Abstract
This paper describes an approach for making use of the components of the experimentally determined rotational diffusion tensor derived from NMR relaxation measurements in macomolecular structure determination. The parameters of the rotational diffusion tensor describe the shape and size of the macromolecule or macromolecular complex and are therefore complimentary to traditional NMR restraints. The structural information contained in the rotational diffusion tensor is not dissimilar to that present in the small angle region of the solution X-ray scattering profiles. We demonstrate the utility of rotational diffusion tensor restraints for protein structure refinement using the N-terminal domain of enzyme I (EIN) as an example and validate the results by solution small angle X-ray scattering. We also show how rotational diffusion tensor restraints can be used for docking complexes using the dimeric HIV-1 protease and the EIN-HPr complexes as examples. In the former case, the rotational diffusion tensor restraints are sufficient in their own right to determine the position of one subunit relative to another. In the latter case, rotational diffusion tensor restraints complemented by highly ambiguous distance restraints derived from chemical shift pertubation mapping and a hydrophobic contact potential are sufficient to correctly dock EIN to HPr. In each case, the cluster containing the lowest energy structure corresponds to the correct solution.
Introduction
NMR structure determination is primarily based upon short range interactions in the form of nuclear Overhauser enhancement (NOE)-derived short (< 6Å) interproton distance restraints, supplemented by torsion angle restraints derived from 3J couplings and chemical shift restraints.1–3 Residual dipolar couplings measured in weakly aligning media can be used to obtain orientational information,4,5 and paramagnetic NMR in the form of either paramagnetic relaxation enhancement or pseudo-contact shifts can be used to derive long-range (15–35 Å) distance restraints between protons and a paramagnetic center.6,7 The target function that is minimized includes not only terms describing the experimental restraints but terms defining the covalent geometry, stereochemistry and non-bonded interactions.3 The latter can range from a simple hard sphere van der Waals repulsion term to prevent atomic overlap8 to a full scale empirical energy function including a Lennard-Jones potential, electrostatic contributions and explicit solvent molecules.9,10 These terms can also be supplemented by various knowledge-based potentials of mean force, including database-derived multidimensional torsion angle11,12 and hydrogen bonding13 potentials, as well as more global packing potentials describing, for example, the radius of gyration14 or gyration volume.15 As the systems studied become larger and more complex, the relative number for restraints that can be extracted from NOE spectra tends to become sparser on account of extensive spectral overlap and line broadening. Under these circumstances, additional information on the overall shape and size of the system under consideration can become invaluable.
Low resolution information on the overall molecular shape of macromolecules and their complexes is directly encoded in small-angle X-ray scattering (SAXS) profiles,16,17 and indeed direct incorporation of SAXS data into NMR structure calculations has proved extremely useful.18–22 An alternative source of molecular shape information can be obtained by using the components of the overall rotational diffusion tensor as global shape restraints. The principal values of the rotational diffusion tensor describe rates of anisotropic tumbling of a protein or protein complex in solution and depend upon protein size and shape: larger proteins tumble more slowly than smaller ones, while asymmetrically shaped proteins tumble around the different rotation axes with different rates.23–26 The components of the rotational diffusion tensor are readily accessible from 15NR1/R2 ratios obtained from 15N-relaxation measurements, in conjunction with an available structure.27–29 The latter can be a preliminary NMR structure obtained by conventional means, or the known structures (either NMR or X-ray) of the free proteins in the context of a structure determination of a protein-protein complex.
The recent development of a fast and reliable method for calculation of the rotational diffusion tensor of a protein30 makes it possible to incorporate the information encoded in the diffusion tensor directly into NMR structure calculations. This information can be used either for refining structures of globular proteins or for assembling structures of multi-domain proteins and protein-protein complexes. Initial work using this approach described only a search algorithm for protein domain positioning with translational degrees of freedom when the domain orientations were already known from independent measurements.31
In this paper we present the implementation of a pseudo-potential term to minimize the difference between calculated and target values of the components of the rotational diffusion tensor within the Xplor-NIH NMR structure determination package,8,32 and describe simulated annealing protocols for protein structure refinement and for docking of complexes where the structures of the individual components of the complex are known. We illustrate the performance of the rotational diffusion tensor potential with respect to refinement of the structure of the N-terminal domain of enzyme I (EIN) in conjunction with NOE data, to the docking of the subunits of the HIV-1 protease dimer in the absence of any additional experimental data, and to the docking of EIN and the histidine phosphocarrier protein HPr in conjunction with NMR chemical shift perturbation data.
Theory and Computational Methods
The rotational diffusion tensor potential
Restraints on the protein rotational diffusion tensor were implemented in a potential energy term Ediff designed to minimize the sum of squares difference between the experimentally determined components of the protein rotational diffusion tensor, , and those calculated from the molecular structure, :
| (1) |
where kdiff is a force constant, and the indices i and j range over the 6 unique components of the two symmetrical 3×3 rotational diffusion tensors.
To calculate the diffusion tensor given a molecular structure, we represent the surface of the protein by an equivalent ellipsoid,30 and then apply Perrin’s equations23,24 to calculate the diffusion tensor using the ellipsoid’s dimensions and orientation. The key computational feature of this approach lies in an efficient method for mapping the protein’s surface using a fast triangulation algorithm, which is then used for building the equivalent ellipsoid.33 Once the coordinates of the surface are available the calculation of the principal values of the rotational diffusion tensor, , can be expressed in closed form as:
| (2) |
where kB is the Boltzmann constant; l = x, y, z; Tdiff is absolute temperature in Kelvin, and
| (3) |
fy and fz are obtained from Eq. (3) by appropriate permutation of the indices. In Eq. (3) η is the solvent viscosity, and Pl are parameters describing the protein geometry given by:
| (4) |
(with Py and Pz given by cyclic permutations of the indices). The semi-axes of the equivalent ellipsoid, ax, ay and az (in general ax ≠ ay ≠ az) in Eqs. (3–4) can be obtained directly from the eigenvalues, El, of the covariance matrix:
| (5) |
where are the coordinates of the protein surface representation, l,k = x, y, z, and Np is the number of points used to represent the protein surface. The values of the semi-axes of the equivalent ellipsoid are then given by:
| (6) |
Accordingly, the principal vectors of the diffusion tensor coincide with the corresponding eigenvectors of the covariance matrix, vx, vy and vz. Thus, given and vl the full rotational diffusion tensor can be reconstructed.
The gradients of Ediff with respect to all atomic displacements can be evaluated in closed form. However, a finite atomic displacement requires recalculation of the protein surface which is computationally expensive. So in practice we recalculate the protein surface when the backbone atomic root mean square (rms) difference between the previously triangulated and current protein structure is greater than 0.5 Å, or after 30 gradient evaluations, whichever comes first. By default we assume that the protein is tumbling in water. Consequently, we use a series approximation derived from the tabulated values of the water viscosity η given in the CRC Handbook of Chemistry and Physics,34 given by η(Tdiff) = 1.7753 − 0.0565×(Tdiff − 273) + 1.0751×10−3(Tdiff − 273)2 − 9.222×10−6×(Tdiff − 273)3. However, other values of η can be specified explicitly. Two other critical parameters that affect the calculation of and, therefore, the value of Ediff, are the temperature, Tdiff, and the thickness of the hydration layer used to construct the solvated protein. For the latter parameter we assume a uniform value of 2.8 Å which resembles a monolayer of water molecules.30
In many real experimental situations the viscosity and temperature of the sample may not be known precisely. For example, there may be sample heating as a consequence of the application of strong radiofrequency pulses during the NMR experiment. In addition, the use of concentrated (>0.5 mM) solutions of large proteins may markedly change the viscosity of the sample. Further, the details of the hydration layer are expected to change from protein to protein such that, for example, it is common to assign a fitting parameter to this layer when comparing calculated and experimental small angle X-ray scattering (SAXS) data. {Svergun, 1995 #56} Uncertainties in the experimental viscosity and temperature coupled with approximations inherent in the hydration layer model require a procedure to account for these effects. Since the primary result of uncertainties from these three sources is a scaling of the diffusion tensor Dcalc, we chose to compensate for these three factors by adjusting only the apparent diffusion tensor temperature . We reiterate that the value of is no longer a physical temperature, but rather a fitting parameter to collect errors in temperature, viscosity and hydration layer description. In this work we calculated structures using a series of different values. The value of that results in the lowest values of the total target energy is then chosen as the best match for the experimental and sample conditions. We have also implemented a procedure whereby is optimized during the course of the calculations (see below).
Simulated annealing protocols
Two different protocols were employed, one for refinement of globular proteins and the other for docking protein-protein complexes, as described below and in the Supplementary Information.
In the refinement protocol, the rotational diffusion tensor potential, Ediff, was simply added to the standard Xplor-NIH8 structure refinement protocol in torsion angle space.35 In addition to the Ediff term, the minimized target energy function comprises square-well potentials for the experimental NMR restraints, harmonic potentials for the covalent geometry (bonds, angles and improper torsions), a quartic van der Waals repulsion term to prevent atomic overlap,36 and a multidimensional torsion angle database potential of mean force.12 Details of the protocol are provided in Supplementary Information. The experimental NMR data comprise NOE-derived approximate interproton distance restraints and backbone and sidechain torsion angle restraints for the N-terminal domain of enzyme I (EIN).37 The components of the diffusion tensor were determined from the published 15NR1 and R2 data for EIN38 in conjunction with the NMR coordinates of EIN (1EZA)37 using well-established procedures.25,26
The refinement protocol can be used in two modes. In the first mode the value of used to calculate the components of the rotational diffusion tensor is held fixed. In the second mode, the value of is optimized during the course of the calculation. The latter is carried out by introducing three pseudo atoms, O, X and Y, with the XOY angle (θXOY) directly mapped onto :
| (7) |
where is the nominal experimental temperature, and the value of can vary within a range of . Aside from Ediff, the three pseudo atoms are coupled to the rest of the protein structure only through interaction with the thermal bath. The same approach was previously used to optimize the magnitude of the dipolar coupling alignment tensor.39
The docking protocol involves the application of conjoined rigid body/torsion angle dynamics and minimization35 and was designed for protein-protein docking where the backbone structures of the individual proteins or subunits are known. Conceptually, this protocol consists of two parts: rough positioning of the proteins within the complex, followed by simulated annealing refinement of the complex. Specifically, the protocol starts with one protein or subunit held fixed, and the position and orientation of the second protein randomized around the first. The second protein is then translated and rotated as a rigid body such that the energies corresponding to the experimental NMR restraints (i.e. the rotational diffusion tensor and, if available, supplementary data such as highly ambiguous intermolecular distance restraints from chemical shift perturbation mapping40) and van der Waals repulsion term are minimized using conjugate gradient minimization. This procedure is repeated 10 times and the resulting lowest energy structure is used as the starting point for molecular dynamics simulated annealing optimization. Simulated annealing starts at 500 K and then performs relatively short runs of conjoined rigid body/torsion angle dynamics in 10 K steps down to a temperature of 10 K with the backbone of the first protein held fixed, the backbone of the second protein allowed to rotate and translate as a rigid body, and the surface exposed sidechains of both proteins given torsional degrees of freedom. (Note that in the case of a symmetric homodimer for which C2 symmetry restraints are applied, both subunits are allowed to rotate and translate during simulated annealing.) In addition to terms representing the experimental restraints and the quartic van der Waals repulsion term, the target energy includes a term for the radius of gyration (calculated from the total number of residues, N, in the complex given by Rgyr = 2.2 N0.38)14 to ensure reasonable intermolecular packing density, a multidimensional torsion angle database potential of mean force to ensure that sidechain torsion angles populate physically realistic rotamers,12 and a new hydrophobic contact potential described below. The force constants for most of the potential terms are set low at the higher temperatures and gradually increased during the course of simulated annealing as the temperature is lowered. Further details of the docking protocol are provided in Supplementary Information.
The value of can also be optimized in the docking protocol. However, for the docking protocol we found that optimization strategy described above for the refinement protocol was unsuccessful. This is largely due to the fact that variations in the value of correspond essentially to changes in the trace of the calculated diffusion tensor, and, therefore, to alterations in the overall size of the complex. As a result, variation of during the course of the docking calculation can lead to instabilities that severely reduce the convergence properties of the protocol. We therefore employed an alternative strategy based on random sampling: for every run of initial rigid body domain positioning (by minimization) was fixed to a random value within a specified range (usually ±5 K), and the number of repetitions was increased from 10 to 50. The lowest energy structure was then used as the starting coordinates for the simulated annealing phase of the docking protocol.
The hydrophobic contact potential
A knowledge-based, low-resolution hydrophobic contact potential comprising interaction strengths calibrated using known protein structures41 was added to Xplor-NIH and found to be useful in the docking calculations. Unlike a full, realistic nonbonded description including electrostatics and discrete solvent, this contact term is computationally inexpensive. Further, unlike the all-atom description, the contact potential is quite insensitive to sidechain conformations, information about which is usually lacking in docking calculations.
The contact potential depends linearly on the distance di,j between residues i and j, defined as:
| (8) |
where there are sums over qki, the positions of the atoms in residue i, and over qkj the positions of the atoms in residue j. The contact energy term, Econtact, is given by:
| (9) |
where kcontact is a force constant, the sum is over all pairs of residues, Mi,j is the interaction strength between the residue types as determined by Miyazawa et al.41, and Vp is the following continuous piecewise linear potential:
| (10) |
where γ = 2/[dsw (d> − d<)] and . In this work we chose d< = 0 Å, d> = 10Å and dsw = 0.5 Å. Thus, the Econtact potential is fully enabled when the residues are touching, and disabled for residues separated by more than 10 Å. The Econtact potential proved useful in discriminating against docked structures containing bad hydrophobic interactions which otherwise met the various other restraints.
Cluster analysis
For the docking calculations, 512 independent structure calculations were carried out. To characterize the efficacy of the structure determination algorithm, it is essential to group the calculated structures together in clusters that are similar to one another by an appropriate metric. For this purpose we used the agglomerative algorithm outlined by Ward.42 Initially, each structure is placed in a separate cluster and the inter-cluster distance is computed as the rms distance between the Cα atoms in any two clusters. The clusters are then combined as follows: (a) the two closest clusters are found and combined; (b) a new distance matrix is generated in which the distance between clusters is the shortest structure-structure distance for these clusters; (c) this process is repeated until the shortest distance is larger than a specified tolerance, which in this work was set to 1 Å. This algorithm detects clusters of structures that are well-separated from each other in atomic rms displacement space, but does not guarantee that structures within a given cluster are similar. A measure of this similarity can be calculated as the Cα atomic rms difference to the mean structure after the clusters have been determined. This clustering algorithm is available as the findClusters helper script included in the Xplor-NIH package (version 2.22 or later).
Experimental methods
Sample preparation
EIN and HPr from E. coli were expressed and purified as described previously.43,44 The EIN construct employed in the current work consists of residues 1–249 of Enzyme I. The original NMR37 and X-ray45 work on EIN made use of a construct comprising residues 1–259 of Enzyme I. However, residues 250–259 of EIN are disordered in solution and invisible in the X-ray derived electron density map. NMR samples were prepared in 20 mM Tris buffer, pH 7.4, 90% H2O/10% D2O (v/v). Two samples were used for 15Nrelaxation measurements: the first comprised 0.5 mM U-[15N/2H]-EIN and 0.8 mM HPr at natural isotopic abundance; the second consisted of 0.5 mM U-[15N/2H]-HPr and 0.8 mM EIN at natural isotopic abundance.
NMR spectroscopy
NMR spectra were recorded at 313 K on a DRX Bruker 600 MHz spectrometer equipped with a z-shielded gradient triple resonance cryoprobe. 15NR1 and R1ρ relaxation measurements were carried out using pulse schemes described previously,46,47 and recorded in an interleaved manner using seven different relaxation delays. Delays of 103.3, 151.3, 391.3, 691.3, 1291.3, 1891.3, and 2491.3 ms were used for the R1 relaxation measurements, and delays of 6.7, 16.3, 25.9, 32.3, 48.3, 64.3, 96.3 ms were used for R1ρ relaxation measurement, with a recycle delay of 5 s for both measurements. R1ρ relaxation was obtained using a 15Nspin-lock field strength of 1.8 kHz. Spectra were processed using NMRPipe,48 and peak intensity values were fit to a single-exponential decay. 15NR2 relaxation rates were calculated from the 15NR1 and 15NR1ρ relaxation rates using the following equation:49
| (11) |
where θ = arctan(ΩN/γNB1), and ΩN is the resonance offset and γNB1 the spin-lock field strength.
SAXS measurements
SAXS data for EIN were collected on a SAXSess instrument from Anton Paar, which is configured as a Kratky camera coupled with high-flux multilayer monochromator optics. X-ray radiation from a sealed fine-focus tube source (Princeton Instruments), operating at 40 kV and 50 mA, was monochromated at the Cu Kα wavelength (1.542 Å) and incident on the sample in a 1-mm inner diameter quartz capillary of 24 mm length, thermostated at 25 °C. A line-shaped X-ray beam 20 mm in length was used to maximize the incident flux. Sample buffer conditions were the same as those used for NMR measurements described above, except that 150 mM NaCl was used to suppress the effects of inter-particle correlations (structure factor). Data were collected as a series of sequential 1 hr acquisitions with the protein sample, followed immediately by the dialysis buffer. Due to signal relaxation, the imaging plates were read out with a 5 min delay at the end of each acquisition session. Data at 50% dilution were collected to investigate the magnitude of the inter-particle structure factor. Wide-angle scattering data were collected within a q-range of ~0.02 Å−1 to ~2.80Å−1. Here, q = 4 π sinθ/λ, where 2θ is the scattering angle and λ the wavelength of the incident radiation. The recorded two-dimentional (2D) images were converted to one-dimensional (1D) scattering profiles by radial integration within 15 mm strips aligned at the center of the incident beam. 1D profiles were then mapped onto the q-axis by reference to the position of the primary beam attenuated by the semi-transparent beam stop of the instrument. The converted profiles were corrected for the readout noise of the imaging plate scanner and normalized to the recorded intensities of the transmitted primary beam. The scattering curves from the buffer were then subtracted from the scattering curve of the protein sample. The final scattering data were averaged over three independent sample/buffer data acquisitions of 1 hr each. The line-collimation 1D profiles were desmeared using the GNOM software,50 taking into account the recorded length profile of the incident beam. A maximum dimension of 70 Å was obtained by application of GNOM’s regularized Fourier transform procedure. The resulting point collimation-like data were used for the subsequent structural analysis in the q interval from 0.03 Å−1 to 0.60 Å−1 (crystallographic resolutions between ~300Å and ~10 Å). Evaluation of the quality of the fit of the scattering data to the various structural models was carried out with CRYSOL version 2.5.16
Results and Discussion
Refinement of a globular protein structure
To demonstrate the impact of rotational diffusion tensor restraints on structure refinement of globular proteins we made use of the N-terminal domain of enyzme I (EIN). EIN is an elongated molecule of approximately 30 kDa with a ratio of 3:3:1 for the principal components of the inertia tensor.37,38 The initial structure used for refinement is the NMR structure (specifically the restrained regularized mean coordinates; PDB code 1EZA) based on NOE, torsion angle, 3JHNα coupling constant and 13Cα/13Cβ restraints.37 The target values for the tensor components of the rotational diffusion tensor were obtained by least-squares minimization between the observed R1/R2 ratios (recorded at 313 K on a sample containing 1.1 mM EIN)38 and those calculated from the 1EZA coordinates (see Supplementary Information). Only data in regular secondary structure elements were employed to exclude potential errors arising from any large scale motions in the picosecond to nanosecond timescale or from exchange line broadening, and to minimize the impact of inaccuracies in the coordinates. The resulting tensor components for an axially symmetric diffusion model (see Supplementary Information) were then used as target values in a simulated annealing refinement protocol incorporating the rotational diffusion tensor potential Ediff. The other experimental NMR data included in the refinement were a set of distance restraints derived from NOE measurements (2818 interproton distance restraints) and backbone hydrogen bond analysis (230 distance restraints for 115 backbone hydrogen bonds) and 571 torsion angle restraints.37 The target function also included a quartic van der Waals repulsion term and a multidimensional torsion angle database potential of mean force.12 The latter is an improved version relative to that used in the original NMR structure determination.11
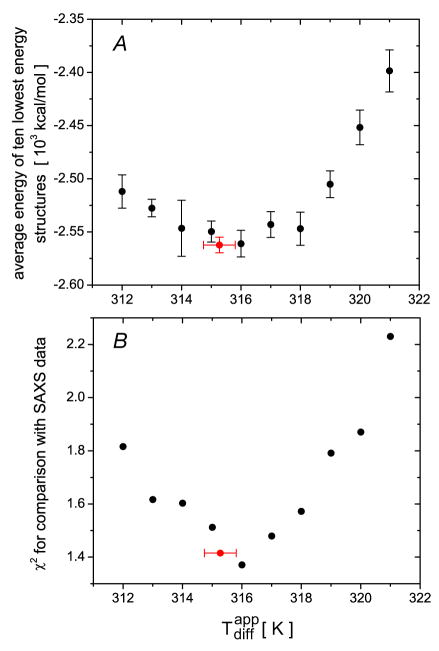
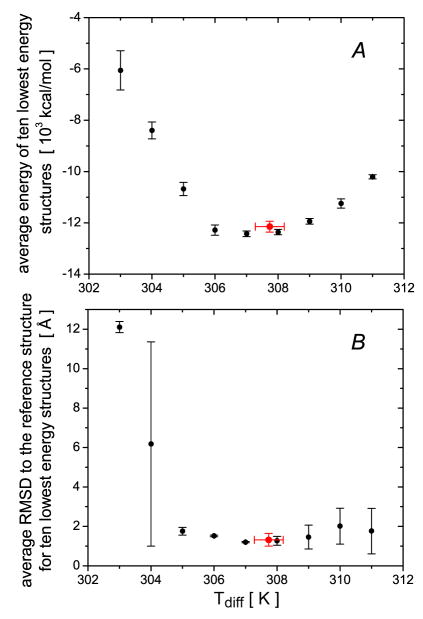
The values of the total Xplor-NIH energies averaged over the 10 lowest energy structures obtained at a series of values are plotted in Fig. 1A. The minimum in the Xplor-NIH energy is observed at compared to a nominal experimental temperature of 313 K. Independent validation of these results is obtained from the corresponding agreement of these structures with experimental SAXS data where the minimum χ2 value is also found for the structures calculated at (Fig. 1B). Thus, the agreement between the locations of the minima in the dependence of the total Xplor-NIH energy and the independently validated χ2-fit to the SAXS data strongly suggest that the use of the rotational diffusion tensor for protein structure refinement provides structures that are not only consistent with the experimental NMR restraints used in the refinement but also with other independent experimental information. When the force constant for the diffusion tensor potential is increased five-fold, the minimum in the total Xplor-NIH energy is found at compared to a minimum at 316 K for the χ2-fit to the SAXS data (Fig. S1, Supplementary Information). This difference is sufficiently small that the procedure of choosing the value that minimizes the overall Xplor-NIH energy seems to be a reasonable strategy and leads to a degree of self-consistency. Automated optimization of during simulated annealing (within a range of 313±5 K) also indicates that the lowest energy structures correspond to the optimal structures at 315.3 ± 0.5 K (see Fig. 1).
Figure 1.
Results of refinement of the structure of EIN with rotational diffusion tensor restraints as a function of the value used in the calculations. (A) Total Xplor-NIH energies, averaged over the ten lowest energy structures (errors bars, 1 s.d.). (B) χ2-fit16 of the calculated structures to the SAXS data. The structures used for fitting the SAXS data are the restrained regularized averages over the ten lowest energy structures calculated at each value of . The back symbols correspond to the structures obtained using a grid search for the settings; the red symbols are the results of automated optimization of .
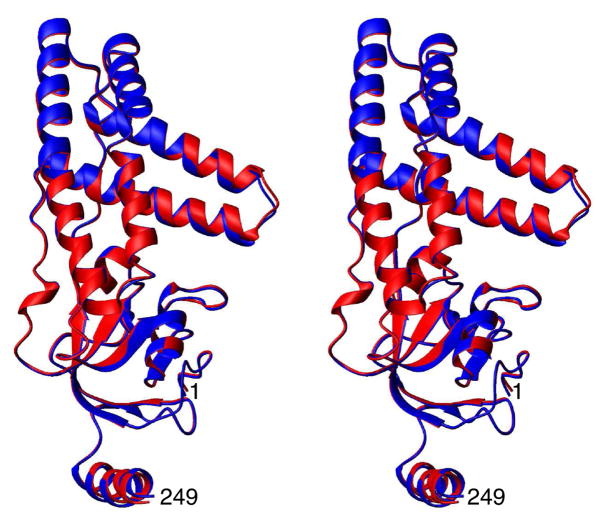
Comparison of previously determined X-ray (1ZYM45) and NMR (1EZA37) structures with the structures refined using the current refinement protocol with and without inclusion of the Ediff term is presented in Table 1. Although both sets of structures calculated with the current protocol are quite similar to each other (Cα atomic rms difference of 0.4 Å; Fig. 2) and exhibit approximately the same Cα atomic rms difference to the 1ZYM and 1EZA structures, the addition of the Ediff term does lead to a significant improvement in agreement with the SAXS data (Table 2, and Fig. S2, Supplementary information). Indeed, the χ2-fit to the SAXS data is as good for the current NMR structure refined with the Ediff term as the 1ZYM X-ray structure.
Table 1.
Cα atomic rms differences for the refined EIN structures
| Cα rms difference (Å) | ||
|---|---|---|
| Refinement with Ediff | Refinement without Ediff | |
| Precisiona | 0.93 | 1.02 |
| vs 1EZA NMR structureb | 1.26 | 1.21 |
| vs 1ZYM X-ray structureb | 1.37 | 1.36 |
| vs each otherb | 0.43 | |
Defined as the Cα rms difference to the mean coordinates averaged over the 10 lowest energy structures
Cα atomic rms difference to the mean coordinates averaged over the 10 lowest energy structures
Figure 2.
Stereoview showing a best-fit superposition of the two restrained regularized average structures obtained from refinement with (red) and without (blue) the rotational diffusion tensor restraints. (The value of was 316 K.)
Table 2.
Comparison of EIN structures with SAXS dataa
| SAXS χ2 | |
|---|---|
| 1EZA (original NMR) | 2.03 |
| 1ZYM (X-ray) | 1.38 |
| Refinement without Ediff | 1.58 |
| Refinement with Ediff | 1.37 |
Analysis of the SAXS data was performed using CRYSOL version 2.5.16 The SAXS data were acquired on EIN(1-249), and therefore the analysis was performed using residues 1-249. The NMR and X-ray data were acquired on EIN(1-259). However, residues 250–259 are disordered in solution37,38 and invisible in the electron density map.45
Protein-protein docking
In the two following sections we examine the impact of rotational diffusion tensor restraints on protein-protein docking using two extreme examples. The first consists of assembling the two identical subunits of HIV-1 protease into the homodimer on the basis of only rotational diffusion tensor restraints. The second involves assembly of the EIN-HPr complex from the X-ray structures of the free proteins based upon the rotational diffusion tensor restraints combined with highly ambiguous distance restraints derived from chemical shift perturbation mapping. In most instances additional experimental data would be available in the form of intermolecular distance restraints (e.g. from NOE, paramagnetic relaxation enhancement, FRET or DEER measurements), possibly supplemented by orientational restraints (from dipolar couplings or paramagnetic-induced pseudo-contact shifts).
Application to the HIV-1 protease dimmer
HIV-1 protease is a symmetric dimer of identical subunits. Here we demonstrate the utility of rotational diffusion tensor restraints alone to correctly assemble a homodimer. In the context of an NMR structure determination of a homodimer, this type of docking calculation would be useful in cases where the structure of an individual subunit is determined with confidence but no reliable intersubunit NOE information could be obtained (e.g. due to spectral overlap, exchange line broadening of interfacial residues, inability to make mixed isotopically labeled samples, etc….).
Calculations were performed using the X-ray coordinates for the individual subunits (PDB accesion code 2NPH51). The only experimental restraints employed in the docking protocol were the components of the rotational diffusion tensor calculated from 15Nrelaxation data acquired at 300 K (see Supplementary Information).47 More specifically, we used the experimental data recorded at a spectrometer frequency of 600 MHz and derived the diffusion tensor using the coordinates of the first subunit of the 2PNH structure. Although a fully anisotropic diffusion tensor model yields a lower χ2 than the axially symmetric model, the statistical F-test criterion does not justify the use of the fully anisotropic model (see Supplementary Information for details). Trials with Dcalc computed using different values show that a value of 303 K results in structures with the lowest total Xplor-NIH energies (Fig. S3, Supplementary Information).
Since HIV-1 protease is a natural homodimer, only the coordinates of one of the subunits of 2NPH51 were used in the calculations. One copy of this subunit was held fixed during the initial stage of rigid body minimization while the position and orientation of the second copy was randomized around the center of gravity of the first copy. In the crystal structure, the sidechains at the dimer interface are in their optimal configurations to yield good packing. To mimic the more general situation in which the docking protocol starts with undocked subunits or proteins, the sidechain conformations of the individual subunits were randomized by running 1 ps of torsion angle molecular dynamics at 500 K with the backbone coordinates held fixed. The resulting structures were then subjected to the docking protocol described in the Computational Methods Section and Supplementary Information.
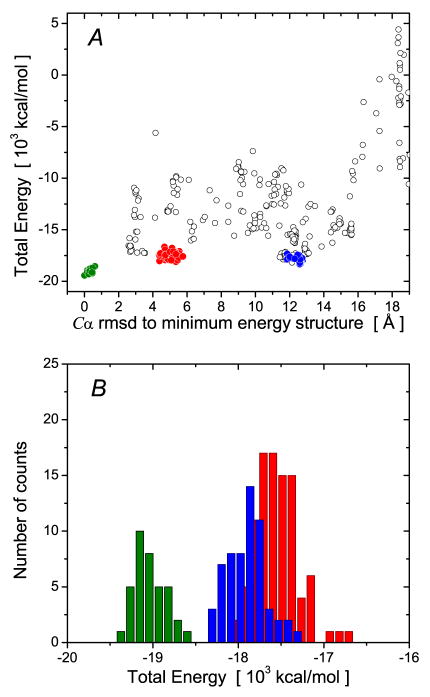
The results of cluster analysis of the 512 calculated structures is shown in Fig. 3. There are 3 major clusters of solutions, comprising 19, 11 and 7% of the structures, respectively. The third cluster contains the lowest energy structures. The rest of the structures are distributed among even smaller clusters, the majority of which include only a single member. We compared the 10 lowest energy structures (Fig. 4A), all of which are members of the same cluster, to the reference X-ray structure. (Note, in the reference structure the two subunits are identical and the dimer coordinates were created by superimposing the coordinates of the first subunit onto the second subunit of the 2NPH structure). The average Cα rms difference beween the 10 lowest energy dimer structures and the reference structure is 0.35 ± 0.09 Å; the Cα rms difference between the regularized mean coordinates and the reference structure is 0.31 Å (Fig. 4B). Thus the rotational diffusion tensor-based docking protocol coupled with clustering analysis is sufficient in its own right to define the subunit arrangement of the HIV-1 protease homodimer.
Figure 3.
Statistical properties of the largest clusters of docking solutions for HIV-1 protease. (A) Dependence of the total Xplor-NIH energy on the Cα rms difference to the minimum energy structure. Structures in the first, second and third largest clusters are indicated in red, blue and green filled-in circles, respectively. The remaining structures, the majority of which are in single member clusters, are shown as open circles. (B) Histograms of the distribution of total Xplor-NIH energy values with the same color-coding scheme as in panel A. The value of used was 303 K.
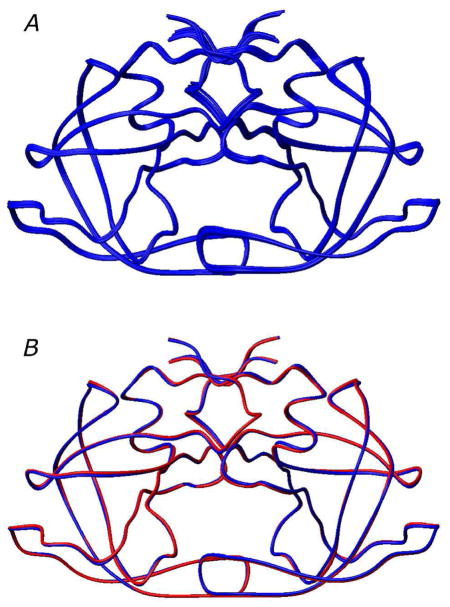
Figure 4.
Lowest energy HIV-1 protease dimer structures. (A) Best-fit superposition of the 10 lowest energy structures from the cluster containing the lowest energy structure. (B) Comparison of the restrained regularized mean structure (blue), derived from the 10 lowest energy structures, with the subunit arrangement in the X-ray structure51 (red). ( .)
Application to the EIN-HPr complex
The structure of the 40 kDa EIN-HPr complex was previously determined by NMR based on NOE and residual dipolar coupling data,52 and has been used as a test system for docking based on highly ambiguous intermolecular distances derived from chemical shift perturbation mapping either alone53 or in combination with residual dipolar coupling data.40 Here we demonstrate the use of rotational diffusion tensor restraints in conjunction with highly ambiguous intermolecular distance restraints to dock the EIN-HPr complex using the crystal structures of free EIN (1ZYM45) and HPr (1POH54) as starting coordinates.
The 15NR1 and R2 relaxation rates for the EIN-HPr complex were acquired as described in the Experimental Methods section and used to calculate the components of the rotational diffusion tensor making use of only the coordinates of N-H bond vectors in regions of regular secondary structure. The eigenvalues of the diffusion tensor were assumed to be the same for the EIN and HPr partners in the complex. However, the orientations of the principal axis frame (PAF) of the diffusion tensor with respect to the molecular frames of 1ZYM45 and 1POH54 were fit independently. The relaxation data were analyzed using two different models for the rotation diffusion tensor: axially symmetric and fully anisotropic. Comparison of the normalized χ2 values for these models using the F-test55 criterion (see Supplementary Information) indicates that the fully anisotropic model does not provide a statistically significant improvement in the description of the relaxation data. We therefore used the tensor components of the diffusion tensor obtained from the axially symmetric model in the docking calculations. In addition to the rotational diffusion tensors the docking protocol used highly ambiguous distance restraints obtained from chemical shift perturbation mapping,40 a hydrophobic contact potential (see Computational Methods section), a radius of gyration (Rgyr) potential with the target Rgyr value set to 20 Å,14 and a multidimensional torsion angle database potential of mean force for the sidechains.12
Unlike the identical subunits of the HIV-1 protease homodimer, EIN (249 residues) and HPr (85 residues) are markedly different in size and shape and thus represent a more general situation. In the generic simulated annealing docking protocol described in the Computational Methods section one of the partners of the complex is held fixed in space together with the corresponding orientation of the diffusion tensor. Note that the diffusion tensor not only provides shape restraints but also contains information about its orientation with respect to a molecular reference frame. With HPr held fixed, and EIN free to rotate and translate, convergence to the correct structure is achieved. If, on the other hand, EIN is held fixed and HPr is free to rotate and translate, HPr is placed at the correct binding site but the correct orientation of HPr cannot be determined. This is because reorientation of HPr does not significantly affect the overall shape of the complex, and hence the calculated diffusion tensor, owing to the relatively small size and almost spherical shape of HPr. When HPr, on the other hand, is held fixed, and EIN is allowed to rotate and translate, the simulated annealing docking protocol correctly determines the structure of the complex since the structure of EIN is quite asymmetric and much larger than that of HPr.
In addition to calculations with the experimental 15Nrelaxation data, calculations with several sets of synthetic relaxation data were carried out to assess the robustness of the docking protocol. To this end we calculated the diffusion tensor for a synthetic reference structure assembled from the X-ray coordinates of EIN (1ZYM45) and HPr (1POH54) superimposed onto the NMR structure of the EIN-HPr complex (3EZA52). This diffusion tensor together with the structures of EIN and HPr were used to calculate simulated 15NR1 and R2 relaxation rates to which random Gaussian noise ranging from 0 to 10% was added. The simulated data were then processed to calculate synthetic diffusion tensors. Trials with different values using the diffusion tensor derived from the experimental 15Nrelaxation data yielded an optimal value of (compared to the nominal experimental temperature of 313 K) resulting in both the lowest overall Xplor-NIH target energy and the smallest Cα atomic rms difference to the reference structure (Fig. 5). Automated optimization of (within a range 313±8 K) leads to a very similar value of 307.7 ± 0.5 K (Fig. 5).
Figure 5.
Results of several runs of the docking protocol for the EIN-HPr complex using different values of . The data for the two panels are averaged over the 10 lowest energy structures (errors bars = 1 s.d.). The total target energy is shown in (A) and the Cα atomic rms difference to the reference structure in (B). In both panels, the black symbols correspond to the grid search results obtained with a fixed value of ; the red symbols are the results using automated optimization of .
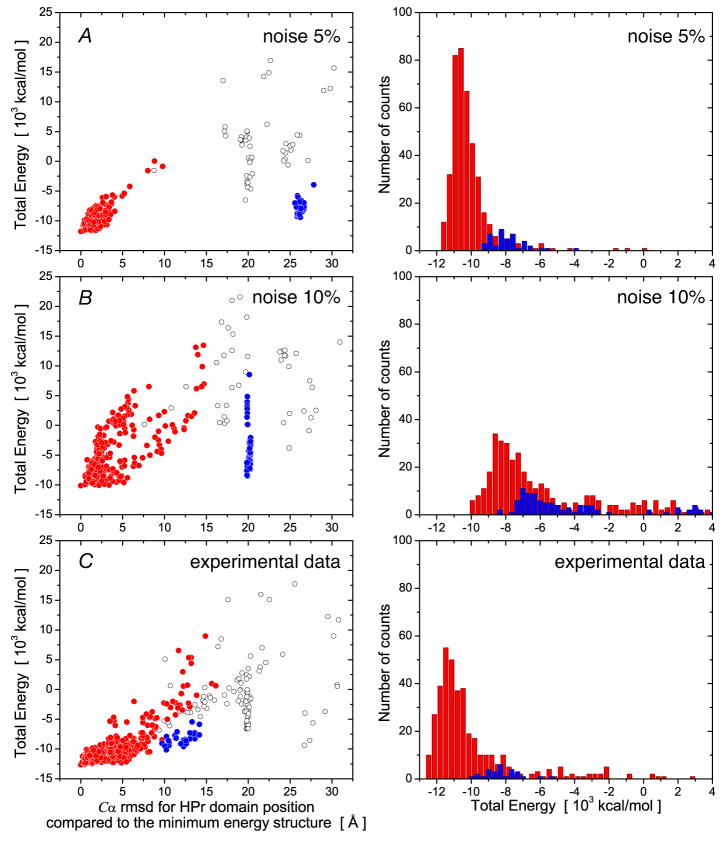
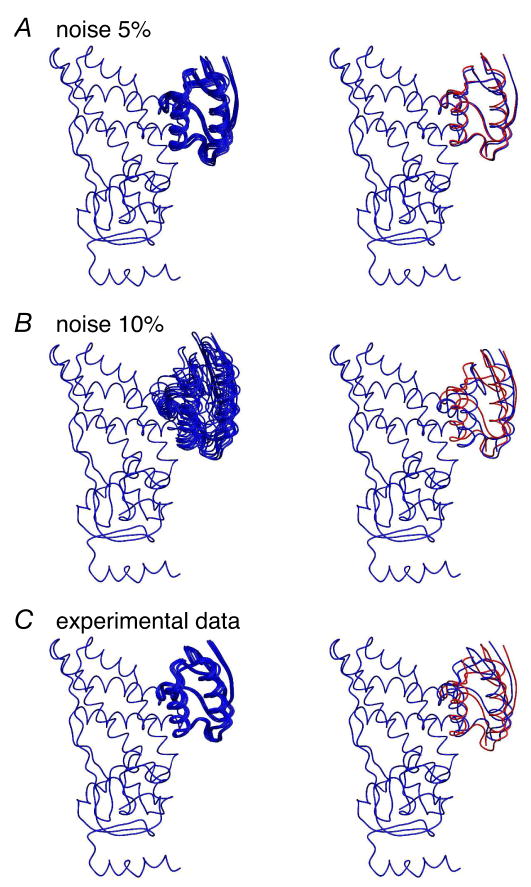
The performance of the diffusion tensor-based docking protocol for the EIN-HPr complex is summarized in Table 3 and Figs. 6 and 7. For both simulated and experimental target diffusion tensors the resulting sets of calculated structures always exhibit significant clustering with the cluster containing the lowest energy structure comprising more than 75% of all calculated structures. The trials with simulated diffusion tensors presented in Table 3 suggest that the docking protocol demonstrates reasonable accuracy and precision up to a level of 10% noise in the simulated R1 and R2 relaxation data. Inclusion of the hydrophobic contact potential energy term does not significantly affect the accuracy of the computed structures, but improves both convergence and precision. Using the experimentally derived components of the rotational diffusion tensor yields good agreement between the docked and reference structures (Table 3 and Figs. 6 and 7). Fig. 6 presents more detailed statistics for the distributions of Xplor-NIH energy values obtained for the docking calculations with both simulated (with 5 and 10% Gaussian noise) and experimental data. The corresponding lowest energy structures calculated from these input data sets are displayed and compared to the reference structure in Fig. 7.
Table 3.
Clustering statistics and Cα atomic rms differences to the reference structure for the EIN-HPr docking calculations.
| Relaxation Data | % structures in cluster containing lowest energy structure | C α rms difference to reference structure (Å)a | |
|---|---|---|---|
| Overall | HPr with EIN superimposed | ||
| Experimental data | 76 | 1.20±0.03 | 2.79±0.12 |
| Synthetic data | |||
| 0% noiseb | 83 | 0.51±0.27 | 1.32±0.76 |
| 0% noise | 83 | 0.48±0.19 | 1.35±0.39 |
| 1% noise | 80 | 0.59±0.15 | 1.65±0.32 |
| 2% noise | 89 | 0.79±0.17 | 2.04±0.37 |
| 5% noise | 82 | 0.48±0.15 | 1.27±0.43 |
| 10% noise | 72 | 1.82±0.35 | 4.08±1.02 |
The reference structure of the complex was generated by best-fitting the X-ray structures of free EIN (1ZYM)45 and HPr (1POH)54 on to the NMR structure of the EIN-HPr complex (3EZA).52 This ensures that the reported rms differences do not reflect internal coordinate differences between the NMR and X-ray coordinates of EIN and HPr.40
The hydrophobic contact potential was not included in these calculations.
Figure 6.
Statistical properties of the two largest clusters of docking solutions for the EIN-HPr complex using rotational diffusion tensors derived from simulated (5 and 10% added noise) and experimental 15NR1 and R2 relaxation data. (A) and (B), synthetic diffusion tensor derived from simulated relaxation data with 5 and 10% added Gaussian noise, respectively; (C) experimentally determined diffusion tensor. The left-hand panels display the Cα atomic rms difference of HPr relative to the minimum energy structure with the coordinates of EIN superimposed; the right-hand panels present a histogram of the distribution of total Xplor-NIH energies for the two largest clusters of calculated structures. The filled-in red and blue circles represent the first and second largest clusters of docked structures, respectively; the open circles in the left-hand panels correspond to structures in the remaining smaller clusters.
Figure 7.
Lowest energy docked structures obtained from rotational diffusion tensors derived from (A and B) simulated (5 and 10% added Gaussian noise) and (C) experimental 15NR1 and R2 relaxation data. In all panels the coordinates of EIN are superimposed. The left-hand panels show a superposition of the 10 lowest energy structures; the right-hand panels show the comparison of the restrained regularized mean structures (blue) derived from the 10 lowest energy structures with the reference structure (red).
In the above calculations the diffusion tensor was derived from relaxation data acquired for both components of the EIN-HPr complex. In principle, however, relaxation data need only be acquired for one of the components of a complex, since the resulting diffusion tensor will contain all the relevant information on the rotational diffusion for the entire complex. (Note that a reduction in the number of R1/R2 data points will lead to a decrease in the accuracy with which the components of the diffusion tensor are determined).56 Such an approach could be especially useful for complexes in which one of the components is large and hence more difficult to analyze data for due to spectral overlap. The use of relaxation data for the smaller component may then still be sufficient to permit reasonably accurate docking of the complex. In the case of the EIN-HPr complex, using only the relaxation data from HPr to derive the components of the rotational diffusion tensor results in an average Cα rms difference between the 10 lowest energy docked structures and the reference structure of 2.12 ± 0.28 Å compared to 1.20 ± 0.03 Å and 1.82 ± 0.35 Å when the experimental and synthetic (with 10% added noise) relaxation data, respectively, for both EIN and HPr are employed.
Concluding Remarks
In this paper we have shown that the components of the rotational diffusion tensor provide effective restraints on overall molecular shape and size. The incorporation of rotational diffusion restraints into protein structure refinement leads to increased accuracy in the resulting shape and size, as judged by improvements in the agreement with independent SAXS data. The rotational diffusion tensor restraints can also be used to efficiently assemble complexes comprising proteins of known structure either in their own right or in conjunction with minimal additional experimental information such as highly ambiguous intermolecular distance restraints derived from chemical shift perturbation mapping. The latter are required when the shape of one or more components of the complex is close to spherical or when one of the components of the complex is significantly smaller than the other. Although there are uncertainties in sample temperature, viscosity and the representation of the protein hydration shell, these can be readily compensated for by optimization of the total target function energy with respect to , either using a grid search or by optimizing during the course of the calculations. The fact that the optimal value of is close to that of the nominal experimental temperature indicates that the approximations used in the representation of the protein hydration shell (possibly the largest source of uncertainty, the details of which are expected to vary from protein to protein),16 including the assumption of a uniform thickness of 2.8 Å, is remarkably good, and similarly the estimate of sample viscosity based on the viscosity of water as a function of temperature,34 is reasonably accurate.
Supplementary Material
Acknowledgments
We thank Dan Garrett and John Kuszewski for useful discussions. Y.R. acknowledges a National Research Council Research Associateship (Award Number 0710430). This work was supported by the NIDDK Intramural Research Program of the NIH (to G.M.C.), the AIDS Targeted Antiviral Program of the Office of the Director of the NIH (to G.M.C.), and the CIT intramural research program of the NIH (to C.D.S.)
References
- 1.Wüthrich K. NMR of Proteins and Nucleic Acids. John Wiley & Sons; New York: 1986. [Google Scholar]
- 2.Clore GM, Gronenborn AM. Crit Rev Biochem Mol Biol. 1989;24:479–564. doi: 10.3109/10409238909086962. [DOI] [PubMed] [Google Scholar]
- 3.Clore GM, Gronenborn AM. Proc Natl Acad Sci U S A. 1998;95:5891–5898. doi: 10.1073/pnas.95.11.5891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bax A, Kontaxis G, Tjandra N. Methods Enzymol. 2001;339:127–174. doi: 10.1016/s0076-6879(01)39313-8. [DOI] [PubMed] [Google Scholar]
- 5.Prestegard JH, al-Hashimi HM, Tolman JR. Q Rev Biophys. 2000;33:371–424. doi: 10.1017/s0033583500003656. [DOI] [PubMed] [Google Scholar]
- 6.Iwahara J, Schwieters CD, Clore GM. J Am Chem Soc. 2004;126:5879–5896. doi: 10.1021/ja031580d. [DOI] [PubMed] [Google Scholar]
- 7.Pintacuda G, John M, Su XC, Otting G. Acc Chem Res. 2007;40:206–212. doi: 10.1021/ar050087z. [DOI] [PubMed] [Google Scholar]
- 8.Schwieters CD, Kuszewski JJ, Clore GM. Progr Nucl Magn Reson Spectros. 2006;48:47–62. [Google Scholar]
- 9.Brooks BR, Bruccoleri RE, Olafson BD, States DJ, Swaminathan S, Karplus M. J Comput Chem. 1983;4:187–217. [Google Scholar]
- 10.Linge JP, Williams MA, Spronk CA, Bonvin AM, Nilges M. Proteins Struct Funct Genet. 2003;50:496–506. doi: 10.1002/prot.10299. [DOI] [PubMed] [Google Scholar]
- 11.Kuszewski J, Gronenborn AM, Clore GM. Prot Sci. 1996;5:1067–1080. doi: 10.1002/pro.5560050609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clore GM, Kuszewski J. J Am Chem Soc. 2002;124:2866–2867. doi: 10.1021/ja017712p. [DOI] [PubMed] [Google Scholar]
- 13.Grishaev A, Bax A. J Am Chem Soc. 2004;126:7281–7292. doi: 10.1021/ja0319994. [DOI] [PubMed] [Google Scholar]
- 14.Kuszewski J, Gronenborn AM, Clore GM. J Am Chem Soc. 1999;121:2337–2338. [Google Scholar]
- 15.Schwieters CD, Clore GM. J Phys Chem B. 2008;112:6070–6073. doi: 10.1021/jp076244o. [DOI] [PubMed] [Google Scholar]
- 16.Svergun D, Barberato C, Koch MHJ. J Appl Crystallogr. 1995;28:768–773. [Google Scholar]
- 17.Svergun DI, Koch MH. Curr Op Struct Biol. 2002;12:654–660. doi: 10.1016/s0959-440x(02)00363-9. [DOI] [PubMed] [Google Scholar]
- 18.Grishaev A, Wu J, Trewhella J, Bax A. J Am Chem Soc. 2005;127:16621–16628. doi: 10.1021/ja054342m. [DOI] [PubMed] [Google Scholar]
- 19.Schwieters CD, Clore GM. Biochemistry. 2007;46:1152–1166. doi: 10.1021/bi061943x. [DOI] [PubMed] [Google Scholar]
- 20.Grishaev A, Ying J, Canny MD, Pardi A, Bax A. J Biomol NMR. 2008;42:99–109. doi: 10.1007/s10858-008-9267-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grishaev A, Tugarinov V, Kay LE, Trewhella J, Bax A. J Biomol NMR. 2008;40:95–106. doi: 10.1007/s10858-007-9211-5. [DOI] [PubMed] [Google Scholar]
- 22.Gabel F, Simon B, Nilges M, Petoukhov M, Svergun D, Sattler M. J Biomol NMR. 2008;41:199–208. doi: 10.1007/s10858-008-9258-y. [DOI] [PubMed] [Google Scholar]
- 23.Perrin F. J Phys Radium. 1934;5:497–511. [Google Scholar]
- 24.Perrin F. J Phys Radium. 1936;7:1–11. [Google Scholar]
- 25.Favro DL. Phys Rev. 1960;119:53–62. [Google Scholar]
- 26.Woessner DE. J Chem Phys. 1962;37:647–654. [Google Scholar]
- 27.Tjandra N, Feller SE, Pastor RW, Bax A. J Am Chem Soc. 1995;117:12562–12566. [Google Scholar]
- 28.Fushman D, Xu R, Cowburn D. Biochemistry. 1999;38:10225–10230. doi: 10.1021/bi990897g. [DOI] [PubMed] [Google Scholar]
- 29.Dosset P, Hus JC, Blackledge M, Marion D. J Biomol NMR. 2000;16:23–28. doi: 10.1023/a:1008305808620. [DOI] [PubMed] [Google Scholar]
- 30.Ryabov YE, Geraghty C, Varshney A, Fushman D. J Am Chem Soc. 2006;128:15432–15444. doi: 10.1021/ja062715t. [DOI] [PubMed] [Google Scholar]
- 31.Ryabov Y, Fushman D. J Am Chem Soc. 2007;129:7894–7902. doi: 10.1021/ja071185d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schwieters CD, Kuszewski JJ, Tjandra N, Clore GM. J Magn Reson. 2003;160:65–73. doi: 10.1016/s1090-7807(02)00014-9. [DOI] [PubMed] [Google Scholar]
- 33.Varshney A, Brooks FP, Wright WV. IEEE Comp Graphics App. 1994;14:19–25. [Google Scholar]
- 34.Weast RC. CRC Handbook of Chemistry and Physics. CRC Press; Boca Raton, FL: 1988. 1st Student. [Google Scholar]
- 35.Schwieters CD, Clore GM. J Magn Reson. 2001;152:288–302. doi: 10.1006/jmre.2001.2413. [DOI] [PubMed] [Google Scholar]
- 36.Nilges M, Gronenborn AM, Brunger AT, Clore GM. Protein Eng. 1988;2:27–38. doi: 10.1093/protein/2.1.27. [DOI] [PubMed] [Google Scholar]
- 37.Garrett DS, Seok YJ, Liao DI, Peterkofsky A, Gronenborn AM, Clore GM. Biochemistry. 1997;36:2517–2530. doi: 10.1021/bi962924y. [DOI] [PubMed] [Google Scholar]
- 38.Tjandra N, Garrett DS, Gronenborn AM, Bax A, Clore GM. Nature Struct Biol. 1997;4:443–449. doi: 10.1038/nsb0697-443. [DOI] [PubMed] [Google Scholar]
- 39.Clore GM, Schwieters CD. J Am Chem Soc. 2004;126:2923–38. doi: 10.1021/ja0386804. [DOI] [PubMed] [Google Scholar]
- 40.Clore GM, Schwieters CD. J Am Chem Soc. 2003;125:2902–2912. doi: 10.1021/ja028893d. [DOI] [PubMed] [Google Scholar]
- 41.Miyazawa S, Jernigan RL. Proteins Struct Funct Genet. 1999;34:49–68. doi: 10.1002/(sici)1097-0134(19990101)34:1<49::aid-prot5>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 42.Ward JH. J Am Stat Ass. 1963;58:236–244. [Google Scholar]
- 43.Garrett DS, Seok YJ, Peterkofsky A, Clore GM, Gronenborn AM. Biochemistry. 1997;36:4393–4398. doi: 10.1021/bi970221q. [DOI] [PubMed] [Google Scholar]
- 44.Suh JY, Cai ML, Clore GM. J Biol Chem. 2008;283:18980–18989. doi: 10.1074/jbc.M802211200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liao DI, Silverton E, Seok YJ, Lee BR, Peterkofsky A, Davies DR. Structure. 1996;4:861–872. doi: 10.1016/s0969-2126(96)00092-5. [DOI] [PubMed] [Google Scholar]
- 46.Farrow NA, Zhang OW, Formankay JD, Kay LE. J Biomol NMR. 1994;4:727–734. doi: 10.1007/BF00404280. [DOI] [PubMed] [Google Scholar]
- 47.Tjandra N, Wingfield P, Stahl S, Bax A. J Biomol NMR. 1996;8:273–284. doi: 10.1007/BF00410326. [DOI] [PubMed] [Google Scholar]
- 48.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 49.Davis DG, Perlman ME, London RE. J Magn Reson Series B. 1994;104:266–275. doi: 10.1006/jmrb.1994.1084. [DOI] [PubMed] [Google Scholar]
- 50.Semenyuk AV, Svergun DI. J Appl Crystallogr. 1991;24:537–540. [Google Scholar]
- 51.Das A, Prashar V, Mahale S, Serre L, Ferrer JL, Hosur MV. Proc Natl Acad Sci U S A. 2006;103:18464–18469. doi: 10.1073/pnas.0605809103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Garrett DS, Seok YJ, Peterkofsky A, Gronenborn AM, Clore GM. Nature Struct Biol. 1999;6:166–173. doi: 10.1038/5854. [DOI] [PubMed] [Google Scholar]
- 53.Dominguez C, Boelens R, Bonvin AM. J Am Chem Soc. 2003;125:1731–1737. doi: 10.1021/ja026939x. [DOI] [PubMed] [Google Scholar]
- 54.Jia ZC, Quail JW, Waygood EB, Delbaere LTJ. J Biol Chem. 1993;268:22490–22501. doi: 10.2210/pdb1poh/pdb. [DOI] [PubMed] [Google Scholar]
- 55.Snedecor GW, Cochran WG. Statistical methods. 8. Iowa State University Press; Ames: 1989. [Google Scholar]
- 56.Zweckstetter M, Bax A. J Biomol NMR. 2002;23:127–137. doi: 10.1023/a:1016316415261. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.