | 2.7 |
| Lys34 | 10.1 | 11.7 | 17.3 | 7.1 | 5.6 | 10.1 | 0.0 | 1.6 | 13.8 | 3.7 | 2.1 |
| His52 | 7.5 | 6.2 | 10.2 | 2.7 | 4.0 | 11.7 | 4.2 | 5.5 | 9.1 | 1.6 | 2.9 |
| Lys55 | 11.1 | 11.3 | 16.0 | 4.9 | 4.7 | 10.1 | 1.0 | 1.2 | 11.1 | 0.0 | 0.2 |
| avg. err. | 6.5 | 6.2 | 2.1 | 2.8 | 3.8 | 3.7 | |||||
Reference 6.
Table 2.
pKa values for basic residues of OMTKY3 compared with the experimental and the NMR-calculated ones – using advanced nonpolar solvation energy term.
| with advanced nonpolar solution term | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SGB/OPLS | PBF/OPLS | PBF/PFF | |||||||||||
| Expt.a | NMRa | pKa | Errexp | Errnmr | pKa | Errexp | Errnmr | pKa | Errexp | Errnmr | |||
| Lys13 | 9.9 | 11.2 | 16.7 | 6.9 | 5.5 | 9.0 | 0.8 | 2.2 | 10.4 | 0.5 | 0.8 | ||
| Arg21 | 12.8 | 15.0 | 2.2 | 12.4 | 0.4 | 12.3 | 0.5 | ||||||
| Lys29 | 11.1 | 11.2 | 23.4 | 12.3 | 12.2 | 17.2 | 6.0 | 6.0 | 12.9 | 1.7 | 1.7 | ||
| Lys34 | 10.1 | 11.7 | 16.3 | 6.2 | 4.6 | 9.9 | 0.3 | 1.8 | 9.9 | 0.3 | 1.8 | ||
| His52 | 7.5 | 6.2 | 11.8 | 4.3 | 5.6 | 11.2 | 3.7 | 5.0 | 7.6 | 0.1 | 1.4 | ||
| Lys55 | 11.1 | 11.3 | 15.7 | 4.6 | 4.4 | 9.5 | 1.6 | 1.8 | 10.1 | 1.0 | 1.2 | ||
| avg. err. | 6.1 | 5.8 | 2.2 | 2.9 | 0.7 | 1.2 | |||||||
Reference 6.
Histidine 52 Residue
Histidine seemed to be the most difficult residue for which to calculate the relative pKa value. The other residues involved alkanes while histidine had a five-membered imidazole ring. Lysine and arginine had similar free energies for the deprotonated residues; His52, on the other hand, had a lower free energy when the hydrogen was covalently bonded to Nε. When simulated using PBF/PFF with the nonpolar solvation term, the relative pKa of His52 was within 0.1 pKa units of the experimental value. The simulation of His52 without the nonpolar term in the continuum solvation energy was within 1.4 pH units of the experimental value, but neither the SGB/OPLS nor the PBF/OPLS simulation could predict a reasonable relative pKa value for His52. Polarizability appears to play a role in the imidazole ring, and because the OPLS-AA force field does not account for this, neither solvent model could achieve a relative pKa value similar to the experimental one.
Arginine 21 Residue
Of all the relative pKa calculations, Arg21 had pKa values less than 3 pH units from the NMR-calculated value for all solvent and force field models except PBF/PFF — if the advanced nonpolar term was not included in the solvation energy calculations. The relative pKa value of Arg21 was not determined experimentally by Forsyth et al. Based upon energetics, the PBF/PFF simulations appear to be more stable than their OPLS-AA counterparts. The deprotonated residue and side chains are actually more stable when simulated with PBF/PFF; the deprotonated residue simulated in SGB/OPLS with the nonpolar effects, however, had a lower energy compared with the PBF/PFF simulation. The polarizability may account for the energetic stability in this case, but arginine has a δ-guanidino group which is affected by the solvent. When the advanced nonpolar term is included, |ΔΔG| drops considerably, and the absolute error follows suit. Both SGB/OPLS simulations have pKa values deviating more than 1 pH unit from the NMR-calculated pKa result. The SGB solvent model has been shown to overstabilize nonbonded interactions41, which explains the high pKa predictions and tends to overestimate the atomic radius, which in turn, affects the electrostatics32.
Arg21 simulations performed with PBF/OPLS offer an accurate estimate to the NMR-calculated pKa value, the acidity constant is similar to that obtained using the PBF/PFF model with nonpolar effects. This similarity suggests that PBF/OPLS can offer good pKa values for arginine residues. There is, however, a conformational dependence of the pKa values. When arginine was simulated using generic, not protein-specific coordinates provided by Impact 3.0, the absolute pKa error was 0.8 pH units for PBF/OPLS, 2.1 units for PBF/OPLS with nonpolar effects, and 0.1 units for PBF/PFF with nonpolar effects. Base on these results, PBF/OPLS either with or without the nonpolar effects will not necessarily provide relative pKa values as close to experimental or NMR-calculated ones as produced by the polarizable force field.
Lysine 29 Residue
Lys29 was the only basic residue that had a potential hydrogen bonding interaction; within proximity to Asp27. When simulated with SGB/OPLS either with or without the nonpolar effect, the relative pKa value was quite high, over 20 pH units. The total free energies for the simulations were lower than the free energies of the other simulations, which indicated that the interactions were stabilized, probably by the hydrogen bonds. As stated in the explanation for Arg21, SGB tends to overestimate the atomic radius thereby affecting the electrostatic interactions and the pKa values.
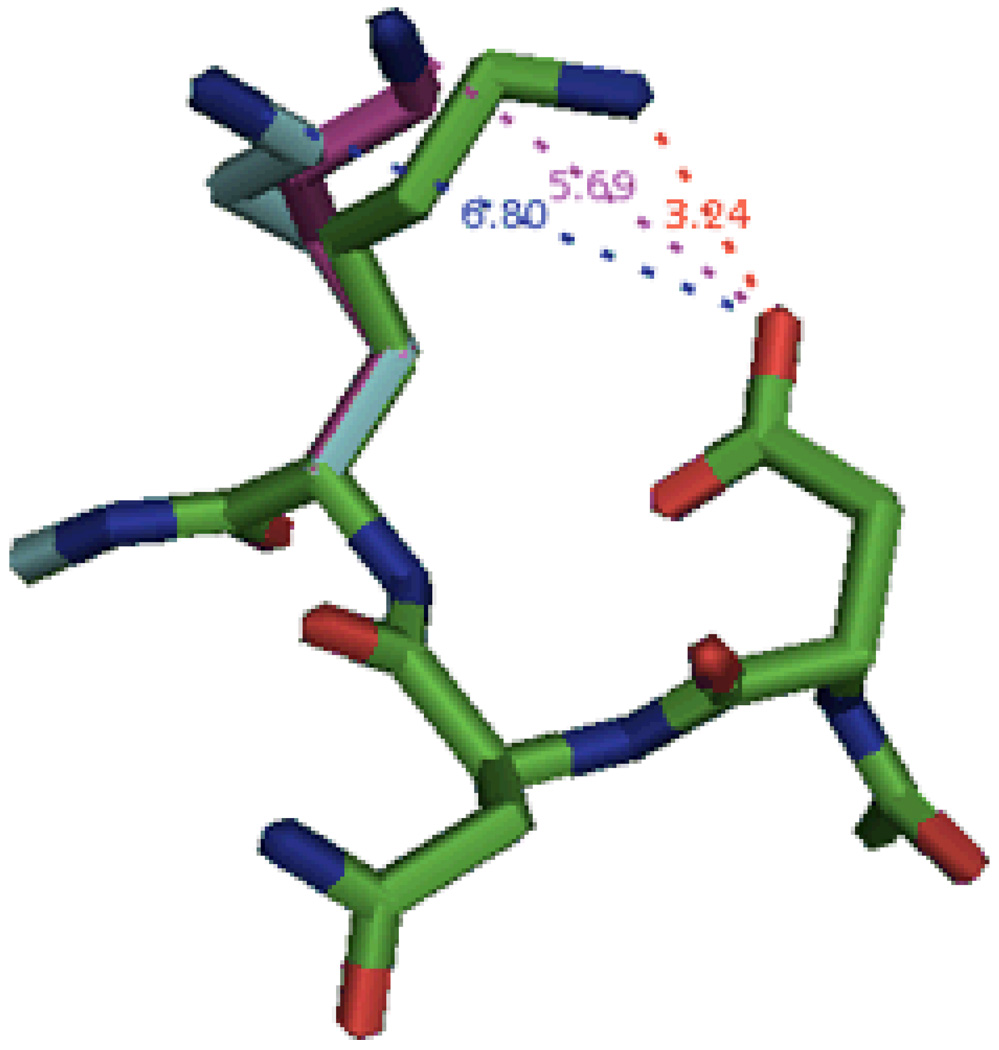
As mentioned in the Methods section, the neutral Lys29 was simulated without constraints upon the side chain. By removing the constraints, the side chain was able to move away from Asp27, which as noted earlier, had a possible interaction with the charged side chain (Figure 5). The interaction distance increased from 3.24 Å to ca. 5.7 – 6.8 Å. This alleviated the strain on the system and permitted a more accurate calculation of the relative pKa value for Lys29. Such a choice makes sense because the PDB structure contains the protonated Lys29 residue, and the deprotonated state will exhibit a noticeably different geometry, as is indeed shown on Figure 5.
Figure 5.
Comparison of Lys29 side chain geometries before and after optimizations. The NMR model of neutral Lys29 (green) was simulated in PBF/PFF with and without nonpolar solvent effects (purple and cyan, respectively) as well as without constraints imposed upon the Lys side chain. The distances between the O and N atoms are listed for the NMR (red), PBF/PFF (purple), and PBF/PFF with nonpolar solvent effects (blue).
Residues Lys13, Lys34, and Lys55
The three remaining residues (Lys13, Lys34, and Lys55) had no hydrogen bonds with other residues withn the protein. Lys34 appeared to be internal compared with Lys13 and Lys55, which were exposed to the solvent. As expected, the three residues had large absolute pKa errors when simulated in either SGB/OPLS models. The pKa error of Lys34 was greater than 3.0 pH units when simulated in PBF/PFF without any consideration for the nonpolar effects, and Lys55 had an error of 1.6 units when simulated in PBF/OPLS with the nonpolar effects enabled. Otherwise, the pKa errors were less than 1.5 pH units for the remaining simulations.
The PBF/PFF error for the Lys34 residue was reduced from 3.7 to 0.3 pH units by using the advanced nonpolar treatment as shown in Table 2, which indicates that the nonpolar term may better represent the internal hydrophobic environment of a protein. Yet, the OPLS results were marginally affected with either the PBF or the SGB solvation model. Overall, the PBF/OPLS performs rather well in this case.
As stated earlier, atomic positions can significantly affect the calculation of relative pKa values. Lysine, for example, offers an absolute pKa error of less than 1.0 pH units for PBF/OPLS, PBF/PFF and PBF/PFF with nonpolar effects when simulated using the generic , non-protein specific coordinates provided by Impact. Yet, the average pKa errors (Arg, His, and Lys) were less than 1.0 pH unit for the residues simulated in PBF/PFF with consideration of nonpolar effects. Overall, while the PBF/PFF combination with the advanced nonpolar term is the best one for the Lys residues, it looks like the three lysine residues which have no hydrogen bonds to the rest of the protein (Lys13, Lys34, and Lys55) can be simulated reasonably well with the nonpolarizable OPLS force field. This is true for the PBF solvation model but not for the SGB solvation model. This suggests that reproducing the solvent polarization is much more important for this residues than the solute one, which is not suprising given that the protonated lysine residues are positively charged.
At this point, we would like to compare the results of this research, as well as of our calculations presented in Reference 17, with those arising from protein pKa calculations described in References 23 and 42. The former addresses explicit and implicit solvent models used with AMBER and CHARMM force fields to evaluate pKa values of aspartic acid residues in thioredoxin. Molecular dynamics simulations were employed, but the force fields did not include explicit electrostatic polarizability. The results are presented in Table 3. It can be seen that the explicit solvent simulations lead to the average pKa errors of 1.9–2.6 pH units, with the maximum deviations of ca. 3.2–4.5 unites, which is not inconsistent with the results presented in this article and in Reference 17 for the fixed-charges force field. The authors of that work themselves admit that such a great magnitude of the error is likely to stem from the absence of the explicit treatment of the electrostatic polarization.23 The results of the continuum solvent simulations are significantly better, even though both the average and the maximum deviations from the experimental results are still not as good as those presented for the PFF in this work and in Reference 17. Moreover, only three residues are covered, while we have simulated a total of eleven. Nevertheless, the importance of the thermodynamic sampling, as suggested by the authors of the work in Reference 23 seems to be clear, and this is a direction which we hope to explore in the future to improve our results for the pKa shifts even further. Furthermore, the continuum solvent model does present a model of polarization. The very fact that this permits to improve the results as compared to the non-polarizable fixed-charges explicit solvent can probably serve as a proof, albeit indirect, of the need to explicitly address the electrostatic polarization in protein pKa calculations.
Table 3.
Relative pKa values for Asp residues of thioredoxin, from Reference 23
| Explicit solvent | Continuum Solvent | Experiment | ||
|---|---|---|---|---|
| AMBER | CHARMM | |||
| Asp14 | −0.66 | 0.81 | −0.81, −1.61 | −1.98 |
| Asp20 | 1.25 | 0.66 | 0.37 | 0.0 |
| Asp26 | 6.67 | 7.99 | 5.21 | 3.52 |
| Average Error | 1.91 | 2.64 | 1.10, 0.81 | |
The authors of the work in Reference 42 used a different approach. They have calculated the Asp20 and Asp26 pKa shifts in the thioredoxin with the molecular dynamics approach and fixed charges, but the chares employed were obtained with the Ppolarized Protein-Specific Charges (PPC) procedure. Briefly, quantum mechanical calculations are run for a protein subsystem, and the electrostatic potential fitting is used to derive the charges. While these charges themselves are fixed in the simulations which follow, they are nevertheless produced for a specific protein structure and environment, and thus can be expected to perform better than generic protein charges which are supposed to be transferable to any protein, for as long as they are used in the correct aminoacid. The error in the Asp26 pKa was 0.15 pH units, and it was 0.73 units for Asp20. While these results themselves are excellent, our PFF not only performs uniformly well on a large set of data, but also has an intrinsic advantage of having the polarizable parameters fitted once and for all. These parameters are taking care of the electrostatic response automatically in the process of the simulations, without any need to derive fixed charges from quantum mechanics for each new protein structure. The quality of our results stems from the adequate physical description of the protein electrostatics and not from any conformation-specific refitting.
Moreover, the errors listed in Reference 42 for the Asp26 and Asp20 pKa values obtained with a fixed-charges AMBER force field were 3.15 and 1.15 pH units, respectively. We believe that this bears witnesses to the importance of using an adjustable electrostatics. Apparently, just like in the work presented in Reference 23, an adequate thermodynamic sampling in itself is not sufficient to correctly reproduce pKa shifts in proteins.
It is also worth mentioning that our assumption that only the immediately neighboring and hydrogen boned residues have to be included in the calculations in order to produce accurate assessment of the protein pKa shifts has been proven to work well again. As shown in Table 2, the relatively small parts of the whole OMTKY3 protein shown on Figure 1 – Figure 4 and employed in our calculations were sufficient to produce the good results. Moreover, the same formalism worked well in our calculations of pKa shifts in the Asp and Glu residues of OMTKY.17 The average error in pKa values was found to be 0.58 pH units, with the maximum deviation of 0.8 units. Furthermore, experimental studies of the OMTKY3 have also demonstrated that mutations of the residues beyond the immediate proximity has little effect upon the pKa shifts of the acidic residues.7 At this point, we cannot be absolutely confident that this hypothesis will remain valid for other proteins, but the consistency of the above results suggests that this might well be the case.
The above notion has yet another positive implication. None of the six residues considered in this work were in an immediate nearness to each other. Therefore, none were explicitly included in calculations of pKs values of another residue. Thus, the combinatorial problem of considering all possible compositions of the protonated and deprotonated residue A, interacting with the protonated and deprotonated residue B, interacting with protonated and deprotonated residue C, etc. did not emerge. Each residue was considered independently. And while the Lys29 case did include the ionazable Asp27 residue in the vicinity, the acidity constant of the latter is so low (pKa = 2.3) that we could simply assume its presence in the deprotonated form.
IV. Conclusions
Following the success of our previous calculations of pKa shifts for the acidic residues of the OMTKT3 protein, we have calculated values of acidity constants for six basic residues (Lys13, Arg21, Lys29, Lys34, His52, and Lys55) of the same protein. A polarizable force field was employed, as well as the fixed-charges OPLS-AA with both the Poisson Boltzmann and the Generalized Born continuum solvation models. Moreover, the effect of utilizing an advanced treatment of nonpolar solvation effects was also tested.
The polarizable force field was demonstrated to produce results superior to the fixed-charges OPLS-AA, and the choice of the continuum solvation model was certainly important in using the OPLS force field. The average pKa error with the PFF was found to be only 0.7 pH units compared with 2.1–2.2 units for the PBF/OPLS model.
The choice of the continuum solvation model is certainly important in using the OPLS force field, as the OPLS/SGB produced a much larger average error of 6.1–6.5 pH units.
Employing the advanced nonpolar term with the PBF solvation model was necessary to represent the hydrophobic protein environment better and to obtain results agreeable with experiment for the PFF technique . However, this term did not seem to play a significant role for the OPLS-based simulations.
One can also make the following observation based on the data presented in this paper. There are two important components which are missing in fixed-charges single-point calculations of protein pKa values. The first is the need for an adequate thermodynamic sampling, as opposed to using a single geometry. The other is the necessity to use adjustable electrostatics (explicit polarization). We have partially addressed the former issue by performing geometry optimizations of parts of the residues involved. At the same time, it appears that the latter issue is much more important. On one hand, our polarizable force field, which was not produced or fitted specifically for pKa calculations, yields results in uniformly good agreement with experiment, even without Monte Carlo or molecular dynamics simulations. On the other hand, results presented in References 23 and 42 demonstrate that fixed charges force fields are not sufficiently adequate in calculating pKa values of protein residues, even when a physically correct thermodynamic sampling is involved. Therefore, while we do plan to add a Monte Carlo component to our simulations in the future, we believe that our work has demonstrated that the functional form of the force field (polarization) is much more critical for the research in hand.
Overall, we have demonstrated that our polarizable force field is suitable for simulating both acidic and basic residues with a chemically significant level of accuracy, and the explicit treatment of polarization was necessary to achieve this agreement with the experimental data.
Figure 2.
Representation of histidine reference system (top) and its residue in the protein (bottom). The geometry of the residue was that found in the actual OMTKY structure, further optimized with Impact. The bold bonds were restrained throughout the simulations while the rest were permitted to move freely. The reference system (4-ethylimidazole) pKa was 7.55 37.
Figure 3.
Representation of lysine reference system (top) and its residue in the protein (bottom). The geometry of the residue was that found in the actual OMTKY structure, further optimized with Impact. The bold bonds were restrained throughout the simulations. The other bonds were permitted to move freely. The reference system (pentylamine) pKa was 10.6338.
Acknowledgments
The project described was supported by Grant Number R01GM074624 from the National Institute of General medical Sciences. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of General Medical Sciences or the national Institutes of Health. The authors express gratitude to Schrödinger, LLC for the Impact software. We also wish to thank Jeff Saunders of Schrodinger, LLC for his technical assistance.
References
- 1.Antosiewicz J, Mccammon JA, Gilson MK. J Mol Biol. 1994;238:415–436. doi: 10.1006/jmbi.1994.1301. [DOI] [PubMed] [Google Scholar]
- 2.Antosiewicz J, McCammon JA, Gilson MK. Biochemistry-Us. 1996;35:7819–7833. doi: 10.1021/bi9601565. [DOI] [PubMed] [Google Scholar]
- 3.Barth P, Alber T, Harbury PB. P Natl Acad Sci USA. 2007;104:4898–4903. doi: 10.1073/pnas.0700188104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dixit SB, Bhasin R, Rajasekaran E, Jayaram B. J Chem Soc Faraday T. 1997;93:1105–1113. [Google Scholar]
- 5.Forsyth WR, Antosiewiez JM, Robertson AD. Proteins. 2002;48:388–403. doi: 10.1002/prot.10174. [DOI] [PubMed] [Google Scholar]
- 6.Forsyth WR, Gilson MK, Antosiewicz J, Jaren OR, Robertson AD. Biochemistry-Us. 1998;37:8643–8652. doi: 10.1021/bi980187v. [DOI] [PubMed] [Google Scholar]
- 7.Forsyth WR, Robertson AD. Biochemistry-Us. 2000;39:8067–8072. doi: 10.1021/bi992967p. [DOI] [PubMed] [Google Scholar]
- 8.Havranek JJ, Harbury PB. P Natl Acad Sci USA. 1999;96:11145–11150. doi: 10.1073/pnas.96.20.11145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jorgensen WL, Briggs JM. J Am Chem Soc. 1989;111:4190–4197. [Google Scholar]
- 10.Kallies B, Mitzner R. J Phys Chem B. 1997;101:2959–2967. [Google Scholar]
- 11.Kaminski GA. J Chem Theory Comput. 2005;1:248–254. doi: 10.1021/ct049880o. [DOI] [PubMed] [Google Scholar]
- 12.Kaminski GA. J Phys Chem B. 2005;109:5884–5890. doi: 10.1021/jp050156r. [DOI] [PubMed] [Google Scholar]
- 13.Khandogin J, York DM. Proteins-Structure Function and Bioinformatics. 2004;56:724–737. doi: 10.1002/prot.20171. [DOI] [PubMed] [Google Scholar]
- 14.Klicic JJ, Friesner RA, Liu SY, Guida WC. J Phys Chem A. 2002;106:1327–1335. [Google Scholar]
- 15.Li H, Hains AW, Everts JE, Robertson AD, Jensen JH. J Phys Chem B. 2002;106:3486–3494. [Google Scholar]
- 16.Li H, Robertson AD, Jensen JH. Proteins-Structure Function and Bioinformatics. 2004;55:689–704. doi: 10.1002/prot.20032. [DOI] [PubMed] [Google Scholar]
- 17.MacDermaid CM, Kaminski GA. J Phys Chem B. 2007;111:9036–9044. doi: 10.1021/jp071284d. [DOI] [PubMed] [Google Scholar]
- 18.Mehler EL, Guarnieri F. Biophys J. 1999;77:3–22. doi: 10.1016/S0006-3495(99)76868-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nielsen JE, Vriend G. Proteins. 2001;43:403–412. doi: 10.1002/prot.1053. [DOI] [PubMed] [Google Scholar]
- 20.Ohno K, Sakurai M. J Comput Chem. 2006;27:906–916. doi: 10.1002/jcc.20372. [DOI] [PubMed] [Google Scholar]
- 21.Sandberg L, Edholm O. Proteins. 1999;36:474–483. doi: 10.1002/(sici)1097-0134(19990901)36:4<474::aid-prot12>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- 22.Schaller W, Robertson AD. Biochemistry-Us. 1995;34:4714–4723. doi: 10.1021/bi00014a028. [DOI] [PubMed] [Google Scholar]
- 23.Simonson T, Carlsson J, Case DA. J Am Chem Soc. 2004;126:4167–4180. doi: 10.1021/ja039788m. [DOI] [PubMed] [Google Scholar]
- 24.Song JK, Laskowski M, Qasim MA, Markley JL. Biochemistry-Us. 2003;42:2847–2856. doi: 10.1021/bi0269512. [DOI] [PubMed] [Google Scholar]
- 25.Zhang MJ, Vogel HJ. J Biol Chem. 1993;268:22420–22428. [PubMed] [Google Scholar]
- 26.Tanford C, Kirkwood JG. Journal Title:Journal of the American Chemical Society. 1957;79:5333–5339. [Google Scholar]
- 27.Harris TK, Turner GJ. Iubmb Life. 2002;53:85–98. doi: 10.1080/15216540211468. [DOI] [PubMed] [Google Scholar]
- 28.Hoogstraten CG, Choe S, Westler WM, Markley JL. Protein Sci. 1995;4:2289–2299. doi: 10.1002/pro.5560041106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bode W, Wei AZ, Huber R, Meyer E, Travis J, Neumann S. Embo J. 1986;5:2453–2458. doi: 10.1002/j.1460-2075.1986.tb04521.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maple JR, Cao YX, Damm WG, Halgren TA, Kaminski GA, Zhang LY, Friesner RA. J Chem Theory Comput. 2005;1:694–715. doi: 10.1021/ct049855i. [DOI] [PubMed] [Google Scholar]
- 31.Kaminski GA, Friesner RA, Tirado-Rives J, Jorgensen WL. J Phys Chem B. 2001;105:6474–6487. [Google Scholar]
- 32.Gallicchio E, Zhang LY, Levy RM. J Comput Chem. 2002;23:517–529. doi: 10.1002/jcc.10045. [DOI] [PubMed] [Google Scholar]
- 33.Impact, version 3.0. New York: Schrödinger, LLC; 2004. [Google Scholar]
- 34.Kaminski GA, Stern HA, Berne BJ, Friesner RA. J Phys Chem A. 2004;108:621–627. [Google Scholar]
- 35.Dunning TH. J Chem Phys. 1989;90:1007–1023. [Google Scholar]
- 36.Kaminski GA, Stern HA, Berne BJ, Friesner RA, Cao YXX, Murphy RB, Zhou RH, Halgren TA. J Comput Chem. 2002;23:1515–1531. doi: 10.1002/jcc.10125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dean JA, Lange NA. Lange's handbook of chemistry. 15th ed. New York: McGraw-Hill; 1999. [Google Scholar]
- 38.Lide DR. CRC handbook of chemistry and physics : a ready-reference book of chemical and physical data; 87th ed. Boca Raton, Fla: CRC Press; 2006. [Google Scholar]
- 39.Ghosh A, Rapp CS, Friesner RA. J Phys Chem B. 1998;102:10983–10990. [Google Scholar]
- 40.Tannor DJ, Marten B, Murphy R, Friesner RA, Sitkoff D, Nicholls A, Ringnalda M, Goddard WA, Honig B. J Am Chem Soc. 1994;116:11875–11882. [Google Scholar]
- 41.Zhou RH, Berne BJ. P Natl Acad Sci USA. 2002;99:12777–12782. doi: 10.1073/pnas.142430099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ji CG, Mei Y, Zhang JZH. Biophys. J. 2008;95:1080–1088. doi: 10.1529/biophysj.108.131110. [DOI] [PMC free article] [PubMed] [Google Scholar]