Abstract
The regenerating (Reg) family comprises an extensive, diversified group of proteins with homology to C-type lectins. Several members of this family are highly expressed in the gastrointestinal tract under normal conditions, and often show increased expression in inflammatory bowel disease. However, little is known about Reg protein function, and the carbohydrate ligands for these proteins are poorly characterized. We report here the first expression and purification of Reg proteins using a bacterial system. Mouse RegIIIγ and its human counterpart, HIP/PAP, were expressed in Escherichia coli, resulting in the accumulation of aggregated recombinant protein. Both proteins were renatured by arginine-assisted procedures and were further purified using cation-exchange chromatography. The identities of the purified proteins were confirmed by SDS–PAGE, N-terminal sequencing, and MALDI-TOF mass spectrometry. Size exclusion chromatography revealed that both proteins exist as monomers, and circular dichroism showed that their secondary structures exhibit a predominance of β-strands which is typical of C-type lectins. Finally, both RegIIIγ and human HIP/PAP bind to mannan but not to monomeric mannose, giving initial insights into their carbohydrate ligands. These studies thus provide an essential foundation for further analyses of human and mouse RegIII protein function.
Keywords: C-type lectins, Regenerating protein family, Carbohydrate binding, Mucosal injury, Inflammatory bowel disease
C-type lectins are proteins that contain carbohydrate recognition domains (CRDs)1 and bind selectively to specific carbohydrate structures, often in a Ca2+-dependent manner. They mediate a variety of functions including cellular adhesion, clearance of circulating proteins, and recognition of microbe-associated molecular patterns (reviewed in [1]). The Reg gene family encodes an extensive group of secreted proteins that contain conserved sequence motifs found in all C-type lectin CRDs. The family is so named because the first member to be identified was cloned from a cDNA library derived from regenerating pancreatic islets [2]. Subsequently, several members of this multigene family have been identified in mice and humans, and are grouped according to homology into four subfamilies: RegI, RegII, RegIII, and RegIV. Despite their similarities to well-characterized C-type lectins, the members of the Reg family have poorly defined functions and their carbohydrate ligands have not been clearly identified.
Members of the RegIII family are constitutively expressed at high levels in mouse and human gastrointestinal tissues. RegIIIα, β, and γ are expressed in mouse small intestine [3], while human hepatocarcinoma–intestine–pan-creas/pancreatitis associated protein (HIP/PAP) is made in human small intestine. RegIIIβ and γ expression levels increase dramatically in response to bacterial colonization and other inflammatory stimuli in mice [4–6]. In addition, HIP/PAP expression is upregulated in the mucosal tissues of patients with inflammatory bowel disease [5,7]. Despite these insights into the forces regulating RegIII protein expression, almost nothing is known about the biological functions of RegIII proteins or their role in disease.
An abundant source of purified recombinant mouse and human RegIII proteins is needed to delineate the role of RegIII proteins in intestinal biology and human disease. Human HIP/PAP has been purified previously from the milk of transgenic mice engineered to express the protein in mammary gland [8], and as a glutathione S-transferase (GST) fusion protein in an Escherichia coli expression system [9]. Although the transgenic approach yielded quantities of protein sufficient for crystallographic analysis [10], this method is technically challenging, time-consuming, and expensive. The recombinant fusion protein procedure produced only microgram quantities of the GST-tagged protein [9]. We therefore wished to develop a system for the rapid expression and purification of recombinant RegIII proteins that is simple, high yield, and readily adaptable to other Reg family members.
In this report, we describe a new method for high level expression of mouse RegIIIγ and HIP/PAP using a bacterial expression system. Initial problems with low HIP/PAP expression were solved by introducing silent mutations into the 5′ end of the gene that were designed to relax local mRNA secondary structure. We present details of a procedure for the refolding and purification of RegIIIγ and HIP/PAP from bacterial inclusion bodies. This simple protocol yields milligram quantities of both proteins, and is the first example of high level Reg protein purification from a bacterial expression system. Finally, we show initial evidence suggesting that both RegIIIγ and HIP/PAP bind polymeric but not monomeric mannose.
Materials and methods
Vectors, strains, and supplies
The expression vector pET3a was from Novagen. E. coli BL21-CodonPlus (DE3)-RIL and E. coli BL21-CodonPlus (DE3)-RILP competent cells were from Stratagene. Oligonucleotides and restriction enzymes were supplied by Invitrogen. Other DNA modifying enzymes and isopropyl-β-d-thiogalactopyranoside (IPTG) were from Roche Molecular Biochemicals. Luria Broth was purchased from VWR. Sephacryl S-100 high resolution gel filtration medium and size exclusion chromatography standards were from GE Healthcare. All other chemicals and reagents were from Sigma.
Construction of the mouse RegIIIγ expression vector
A 474 bp amplicon was generated by RT-PCR from mouse small intestinal RNA using the specific primers 5′-ATTGCGAGGCATATGGAAGTTGCCAAGAAAGATGCCCCAT-3′ (forward primer) and 5′- CTATGGGG ATCCCTAGGCCTTGAATTTGCAGACATAGGGT-3′ (reverse primer). The forward primer contained an NdeI restriction site (underlined) for cloning into pET3a. The reverse primer incorporated the native stop codon followed by an engineered BamHI site (underlined). The resulting amplicon contained a methionine start codon in place of the signal sequence and thus encoded the mature secreted protein. PCR products and vector were digested with NdeI and BamHI, gel-purified, and ligated. The recombinant plasmid (pET3a-RegIIIγ) was sequenced to confirm the absence of mutations, and was transformed into E. coli BL21-CodonPlus (DE3)-RIL for protein expression.
Construction of HIP/PAP expression strains
A 474 bp amplicon was generated by RT-PCR from human small intestinal RNA (Ambion) using the specific primers 5′-ATTGCGAGGCATATGGAAGAACCCCAGAGAGGAACTGC-3′ (forward primer) and 5′-CTATGGTGATCACTAGTCAGTGAACTTGCAGACATAG GGTAA-3′ (reverse primer). The forward primer contained an NdeI restriction site (underlined) for cloning into pET3a. The reverse primer incorporated the native stop codon followed by an engineered BclI site (underlined). The resulting amplicon lacked the HIP/PAP signal sequence and thus encoded the mature secreted protein [8]. The PCR product was digested with NdeI and BclI, ligated into NdeI/BamHI-digested pET3a, and the resulting plasmid (pET3a-HIP/PAP) sequenced to confirm the absence of mutations.
A second expression construct (pET3a-HIP/PAPmut) was generated with silent mutations engineered into the 5′ end of the HIP/PAP coding sequence. Mutations were introduced by redesigning the forward primer that was used to generate the wild-type HIP/PAP construct: 5′- ATTGCGAGGCATATGGAAGAACCACAAAGAGAAA CT GC-3′ (mutant bases are underlined; also see Fig. 1). A 474 bp amplicon was generated by PCR with this mutant primer and the HIP/PAP-specific reverse primer above, using pET3a-HIP/PAP as template. The amplicon was cloned into pET3a as described for pET3a-HIP/PAP. The resulting plasmid was sequenced to confirm incorporation of the silent mutations and the absence of additional mutations. Both pET3a-HIP/PAP and pET3a-HIP/PAPmut were transformed into E. coli BL21-CodonPlus (DE3)-RILP for protein expression.
Fig. 1.
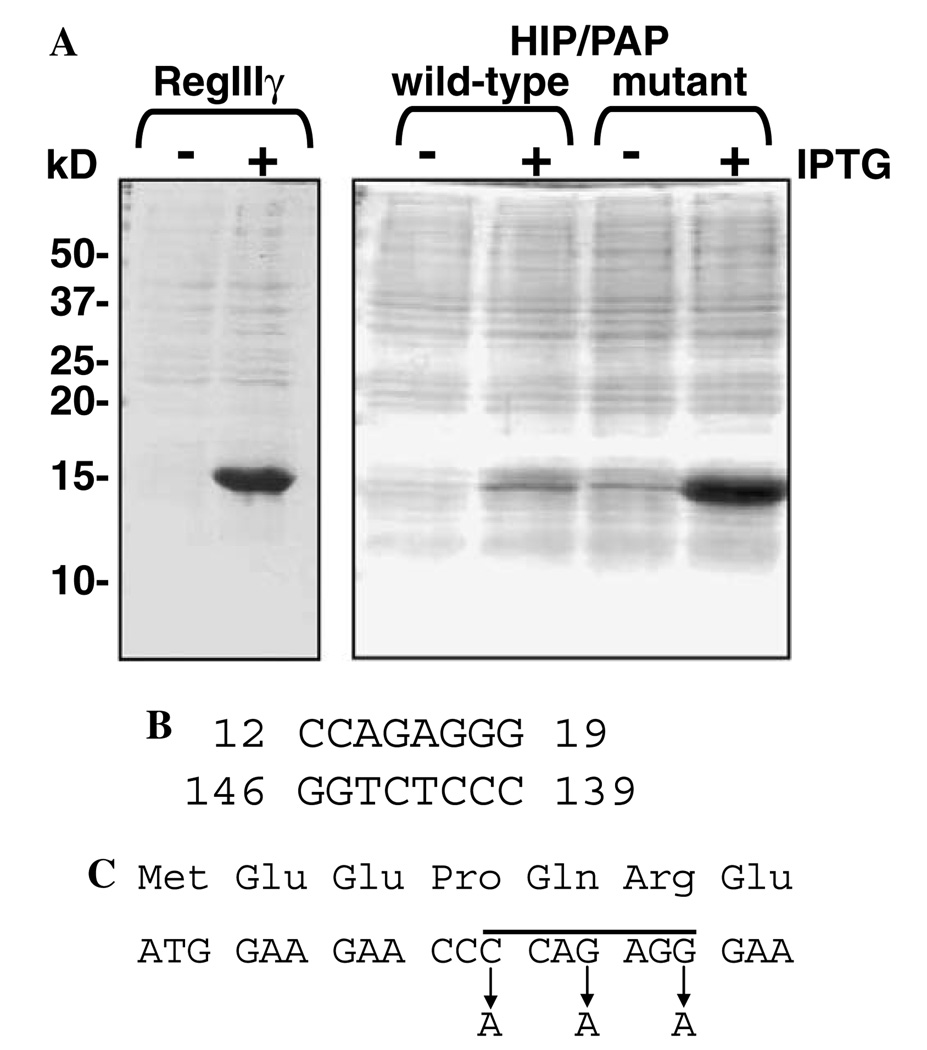
Expression of mouse RegIIIγ and human HIP/PAP in E. coli. (A) Expression of RegIIIγ (pET3a-RegIIIy) was induced by the addition of 0.4 mM IPTG. HIP/PAP expression constructs (wild-type = pET3a-HIP/PAP; mutant = pET3a-HIP/PAPmut) were induced by the addition of 1 mM IPTG. Total E. coli lysates from pre- and post-induction cultures were analyzed by electrophoresis through a 15% SDS–PAGE gel followed by Coomassie blue staining. (B) Predicted stem structure involving residues 12–19 of the HIP/PAP mRNA coding region. The stem was predicted by analyzing the mature HIP/PAP coding sequence using the web-based RNA secondary structure prediction algorithm at www.gene-bee.msu.su. (C) Positions of the silent mutations incorporated into the forward primer used to generate pET3a-HIP/PAPmut. The residues corresponding to the predicted stem are indicated by a line.
Expression and purification of RegIIIγ
Escherichia coli BL21-CodonPlus (DE3)-RIL harboring pET3a-RegIIIγ were grown at 37 °C in 500ml of LB medium supplemented with 0.1 mg/ml ampicillin to an absorbance of 0.6–1.0 (mid-log phase) at 600 nm. Protein expression was induced by the addition of 0.4 mM IPTG, and the culture was incubated for another 3 h at 37 °C with good aeration. Cells were collected by centrifugation at 6500g for 15 min at 4°C, and the pellet resuspended in 1/20 culture volume (25 ml) of Inclusion Body (IB) Wash Buffer (20 mM Tris–HCl, 10mM EDTA, 1% Triton X-100, pH 7.5). The cells were divided into five equal 5 ml aliquots and ruptured by sonication in two 1 min bursts at setting four using a Misonix XL-2020 Sonicator fitted with a 4.8 mm tapered probe. The lysate was centrifuged at 10,000g for 10 min, and the insoluble fraction was resuspended in 50 ml of IB Wash Buffer using a Dounce homogenizer. Centrifugation and resuspension were repeated, and the final insoluble inclusion body preparation was collected by centrifugation at 10,000g for 10 min followed by dispersion in 10 ml of Resuspension Buffer (7 M guanidine–HCl, 0.15 M reduced glutathione, 0.1 M Tris–HCl, 2mM EDTA, pH 8.0) and rotation for 2 h at room temperature. The resuspended inclusion bodies were added dropwise to a total of 500 ml of RegIIIγ Refolding Buffer (0.5 M arginine–HCl, 0.6 mM oxidized glutathione, 50mM Tris–HCl, pH 8.0) and left to stand for 24 h. The solution was clarified by centrifugation at 10,000g for 30 min, dialyzed overnight against 10 volumes of Dialysis Buffer 1 (25 mM Tris–HCl, 25 mM NaCl, 2mM CaCl2, pH 7), followed by a second overnight dialysis against 10 volumes of Dialysis Buffer 2 (25 mM MES, 25 mM NaCl, 2mM CaCl2, pH 6). The dialysate was centrifuged at 10,000g for 30 min, and RegIIIγ was captured by passage over a 5 ml column of SP-Sepharose Fast Flow cation-exchange resin (Sigma) equilibrated in Dialysis Buffer 2. After washing in 10 column volumes of Dialysis Buffer 2, the protein was batch-eluted in 5 column volumes of 0.4 M NaCl in Dialysis Buffer 2. Fractions containing protein were identified by the Bradford method [11] using the Bio-Rad Protein Assay Dye Reagent Concentrate (Bio-Rad).
Expression and purification of HIP/PAP
Recombinant HIP/PAP was expressed from pET3a-HIP/PAPmut. The expression and purification protocol was similar to that of RegIIIγ, with some significant changes. First, the IPTG concentration used for protein induction was 1 mM, and induction proceeded for 2h. HIP/PAP-containing inclusion bodies were refolded in HIP/PAP Refolding Buffer (50 mM Tris–HCl, pH 8.0, l0mM KCl, 240 mM NaCl, 2mM MgCl2, 2mM CaCl2, 0.5 M guanidine–HCl, 400 mM sucrose, 500 mM arginine–HCl, 1 mM reduced glutathione, and 0.1 mM oxidized glutathione). Subsequent dialysis and ion-exchange chromatography steps were performed as described for RegIIIγ, except the SP-Sepharose column was batch-eluted in 0.6 M NaCl in Dialysis Buffer 2.
Characterization of recombinant proteins
Purity of recombinant protein preparations was evaluated by SDS–PAGE through 15% gels, N-terminal sequencing on an ABI494 sequencer (PE Biosystems), and Matrix Assisted Laser Desorption/Ionization Time of Flight (MALDI-TOF) mass spectrometry on a Micromass spectrometer in the UT Southwestern Protein Chemistry Technology Center.
Size exclusion chromatography was performed using a 1.5 × 63 cm Sephacryl S-100 column, at a flow rate of 22 ml/h. The column was equilibrated in 25 mM Tris–HCl, pH 7.5, 25 mM NaCl, 2mM CaCl2. One milliliter of protein at 2 mg/ml was applied and eluted in equilibration buffer, and eluted fractions were monitored for protein at 280 nm. Molecular weights were determined in comparison to the standards provided in the Amersham Low Molecular Weight Calibration Kit (GE Healthcare).
Circular dichroism (CD) analysis was performed on an Aviv 62DS spectropolarimeter with a 1 mm cell length. Spectra of purified RegIIIγ and HIP/PAP were recorded in 25 mM Tris–HCl, pH 7.5, at a protein concentration of 10 µM. Three spectra were recorded for each condition from 190 to 260 nm in 1 nm increments, averaged, and the background spectrum of buffer without protein was subtracted from the protein-containing spectra. CD spectra were initially analyzed by the software accompanying the spectropolarimeter. Analysis of spectra to extrapolate secondary structures was performed by Dichroweb [12] (website is found at http://www.cryst.bbk.ac.uk/cdweb/html/home.html) using the K2D and Selcon 3 analysis programs [13,14].
Polyclonal antibody generation and Western blot analysis
Purified RegIIIγ was submitted to Cocalico Biologicals for polyclonal antibody generation in rabbits. Protein extracts for Western blot analysis were generated from mouse small intestine (jejunum). A 2 cm piece of freshly isolated intestinal tissue was flushed, lyophilized overnight, and pulverized under liquid N2. The pulverized tissue was resuspended in 1 ml of Extraction Buffer (8 M urea, 1% SDS, 0.15 M Tris–HCl, pH 7.5) and lysed by passing the suspension through an 18 gauge needle 3–5 times, followed by 3–5 passages through a 21 gauge needle. Total protein was quantitated with the Bio-Rad Detergent Compatible (DC) protein assay (Bio-Rad). Tissue protein and recombinant RegIIIγ were subjected to SDS–PAGE in a 15% gel and transferred to PVDF (Millipore). Membranes were blocked with 5% nonfat milk and incubated with polyclonal antiserum or preimmune serum followed by horseradish peroxidase-conjugated goat anti-rabbit IgG (Jackson ImmunoResearch). Immunoreactivity was detected using the Pierce SuperSignal West Pico Chemiluminescent detection kit.
Carbohydrate binding assays
Yeast mannan (Sigma) and dextran (Sigma) were coupled to Sepharose 6B using a previously published protocol [15]. Mannose–agarose was purchased from Sigma. 25 µl of each resin was washed extensively in Binding Buffer (25 mM MES, pH 6.0, 25 mM NaCl, 1% BSA), and 50 µg recombinant protein was added to each resin in a total volume of 1 ml of Binding Buffer. After rotation for 2 h at 4 °C, the beads were washed twice with 1 ml of Wash Buffer (25 mM MES, pH 6.0, 25 mM NaCl). Bound protein was eluted by boiling the beads in SDS–PAGE buffer (10% glycerol, 5% β-mercaptoethanol, 2% SDS, 62.5 mM Tris– HCl, 0.003% bromophenol blue, pH 6.8) and resolved by SDS–PAGE through a 15% acrylamide gel.
Ten milliliter mannose–Sepharose or mannan– Sepharose columns were run in 25 mM MES, pH 6.0, 25 mM NaCl, and were eluted in 25 mM MES, pH 6.0, 25 mM NaCl containing either l0mM CaCl2 or 20 mM CaCl2. Collected fractions were analyzed for protein content by spectrophotometry at 280 nm.
Results
Expression of recombinant mouse RegIIIγ and human HIP/PAP in E. coli
The open reading frame corresponding to mature mouse RegIIIγ (lacking the N-terminal signal peptide) was ligated into the bacterial expression vector pET3a to yield pET3a-RegIIIγ. This construct allows IPTG inducible expression from a T7 promoter. We chose to express RegIIIγ in the E. coli expression strain BL21-CodonPlus(DE3)-RIL, which is genetically modified to express tRNAs corresponding to specific Arg, Ile, and Leu codons that are normally rare in E. coli. To assess protein expression levels, IPTG was added to log-phase cultures, and pre- and post-induction cell lysates were analyzed by SDS–PAGE. Coomassie blue staining of gels revealed robust induction of RegIIIγ expression (Fig. 1A).
The mature human HIP/PAP open reading frame was also cloned into pET3a to yield pET3a-HIP/PAP. However, we could detect very little HIP/PAP expression following the addition of IPTG to growing cultures (Fig. 1A). These results are consistent with those of other investigators who have expressed HIP/PAP in E. coli [16]. Attempts to improve protein levels by altering induction conditions (IPTG concentration, induction time, induction temperature) were unsuccessful. We were also unsuccessful at improving protein expression by using an E. coli strain, BL21-CodonPlus(DE3)-RILP, that harbors an additional rare Pro tRNA.
We hypothesized that translation initiation from the pET3a-HIP/PAP construct might be impaired, resulting in poor induction of recombinant HIP/PAP expression. Analysis of the mature HIP/PAP coding sequence (including the engineered start codon), using the RNA secondary structure prediction algorithm at www.genebee.msu.su, revealed the presence of a predicted stem involving nucleotides 12–19, with a free energy of −17.5 kcal/mol (Fig. 1B). By comparison, the mature RegIIIγ coding sequence contains a predicted stem encompassing nucleotides 8–11. This stem has a free energy of −7.1 kcal/mol, indicating a much less stable structure. Based on this analysis, we reasoned that the presence of a stable stem close to the 5′ end of the HIP/PAP mRNA could interfere with ribosome binding and subsequent translation. To test this idea, we engineered three silent mutations into the forward primer used to amplify the HIP/PAP coding sequence (Fig. 1C), and cloned the resulting amplicon into pET3a to generate pET3a-HIP/PAPmut. The mutations were designed to abolish the predicted stem by substituting A for G or C, thus reducing the stability of the base pair interactions. Indeed, re-analysis of the altered sequence via the web-based algorithm confirmed the absence of the predicted stem. Consistent with this, the mutant HIP/PAP construct resulted in a remarkable increase in protein expression relative to the wild-type construct (Fig. 1A). Thus, all further HIP/PAP expression/purification studies were done using recombinant protein derived from pET3a-HIP/PAPmut.
Purification of mouse RegIIIγ and human HIP/PAP
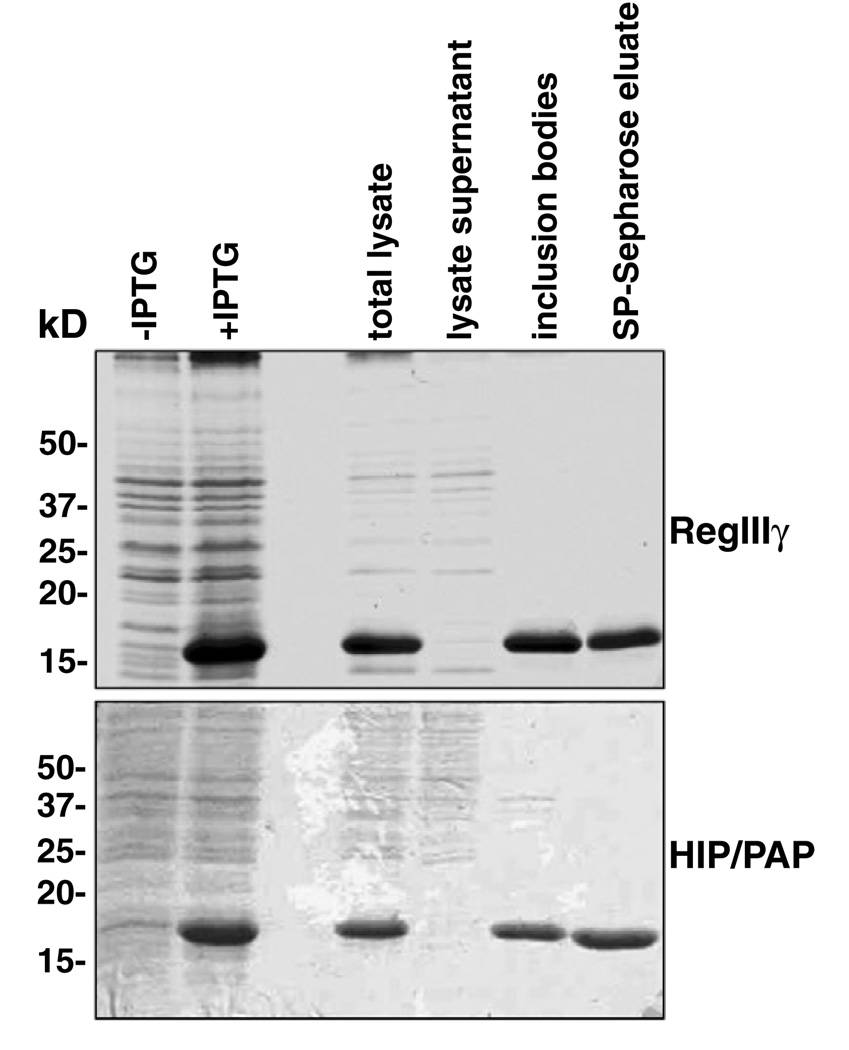
Escherichia coli BL21 expression strains such as BL21-CodonPlus(DE3)-RIL and BL21-CodonPlus(DE3)-RILPlack the ability to generate disulfide bonds between cysteine residues in proteins. As RegIIIγ and HIP/PAP both contain three predicted disulfide bonds, we expected that the recombinant proteins would be misfolded and targeted to bacterial inclusion bodies. As shown in Fig. 2, both proteins were absent from the soluble fraction of E. coli lysates and were found in purified inclusion bodies. In both cases, recombinant protein represented the vast majority of inclusion body protein.
Fig. 2.
SDS–PAGE analysis of samples taken during the purification of RegIIIγ and HIP/PAP. E. coli cells overexpressing recombinant RegIIIγ or HIP/PAP were collected before and after induction with IPTG (+ and −IPTG). The lanes containing total lysate and lysate supernatant samples were loaded with 10 µg total protein. Inclusion body and SP-Sepharose column eluate sample lanes contain 5 µg of protein. Proteins were resolved on a 15% Polyacrylamide gel and stained with Coomassie brilliant blue.
RegIIIγ and HIP/PAP from isolated inclusion bodies could be solubilized in 7 M guanidine hydrochloride under reducing conditions. However, both proteins required refolding and reoxidation prior to purification. In the case of RegIIIγ, we refolded the protein using an approach similar to that used previously to obtain native Angiogenin-4 [17]. This procedure involves the dropwise addition of the solubilized inclusion body protein to a solution containing 0.5 M arginine and oxidized glutathione at pH 8. It has been proposed that arginine inhibits protein aggregation during refolding, while the oxidized glutathione promotes the formation of disulfide bonds [18]. This buffer resulted in good recovery of RegIIIγ (27% of total inclusion body protein) following removal of arginine by dialysis (Table 1).
Table 1.
Purification of recombinant mouse RegIIIγ and human HIP/PAP from overexpressing Escherichia coli
| Purification step | RegIIIγ | HIP/PAP | ||
|---|---|---|---|---|
| Total protein (mg) | Protein yield (%) | Total protein (mg) | Protein yield (%) | |
| Cell lysate | 134 | 100 | 176 | 100 |
| Solubilized inclusion bodies | 34 | 25 | 28 | 16 |
| Post-refolding dialysate | 9 | 6.7 | 22 | 12.5 |
| SP-Sepharose eluate | 7.9 | 6.0 | 12 | 6.8 |
Results are derived from 500 ml cultures of E. coli expressing the recombinant proteins.
Total protein was estimated by the method of Bradford (11).
Attempts to refold HIP/PAP using the RegIIIγ refolding buffer resulted in a large amount of aggregation and the recovery of negligible soluble HIP/PAP. We therefore screened a variety of refolding conditions including cations (Ca2+, Mg2+), chelator (EDTA), salt (NaCl, KCl), pH, and additives such as l-arginine and sucrose. As detailed in Materials and methods, we obtained the best recoveries of soluble HIP/PAP using a solution containing KCl, NaCl, cations, guanidine, sucrose, arginine, and a mixture of reduced and oxidized glutathione. Following dialysis, the yield of soluble HIP/PAP was 78% of total inclusion body protein.
RegIIIγ and HIP/PAP both have a basic predicted pI (8.5 for RegIIIγ and 7.8 for HIP/PAP, as calculated by the algorithm found at the ExPASy website (http://au.expasy.org/tools/pi_tool.html)). Thus, we predicted that both recombinant proteins would bind to a cation exchange resin. Furthermore, recombinant HIP/PAP produced in transgenic mice has previously been shown to bind to a Mono S cation-exchange column [8]. Following refolding, the proteins were dialyzed into a low ionic strength buffer at pH 6 and were bound to SP-Sepharose. RegIIIγ was eluted in 0.4 M NaCl (Fig. 2), and yielded ~16mg/l of culture (Table 1). HIP/PAP required 0.6 M NaCl for elution (Fig. 2), and yielded ~24mg/l of culture (Table 1).
Characterization of recombinant proteins
The purities of the recombinant proteins were assessed initially by SDS–PAGE. Following elution from SP-Sepharose, RegIIIγ and HIP/PAP migrated as single species (Fig. 2). To further confirm their identities, the purified proteins were analyzed by MALDI-TOF mass spectrometry. RegIIIγ yielded a single peak corresponding to a molecular mass of 16.6kDa, in good agreement with the predicted molecular mass of the mature protein (16.5 kDa). Likewise, analysis of purified HIP/PAP gave a single peak indicating a molecular mass of 16.4 kDa, in agreement with its predicted molecular mass (16.7 kDa). Furthermore, Edman N-terminal sequencing of recombinant RegIIIγ yielded MEVAK for RegIIIγ, the expected amino terminus of the mature [Met−1] protein. Likewise, analysis of the HIP/PAP N-terminus yielded the sequence MEEPQ, which corresponds to the predicted N-terminus of the recombinant mature [Met−1] protein.
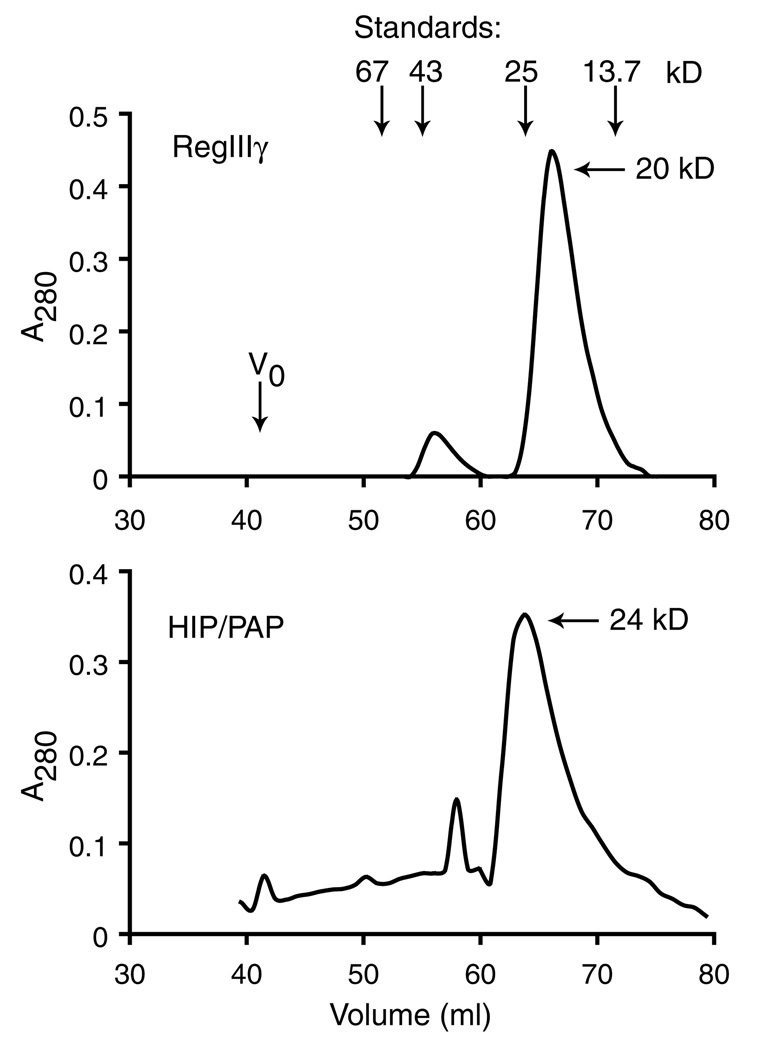
Size exclusion chromatography was performed to determine whether the recombinant proteins formed oligomers in solution. Chromatography through Sephacryl S-100 revealed that RegIIIγ elutes at a molecular mass of 20 kDa (Fig. 3). While this corresponds to a molecular weight slightly greater than the predicted molecular mass of mono-meric protein (16 kDa), dimer would likely elute at a molecular weight in excess of 32kDa. Although a minor protein peak was observed at a molecular weight approximating that of dimer, our results suggest that the majority of RegIIIγ is a monomer in solution. Similarly, HIP/PAP exhibits a major elution peak at 26kDa, suggesting that it also is predominantly monomeric in solution.
Fig. 3.
Size exclusion chromatography of RegIIIγ and HIP/PAP. Two milligram samples of pure RegIIIγ and HIP/PAP were applied to a Sephacryl S-100 column and eluted with detection at 280 nm. Positions of void volume (V0) and standards are indicated: albumin (67 kDa), ovalbumin (43 kDa), chymotrypsinogen (25 kDa), and ribonuclease (13.7 kDa).
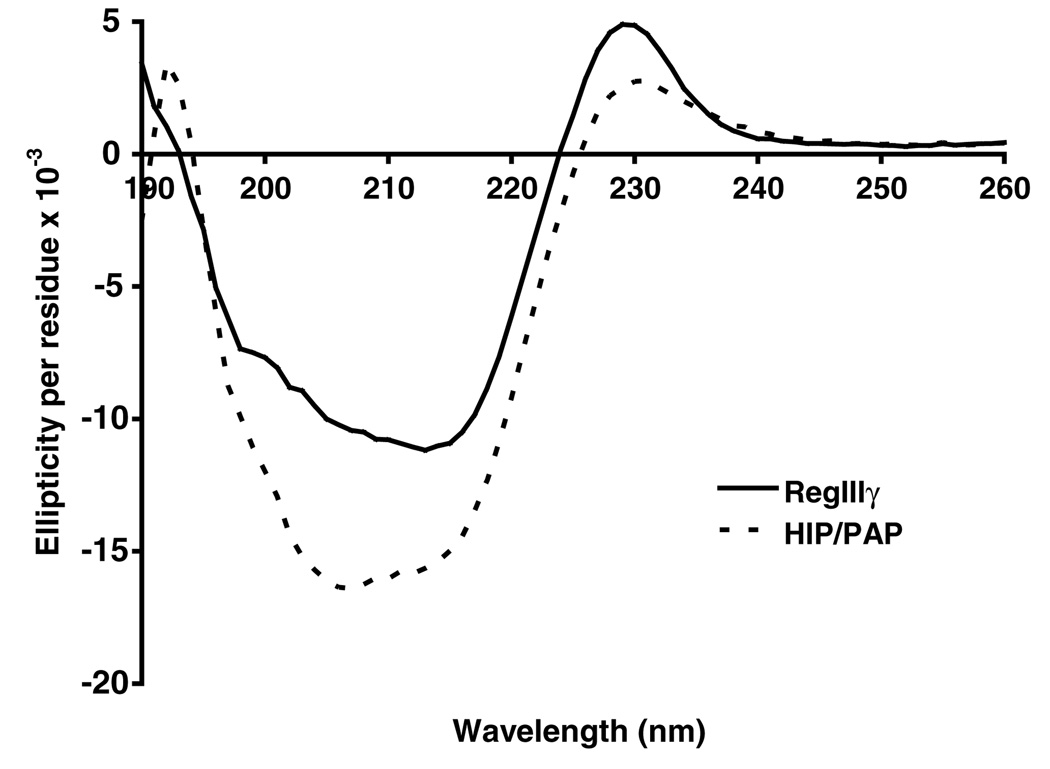
Previous crystallographic analysis of HIP/PAP has elucidated a secondary structure that is composed of 9 β-strands and 2 α-helices [10]. This structure is very similar to those of virtually all other C-type lectin CRDs characterized [19]. We performed circular dichroism spectroscopy to characterize the secondary structures of purified recombinant RegIIIγ and HIP/PAP. The results for both proteins reveal maximal negative ellipticity in the range of 205–215 nm (Fig. 4), and the spectra were similar overall to those derived from other C-type lectin family members, including langerin [20], surfactant protein A [21], and mannose binding lectin [22]. Indeed, analysis of the spectra by Dichroweb [12] using the K2D and Selcon 3 analysis programs [13,14] indicate that RegIIIγ and HIP/PAP are both predominantly comprised of β-sheet structure, while α-helix structure is not as prevalent. Our findings are thus consistent with the secondary structures revealed by the HIP/PAP crystal structure as well as those other C-type lectin CRDs. These results indicate that purified recombinant RegIIIγ and HIP/PAP have acquired their expected secondary structures and are thus correctly refolded.
Fig. 4.
Circular dichroism spectra of RegIIIγ and HIP/PAP. Results show that RegIIIγ and HIP/PAP are composed predominantly of β-sheet structure, which is consistent with correct refolding.
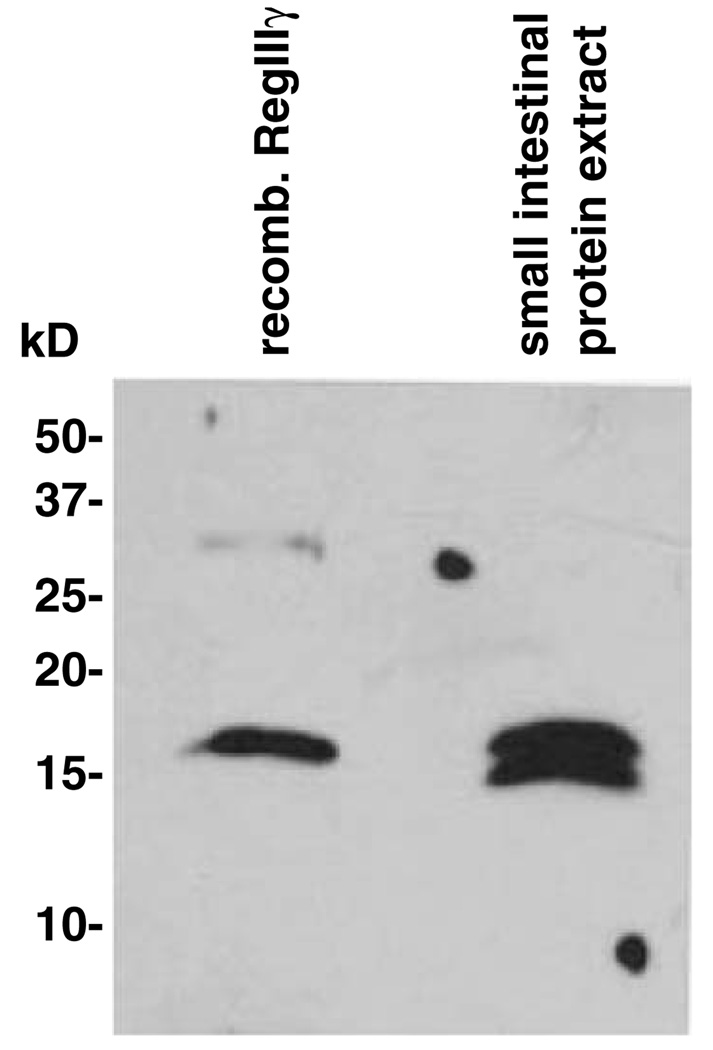
Recombinant mammalian proteins expressed in E. coli generally lack the post-translational modifications present on their endogenous counterparts. Such differences can pose difficulties for functional and biochemical analysis of recombinant proteins. Previous studies have demonstrated the existence of an O-glycosylated form of another Reg family member, human RegIα [23]. Analysis of the RegIIIγ primary sequence indicates that the protein does not harbor a consensus sequence for N-glycosylation (Asn-Xaa-Ser/Thr). However, there is at least one potential O-glycosylation site as determined by the NetOGlyc algorithm at Expasy (http://www.cbs.dtu.dk/services/NetOGlyc/) [24]. To determine whether the endogenous protein is post-translationally modified by glycosylation or another modification, we compared the molecular weight of the endogenous protein with that of recombinant RegIIIγ. Western blot analysis using a polyclonal antibody raised against purified RegIIIγ revealed that the recombinant protein migrates at the same molecular weight as protein from intestinal tissue homogenates (Fig. 5). The lower band detected in the endogenous sample is identical in molecular weight to a cleaved form that we observe following exposure of recombinant RegIIIγ to exogenous proteases, suggesting that RegIIIγ is processed in vivo by endogenous intestinal proteases. These results are thus consistent with the conclusion that RegIIIγ is not modified by glycosylation. However, it is still possible that endogenous RegIIIγ harbors other post-translational modifications that are undetectable by SDS–PAGE analysis.
Fig. 5.
Western blot comparison of recombinant and endogenous mouse RegIIIγ. Total mouse small intestinal protein was prepared as described in Materials and methods. Five nanograms of purified recombinant RegIIIγ and 20 µg of total intestinal protein were separated by SDS-PAGE and transferred to PVDF. The blot was probed with rabbit antiserum raised against recombinant RegIIIγ.
Characterization of RegIIIγ and HIP/PAP carbohydrate binding activity
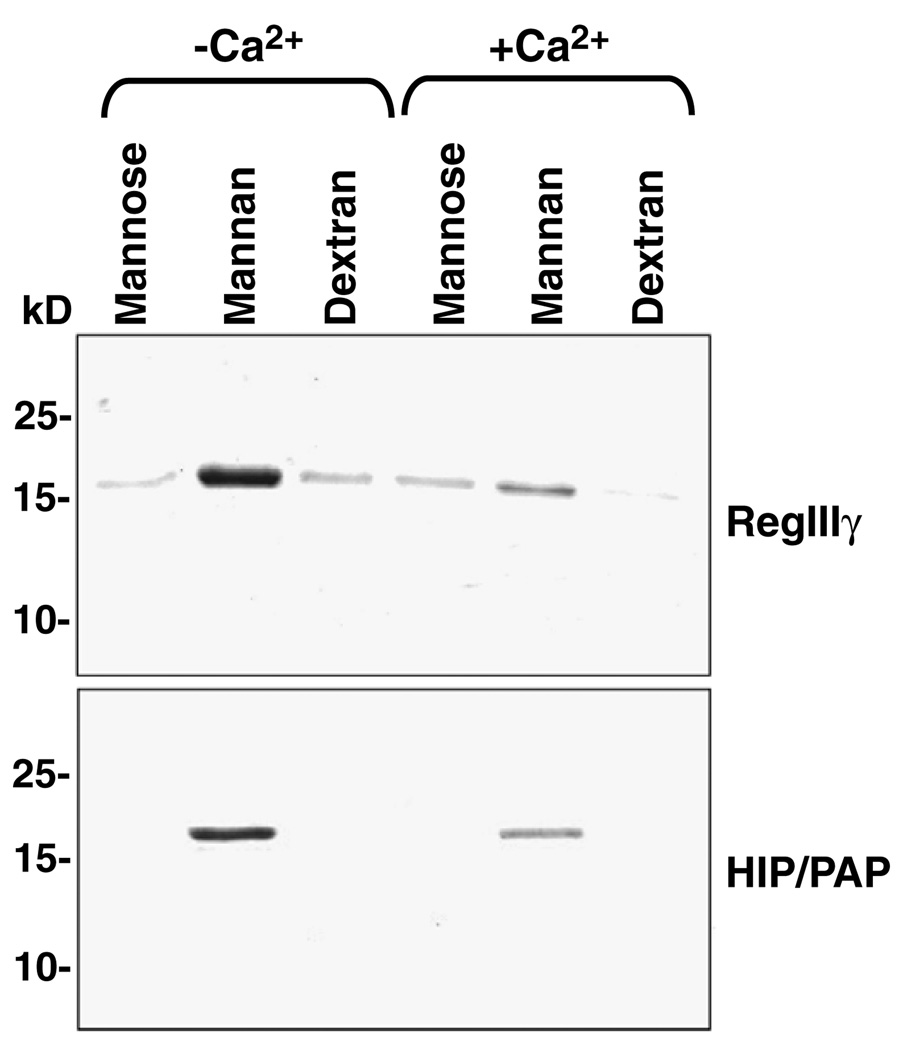
The primary amino acid sequences of both RegIIIγ and HIP/PAP are composed almost entirely of a conserved C-type lectin carbohydrate recognition domain (CRD). According to the classification scheme proposed by Drickamer and Fadden, both proteins are members of the type VII C-type lectin subfamily [25]. Although members of the other subfamilies have well-characterized carbohydrate ligands, the ligands bound by type VII lectins, including RegIIIγ and HIP/PAP, are poorly characterized. To gain insight into the carbohydrate binding specificity of these proteins, we covalently coupled various mono-, di-, and polysaccharides to Sepharose 6B resin and assayed for binding of purified RegIIIγ and HIP/PAP. None of the immobilized monosaccharides tested (glucose, galactose, N-acetylglucosamine, N-acetylgalactosamine, mannose, and fucose) supported binding of either lectin (data not shown). In addition, we did not observe binding to the disaccharide lactose (data not shown). This is in contrast to a previous report demonstrating that lactose is a ligand for GST-tagged HIP/PAP [9]. However, we found that RegIIIγ and HIP/PAP both bound to immobilized mannan (Fig. 6), a polysaccharide composed of polymerized mannose. By contrast, neither protein bound to dextran, a polysaccharide composed of α1,6- and αl,3-linked glucose, suggesting that both lectins are specific for mannose polysaccharides. Moreover, we did not detect binding of either protein to mannose (Fig. 6), suggesting that RegIIIγ and HIP/PAP bind polymeric but not monomeric mannose.
Fig. 6.
RegIIIγ and HIP/PAP binding to immobilized saccharides. 50 µg of each protein was bound to 25 µl of immobilized mono- or polysaccharide for 2 h at 4 °C. After washing, bound proteins were released by boiling the Sepharose beads in SDS–PAGE sample buffer followed by electrophoresis through 15% Polyacrylamide gels and Coomassie blue staining.
The C-type lectin family includes members whose ligand binding is calcium-dependent. However, previous studies have shown that at least one other Reg family member, RegIα, does not bind Ca2+ [23]. Furthermore, crystallographic analysis of RegIα revealed significant alterations in the polypeptide loop that binds Ca2+ in other C-type lectins [23]. We therefore asked whether RegIIIγ and HIP/PAP binding to mannan is influenced by Ca2+. Unexpectedly, our results revealed that 10mM CaCl2 reduces binding of both RegIIIγ and HIP/PAP to mannan (Fig. 6). These results thus suggest that carbohydrate ligand binding to RegIIIγ and HIP/PAP is indeed modulated by Ca2+, but in a way that is distinct from other C-type lectins.
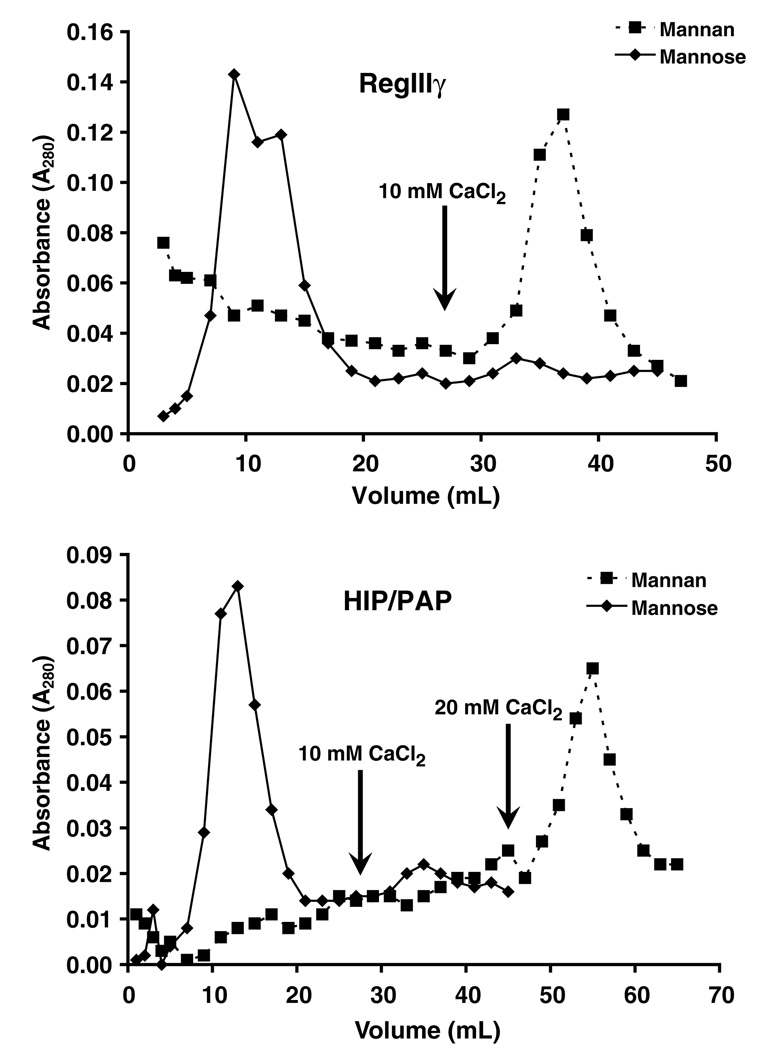
We next wished to determine whether the entire purified RegIIIγ and HIP/PAP protein populations are capable of binding to mannan. We therefore applied purified recombinant protein to a column of mannose– or mannan–conjugated Sepharose beads. As expected, both proteins passed through the mannose–Sepharose column (Fig. 7). In contrast, RegIIIγ and HIP/PAP bound quantitatively to the mannan–Sepharose column (Fig. 7), indicating that each purified protein population is refolded to a functionally active state in its entirety. Furthermore, we eluted both mannan bound proteins with CaCl2, confirming that both lectin–carbohydrate interactions are inhibited by Ca2+.
Fig. 7.
Chromatography of RegIIIγ and HIP/PAP on mannose- and man-nan–Sepharose. 0.25 mg of each purified protein was applied to a 10 ml mannose– or mannan–Sepharose column. Two milliliter fractions were collected and protein was detected by spectrophotometry at 280 nm. Protein bound to mannan–Sepharose was eluted with 10 or 20 mM CaCl2 as indicated.
Discussion
A number of prior studies have suggested that the Reg protein family plays critical roles in intestinal biology. Several members of the Reg family, including mouse RegIIIγ and human HIP/PAP, are highly expressed in the intestine as compared to other tissues [3,16,26]. Additionally, dysregulated expression of these proteins has been associated with intestinal diseases such as inflammatory bowel disease [5,7,27]. Based on their initial identification in regenerating tissue, certain family members have been ascribed a role in tissue repair [28]. However, the exact functions of Reg family proteins are still poorly understood. Moreover, although all are members of the C-type lectin family, their carbohydrate ligands remain poorly characterized. As a first step in elucidating the biological functions and ligands of the RegIII family, we have developed a simple protocol for purification of RegIIIγ and HIP/PAP using a bacterial overexpression system.
Two distinct heterologous expression systems have been used previously to obtain milligram quantities of pure Reg proteins. HIP/PAP has been isolated from lactating transgenic mice expressing the recombinant protein under the control of a mammary gland-specific promoter [8]. Additionally, RegIV has been purified following overexpression in a high density fermentation system using the yeast Pichia pastoris [29]. However, both methods require specialized equipment and techniques that can be expensive and time-consuming. In this report, we have described a relatively simple method for obtaining milligram quantities of RegIIIγ and HIP/PAP without the use of protein tags such as GST or Histidine. By overexpressing the proteins in E. coli, isolating inclusion bodies, and refolding the proteins to their native conformations, we obtained milligram quantities of both RegIIIγ and HIP/PAP. This method is likely to be easily adaptable to other Reg family members by screening for refolding conditions specific to each protein.
Initial attempts to induce production of HIP/PAP in E. coli resulted in very low expression levels in contrast to the robust expression seen with RegIIIγ. To improve HIP/PAP expression, we incorporated silent mutations designed to alleviate predicted RNA secondary structures at the ribosome binding site. This resulted in greatly improved HIP/PAP expression, suggesting that a similar approach could be employed to increase expression of other Reg family members that exhibit low expression levels in E. coli. In fact, an analogous approach has been used previously to amplify E. coli expression of porcine liver cytochrome P-450 reductase [30].
Three lines of evidence indicate that our procedures yield correctly refolded HIP/PAP and RegIIIγ. First, unfolded proteins tend to aggregate. However, our size exclusion chromatography experiments revealed that both recombinant proteins exist primarily as monomers in solution, suggesting that they were not entirely misfolded. Second, circular dichroism analysis shows that the refolded proteins have a secondary structure that is predominantly β-sheet, which is typical of C-type lectins. Third, the fact that RegIIIγ and HIP/PAP bind quantitatively to mannan– Sepharose indicates that they are fully refolded to a functionally active state.
The carbohydrate binding data presented here are among the first to suggest a potential ligand for Reg family members. Our results suggest that the binding specificities of RegIIIγ and HIP/PAP are similar to that of mannose binding lectin, which binds to mannose residues on bacterial surfaces and initiates recruitment of complement components that carry out microbial killing [31]. However, there are two key differences. First, although RegIIIγ and HIP/PAP both interact with mannan, they do not bind monomeric mannose, suggesting a requirement for a highly polymeric ligand. Second, whereas mannose binding lectin requires Ca2+ for ligand binding [32], the binding of RegIIIγ and HIP/PAP to mannan is inhibited by Ca2+. Further biochemical studies will be required to determine the mechanism of RegIII carbohydrate binding and to gain a precise understanding of how Ca2+ modulates this binding. Moreover, once the biological functions of RegIII proteins are better understood, then it will be possible to evaluate the functional significance of this modulation.
Previous studies by other investigators have suggested that RegIIIγ and HIP/PAP are secreted from gut epithelial cells [16,33]. If so, then these proteins are likely targeted to the intestinal lumen which is inhabited by large populations of microbes. The fact that mannan is a yeast-derived polysaccharide raises the possibility that RegIIIγ and HIP/PAP could interact with fungal species that are present in the intestine. However, their mannan binding ability also suggests a binding activity similar to mannose binding lectin, which has been shown to adhere to bacterial cell surfaces [31]. The ability to produce milligram quantities of these proteins in an E. coli overexpression system will allow these hypotheses to be tested, and will facilitate further detailed analyses of the ligands and functions of the RegIII family.
Acknowledgments
This work was supported by the Crohn’s and Colitis Foundation of America, the NIH (R01 DK070855 to L.V.H.), and a Burroughs Wellcome Career Award in the Biomedical Sciences to L.V.H. We thank Karen Lewis and Dr. Philip Thomas for help with circular dichroism studies and members of the Hooper laboratory for constructive criticism of the manuscript.
Footnotes
Abbreviations used: CRD, carbohydrate recognition domain; HIP/PAP, hepatocarcinoma-intestine-pancreas/pancreatitis associated protein; GST, glutathione S-transferase; IPTG, isopropyl-β-d-thiogalactopyranoside; IB, inclusion body; MALDI-TOF, matrix assisted laser desorption/ionization time of flight; CD, circular dichroism; DC, detergent compatible.
References
- 1.Drickamer K, Taylor ME. Biology of animal lectins. Annu. Rev. Cell Biol. 1993;9:237–264. doi: 10.1146/annurev.cb.09.110193.001321. [DOI] [PubMed] [Google Scholar]
- 2.Terazono K, Yamamoto H, Takasawa S, Shiga K, Yonemura Y, Tochino Y, Okamoto H. A novel gene activated in regenerating islets. J. Biol. Chem. 1988;263:2111–2114. [PubMed] [Google Scholar]
- 3.Narushima Y, Unno M, Nakagawara K, Mori M, Miyashita H, Suzuki Y, Noguchi N, Takasawa S, Kumagai T, Yonekura H, Okamoto H. Structure, chromosomal localization and expression of mouse genes encoding type III Reg, RegIIIα, RegIIIβ, RegIIIγ. Gene. 1997;185:159–168. doi: 10.1016/s0378-1119(96)00589-6. [DOI] [PubMed] [Google Scholar]
- 4.Ogawa H, Fukushima K, Sasaki I, Matsuno S. Identification of genes involved in mucosal defense and inflammation associated with normal enteric bacteria. Am. J. Physiol. Gastrointest. Liver Physiol. 2000;279:G492–G499. doi: 10.1152/ajpgi.2000.279.3.G492. [DOI] [PubMed] [Google Scholar]
- 5.Ogawa H, Fukushima K, Naito H, Funayama Y, Unno M, Takahashi K, Kitayama T, Matsuno S, Ohtani H, Takasawa S, Okamoto H, Sasaki I. Increased expression of HIP/PAP and regenerating gene III in human inflammatory bowel disease and a murine bacterial reconstitution model. Inflamm. Bowel Dis. 2003;9:162–170. doi: 10.1097/00054725-200305000-00003. [DOI] [PubMed] [Google Scholar]
- 6.Keilbaugh SA, Shin ME, Banchereau RF, McVay LD, Boyko N, Artis D, Cebra JJ, Wu GD. Activation of RegIIIβ/γ and interferon γ expression in the intestinal tract of SCID mice: an innate response to bacterial colonisation of the gut. Gut. 2005;54:623–629. doi: 10.1136/gut.2004.056028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dieckgraefe BK, Crimmins DL, Landt V, Houchen C, Anant S, Porche-Sorbet R, Ladenson JH. Expression of the regenerating gene family in inflammatory bowel disease mucosa: Reg Iα upregulation, processing, and antiapoptotic activity. J. Investig. Med. 2002;50:421–434. doi: 10.1136/jim-50-06-02. [DOI] [PubMed] [Google Scholar]
- 8.Christa L, Pauloin A, Simon MT, Stinnakre MG, Fontaine ML, Delpal S, Ollivier-Bousquet M, Brechot C, Devinoy E. High expression of the human hepatocarcinoma-intestine-pancreas/pan-creatic-associated protein (HIP/PAP) gene in the mammary gland of lactating transgenic mice. Secretion into the milk and purification of the HIP/PAP lectin. Eur. J. Biochem. 2000;267:1665–1671. doi: 10.1046/j.1432-1327.2000.01159.x. [DOI] [PubMed] [Google Scholar]
- 9.Christa L, Felin M, Morali O, Simon MT, Lasserre C, Brechot C, Seve AP. The human HIP gene, overexpressed in primary liver cancer encodes for a C-type carbohydrate binding protein with lactose binding activity. FEBS Lett. 1994;337:114–118. doi: 10.1016/0014-5793(94)80640-3. [DOI] [PubMed] [Google Scholar]
- 10.Abergel C, Chenivesse S, Stinnakre MG, Guasco S, Brechot C, Claverie JM, Devinoy E, Christa L. Crystallization and preliminary crystallographic study of HIP/PAP, a human C-lectin overexpressed in primary liver cancers. Acta Crystallogr. D. Biol. Crystallogr. 1999;55(Pt 8):1487–1489. doi: 10.1107/s0907444999007969. [DOI] [PubMed] [Google Scholar]
- 11.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 12.Lobley A, Whitmore L, Wallace BA. DICHROWEB: an interactive website for the analysis of protein secondary structure from circular dichroism spectra. Bioinformatics. 2002;18:211–212. doi: 10.1093/bioinformatics/18.1.211. [DOI] [PubMed] [Google Scholar]
- 13.Bohm G, Muhr R, Jaenicke R. Quantitative analysis of protein far UV circular dichroism spectra by neural networks. Protein Eng. 1992;5:191–195. doi: 10.1093/protein/5.3.191. [DOI] [PubMed] [Google Scholar]
- 14.Sreerama N, Woody RW. Protein secondary structure from circular dichroism spectroscopy. Combining variable selection principle and cluster analysis with neural network, ridge regression and self-consistent methods. J. Mol. Biol. 1994;242:497–507. doi: 10.1006/jmbi.1994.1597. [DOI] [PubMed] [Google Scholar]
- 15.Taylor ME, Drickamer K. Structure–function analysis of C-type animal lectins. Methods Enzymol. 2003;363:3–16. doi: 10.1016/S0076-6879(03)01039-5. [DOI] [PubMed] [Google Scholar]
- 16.Christa L, Carnot F, Simon MT, Levavasseur F, Stinnakre MG, Lasserre C, Thepot D, Clement B, Devinoy E, Brechot C. HIP/PAP is an adhesive protein expressed in hepatocarcinoma, normal Paneth, and pancreatic cells. Am. J. Physiol. 1996;271:G993–G1002. doi: 10.1152/ajpgi.1996.271.6.G993. [DOI] [PubMed] [Google Scholar]
- 17.Holloway DE, Hares MC, Shapiro R, Subramanian V, Acharya KR. High-level expression of three members of the murine angioge-nin family in Escherichia coli and purification of the recombinant proteins. Protein Expr. Purif. 2001;22:307–317. doi: 10.1006/prep.2001.1434. [DOI] [PubMed] [Google Scholar]
- 18.De Bernardez Clark E, Schwarz E, Rudolph R. Inhibition of aggregation side reactions during in vitro protein folding. Methods Enzymol. 1999;309:217–236. doi: 10.1016/s0076-6879(99)09017-5. [DOI] [PubMed] [Google Scholar]
- 19.Drickamer K. C-type lectin-like domains. Curr. Opin. Struct. Biol. 1999;9:585–590. doi: 10.1016/s0959-440x(99)00009-3. [DOI] [PubMed] [Google Scholar]
- 20.Stambach NS, Taylor ME. Characterization of carbohydrate recognition by langerin, a C-type lectin of Langerhans cells. Glycobiology. 2003;13:401–410. doi: 10.1093/glycob/cwg045. [DOI] [PubMed] [Google Scholar]
- 21.Taneva S, Voelker DR, Keough KM. Adsorption of pulmonary surfactant protein D to phospholipid monolayers at the air-water interface. Biochemistry. 1997;36:8173–8179. doi: 10.1021/bi963040h. [DOI] [PubMed] [Google Scholar]
- 22.Weis WI, Drickamer K. Trimeric structure of a C-type mannose-binding protein. Structure. 1994;2:1227–1240. doi: 10.1016/S0969-2126(94)00124-3. [DOI] [PubMed] [Google Scholar]
- 23.Bertrand JA, Pignol D, Bernard JP, Verdier JM, Dagorn JC, Fontecilla-Camps JC. Crystal structure of human lithostathine, the pancreatic inhibitor of stone formation. EMBO J. 1996;15:2678–2684. [PMC free article] [PubMed] [Google Scholar]
- 24.Julenius K, Molgaard A, Gupta R, Brunak S. Prediction, conservation analysis, and structural characterization of mammalian mucin-type O-glycosylation sites. Glycobiology. 2005;15:153–164. doi: 10.1093/glycob/cwh151. [DOI] [PubMed] [Google Scholar]
- 25.Drickamer K, Fadden AJ. Genomic analysis of C-type lectins. Biochem. Soc. Symp. 2002:59–72. doi: 10.1042/bss0690059. [DOI] [PubMed] [Google Scholar]
- 26.Nata K, Liu Y, Xu L, Ikeda T, Akiyama T, Noguchi N, Kawaguchi S, Yamauchi A, Takahashi I, Shervani NJ, Onogawa T, Takasawa S, Okamoto H. Molecular cloning, expression and chromosomal localization of a novel human REG family gene, REG III. Gene. 2004;340:161–170. doi: 10.1016/j.gene.2004.06.010. [DOI] [PubMed] [Google Scholar]
- 27.Dieckgraefe BK, Stenson WF, Korzenik JR, Swanson PE, Harrington CA. Analysis of mucosal gene expression in inflammatory bowel disease by parallel oligonucleotide arrays. Physiol. Genomics. 2000;4:1–11. doi: 10.1152/physiolgenomics.2000.4.1.1. [DOI] [PubMed] [Google Scholar]
- 28.Okamoto H. The Reg gene family and Reg proteins: with special attention to the regeneration of pancreatic β-cells. J. Hepatobiliary Pancreat. Surg. 1999;6:254–262. doi: 10.1007/s005340050115. [DOI] [PubMed] [Google Scholar]
- 29.Li A, Crimmins DL, Luo Q, Hartupee J, Landt Y, Ladenson JH, Wilson D, Anant S, Dieckgraefe BK. Expression of a novel regenerating gene product, Reg IV, by high density fermentation in Pichia pastoris: production, purification, and characterization. Protein Expr. Purif. 2003;31:197–206. doi: 10.1016/s1046-5928(03)00164-5. [DOI] [PubMed] [Google Scholar]
- 30.Kimura S, Iyanagi T. High-level expression of porcine liver cytochrome P-450 reductase catalytic domain in Escherichia coli by modulating the predicted local secondary structure of mRNA. J. Biochem. (Tokyo) 2003;134:403–413. doi: 10.1093/jb/mvg158. [DOI] [PubMed] [Google Scholar]
- 31.Ezekowitz RA. Role of the mannose-binding lectin in innate immunity. J. Infect. Dis. 2003;187(Suppl 2):S335–S339. doi: 10.1086/374746. [DOI] [PubMed] [Google Scholar]
- 32.Weis WI, Drickamer K, Hendrickson WA. Structure of a C-type mannose-binding protein complexed with an oligosaccharide. Nature. 1992;360:127–134. doi: 10.1038/360127a0. [DOI] [PubMed] [Google Scholar]
- 33.Lechene de la Porte P, de Caro A, Lafont H, Sarles H. Immunocyto-chemical localization of pancreatic stone protein in the human digestive tract. Pancreas. 1986;1:301–308. doi: 10.1097/00006676-198607000-00002. [DOI] [PubMed] [Google Scholar]