Abstract
Cryptochromes (CRYs) are blue-light photoreceptors with known or presumed functions in light-dependent and light-independent gene regulation in plants and animals. Although the photochemistry of plant CRYs has been studied in some detail, the photochemical behavior of animal cryptochromes remains poorly defined in part because it has been difficult to purify animal CRYs with their flavin cofactors. Here we describe the purification of type 4 CRYs of zebrafish and chicken as recombinant proteins with full flavin complement and compare the spectroscopic properties of type 4 and type 1 CRYs. In addition, we analyzed photoinduced proteolytic degradation of both types of CRYs in vivo in heterologous systems. We find that even though both types of CRYs contain stoichiometric flavin, type 1 CRY is proteolytically degraded by a light-initiated reaction in Drosophila S2, zebrafish Z3, and human HEK293T cell lines, but zebrafish CRY4 (type 4) is not. In vivo degradation of type 1 CRYs does not require continuous illumination, and a single light flash of 1 ms duration leads to degradation of about 80% of Drosophila CRY in 60 min. Finally, we demonstrate that in contrast to animal type 2 CRYs and Arabidopsis CRY1 neither insect type 1 nor type 4 CRYs have autokinase activities.
Cryptochromes are photolyase-like flavoproteins that are known or suspected to function as sensory photoreceptors (1−3). Despite extensive work on the photochemical and photobiological properties of animal CRYs,1 at present, only Drosophila cryptochrome (DmCRY) and some insect CRYs closely related to DmCRY have been shown to function as photosensors (4,5). In an effort to identify other CRYs that have photosensory functions and establish a universal reaction mechanism for all CRYs, we have been isolating and characterizing CRYs from diverse sources.
Phylogenetic analyses have divided the cryptochrome/photolyase family into several classes (3−5) including the CPD (cyclobutane pyrimidine dimer) photolyases and the single-stranded DNA-specific photolyases, which use light energy to repair CPDs in DNA, plant CRYs, which regulate growth and development in response to light, and the animal CRYs. Animal CRYs, of which there are several types, are related to the (6-4) photolyases, which use light energy to repair (6-4) photoproducts in DNA. Type 1 CRYs, which include DmCRY, are degraded in vivo in response to exposure to light, and are thought to regulate the circadian clock as a result of their effects on protein stability. Type 2 CRYs are essential components of the circadian clock, in which they function independently of light as repressors of transcription. Type 2 CRYs have failed to show definitive evidence for a photoreceptor function (3). Type 4 CRYs have not been critically examined. However, based on evolutionary considerations as well as expression patterns, it has been proposed that type 4 CRYs function as potential circadian photoreceptors in zebrafish and in the chicken pineal gland (6,7). Indeed, photoentrainment of peripheral organs of zebrafish and of the circadian rhythm of the zebrafish embryonic cell line Z3 by light (8) and in chicken the presence of a vitamin A-independent pineal photosensor (9) and constriction of the embryonic chick pupil by light, independent of opsins (10), have been considered as evidence for cryptochrome-mediated circadian photoreception. Hence, we decided to purify zebrafish and chicken type 4 CRYs and examine their putative photoreceptor activity using type 1 CRYs, which are known to function as photoreceptors, as reference proteins. In this paper, we describe the purification of type 4 CRYs of zebrafish (Danio rerio) (ZfCRY4) and of chicken (Gallus gallus) (GgCRY4) with stoichiometric amounts of the FAD cofactor. In addition, we compare the spectroscopic, photophysical, and photobiological properties of DmCRY and ZfCRY4 as representatives of animal type 1 and type 4 cryptochromes. We find that the two types of CRYs exhibit distinct excited-state dynamic and photobiological properties. Strikingly, we find that while a light flash of 1 ms duration is sufficient to cause nearly complete proteolysis of DmCRY and that DmCRY is subject to light-induced proteolysis in Drosophila, zebrafish, and mammalian cell lines, ZfCRY4 does not undergo light-induced proteolysis by flash or continuous illumination in either orthologous or heterologous hosts. These results suggest that animal type 1 CRYs which are known to function as photoreceptors and type 4 CRYs which are thought to be photoreceptors have potentially different photosignaling mechanisms.
Materials and Methods
Plasmids and Baculoviruses
Plasmids and viral constructs used in this study are listed in Table 1. The pAc5.1-dCRY-V5/HisA (expressing DmCRY) and pAc5.1-β-Gal-V5/HisA and pAc5.1-DpCRY2-V5/HisA (expressing Danaus plexippus (monarch butterfly) -DpCRY2) vectors have been described previously (5,11). pMal-EcPhr expression vector (Escherichia coli photolyase) (12) and pMal-AgCRY1 (C413N) (13) have been described previously. pcDNA4.ZfCRY4 was constructed by inserting ZfCRY4 cDNA, which was amplified by RT-PCR from total RNA isolated from the zebrafish embryonic Z3 cell line (8), into pcDNA4/myc-his (Invitrogen) in frame with a Flag tag at the C-terminus. The pAc5.1-ZfCRY4-V5/HisA was constructed by inserting ZfCRY4 cDNA from pcDNA4.ZfCRY4 into pAc5.1-V5/HisA (Invitrogen). pcDNA3-dCRY-V5/HisA and pcDNA3-β-gal-V5/HisA were constructed by inserting dCRY-V5/HisA and β-gal-V5/HisA, respectively, into pcDNA3 (Invitrogen).
Table 1. Plasmids and Viral Constructs Used in This Study.
| use | construct | host cells | protein | tag (terminus) | ref |
|---|---|---|---|---|---|
| protein purification | pMalEcPhr | E. coli | Ec photolyase | MBP (N) | (12) |
| pMalAgCRY1 (C413N) | E. coli | AgCRY1-C413N | MBP (N) | (13) | |
| virus | Sf21 | DmCRY | Flag, His (N) | (13) | |
| virus | Sf21 | AtCRY1 | His (N) | (14) | |
| virus | Sf21 | GgCRY4 | Flag (N) | this study | |
| virus | Sf21 | ZfCRY4 | Flag (C) | this study | |
| virus | Sf21 | DpCRY2 | Flag (N) | this study | |
| protein expression | pAc5.1-ZfCRY4-V5/HisA | S2 | ZfCRY4 | V5, His (C) | this study |
| pAc5.1-dCRY-V5/HisA | S2 | DmCRY | V5, His (C) | (5,11) | |
| pAc5.1-β-Gal-V5/HisA | S2 | β-galactosidase | V5, His (C) | (5,11) | |
| pcDNA4.ZfCRY4 | mammalian, Z3 | ZfCRY | Flag, Myc, His (C) | this study | |
| pcDNA3-dCRY-V5/HisA | mammalian, Z3 | DmCRY | V5, His (C) | this study |
Baculoviruses for DmCRY (13) and AtCRY1 (14) have been described previously. The GgCRY4 coding sequence was amplified from chicken brain RNA (Clontech) and cloned into pFastBac1 in frame with a Flag tag at the N-terminus. The DpCRY2 coding sequence was amplified from pAc5.1-DpCRY2-V5/HisA and cloned into pFastBac1 in frame with a Flag tag at the N-terminus using the Gibco BRL Bac-to-Bac baculovirus expression system (Invitrogen). Briefly, pFastBac1 plasmids containing target coding sequences were transformed into the E. coli DH10Bac, and recombinant bacmids were isolated from 1 mL of bacterial cultures grown from colonies of transformants. For recombinant virus generation, Sf21 cells in six-well plates were transfected with 1 μg of bacmids using 6 μL of Cellfectin reagent (Invitrogen), and parental virus was collected after 72 h. After three more 72 h amplification steps, the fourth passage (P4) high titer stock was obtained and used to infect Sf21 cells for large-scale expression of protein.
Cell Lines
The Sf21 insect cell line used for protein purification was obtained from Stratagene. The Drosophila Schneider S2 cell line was obtained from Steven M. Reppert (University of Massachusetts), and the HEK293 human embryonic cell line was obtained from ATCC. The rat retinal ganglion cell line RGC5 (15) was a kind gift of Dr. N. Agarwal (The University of North Texas Health Science Center).
Transfection and Cell Culture
The Drosophila S2 cells were cultured and transfected as described previously (11). The mammalian cells (RGC5 and HEK293T) were transfected using Fugene 6 reagent (Roche), and the cells were maintained in DMEM (Dulbecco’s modified Eagle’s medium; Sigma) supplemented with 10% FBS (Sigma) and 1× penicillin/streptomycin (Gibco). The zebrafish Z3 cell line (8) was cultured in L-15 medium (Cellgro) supplemented with 10% FBS (fetal bovine serum; Sigma) and 1× penicillin/streptomycin (Gibco). The S2, Sf21, and Z3 cells were incubated at 27 °C. The mammalian cell lines, HEK293T and RGC5 (15), were grown in a 37 °C humidified chamber containing 5% CO2. Anti-Flag (Sigma), anti-GAPDH (Cell Signaling), and anti-V5 (Invitrogen) antibodies were obtained from commercial sources. The anti-mCRY1 monoclonal antibody was generated in our laboratory and has been described previously (16).
Protein Purification
The maltose-binding protein (MBP)-Phr (12) and MBP-AgCRY1C413N (a C413N mutant of mosquito CRY1) fusion proteins (13) were expressed in E. coli BL21 strain (Stratagene) and purified as described previously for DpCRY1 (17). Typical yields were about 2 mg of MBP-CRY and 2 mg of MBP-Phr from 12 and 2 L cultures, respectively. The purified proteins were kept at −80 °C in storage buffer containing 50 mM Tris-HCl, pH 7.5, 100 mM KCl, 5 mM dithiothreitol, and 50% (v/v) glycerol. Purification and handling of CRYs were carried out at 4 °C and under dim yellow light (>550 nm) to prevent accidental photoreduction.
To purify Flag-tagged fusion proteins (DmCRY, DpCRY2, ZfCRY4, GgCRY4) from insect cells, Sf21 cells (106 per mL) growing at 27 °C in spinner flasks were infected with the P4 high titer virus at 1:100 (v/v) ratio, the cells were harvested 2 days later and lysed as described previously (14), and the proteins were purified using anti-FLAG M2-agarose beads from Sigma. Briefly, 400 μL of Flag-agarose beads was incubated with 30 mL of cleared cell lysate for 2 h and then washed three times with 15 mL of 1× TBS to remove unbound proteins. Recombinant proteins were eluted from Flag-agarose beads in 4 mL of 1× TBS containing 100 μg/mL Flag peptide (Sigma). Eluted protein samples were dialyzed against storage buffer containing 50 mM Tris-HCl, pH 7.5, 100 mM NaCl, 5 mM dithiothreitol, and 50% (v/v) glycerol. AtCry1 was purified as described, using nickel affinity chromatography (14). Typical yields were about 2 mg of CRY from 1 L of culture. The purified proteins contained an essentially stoichiometric amount of FAD as determined by the absorbance at 440 nm/absorbance at 280 nm ratio as described previously (18).
Excited-State Dynamics
Detailed experimental procedures for excited-state dynamics have been described previously (19).
Light-Induced in Vivo Proteolysis of CRYs
Drosophila S2 and zebrafish Z3 cells were transfected with appropriate plasmids in 60 mm dishes (Z3 cells) or T25 flasks (S2 cells). The cells were split into 35 mm dishes 6 h after transfection and 48 h after transfection were exposed to 366 nm light at a fluence rate of 2 mW cm−2 for the indicated time using a F15T8-BLB black light (General Electric) lamp as a light source. The extent of photoinduced proteolysis was determined by Western blotting using appropriate antibodies, and the level of CRY proteolytic degradation was quantified using ImageQuant software (GE Healthcare) and expressed relative to internal control β-galactosidase. Mammalian cells transfected with appropriate plasmids were processed similarly except that cells were kept in humidified chambers with 5% CO2 and at 37 °C for 48 h, and light was applied from outside of the incubator and filtered through one glass plate. Light intensities were measured with a UVX digital radiometer (UVP Inc.), and they represent the intensities recorded after passage through the culture dish.
Camera Flash Photolysis
S2 cells were transfected with appropriate plasmids and incubated at 27 °C in the dark for 48 h. For flash exposure experiments, cells were equilibrated to room temperature and then were exposed to a single flash from a commercial camera flash attachment (Vivitar). The flash of bright white light lasts ∼1 ms at intensity of 1 W cm−2. Cells were in growth media at the time of the flash and were exposed through a glass plate and the lid of the culture dish. The media (1.5 mL in a 35 mm diameter dish) is a complex mixture with no pH indicator and which has an absorbance of 0.23 at 400 nm. The dish was at a distance of 6 cm from the flash unit. Cells were kept at room temperature, and at various times following the camera flash they were harvested, pelleted, and resuspended in SDS−PAGE sample loading buffer. The proteins were separated on 8% or 9% SDS−PAGE, and the CRY and β-galactosidase (control) levels were determined by immunoblotting with anti-V5 antibodies and quantified using the ImageQuant software (GE Healthcare) system.
Kinase Assay
Purified proteins in the range of 0.5−2 μg were dissolved in 20 μL of the kinase buffer containing 50 mM Tris-HCl at pH 7.4, 5 mM MgCl2, 100 μM ATP (plus 1 μCi of [γ-32P]ATP (3000 Ci/mmol)), and 10 mM β-mercaptoethanol. Irradiation of samples in Eppendorf tubes was carried out at room temperature for 30 min at a fluence rate of 2 mW cm−2 (after passing through Eppendorf tubes) under a F15T8-BLB black light (General Electric) fluorescent lamp. For dark controls, the Eppendorf tubes were covered with aluminum foil and incubated alongside the light condition samples.
Results
Purification and Spectroscopic Properties of Type 1 and Type 4 Animal Cryptochromes
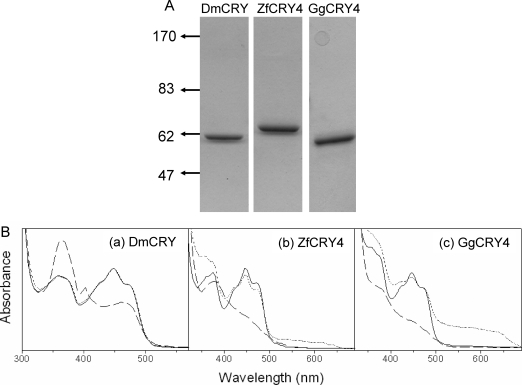
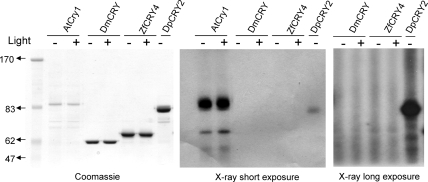
Previously, we reported the purification of type 1 CRYs of several insects using baculovirus/insect cell and bacterial expression systems (13,17). These preparations contained stoichiometric flavin and were amenable to photochemical and photophysical analyses (19,20). In contrast, previous attempts to purify vertebrate CRYs from natural sources have not been successful, and recombinant vertebrate CRYs purified from heterologous sources have contained grossly substoichiometric FAD (14,21). However, these efforts to purify vertebrate CRYs were aimed at obtaining type 2 CRYs which function as transcriptional repressors in addition to any potential photoreceptive activity (22,23) and apparently do not require the FAD cofactor for their repressor activity (24). Hence in this study, in an effort to test for a photoreceptor function of vertebrate CRYs, we concentrated on two type 4 CRYs from zebrafish (ZfCRY4) and chicken (GgCRY4) because some photobiological data suggest that these vertebrate CRYs might function as non-opsin photosensory pigments (9,10). Drosophila CRY (DmCRY) was used as a positive control. We used the baculovirus/insect cell vector/host system for expression, and the proteins were isolated to high levels of purity by affinity chromatography. The three proteins were analyzed by SDS−PAGE and Coomassie staining (Figure 1A). The absorption spectra of the purified proteins show that they contain flavin in the two-electron-oxidized FADox state (Figure 1B). As reported previously, exposure of type 1 CRYs to blue light causes photoreduction of the flavin to the FAD•− anion radical (17,25), which most likely represents the in vivo dark (ground) state of the cofactor (13). In contrast, exposure of type 4 CRYs leads to the formation of two-electron-reduced FADH2 (or FADH−) without a detectable intermediate of an FAD•− anion radical. Interestingly, incubation of DmCRY (type 1) at 4 °C under aerobic conditions and in the dark for 1 h results in complete reoxidation of the cofactor to the FADox form without a detectable neutral radical intermediate. In contrast, incubation of reduced type 4 CRYs under the same condition generates a rather stable FADH• blue neutral radical (Figure 1B), in a manner similar to that of Arabidopsis CRY1 and of photolyases from a variety of sources (19,26). While the functional significance of the differential behaviors of type 1 and type 4 CRYs is unclear at present, for the first time a vertebrate cryptochrome with full complement of FAD has now become available for photophysical, photochemical, and biochemical analyses. Because of the nearly identical steady-state spectroscopic properties of ZfCRY4 and GgCRY4 we conducted the rest of our CRY4 studies with ZfCRY4 for convenience and because of the availability of a light-sensitive zebrafish cell line, in which ZfCRY4 is thought to function as a photoreceptor (8) and could be used in our experiments.
Figure 1.
Spectroscopic properties of type 1 and type 4 animal cryptochromes. (A) Analysis of purified proteins by 8% SDS−PAGE and Coomassie staining. The numbers to the left of the panel indicate the positions of molecular mass markers in kilodaltons. Approximately 5 μg of each cryptochrome was loaded. (B) Absorption spectra of (a) Drosophila CRY, (b) zebrafish CRY4, and (c) chicken CRY4. Solid line: spectra of CRYs purified under yellow light. Dashed line: spectra after exposure to 2 mW cm−2 of 366 nm light for 10 min which reduces FAD in DmCRY to the anion radical and reduces FAD in the type 4 CRYs to the two-electron-reduced FADH− (or FADH2) form without a detectable radical intermediate. Dotted line: absorption spectra of the photoreduced samples after incubating in dark under aerobic conditions for 30 min for DmCRY and for 5 h for ZfCRY4 and GgCRY4. As is apparent DmCRY oxidizes rapidly without a detectable radical intermediate whereas ZfCRY4 and GgCRY4 reoxidize slowly and generate a very stable FAD• neutral radical.
Comparison of Excited-State Dynamics of ZfCRY4 with Dynamics of Photolyase and a Type 1 CRY
Excited-state dynamics of flavoproteins has the potential of providing significant information about their photochemical and biochemical reactions (27). Hence, we carried out a systematic analysis of ZfCRY4 excited-state dynamics using E. coli photolyase (EcPHR) and Anopheles gambiae (mosquito) CRY1 (AgCRY1), both of which are known to carry out photochemical reactions (5) as reference proteins of the photolyase/cryptochrome family. AgCRY1 was used as a representative of type 1 CRYs because of its ready availability and DmCRY-like photochemical and biological properties (5). Similarly, ZfCRY4 was used as a representative of type 4 CRYs because it exhibits 62% sequence identity to GgCRY4 (7), and an orthologous light-sensitive zebrafish cell line (Z3) (8) is available for the expression of ZfCRY4 and photobiological testing. Figure 2A shows the fluorescence emission spectra of ZfCRY4 after excitation at different wavelengths, and Figure 2B shows the lifetime measurements of ZfCRY4, folate-depleted E. coli photolyase, and AgCRY1. Note that in this experiment we used a mutant of AgCRY1 (AgCRY-C413N) that like ZfCRY4 and EcPHR goes through a FADH• neutral radical intermediate during oxidation from the FADH− form (13,19). Because the FADH− form is known to be the active form of photolyase (28,29) and suspected to be the active form of type 4 CRYs, all three proteins were reduced to this form photochemically (19,20) and used in the excited-state dynamic study.
Figure 2.
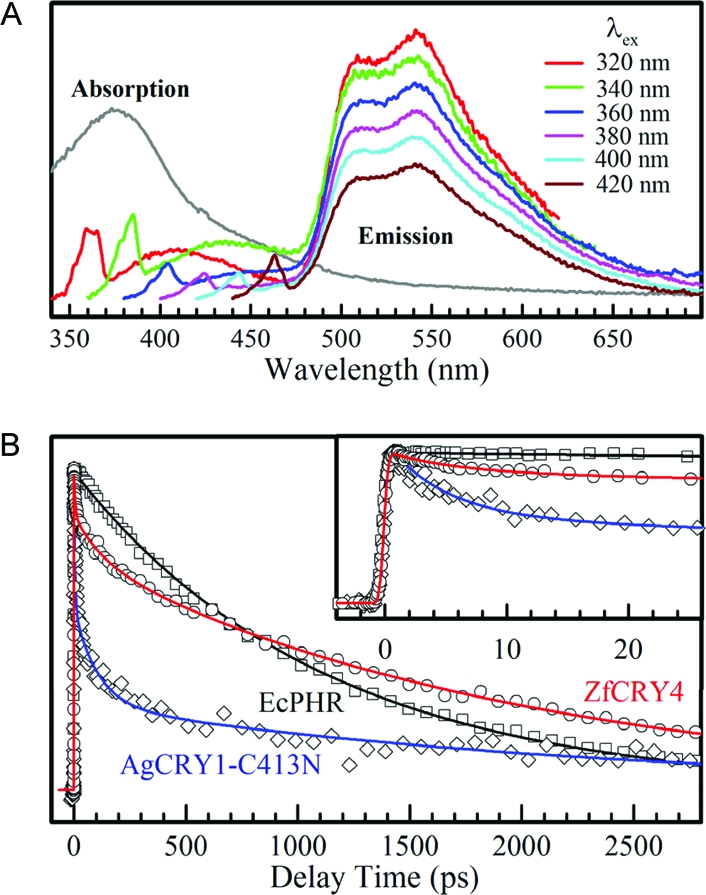
Excited-state dynamics of ZfCRY4. (A) Absorption and emission spectra of ZfCRY4 (FADH−). The emission spectra are excitation wavelength independent. The small shoulders and sharp peaks at short wavelengths (350−470 nm) are potential second chromophore emission and Raman Scattering, respectively. (B) Comparison of the fluorescence transients of ZfCRY4 and EcPHR and mosquito AgCRY1. A short-time range is shown in the inset. All three transients exhibit some unique properties, indicative of different functional behaviors.
Two conclusions can be made from the data in Figure 2. First, the emission spectra of ZfCRY4 with different excitation wavelengths are all the same (Figure 2A) as was observed for E. coli photolyase (20,30) because as expected from Kasha’s law fluorescence emission at all excitation wavelengths is from the lowest excited state. This contrasts with type 1 CRYs whose emission spectra of FAD−* are excitation wavelength-dependent (19). Second, the lifetime of the fluorescence transient of ZfCRY4 stands between that of photolyase and those of type 1 CRYs. E. coli photolyase flavin adopts a rather planar structure and has a single-exponential decay with a lifetime of 1.3 ns that enables the enzyme to maximize its electron-transfer reaction. In contrast, insect type 1 CRYs have multiple deactivation time scales from a few picoseconds up to a few nanoseconds, which is another manifestation of excitation wavelength-dependent fluorescence emission maxima. The flavin moiety of FAD is believed to be flexible in insect type 1 CRYs and experiences a butterfly bending motion to give rise to multiple deactivation pathways and hence multiple lifetimes (19). In the case of AgCRY1-C413N, the transient is best fitted with a three exponential with the two predominant ones in the picosecond range: 5 ps (47%), 105 ps (28%), and 2.3 ns (25%) (Figure 2B) (19). The fluorescence transient signal for ZfCRY4, like AgCRY1, is best fitted with multiple decays but with one important difference in that by far the most predominant species has a photolyase-like lifetime: 6 ps (17%), 118 ps (15%), and 1.85 ns (68%). Thus, it appears that the excited-state dynamics of ZfCRY4 has similarities to both photolyase and insect type 1 CRYs, and probably it represents another type of photoreceptor behavior with its own excited-state photophysics.
Photoinduced Proteolysis
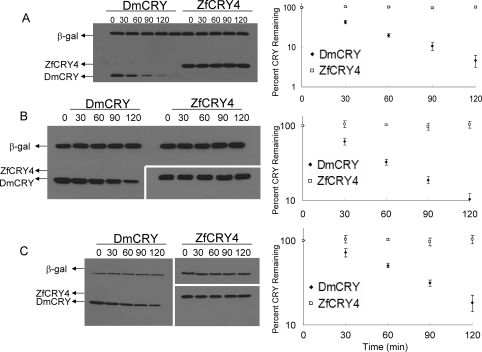
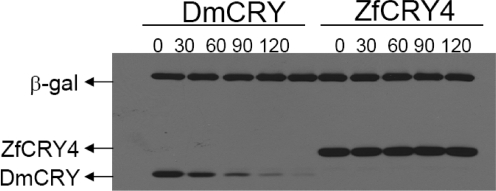
Exposure of Drosophila to light causes degradation of DmCRY (4). The finding that light-induced proteolytic degradation of DmCRY could be achieved in the Drosophila S2 cell line (5,31−33) provided a simple experimental system to study DmCRY, other type 1 CRYs (5,11), and potentially other animal CRYs such as type 4 CRYs that may function as photoreceptors. Hence, we wished to use this system to determine if type 4 CRYs also undergo photoinitiated and proteasome-mediated proteolytic degradation similar to that of DmCRY (34) and all other type 1 CRYs tested so far (5,11). In Figure 3A, we present the results of such an experiment. The S2 cells were transfected with vectors expressing DmCRY or ZfCRY4 as well as β-galactosidase as an internal control. Transfected cells were exposed to increasing doses of 366 nm light and then were lysed, and photoinduced proteolysis was quantified by immunoblotting (Figure 3A). DmCRY is degraded by 366 nm in a dose-dependent manner, in agreement with other reports (11,31,32). In contrast, no degradation of ZfCRY4 is detected within the dose range used (Figure 3A). A recent report concluded that nearly 20 Drosophila proteins including redox mediators (thioredoxin, glutathione S-transferase, and a cytochrome p450), phosphatases, ubiquitin ligases, and several subunits of proteasome are involved in light-induced proteolysis of DmCRY (35). However, another study concluded that the Jetlag E3 ligase is responsible for light-induced ubiquitylation and ultimate proteolysis of DmCRY (36). Hence, it was possible that the lack of photoinduced proteolysis of ZfCRY4 was due to species specificity of the enzymes necessary for modification/proteolysis of type 4 cryptochromes. Also, it was conceivable that ZfCRY4 did not acquire its putative second chromophore in S2 cells, which would decrease its efficiency of photodegradation. Therefore, we tested ZfCRY4 as well as DmCRY for photoinduced proteolysis in the zebrafish Z3 cell line (8). We find that the zebrafish CRY4 is not degraded by light even in this orthologous cell line (Figure 3B). In contrast, DmCRY is degraded by light in Z3 cells almost as efficiently as in the S2 cell line, indicating that the light-activated cryptochrome modification/proteolysis apparatus is present in organisms even in the absence of any indication that such a reaction occurs with those organisms’ intrinsic cryptochromes. Finally, we find that even a human embryonic kidney cell line (HEK293T) is capable of photoinduced proteolysis of DmCRY. As in Z3 cells, ZfCRY4 is not degraded in this vertebrate cell line (Figure 3C). We conclude that vertebrates possess the enzymatic mechanisms for photoinduced proteolysis of cryptochromes, but that if type 4 CRYs function as photoreceptors, photoinduced proteolysis is not a feature of their photocycle, and that type 4 CRYs operate by a mechanism dissimilar to that of type 1 CRYs.
Figure 3.
Photoinduced proteolysis of CRYs in homologous and heterologous systems. Cells were transfected with plasmids encoding the indicated cryptochromes, and β-galactosidase as a negative control, and 48 h later were irradiated with 366 nm light at a fluence rate of 2 mW cm−2 for the indicated times. The cells were then collected, and the level of CRYs and β-galactosidase was analyzed by Western blotting using anti-V5 antibody for DmCRY in S2, Z3, and HEK293T cells and ZfCRY4 in S2 cells and anti-Flag antibody for ZfCRY4 in Z3 and HEK293T cells. Left panels show representative blots, and right panels show quantitative analysis of light-induced proteolysis as a function of light dose. Averages from three experiments including the ones shown in the left panel are included. Bars indicate standard errors of the mean. The host cells were (A) S2 cells, (B) Z3 cells, and (C) HEK293T cells.
Flash Photolysis
Commercial camera flash units deliver sufficient energy in approximately 1 ms to repair all thymine dimers in photolyase−DNA complexes (29,37). This “flash photolysis” technique has been used extensively to characterize the photochemical reaction carried out by photolyase. We wished to employ this method to gain some insight into the photochemical reaction carried out by type 1 CRYs and possibly detect photoinduced proteolysis with a type 4 CRY that was not degraded with the lower fluence rate light employed in our continuous illumination experiments.
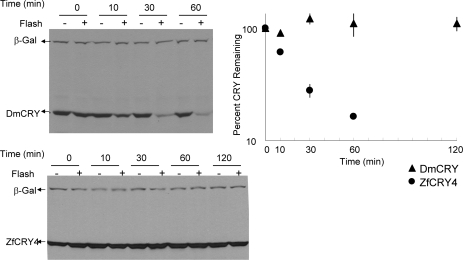
In fact, in a study aimed at determining some basic parameters of light-induced DmCRY degradation it was reported that photoinduced proteolytic degradation of DmCRY requires continuous illumination because under the experimental conditions used in that study cryptochrome degradation stopped when light was turned off (32). At face value, this finding suggests that CRY only in a short-lived photochemically excited state is a target for posttranslational modification that leads to eventual proteolysis. To test this model, we subjected S2 cells expressing recombinant DmCRY to a single camera flash of ∼1 ms duration and then incubated them in the dark at room temperature for various periods of time, and the level of CRY proteolysis was assessed by immunoblotting as a function of time after the flash. Unexpectedly, a single camera flash followed by incubation in the dark results in nearly 80% DmCRY degradation in 1 h (Figure 4). As is apparent from this figure, following the light flash the majority of DmCRY is degraded in the dark with a first-order rate constant of k1 = 5.8 × 10−4 s−1 (t1/2 = 20 min). A second population of DmCRY consisting of about 20% of the total is resistant to proteolysis because under the experimental conditions used DmCRY continues to be synthesized after the flash as observed under conditions of continuous illuminations (32). Importantly, these results unambiguously show that continuous illumination is not necessary for light-initiated DmCRY degradation. Furthermore, this finding suggests that light exposure creates a CRY metastable state (we prefer this term to “excited state” or “transition state” as these terms have precise meanings in photophysics and kinetics, respectively) that lasts long enough to enable protein kinases and E3 ligases to act on CRY to make it a target for the proteasome. In any event, these results place certain limitations on the possible sequence of events that occur from absorption of a photon to posttranslational modification and ultimate proteolysis of type 1 CRYs.
Figure 4.
Effect of a single flash of light proteolysis of a type 1 and a type 4 CRY. S2 cells were transfected with plasmids expressing V5-tagged β-galactosidase and either DmCRY or ZfCRY4 and 48 h after transfection cells were exposed to a single camera flash and incubated in the dark at room temperature. At the indicated time points cells were harvested, homogenates were prepared, and proteins were separated on SDS−PAGE and immunoblotted using anti-V5 antibody. Representative blots are shown in the left panel, and quantitative analyses of two experiments are plotted in the right panel (means and standard deviations where larger than the data points).
Because the fluence rate of a multiunit camera flash is much higher than the conventional light sources used in our continuous illumination experiments (38), we considered the possibility that type 4 CRYs that do not undergo light-induced proteolysis with continuous illumination might do so with the higher fluence rate of flash photolysis. Hence, we carried out the same camera flash experiments on ZfCRY4. There was no degradation of this type 4 CRY even under this excitation condition (Figure 4). We conclude that photoinduced proteolysis is not a part of the photosignaling or of the downregulation of the photosignal initiated by type 4 CRYs, if indeed these CRYs function as photosensory photoreceptors.
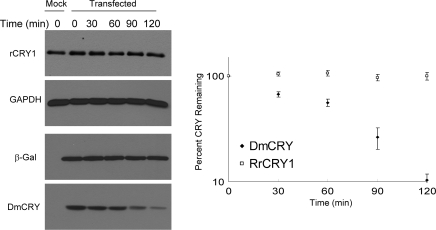
Finally, even though there is no evidence that mammalian cryptochromes are degraded by light in their natural hosts, it was recently reported that human CRY1 (a type 2 CRY) expressed in Drosophila was degraded upon exposure of the flies to light (39). We were unable to observe any light-induced degradation of HsCRY1 in Drosophila S2 cells. Hence, we decided to test the photoinduced proteolysis of a type 2 CRY in its native host. We chose to use the RGC5 rat ganglion cell line (15) for several reasons. First, the endogenous RrCRY1 (Rattus rattus CRY1) is expressed at a high level in these cells and can be readily detected using our anti-mCRY1 antibodies (16). Also, RGC5 cells are more likely to have a CRY photosignaling machinery including proteolysis because of its origin from a photosensory tissue. We find that RrCRY1 does not undergo photoinduced proteolysis in RGC5. In contrast, DmCRY, as was observed in all other insect, zebrafish, and human cell lines tested, is subject to photoinduced proteolysis in RGC5 cells as well (Figure 5). We conclude that neither type 2 nor type 4 CRYs are subject to light-activated proteolysis in any cryptochrome/host cell system tested even under the relatively high blue light dose used in our study.
Figure 5.
Light-induced proteolysis of Drosophila CRY but not of rat CRY1 in the RGC5 rat ganglion cell line. Proliferating RGC5 cells in 60 mm dishes were transfected with pcDNA3-DmCRY-V5/HisA and pcDNA3-β-gal-V5/HisA (1:1 ratio) or pcDNA3 vector. Cells were split into 35 mm dishes and kept in the dark at 37 °C for 48 h. Light at 2 mW cm−2 fluence rate was applied for the indicated times in a sealed incubator. Cells were collected and lysed, and the levels of DmCRY were determined by Western blotting using anti-V5 antibody. For RrCRY1, cells were subjected to the same growth conditions and irradiation regimen but were not transfected. Following lysis, the status of RrCRY1 was probed using anti-mCRY1 monoclonal antibodies and anti-GAPDH as a loading control. Left panel: A representative blot. Right panel: Quantitative analysis of data from two experiments. Bars indicate the standard deviations. Circles, DmCRY; diamonds, RrCRY1.
Autokinase Activities of Cryptochromes
Many photosensory proteins including phytochrome and phototropin are autophosphorylating kinases. It has been reported that AtCRY1, HsCRY1, and HsCRY2 but not AtCRY2 have autokinase activities (21,40,41). However, there is disagreement on whether or not the CRY kinase activity is stimulated by light (21,40). The fact that AtCRY2, which is a bona fide blue-light photoreceptor, lacks kinase activity suggests that autokinase function is not a sine qua non of sensory photoreceptors. With this consideration in mind, we tested both a type 4 CRY (ZfCRY4), which contains stoichiometric FAD, and an insect type 2 CRY (DpCRY2), which contains only trace FAD when purified as recombinant protein for kinase activity (Figure 6). AtCRY1, which contains stoichiometric FAD, as expected, is an autokinase in agreement with our previous report, and under our assay conditions the kinase activity is not stimulated by light (21). Interestingly, we find that the butterfly (D. plexippus) type 2 CRY (DpCRY2) which phylogenetically segregates with mammalian CRYs (5) also has autokinase activity. Importantly, DmCRY (a type 1 CRY) which is known to function as a photoreceptor has no kinase activity, and ZfCRY4 (a type 4 CRY) which is suspected to be a photoreceptor lacks autokinase activity both in light and in the dark.
Figure 6.
Kinase activities of AtCRY1 and animal type 1, type 2, and type 4 CRYs. Reaction mixtures in kinase buffer with [γ-32P]ATP label were incubated for 30 min either in dark or under 366 nm light at 2 mW cm−2 for 30 min and analyzed by SDS−PAGE. Left, Coomassie blue stain; right, autoradiogram.
Conclusions
In this study, we compared the photochemical/photobiological properties of type 1 and type 4 CRYs with the overall goal of gaining further insight into the cryptochrome photosignaling mechanism. Several facts salient to the photosensory properties of cryptochrome have emerged from this study.
First, in both orthotopic and heterotopic hosts DmCRY (a type 1 CRY) is subject to photoinitiated proteolysis, indicating that the molecular machinery of photoinduced CRY proteolysis, which encompasses specific phosphorylation, ubiquitination, and proteolysis by the proteasome, is conserved in all animals tested including Drosophila, zebrafish, rat, and human. Cell lines from some of these organisms (insect, zebrafish) are known to be photoresponsive; others (rat, human) are not photoresponsive. Hence, the lack of photoinitiated proteolysis of type 4 CRYs in any of these cell lines provides reasonable evidence that these cryptochromes are not subject to light-induced proteolysis under physiological conditions.
Second, DmCRY is degraded in the dark nearly completely in 60 min following exposure to a camera flash of ∼1 ms duration and of 1 W cm−2 fluence rate. Using the known extinction coefficient of the cryptochrome, the number of photons absorbed by CRY within this period can be calculated from the formula:
where n = photons absorbed by CRY per second, ε = molar extinction coefficient of CRY (∼104 M−1 cm−1),and Nhυ = photons cm−2 s−1 (fluence rate).
At a fluence rate of 1 W cm−2 of 400−600 nm photons delivered by the camera flash, it is estimated that about 106 photons are absorbed by a CRY molecule per second or 103 photons absorbed per millisecond. Because the average lifetime of the excited state of a type 1 CRY is about 0.5 ns (19), the probability of a biphotonic reaction (absorption of a photon by a molecule in a photochemically excited state) for type 1 is negligible (0.5 × 10−9 × 103 = 0.5 × 10−6). Therefore, any model proposed to explain the action mechanism of type 1 CRYs must take into account the fact that the photophysical/photochemical reaction necessary to initiate the proteolysis of type 1 CRYs must be completed within <1 ns. This time is far too short for a diffusion-controlled bimolecular reaction to take place even when the reactants are at millimolar concentrations. These considerations lead to the conclusion that the primary photochemical reaction of CRY is either intramolecular (conformational change or redox reaction) or intermolecular (energy or electron transfer) between cryptochrome and a highly abundant substrate that has high enough affinity for CRY such that at any given moment at least 80% of CRY must be in complex with its substrate. Considering the unlikelihood of having a reactant at high concentration to saturate the grossly overproduced CRY in transfected S2 cells, we conclude that the primary photochemical reaction in type 1 CRYs is most likely intramolecular.
Third, the type 4 CRYs are, so far, the only vertebrate CRYs purified with a stoichiometric amount of flavin and as such provide a unique opportunity to analyze vertebrate CRY photochemistry in vitro. From a photophysical standpoint, we find that type 4 CRYs exhibit characteristics more similar to photolyase than type 1 CRYs in that the excited state is dominated by a long-lived singlet with a lifetime of about 1 ns. Along these lines, while the FADox form of type 1 CRYs is photoreduced to FAD•−, the type 4 CRYs are photoreduced to FADH2 (or FADH−). Furthermore, when the FADH− form of type 4 CRY is incubated under aerobic conditions, it is first oxidized to the rather stable FADH• blue neutral radical before conversion to the two-electron-oxidized FADox form.
Lastly, type 4 CRYs do not undergo photoinduced proteolysis during their photocycle in vivo. In this regard type 4 CRYs are similar to Arabidopsis CRY1 which contains FADox after purification, is photoreduced to FADH−, and is reoxidized under aerobic conditions to FADH• first, before full oxidation to the FADox form. Importantly, Arabidopsis CRY1, like type 4 CRYs, does not undergo photoinduced proteolysis during its photosensory photocycle.
In Table 2, we compare some of the plant and animal CRYs that have been characterized to date with respect to their photosensory and transcriptional repressor functions, light-induced proteolytic degradation, and autokinase activity. Some generalizations can be made from this table. First, photoinduced proteolysis is not a general property of photosensory CRYs as AtCRY2 and DmCRY undergo light-induced proteolysis, but AtCRY1 whose photosensory activity is well characterized does not. Second, CRYs whose transcriptional repressor activities are well established such as HsCRY1 and HsCRY2 and type 2 insect CRYs have kinase activities, but some of the primarily photosensory CRYs such as AtCRY1 do while AtCRY2 does not. Third, it appears that the FAD cofactor is not needed for the kinase function because type 2 CRYs, which function as transcriptional inhibitors when expressed in Sf21 or S2 cells, do not bind measurable amounts of FAD but function as potent inhibitors of Clock:BMal1 complex (5). Finally, type 4 CRYs are unique among all of the animal CRYs characterized so far because they neither repress Clock:BMal1 nor undergo photoinduced proteolysis. The full complement of FAD in the type 4 CRYs we have analyzed and the unique expression patterns of these CRYs in zebrafish and chicken tissues that are photosensitive (6−10) are strong circumstantial evidence for a circadian photoreceptor function of these cryptochromes. It is expected that further work on these CRYs will contribute significantly to the development of a unified model for the cryptochrome photocycle.
Table 2. Comparison of Animal Cryptochromes Along with Arabidopsis CRYs for Photosensory Function, Presence of Stoichiometric FAD in Purified Recombinant Proteins, Inhibition of Clock:BMal1 Transactivation, Light-Induced Proteolysis, and Autokinase Activity.
| property | AtCRY1a | AtCRY2a | type 1 (DmCRY) | type 2 (DpCRY2) | type 4 (ZfCRY4) |
|---|---|---|---|---|---|
| photosensory function | + | + | + | ? | ? |
| stoichiometric flavin when purified from Sf21 cells | + | + | + | − | + |
| flavin radical by photoreduction | FADH0 | FADH0 | FAD0 | NAc | FADH0 |
| light-induced proteolysis | − | + | + | − | − |
| inhibition of Clock:BMal1 transactivation | + | − | ∓b | + | − |
| autokinase activity | + | − | − | + | − |
Arabidopsis cryptochromes, AtCRY1 and AtCRY2, are phylogenetically closer to the type 2 animal cryptochromes (5).
It appears that DmCRY functions as a repressor of Clock:Cycle (BMal1) in peripheral organs but not in the brain of Drosophila(42).
NA: not applicable since appreciable amounts of flavin are not bound to purified DpCRY2.
Acknowledgments
We thank N. Ozlem Arat (University of North Carolina) for help with the purification of GgCRY4, Steven M. Reppert (University of Massachusetts Medical School) for providing the plasmids carrying the cDNAs of DmCRY, DpCRY2, and the lac Z (β-galactosidase gene) in the pAc5.1 V5/HisA, and Carrie Partch (University of North Carolina School of Medicine) for the pcDNA4.ZfCRY4 vector used in our study.
This work was supported by NIH Grant GM 31082.
Funding Statement
National Institutes of Health, United States
Footnotes
Abbreviations: FAD, flavin adenine dinucleotide; MTHF, methenyltetrahydrofolate; CRY, cryptochrome; Phr, photolyase.
References
- Cashmore A. R. (2003) Cryptochromes: enabling plants and animals to determine circadian time. Cell 114, 537–543. [PubMed] [Google Scholar]
- Lin C.; Shalitin D. (2003) Cryptochrome structure and signal transduction. Annu. Rev. Plant Biol. 54, 469–496. [DOI] [PubMed] [Google Scholar]
- Sancar A. (2004) Regulation of the mammalian circadian clock by cryptochrome. J. Biol. Chem. 279, 34079–34082. [DOI] [PubMed] [Google Scholar]
- Stanewsky R.; Kaneko M.; Emery P.; Beretta B.; Wager-Smith K.; Kay S. A.; Rosbash M.; Hall J. C. (1998) The cryb mutation identifies cryptochrome as a circadian photoreceptor in Drosophila. Cell 95, 681–692. [DOI] [PubMed] [Google Scholar]
- Yuan Q.; Metterville D.; Briscoe A. D.; Reppert S. M. (2007) Insect cryptochromes: gene duplication and loss define diverse ways to construct insect circadian clocks. Mol. Biol. Evol. 24, 948–955. [DOI] [PubMed] [Google Scholar]
- Kobayashi Y.; Ishikawa T.; Hirayama J.; Daiyasu H.; Kanai S.; Toh H.; Fukuda I.; Tsujimura T.; Terada N.; Kamei Y.; Yuba S.; Iwai S.; Todo T. (2000) Molecular analysis of zebrafish photolyase/cryptochrome family: two types of cryptochromes present in zebrafish. Genes Cells 5, 725–738. [DOI] [PubMed] [Google Scholar]
- Kubo Y.; Akiyama M.; Fukada Y.; Okano T. (2006) Molecular cloning, mRNA expression, and immunocytochemical localization of a putative blue-light photoreceptor CRY4 in the chicken pineal gland. J. Neurochem. 97, 1155–1165. [DOI] [PubMed] [Google Scholar]
- Cermakian N.; Whitmore D.; Foulkes N. S.; Sassone-Corsi P. (2000) Asynchronous oscillations of two zebrafish CLOCK partners reveal differential clock control and function. Proc. Natl. Acad. Sci. U.S.A. 97, 4339–4344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zatz M. (1994) Photoendocrine transduction in cultured chick pineal cells: IV. What do vitamin A depletion and retinaldehyde addition do to the effects of light on the melatonin rhythm?. J. Neurochem. 62, 2001–2011. [DOI] [PubMed] [Google Scholar]
- Tu D. C.; Batten M. L.; Palczewski K.; Van Gelder R. N. (2004) Nonvisual photoreception in the chick iris. Science 306, 129–131. [DOI] [PubMed] [Google Scholar]
- Zhu H.; Yuan Q.; Briscoe A. D.; Froy O.; Casselman A.; Reppert S. M. (2005) The two CRYs of the butterfly. Curr. Biol. 15, R953–R954. [DOI] [PubMed] [Google Scholar]
- Selby C. P.; Sancar A. (2006) A cryptochrome/photolyase class of enzymes with single-stranded DNA-specific photolyase activity. Proc. Natl. Acad. Sci. U.S.A. 103, 17696–17700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozturk N.; Song S. H.; Selby C. P.; Sancar A. (2008) Animal type 1 cryptochromes. Analysis of the redox state of the flavin cofactor by site-directed mutagenesis. J. Biol. Chem. 283, 3256–3263. [DOI] [PubMed] [Google Scholar]
- Ozgur S.; Sancar A. (2006) Analysis of autophosphorylating kinase activities of Arabidopsis and human cryptochromes. Biochemistry 45, 13369–13374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnamoorthy R. R.; Agarwal P.; Prasanna G.; Vopat K.; Lambert W.; Sheedlo H. J.; Pang I. H.; Shade D.; Wordinger R. J.; Yorio T.; Clark A. F.; Agarwal N. (2001) Characterization of a transformed rat retinal ganglion cell line. Brain Res. Mol. Brain Res. 86, 1–12. [DOI] [PubMed] [Google Scholar]
- Gauger M. A.; Sancar A. (2005) Cryptochrome, circadian cycle, cell cycle checkpoints, and cancer. Cancer Res. 65, 6828–6834. [DOI] [PubMed] [Google Scholar]
- Song S. H.; Ozturk N.; Denaro T. R.; Arat N. O.; Kao Y. T.; Zhu H.; Zhong D.; Reppert S. M.; Sancar A. (2007) Formation and function of flavin anion radical in cryptochrome 1 blue-light photoreceptor of monarch butterfly. J. Biol. Chem. 282, 17608–17612. [DOI] [PubMed] [Google Scholar]
- Worthington E. N.; Kavakli I. H.; Berrocal-Tito G.; Bondo B. E.; Sancar A. (2003) Purification and characterization of three members of the photolyase/cryptochrome family glue-light photoreceptors from Vibrio cholerae. J. Biol. Chem. 278, 39143–39154. [DOI] [PubMed] [Google Scholar]
- Kao Y. T.; Tan C.; Song S. H.; Ozturk N.; Li J.; Wang L.; Sancar A.; Zhong D. (2008) Ultrafast dynamics and anionic active states of the flavin cofactor in cryptochrome and photolyase. J. Am. Chem. Soc. 130, 7695–7701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao Y. T.; Saxena C.; He T. F.; Guo L.; Wang L.; Sancar A.; Zhong D. (2008) Ultrafast dynamics of flavins in five redox states. J. Am. Chem. Soc. 130, 13132–13139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozgur S.; Sancar A. (2003) Purification and properties of human blue-light photoreceptor cryptochrome 2. Biochemistry 42, 2926–2932. [DOI] [PubMed] [Google Scholar]
- Kume K.; Zylka M. J.; Sriram S.; Shearman L. P.; Weaver D. R.; Jin X.; Maywood E. S.; Hastings M. H.; Reppert S. M. (1999) mCRY1 and mCRY2 are essential components of the negative limb of the circadian clock feedback loop. Cell 98, 193–205. [DOI] [PubMed] [Google Scholar]
- Vitaterna M. H.; Selby C. P.; Todo T.; Niwa H.; Thompson C.; Fruechte E. M.; Hitomi K.; Thresher R. J.; Ishikawa T.; Miyazaki J.; Takahashi J. S.; Sancar A. (1999) Differential regulation of mammalian period genes and circadian rhythmicity by cryptochromes 1 and 2. Proc. Natl. Acad. Sci. U.S.A. 96, 12114–12119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceriani M. F.; Darlington T. K.; Staknis D.; Mas P.; Petti A. A.; Weitz C. J.; Kay S. A. (1999) Light-dependent sequestration of timeless by cryptochrome. Science 285, 553–556. [DOI] [PubMed] [Google Scholar]
- Berndt A.; Kottke T.; Breitkreuz H.; Dvorsky R.; Hennig S.; Alexander M.; Wolf E. (2007) A novel photoreaction mechanism for the circadian blue light photoreceptor Drosophila cryptochrome. J. Biol. Chem. 282, 13011–13021. [DOI] [PubMed] [Google Scholar]
- Liu C.; Robertson D. E.; Ahmad M.; Raibekas A. A.; Jorns M. S.; Dutton P. L.; Cashmore A. R. (1995) Association of flavin adenine dinucleotide with the Arabidopsis blue light receptor CRY1. Science 269, 968–70. [DOI] [PubMed] [Google Scholar]
- Zhong D. (2007) Ultrafast catalytic processes in enzymes. Curr. Opin. Chem. Biol. 11, 174–181. [DOI] [PubMed] [Google Scholar]
- Sancar A. (2003) Structure and function of DNA photolyase and cryptochrome blue-light photoreceptors. Chem. Rev. 103, 2203–2237. [DOI] [PubMed] [Google Scholar]
- Sancar A. (2008) Structure and function of photolyase and in vivo enzymology: 50th anniversary. J. Biol. Chem. 283, 32153–32157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao Y. T.; Saxena C.; Wang L.; Sancar A.; Zhong D. (2005) Direct observation of thymine dimer repair in DNA by photolyase. Proc. Natl. Acad. Sci. U.S.A. 102, 16128–16132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naidoo N.; Song W.; Hunter-Ensor M.; Sehgal A. (1999) A role for the proteasome in the light response of the timeless clock protein. Science 285, 1737–1741. [DOI] [PubMed] [Google Scholar]
- Busza A.; Emery-Le M.; Rosbash M.; Emery P. (2004) Roles of the two Drosophila cryptochrome structural domains in circadian photoreception. Science 304, 1503–1506. [DOI] [PubMed] [Google Scholar]
- VanVickle-Chavez S. J.; Van Gelder R. N. (2007) Action spectrum of Drosophila cryptochrome. J. Biol. Chem. 282, 10561–10566. [DOI] [PubMed] [Google Scholar]
- Emery P.; So W. V.; Kaneko M.; Hall J. C.; Rosbash M. (1998) CRY, a Drosophila clock and light-regulated cryptochrome, is a major contributor to circadian rhythm resetting and photosensitivity. Cell 95, 669–679. [DOI] [PubMed] [Google Scholar]
- Sathyanarayanan S.; Zheng X.; Kumar S.; Chen C. H.; Chen D.; Hay B.; Sehgal A. (2008) Identification of novel genes involved in light-dependent CRY degradation through a genome-wide RNAi screen. Genes Dev. 22, 1522–1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peschel N.; Chen K. F.; Szabo G.; Stanewsky R. (2009) Light-dependent interactions between the Drosophila circadian clock factors cryptochrome, jetlag, and timeless. Curr. Biol. 19, 241–247. [DOI] [PubMed] [Google Scholar]
- Harm H.; Rupert C. S. (1968) Analysis of photoenzymatic repair of UV lesions in DNA by single light flashes. I. In vitro studies with Haemophilus influenzae transforming DNA and yeast photoreactivating enzyme. Mutat. Res. 6, 355–370. [DOI] [PubMed] [Google Scholar]
- Li Y. F.; Heelis P. F.; Sancar A. (1991) Active site of DNA photolyase: tryptophan-306 is the intrinsic hydrogen atom donor essential for flavin radical photoreduction and DNA repair in vitro. Biochemistry 30, 6322–6329. [DOI] [PubMed] [Google Scholar]
- Hoang N.; Schleicher E.; Kacprzak S.; Bouly J. P.; Picot M.; Wu W.; Berndt A.; Wolf E.; Bittl R.; Ahmad M. (2008) Human and Drosophila cryptochromes are light activated by flavin photoreduction in living cells. PLoS Biol. 6, e160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouly J. P.; Giovani B.; Djamei A.; Mueller M.; Zeugner A.; Dudkin E. A.; Batschauer A.; Ahmad M. (2003) Novel ATP-binding and autophosphorylation activity associated with Arabidopsis and human cryptochrome-1. Eur. J. Biochem. 270, 2921–2928. [DOI] [PubMed] [Google Scholar]
- Shalitin D.; Yu X.; Maymon M.; Mockler T.; Lin C. (2003) Blue light-dependent in vivo and in vitro phosphorylation of Arabidopsis cryptochrome 1. Plant Cell 15, 2421–2429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanchenko M.; Stanewsky R.; Giebultowicz J. M. (2001) Circadian photoreception in Drosophila: functions of cryptochrome in peripheral and central clocks. J. Biol. Rhythms 16, 205–215. [DOI] [PubMed] [Google Scholar]