Summary
The integrase protein (Int) from bacteriophage lambda is the archetype member of the tyrosine recombinase family, a large group of enzymes that rearrange DNA in all domains of life. Int catalyzes the insertion and excision of the viral genome into and out of the E. coli chromosome. Recombination transpires within higher-order nucleoprotein complexes that form when its amino-terminal domain binds to arm-type DNA sequences that are located distal to the site of strand exchange. Arm-site binding by Int is essential for catalysis, as it promotes Int mediated bridge structures that stabilize the recombination machinery. We have elucidated how Int is able to sequence specifically recognize the arm-sites by determining the solution structure of its amino-terminal domain (IntN, residues Met1 to Leu64) in complex with its P’2 DNA binding site. Previous studies have shown that IntN adopts a rare monomeric DNA binding fold that consists of a three-stranded anti-parallel beta-sheet that is packed against a C-terminal alpha helix. A low resolution crystal structure of the full-length protein also revealed that the sheet is inserted into the major groove of the arm-site. The solution structure presented here reveals how IntN specifically recognizes the arm-site sequence. A novel feature of the new solution structure is the use of an eleven residue tail that is located at the amino terminus. DNA binding induces the folding of a 310 helix in the tail that projects the amino-terminus of the protein deep into the minor groove for stabilizing DNA contacts. This finding reveals the structural basis for the observation that the ‘unstructured’ N-terminus is required for recombination.
Introduction
Bacteriophage lambda employs a tightly regulated set of site-specific DNA recombination reactions that integrate and excise its genome into and out of the E. coli chromosome (reviewed in 1). Phage integration occurs when the attachment site (attP) on the phage and a specific site on the bacterial chromosome (attB) recombine to generate the hybrid recombination sites attL and attR at the phage-host DNA junction. Under the appropriate inducing signals, the attR and attL sites recombine regenerating an excised circular lambda chromosome and a reconstituted attB site on the bacterial chromosome. Although each attachment site contains identical 15 base pair ‘core-type’ sequences that are recombined by the phage-encoded integrase (Int) 2; 3, the directionality of recombination is controlled by protein binding to distally positioned sites within the P and P’ arms (the location of these sites is shown in Fig. S1). The latter interactions promote the assembly of distinct higher-order nucleoprotein complexes called ‘intasomes’, in which the excisive and integrative recombination reactions transpire. Structurally distinct intasomes excise and integrate the phage, and differ in the specific arm-type sites that are occupied by Int, which accessory factors are present, and the specific phage attachment sites that are involved in recombination. Integrative recombination requires Int 4 and the E. coli-encoded integration host factor (IHF) 5; 6, while excisive recombination requires the phage-encoded excise (Xis) protein in addition to Int and IHF 7. The host-encoded factor for inversion stimulation (Fis) is also required for efficient excision in vivo and at limiting concentrations of Xis in vitro 8; 9.
The integrase protein (Int) is the archetypal member of the tyrosine recombinase family, a large group of enzymes that rearrange DNA in archaebacteria, eubacteria, and yeast 1; 10. It belongs to an important subgroup of these enzymes called heterobivalent recombinases, which mobilize bacteriophages and transposons. Heterobivalent recombinases are distinguished by their ability to bind to two distinct DNA sites and by the high degree of control they exert over the directionality of recombination. In the case of Int, its C-terminal catalytic and core domains bind to the core-type sites that bracket the site of strand exchange, while its N-terminal domain interacts with arm-type DNA sequences located 50 to 150 base-pairs from the site of recombination 11. Simultaneous occupancy of the core- and arm-type is essential for recombination because it creates Int-mediated bridges that stabilize the excisive and integrative intasomes 12. Arm-site binding by N-terminal domain also biases the reaction towards product formation and allosterically regulates the activity of the catalytic domain 13; 14.
A NMR structure of the N-terminal domain solved in the absence of DNA revealed that it adopts a three-stranded beta-sheet DNA binding fold 15. In addition, a crystal structure of full-length Int bound to DNA has shown that the sheet in the N-terminal domain interacts with the major groove 16. However, the crystal structure was determined at a resolution of only 4.4 Å and thus yielded limited insight into the origins of nucleotide sequence specific binding. Most importantly, the crystal structure failed to reveal how the amino-terminal tail of Int participates in recombination. The tail precedes the N-terminal domain and is essential for recombination and DNA binding, but clear electron density for its amino acids was missing in the crystal structure 15; 16. In this communication, we report the NMR structure of the N-terminal domain of the lambda integrase protein in complex with DNA. The structure reveals the role of the essential amino-terminal tail in arm-site binding and recombination.
NMR structure determination of the IntN-DNA complex
NMR was used to determine the structure of the N-terminal domain of Int (IntN, residues Met 1 to Leu 64) in complex with a 15-base pair oligonucleotide corresponding to the P’2 arm-type site within the phage (5′- G CAG TCA AAA T C -3′// 3′- C GTC AGT TTT A G -5′, binding site underlined). The NMR spectra of the 14.8 kDa IntN-DNA complex were of good quality enabling the identification of 29 intermolecular NOEs (Fig. 1A). Representative NMR spectra and a summary of the chemical shift, NOE and coupling constant data is also provided as supplemental material (Figs. S2-S4). The solution structure of the complex was calculated using simulated annealing methods from a total of 1,163 experimental derived restraints (1,023 NOE distance, 120 dihedral angle and 20 coupling constant restraints). The coordinate precision of the ensemble of 20 conformers to the average structure is 0.60 ± 0.11 Å for the IntN and DNA backbone atoms (residues Met1 to Leu55 of IntN and base pairs Cyt2 to Ade10) (Fig. 2A). All of the structures exhibit good covalent geometry and are compatible with the NMR data (Table 1). The coordinates and experimental data have been deposited at the protein data bank (accession code 2WCC).
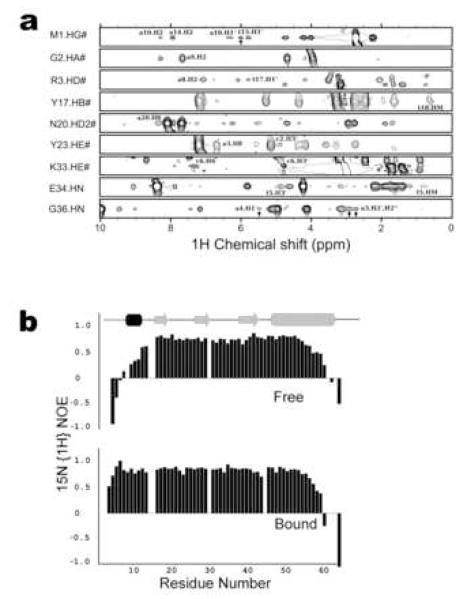
Figure 1. NMR data of the IntN-DNA complex.
(a) Representative intermolecular NOE data that was used to determine the structure of the IntN-DNA complex. Selected panels from NOESY data are shown for amino acids that are in close proximity to the DNA. The amino acids and corresponding hydrogen atoms are indicated on the left side of each panel. Relevant intermolecular NOE crosspeaks to DNA protons are labeled and indicated by an arrow where resonance overlap exists. The top seven panels are taken from a 3D 13C-edited NOESY spectrum of the complex dissolved in deuterated buffer. The bottom two panels are taken from a 3D 15N-edited NOESY spectrum of the complex dissolved in protonated buffer. (b) A comparison of the 15N{1H} heteronuclear NOE data of the DNA-free and -bound forms of IntN showing that its amino-terminal tail becomes ordered upon interacting with DNA. Top and bottom panels correspond to IntN in its DNA free and bound states, respectively. The N-terminal domain of Int (IntN, residues Met 1 to Leu 64) labeled with 13C and 15N was produced as described previously 15. DNA corresponding to the P’2 arm-site (5′- G CAG TCA AAA T C - 3′// 3′- C GTC AGT TTT A G -5′, binding site underlined) was purchased from Biosource and purified by gel electrophoresis. Initially, a 0.1 mM sample of the IntN:DNA complex was formed in the presence of high salt (50 mM HEPES (pH 7), 500 mM NaCl, and 2 mM deuterated DTT). The complex was then concentrated using a centricon (Amersham) and dialyzed into low salt NMR buffer. Two samples were prepared: (a) 0.85 mM IntN:DNA complex dissolved in water containing 7% D2O, 25 mM HEPES (pH 7), 15 mM NaCl, 2 mM deuterated DTT, and 0.01% NaN3 and (b) 0.9 mM IntN:DNA complex dissolved in 100% D2O, 26 mM deuterated TRIS (pH 7), 15 mM NaCl, 5.3 mM deuterated DTT, 0.01% NaN3. NMR spectra were recorded at 37 °C using Bruker Avance-500, -600, and -800 MHz NMR spectrometers. The 1H, 13C, and 15N protein backbone and side chain resonance assignments were obtained using conventional methods and have been described previously 33. The 12C attached 1H resonances of the DNA were assigned by analyzing a 2D F1,F2 13C filtered NOESY spectrum 34. NOE cross-peaks were recorded using 3D 13C- and 15N- edited NOESY spectra recorded with mixing times of 100 ms 35. Intermolecular NOE’s were obtained from 2D F1 13C filtered NOESY, and 15N- and 13C-edited 3D NOESY experiments 36. NMR data were processed and analyzed using the programs NMRPipe 37 and PIPP 38, respectively. The software package MARS was used to automatically assign the backbone resonances of the protein 39.
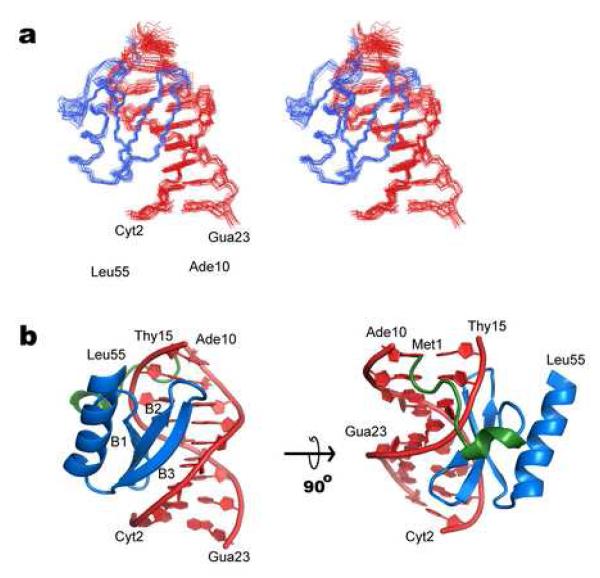
Figure 2. NMR solution structure of the IntN-DNA complex.
(a) A cross-eyed stereo view showing the ensemble of 20 lowest energy structures of the IntN-DNA complex. The protein (amino acids 1 to 55) and DNA backbone (nucleotides Cyt2 to Ade10, and Thy15 to Gua23) are shown in blue and red, respectively. (b) Ribbon drawing of the lowest energy structure of the complex. The strands in the beta-sheet and the helices are labeled. The view in the left image is identical to that shown in panel a. The amino-terminal portion of the protein that becomes ordered upon binding DNA is colored green. The solution structure of the IntN:DNA complex was determined in two stages. The structure of IntN in the complex was determined using the ATNOS/CANDID software package, which identifies NOE distance restraints by automatically assigning the NOESY NMR data 40; 41. Input spectra for the calculations included: a 3D 13C- edited NOESY spectrum recorded using the sample dissolved in 100% D2O and a 15N- edited NOESY spectrum of the sample dissolved in water. Chemical shift assignments for residues Met1-Asp11 were excluded from the ATNOS/CANDID calculations because long range NOE signals from this part of the protein were sparse and the software tended to miss-assign these signals. After seven rounds of calculations CANDID yielded a converged bundle of conformers representing the structure of IntN. In a separate set of calculations the program NIH-XPLOR was used to calculate the structure of the bound DNA molecule. Distance restraints for the DNA were obtained by manually assigning 2D F1,F2 13C filtered NOESY spectra of the complex. In addition, the structures were refined using dihedral angle restraints obtained from the program TALOS and loose DNA dihedral angle restraints for the DNA. The latter, maintained the DNA molecule in a B-form conformation and facilitated convergence, but otherwise did not alter the structure of the complex. NIH-XPLOR was then used to calculate structures of the complex 42. The structure was calculated using the previously determined structure of the DNA molecule and the protein in its unfolded state. The initial docking calculations made use of full-set of distance restraints for the DNA and protein, as well as a limited number of intermolecular NOE’s to orientate the protein and the duplex. The resultant structure was then refined in an iterative manner by manually inspecting the NMR data. During the refinement the program QUEEN was used to sort NOE restraints by decreasing information content 43. The fifty most significant restraints were checked manually and this process (restraint sorting by QUEEN and manual restraint checking) was repeated until all of the most significant restraints were correct. At the final stages of refinement intra-protein hydrogen bonds in regions of regular secondary structure were identified by inspecting NOE data for characteristic patterns. In addition to standard energy terms to maintain appropriate covalent geometry and to account for distance and dihedral angle data, a mean force potential was employed to improve the structure of the DNA molecule 33,44. The final calculations produced 200 structures, 84 of which completely satisfied the experimental data. The program MOLMOL was used to make figures for this paper45.
Table 1. Structural statistics for the solution structure of IntN-DNA complex.
The notation for the NMR structures is as follows: <SA> is the final 20 simulated annealing structures; is the average energy minimized structure. The number of terms for each restraint is given in parentheses.
| <SA> | ||
|---|---|---|
| R.m.s. deviations from NOE restraints (Å) a | ||
| All protein ( 664 ) | ||
| sequential [ | i - j| = 1] (171) | 0.041 ± 0.002 | 0.062 |
| medium range [ | i - j | ≤4] (213) | 0.052 ± 0.002 | 0.082 |
| long range [ | i - j | ≥5 ] (132) | 0.034 ± 0.005 | 0.047 |
| intra-residue (148) | 0.028 ± 0.008 | 0.062 |
| DNA (320) | ||
| sequential (109) | 0.039 ± 0.006 | 0.034 |
| intraresidue (211) | 0.109 ± 0.001 | 0.108 |
| protein/DNA (39) | 0.133 ± 0.007 | 0.111 |
| R.m.s. deviations from hydrogen bonding restraints (Å) b (22) | 0.056 ± 0.007 | 0.042 |
| R.m.s. deviations from dihedral angles restraints (°)c (120) | 0.669± 0.098 | 0.367 |
| R.m.s. deviations from 3JHNα coupling constants (Hz) (20) | 0.598 ± 0.041 | 0.761 |
| Deviation from idealized covalent geometry | ||
| bonds (Å) | 0.003 ± 0.00005 | 0.004 |
| angles (°) | 0.755 ± 0.003 | 0.817 |
| impropers (°) | 0.402 ± 0.007 | 0.483 |
| PROCHECK results (%) d | ||
| most favorable region | 85.9 ± 1.4 | 89.4 |
| additionally allowed region | 14.1 ± 1.4 | 10.6 |
| generously allowed region | 0.0 ± 0.0 | 0.0 |
| disallowed region | 0.0 ± 0.0 | 0.0 |
| Coordinate Precision (Å) e | ||
| Protein backbone | 0.60 ± 0.11 | 0.43 |
| Protein heavy atoms | 1.07 ± 0.10 | 0.61 |
| All heavy atoms | 0.88 ± 0.09 | 0.55 |
None of the structures exhibited distance violations greater than 0.5 Å, dihedral angle violations greater than 5°, or coupling constant violations greater than 2 Hz.
Two distance restraints were employed for each hydrogen bond (rNH⋯O < 2.3 Å and rN⋯O < 3.3 Å).
The experimental dihedral angle restraints were as follows: 53 φ, 53 ψ, 11 χ1, and 4 χ2 angular restraints
Determined using the program PROCHECK described in Ref. 46
The coordinate precision is defined as the average atomic root mean square deviation (rmsd) of the 20 individual SA structures and their mean coordinates. These values are for residues Met1 to Leu64 of IntN. The protein backbone is defined as the N, Cα, and C’ atoms.
Sequence specific recognition of the arm-site
Int recognizes the P’2 arm-site by inserting a three-stranded beta-sheet into the major groove, and by wrapping its amino-terminal tail around the duplex (Fig. 2B). These interactions bury ∼2,220 Å2 of solvent accessible surface area, but only modestly bend the duplex by ∼35°. Similar to previously reported structures, the N-terminal domain consists of a three-stranded anti-parallel beta-sheet (strands B1 (Leu16-Ile18), B2 (Tyr24-Arg27) and B3 (Glu34-Gly38)) that is packed against a C-terminal α-helix (Arg41-Leu55) 15; 16. However, in the structure of the IntN-DNA complex this core domain is supplemented by an ordered amino-terminal extension that has not been observed previously (colored green in Fig. 2B). This appendage immediately precedes the beta-sheet and consists of a 310 helix (Glu8 to Arg10) that rests against the phosphodiester backbone of the duplex, and an amino-terminal tail that is inserted deep into the minor groove. The presence of the 310 helix and other secondary structural elements is substantiated by characteristic NOE patterns in the NOESY data (Fig. S2).
Sequence specific binding is achieved by major groove contacts from amino acids located in the beta-sheet. The side chain atoms in the ensemble are reasonably well defined by the NMR data and have a coordinate precision of 1.07 ± 0.10 Å. Within the major groove the side chains of Arg19 (strand B1) and Glu34 (strand B3) project from opposing sides of the sheet to form a salt-bridge (Fig. 3A). This positions the carboxyl group of Glu34 for interactions with the N4 group of Cyt6 and enables the guanidine group of Arg19 to donate a hydrogen-bond to the O6 atom of Gua19. This interface is stabilized by the side chains of Asn20 and Lys33, which interact with the phosphate groups of Gua19 and Ade7, respectively. All arm-type binding sites contain a conserved Cyt6-Gua19 base pair suggesting that these interactions are required for nucleotide sequence specific binding. A second base specific contact originates from Asn21, whose side chain amide is appropriately positioned to donate and accept hydrogen bonds from the Gua6 O6 atom and the N4 amine group of Cyt21, respectively. Contacts at this interface may explain why Int binds to sites P1 and P1’ approximately 2-3 times more tightly than to sites P2, P’2 and P’3 (Fig. S5) 17. These higher affinity sites contain a guanine base on the 3′ side of this contact interface that likely participate in complementary hydrogen-bonding interactions to the amide group of Asn21. In contrast, all of the weaker binding sites contain an adenine at this position whose N8 amine group presumably precludes these favorable interactions. The structure of the complex also explains why modification of the side chain of Cys25 with the sulfhydryl-directed reagent N-ethylmaleimide (NEM) disrupts Int’s capacity to bind arm-type DNA and, consequently the ability to carry out recombination 18. The thiol side chain of Cys25 is positioned adjacent to the Arg19-Glu34 salt bridge and its modification likely creates a steric clash with the O4 atom of Thy5 that disrupts binding. A list of the intermolecular protein-DNA contacts present in the complex is summarized in Fig. 3C.
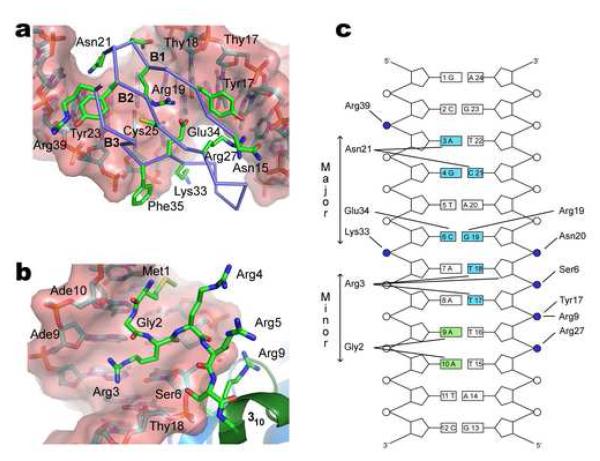
Figure 3. Mechanism of DNA binding.
(a) Expanded view of the major groove interface. Beta-sheet strands B1 (Leu16-Ile18), B2 (Tyr24-Arg27) and B3 (Glu34-Gly38) insert into the major groove. The side chains of Arg19 and Glu34 contact the Cyt6-Gua19 base pair and simultaneously form a salt-bridge. The carboxyl group of Glu34 interacts with the N4 atom of Cyt6 and the guanidine group of Arg19 donates a hydrogen-bond to the O6 atom of Gua19. The side chains of Asn20 and Lys33 contact phosphate groups of Glu19 and Ade7, respectively. The side chain of Asn21 is juxtaposed with the Gua4-Cyt21 base step in the major groove, stabilizing the binding interface. (b) Expanded view of the minor groove interface and role of the amino-terminus in DNA binding. The side chain of Met1, the backbone residues Met1-Gly2 and the side chain of Arg3 are deeply inserted into the minor groove. Gly2 contacts the Ade9-Ade10 base-step and side chain of Arg3 contacts the Thy17-Thy18 base step. Arginine residues 3, 4, 5, 9 and 10 and the N-terminal amino group are favorably positioned for electrostatic interactions with the phosphodiester backbone adjacent to the minor groove interface and the 310 helix. (c) Schematic summarizing the protein-DNA contacts in the structure of the IntN-DNA complex. Phosphodiester linkages are shown as circles, those that are contacting by IntN highlighted in blue. Bases shaded blue and green are contacted by the protein from the major- and minor-groove, respectively. A hydrogen bond is considered to be present when potential donor and acceptor atoms are separated by less than 3 Å. Salt-bridge interactions occur when appropriately charged groups are separated by less than 4.5 Å. Interactions shown in the figure occur in >40% of the structures within the ensemble.
The essential amino-terminal tail of integrase is immobilized in the minor groove
Alteration of the first five amino acids of Int disrupts recombination and DNA binding 15. In the NMR structure of IntN solved the absence of DNA these residues are part of an eleven amino acid disordered tail that precedes strand B1 15. Electron density for these residues is also absent in all previously reported crystal structures of full-length Int 16. Our results indicate that the tail undergoes a disordered-ordered transition upon binding the duplex as result of extensive hydrogen bonding to the minor groove. This is evident by analyzing the 15N{1H} heteronuclear NOE values of the backbone amide groups of the protein in its DNA-free and - bound states (Fig. 1B). In the apo-form, residues preceding strand B1 are unfolded and dynamic as evidenced by small magnitude or negative NOE values. However, upon complex formation the entire N-terminus becomes ordered, as its residues exhibit large NOE values similar to amino acids in the remainder of the domain. Interestingly, other parts of the Int protein also undergo DNA induced folding as recent studies have shown that the core domain is poorly structured in its free form, but folds upon binding to DNA 19.
Inspection of the NMR structure reveals that DNA binding induces the folding of residues Glu8-Arg10 into a 310 helix as a result of stabilizing contacts from the side chains of Ser6 and Arg9 to the phosphates of Thy18 and Thy17, respectively (Fig. 3B). Residues Met1-Gly2-Arg3 preceding the 310 helix then extend into the minor groove enabling the backbone amide of Gly2 to hydrogen bond to the N3 group of Ade10 and the side chain of Met1 to form non-polar interactions with the Ade10-Thy11 base step (Fig. 3B). The tail is further positioned by contacts from by the side chain of Arg3, which is poised to interact with the phosphate of Gua19. Previous studies have investigated the function of residues Arg3, Arg4 and Arg5 in DNA binding and recombination 15. Deleting a single arginine amino acid within this tripeptide has little impact, while protein mutants that eliminate two or more of the arginines are defective in binding and recombination. These results are compatible with the structure, since it is likely that the polypeptide segment preceding the 310 helix can accommodate the removal of a single amino acid without altering its position within the minor groove, while larger deletions would necessitate major changes in the positioning of the Met1-Gly2-Arg3 tripeptide. Because the single amino acid deletion mutant also contains a Gly2Lys substitution, this data suggests that Gly2 can be replaced with lysine without affecting DNA binding. This is compatible with the structure of the complex, since the side chain hydrogen of Gly2 projects into the solvent and is therefore presumably tolerant to mutation. In contrast to the amino-terminal tail, DNA binding causes only modest structural changes in the remainder of the IntN domain as the backbone atoms of residues Leu12 to Leu55 can be superimposed with a rmsd of 0.55 Å when the NMR structures of the DNA-free and -bound forms of IntN are compared 15. Interestingly, in the 4.4 Å crystal structure of full-length Int bound to DNA electron density for residues Met1 to His7 is missing, and residues Glu8 to Arg10 project away from the duplex instead of forming the DNA contacting 310-helix observed in the NMR structure of the complex. The origin of this difference remains unknown, but could be caused by crystal packing effects.
Comparison to other three-stranded beta-sheet DNA binding domains
The three-stranded beta-sheet DNA binding domain present in IntN is very rare and only two other proteins containing this fold have been visualized in complex with DNA: the DNA binding domain from the integrase protein encoded by the Tn916 transposon (IntTn916) 20; 21 and the ethylene responsive factor domain 1 from Arabidopsis thaliana (AtERF1) 22. These proteins share no significant sequence homology with one another, but adopt similar three-dimensional structures that engage the major groove through a three-stranded beta-sheet (Fig. 4). In each protein, the anti-parallel beta-sheet is formed by residues that are contiguous in the primary sequence (B1-B2-B3 topology of secondary structural elements). The sheet is also similarly positioned next to the duplex. In this orientation strand B3 runs from its amino to carboxyl terminus, while the adjacent phosphodiester backbone of the DNA moves from its 3′ to 5′ end. An opposite polarity exists on the other side of the sheet-DNA interface. Here, strand B1 is placed in an amino to carboxyl orientation, while the proximal DNA strand runs from 5′ to 3′. This orientation maximizes the protein-DNA contact surface because the inherent right-handed twist in the beta strands causes the B1-B2 portion of the sheet to form a concave surface that is complementary to the curvature of the duplex 23; 24. In the IntN-DNA complex this enables Arg19 in strand B1, as well as the side chains of Asn20 and Asn21 to make major groove contacts. A comparison of the structures reveals that the sheet in the IntN domain is smaller than the sheets present in the AtERF1 and the IntTn916 proteins because its B2 and B3 strands are shorter. As a result, IntN only projects seven residues from its beta-sheet towards the duplex as compared to nine in the AtERF1 and the IntTn916 proteins. IntN presumably compensates for the reduced size of its sheet-DNA interface by forming additional stabilizing contacts from its amino-terminal tail that are absent in the AtERF1- and the IntTn916-DNA complexes (in IntTn916 and AtERF1 several amino acids precede the DNA binding domain, but they share no significant sequence homology with IntN and do not interact with the minor groove). A similar strategy to increase the size of the DNA contact interface, and thus affinity, is used by helical binding motifs 25. The IntN-DNA NMR structure forms the foundation for future studies to elucidate how the Xis and Fis proteins control the directionality of recombination. Recent studies have shown that these key accessory factors regulate recombination by cooperatively assembling with Int onto attR to form a Int-Xis-Xis-Xis-Fis:DNA complex 26; 27; 28. Moreover, Xis cooperatively binds DNA with Int by interacting with the IntN domain 29; 30; 31. The structure of the IntN-DNA complex serves as a foundation for structural studies of these larger multi protein-DNA complexes that comprise the intasome that mediates phage excision 12; 32.
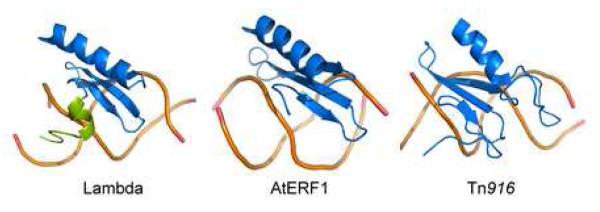
Figure 4. Comparison of three-stranded beta-sheet DNA binding proteins.
The image compares the structures of the DNA complexes of the following proteins: IntN from bacteriophage lambda (left), the ethylene responsive factor domain 1 from Arabidopsis thaliana (AtERF1) 22 and the DNA binding domain from the integrase protein encoded by the Tn916 transposon (IntTn916) 20; 21. Only the backbone of the DNA is shown. Each protein is colored blue, with the exception of the amino-terminal tail of IntN that becomes ordered upon DNA binding, which is colored green. None of the proteins share significant primary sequence homology with one another.
Supplementary Material
Acknowledgements
We thank Dr. Scott Robson for useful discussions and Dr. Robert Peterson for his assistance with NMR experiments. This work was supported by grant from the U.S. Department of Energy (DE-FC-03-87ER60615).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Accession Numbers: Coordinates and structure factors have been deposited in the Protein Data Bank with accession number 2WCC.
References
- 1.Azaro MA, Landy A. Lambda integrase and the lambda Int family. In: Craig NL, Craigie R, Gellert M, Lambowitz AM, editors. Mobile DNA II. ASM Press; Washington DC: 2002. pp. 118–148. [Google Scholar]
- 2.Mizuuchi K, Weisberg R, Enquist L, Mizuuchi M, Buraczynska M, Foeller C, Hsu PL, Ross W, Landy A. Structure and function of the phage lambda att site: size, int-binding sites, and location of the crossover point. Cold Spring Harb Symp Quant Biol. 1981;45(Pt 1):429–37. doi: 10.1101/sqb.1981.045.01.057. [DOI] [PubMed] [Google Scholar]
- 3.Ross W, Landy A. Patterns of lambda Int recognition in the regions of strand exchange. Cell. 1983;33:261–72. doi: 10.1016/0092-8674(83)90355-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kikuchi Y, Nash HA. The bacteriophage lambda int gene product. A filter assay for genetic recombination, purification of int, and specific binding to DNA. J Biol Chem. 1978;253:7149–57. [PubMed] [Google Scholar]
- 5.Nash HA, Robertson CA. Purification and properties of the Escherichia coli protein factor required for lambda integrative recombination. J Biol Chem. 1981;256:9246–53. [PubMed] [Google Scholar]
- 6.Miller HI, Kikuchi A, Nash HA, Weisberg RA, Friedman DI. Site-specific recombination of bacteriophage lambda: the role of host gene products. Cold Spring Harb Symp Quant Biol. 1979;43(Pt 2):1121–6. doi: 10.1101/sqb.1979.043.01.125. [DOI] [PubMed] [Google Scholar]
- 7.Abremski K, Gottesman S. Purification of the bacteriophage lambda xis gene product required for lambda excisive recombination. Journal of Biological Chemistry. 1982;257:9658–62. [PubMed] [Google Scholar]
- 8.Ball CA, Johnson RC. Multiple effects of Fis on integration and the control of lysogeny in phage lambda. J Bacteriol. 1991;173:4032–8. doi: 10.1128/jb.173.13.4032-4038.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thompson JF, de Vargas L. Moitoso, Koch C, Kahmann R, Landy A. Cellular factors couple recombination with growth phase: characterization of a new component in the lambda site-specific recombination pathway. Cell. 1987;50:901–8. doi: 10.1016/0092-8674(87)90516-2. [DOI] [PubMed] [Google Scholar]
- 10.Van Duyne GD. A structural view of tyrosine recombinase site-specific recombination. In: Craig NL, Craigie R, Gellert M, Lambowitz AM, editors. Mobile DNA II. ASM Press; Washington DC: 2002. pp. 93–117. [Google Scholar]
- 11.Ross W, Landy A, Kikuchi Y, Nash H. Interaction of int protein with specific sites on lambda att DNA. Cell. 1979;18:297–307. doi: 10.1016/0092-8674(79)90049-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim S, Landy A. Lambda Int protein bridges between higher order complexes at distant chromosomal loci attL and attR. Science. 1992;256:198–203. doi: 10.1126/science.1533056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sarkar D, Radman-Livaja M, Landy A. The small DNA binding domain of lambda integrase is a context-sensitive modulator of recombinase functions. Embo Journal. 2001;20:1203–1212. doi: 10.1093/emboj/20.5.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Radman-Livaja M, Shaw C, Azaro M, Biswas T, Ellenberger T, Landy A. Arm sequences contribute to the architecture and catalytic function of a lambda integrase-Holliday junction complex. Mol Cell. 2003;11:783–94. doi: 10.1016/s1097-2765(03)00111-4. [DOI] [PubMed] [Google Scholar]
- 15.Wojciak JM, Sarkar S, Landy A, Clubb RT. Arm-site binding by lambda-integrase: Solution structure and functional characterization of its amino-terminal domain. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:3434–3439. doi: 10.1073/pnas.052017999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Biswas T, Aihara H, Radman-Livaja M, Filman D, Landy A, Ellenberger T. A structural basis for allosteric control of DNA recombination by lambda integrase. Nature. 2005;435:1059–66. doi: 10.1038/nature03657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sarkar D, Azaro MA, Aihara H, Papagiannis CV, Tirumalai R, Nunes-Duby SE, Johnson RC, Ellenberger T, Landy A. Differential affinity and cooperativity functions of the amino-terminal 70 residues of lambda integrase. J Mol Biol. 2002;324:775–89. doi: 10.1016/s0022-2836(02)01199-3. [DOI] [PubMed] [Google Scholar]
- 18.Tirumalai RS, Pargellis CA, Landy A. Identification and characterization of the N-ethylmaleimide-sensitive site in lambda-integrase. J Biol Chem. 1996;271:29599–604. doi: 10.1074/jbc.271.47.29599. [DOI] [PubMed] [Google Scholar]
- 19.Kamadurai HB, Foster MP. DNA recognition via mutual-induced fit by the core-binding domain of bacteriophage lambda integrase. Biochemistry. 2007;46:13939–47. doi: 10.1021/bi700974t. [DOI] [PubMed] [Google Scholar]
- 20.Connolly KM, Wojciak JM, Clubb RT. Site-specific DNA binding using a variation of the double stranded RNA binding motif. Nature Structural Biology. 1998;5:546–550. doi: 10.1038/799. [DOI] [PubMed] [Google Scholar]
- 21.Wojciak JM, Connolly KM, Clubb RT. NMR structure of the Tn916 integrase-DNA complex. Nature Structural Biology. 1999;6:366–373. doi: 10.1038/7603. [DOI] [PubMed] [Google Scholar]
- 22.Allen MD, Yamasaki K, Ohme-Takagi M, Tateno M, Suzuki M. A novel mode of DNA recognition by a beta-sheet revealed by the solution structure of the GCC-box binding domain in complex with DNA. Embo Journal. 1998;17:5484–96. doi: 10.1093/emboj/17.18.5484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Suzuki M. DNA recognition by a b-sheet. Protein Engineering. 1995;8:1–43. doi: 10.1093/protein/8.1.1. [DOI] [PubMed] [Google Scholar]
- 24.Connolly KM, Ilangovan U, Wojciak JM, Iwahara M, Clubb RT. Major groove recognition by three-stranded beta-sheets: Affinity determinants and conserved structural features. Journal of Molecular Biology. 2000;300:841–856. doi: 10.1006/jmbi.2000.3888. [DOI] [PubMed] [Google Scholar]
- 25.Clarke ND, Beamer LJ, Goldberg HR, Berkower C, Pabo CO. The DNA binding arm of lambda repressor: critical contacts from a flexible region. Science. 1991;254:267–70. doi: 10.1126/science.254.5029.267. [DOI] [PubMed] [Google Scholar]
- 26.Abbani MA, Papagiannis CV, Sam MD, Cascio D, Johnson RC, Clubb RT. Structure of the cooperative Xis-DNA complex reveals a micronucleoprotein filament that regulates phage lambda intasome assembly. Proc Natl Acad Sci U S A. 2007;104:2109–14. doi: 10.1073/pnas.0607820104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pa0pagiannis CV, Sam MD, Abbani MA, Yoo D, Cascio D, Clubb RT, Johnson RC. Fis targets assembly of the Xis nucleoprotein filament to promote excisive recombination by phage lambda. J Mol Biol. 2007;367:328–43. doi: 10.1016/j.jmb.2006.12.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sun X, Mierke DF, Biswas T, Lee SY, Landy A, Radman-Livaja M. Architecture of the 99 bp DNA-six-protein regulatory complex of the lambda att site. Mol Cell. 2006;24:569–80. doi: 10.1016/j.molcel.2006.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Numrych TE, Gumport RI, Gardner JF. Characterization of the bacteriophage lambda excisionase (Xis) protein: the C-terminus is required for Xis-integrase cooperativity but not for DNA binding. EMBO Journal. 1992;11:3797–3806. doi: 10.1002/j.1460-2075.1992.tb05465.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Swalla BM, Cho EH, Gumport RI, Gardner JF. The molecular basis of co-operative DNA binding between lambda integrase and excisionase. Mol Microbiol. 2003;50:89–99. doi: 10.1046/j.1365-2958.2003.03687.x. [DOI] [PubMed] [Google Scholar]
- 31.Warren D, Sam MD, Manley K, Sarkar D, Lee SY, Abbani M, Wojciak JM, Clubb RT, Landy A. Identification of the lambda integrase surface that interacts with Xis reveals a residue that is also critical for Int dimer formation. Proc Natl Acad Sci U S A. 2003;100:8176–81. doi: 10.1073/pnas.1033041100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Franz B, Landy A. The Holliday junction intermediates of lambda integrative and excisive recombination respond differently to the bending proteins integration host factor and excisionase. Embo Journal. 1995;14:397–406. doi: 10.1002/j.1460-2075.1995.tb07014.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Iwahara J, Iwahara M, Daughdrill GW, Ford J, Clubb RT. The structure of the Dead ringer-DNA complex reveals how AT-rich interaction domains (ARIDs) recognize DNA. Embo Journal. 2002;21:1197–1209. doi: 10.1093/emboj/21.5.1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Iwahara J, Wojciak JM, Clubb RT. Improved NMR spectra of a protein-DNA complex through rational mutagenesis and the application of a sensitivity optimized isotope-filtered NOESY experiment. Journal of Biomolecular Nmr. 2001;19:231–241. doi: 10.1023/a:1011296112710. [DOI] [PubMed] [Google Scholar]
- 35.Muhandiram DR, Farrow NA, Xu G-Y, Smallcombe SH, Kay LE. A gradient 13C NOESY-HSQC experiment for recording 13C-labeled proteins dissolved in H2O. J. Magn. Reson. B. 1993;102:317–321. [Google Scholar]
- 36.Zwahlen C, Legault P, Vincent SJF, Greenblatt J, Konrat R, Kay LE. Methods for measurement of intermolecular NOEs by multinuclear NMR spectroscopy: Application to a bacteriophage lambda N-peptide/boxB RNA complex. Journal of the American Chemical Society. 1997;119:6711–6721. [Google Scholar]
- 37.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 38.Garrett DS, Powers R, Gronenborn AM, Clore GM. A Common Sense Approach to Peak Picking in Two-, Three-, and Four-Dimensional Spectra Using Automated Computer Analysis of Contour Diagrams. Journal of Magnetic Resonance. 1991;95:214–220. doi: 10.1016/j.jmr.2011.09.007. [DOI] [PubMed] [Google Scholar]
- 39.Jung YS, Zweckstetter M. Mars -- robust automatic backbone assignment of proteins. J Biomol NMR. 2004;30:11–23. doi: 10.1023/B:JNMR.0000042954.99056.ad. [DOI] [PubMed] [Google Scholar]
- 40.Herrmann T, Guntert P, Wuthrich K. Protein NMR structure determination with automated NOE-identification in the NOESY spectra using the new software ATNOS. J Biomol NMR. 2002;24:171–89. doi: 10.1023/a:1021614115432. [DOI] [PubMed] [Google Scholar]
- 41.Herrmann T, Guntert P, Wuthrich K. Protein NMR structure determination with automated NOE assignment using the new software CANDID and the torsion angle dynamics algorithm DYANA. J Mol Biol. 2002;319:209–27. doi: 10.1016/s0022-2836(02)00241-3. [DOI] [PubMed] [Google Scholar]
- 42.Schwieters CD, Kuszewski JJ, Tjandra N, Clore GM. The Xplor-NIH NMR molecular structure determination package. J Magn Reson. 2003;160:65–73. doi: 10.1016/s1090-7807(02)00014-9. [DOI] [PubMed] [Google Scholar]
- 43.Nabuurs SB, Spronk CA, Krieger E, Maassen H, Vriend G, Vuister GW. Quantitative evaluation of experimental NMR restraints. J Am Chem Soc. 2003;125:12026–34. doi: 10.1021/ja035440f. [DOI] [PubMed] [Google Scholar]
- 44.Kuszewski J, Schwieters C, Clore GM. Improving the accuracy of NMR structures of DNA by means of a database potential of mean force describing base-base positional interactions. J Am Chem Soc. 2001;123:3903–18. doi: 10.1021/ja010033u. [DOI] [PubMed] [Google Scholar]
- 45.Koradi R, Billeter M, Wuthrich K. MOLMOL: A Program for Display and Analysis of Macromolecular Structures. Journal of Molecular Graphics. 1996;14:51–55. doi: 10.1016/0263-7855(96)00009-4. [DOI] [PubMed] [Google Scholar]
- 46.Laskowski RA, Rullmannn JA, MacArthur MW, Kaptein R, Thornton JM. AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. Journal of Biomolecular Nmr. 1996;8:477–86. doi: 10.1007/BF00228148. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.