SUMMARY
LDL delivers cholesterol to lysosomes by receptor-mediated endocytosis. Exit of cholesterol from lysosomes requires two proteins, membrane-bound Niemann-Pick C1 (NPC1) and soluble NPC2. NPC2 binds cholesterol with its isooctyl side chain buried and its 3ß-hydroxyl exposed. Here, we describe high-resolution structures of the N-terminal domain (NTD) of NPC1 and complexes with cholesterol and 25-hydroxycholesterol. NPC1(NTD) binds cholesterol in an orientation opposite to NPC2: 3ß-hydroxyl buried and isooctyl side chain exposed. Cholesterol transfer from NPC2 to NPC1(NTD) requires reorientation of a helical subdomain in NPC1(NTD), enlarging the opening for cholesterol entry. NPC1 with point mutations in this subdomain (distinct from the binding subdomain) cannot accept cholesterol from NPC2 and cannot restore cholesterol exit from lysosomes in NPC1-deficient cells. We propose a working model: after lysosomal hydrolysis of LDL-cholesteryl esters, cholesterol binds NPC2, which transfers it to NPC1(NTD), reversing its orientation and allowing insertion of its isooctyl side chain into the outer lysosomal membranes.
INTRODUCTION
Over the last 30 years, much has been learned about the receptor-mediated endocytosis of plasma low-density lipoprotein (LDL) in coated pits and its subsequent delivery to endosomes and lysosomes (Brown and Goldstein, 1986; Roth, 2006). Each LDL particle contains ∼500 molecules of free cholesterol and ∼1,500 molecules of esterified cholesterol that are hydrolyzed by acid lipase in the lumen of the lysosome (Goldstein et al., 1975). The liberated cholesterol must then exit the lysosomal compartment in order to reach the plasma membrane and the endoplasmic reticulum (ER) where it performs structural and regulatory roles, respectively (Demel and De Kruyff, 1976; Simons and Ikonen, 2000; Goldstein et al., 2006). A major unanswered question is how this exit process is accomplished.
Insight into two proteins required for cholesterol exit from lysosomes comes from Niemann-Pick Type C (NPC) disease (Pentchev et al., 1995), a fatal hereditary disorder characterized by the accumulation of cholesterol, sphingomyelin, and other lipids in endosomes and lysosomes. Evidence indicates that the primary cause is a failure of LDL-derived cholesterol to exit the lysosomes, which secondarily causes the buildup of other lipids. Mutations in either of two genes underlie NPC disease. Both genes encode lysosomal cholesterol-binding proteins. One gene encodes NPC1, a large 1278-amino acid polytopic membrane protein that is localized to the limiting membranes of endosomes and lysosomes (Carstea et al., 1997; Davies and Ioannou, 2000). The other gene encodes NPC2, a small soluble protein of 132 amino acids that resides in endosomes and lysosomes and is also secreted from the cell (Naureckiene et al., 2000). Homozygous mutations in either gene produce the same pattern of lysosomal lipid accumulation and the same clinical phenotype, providing strong genetic evidence that both proteins are required for cholesterol egress (Sleat et al., 2004).
NPC2 was shown previously to bind with high affinity to cholesterol, but not to cholesterol derivatives with hydrophilic substitutions on the isooctyl side chain, such as 25-hydroxycholesterol (25-HC) and 27-HC (Okamura et al., 1999; Friedland et al., 2003; Ko et al., 2003; Infante et al., 2008b). Xu et al. (2007) used x-ray crystallography to reveal the structure of bovine NPC2 in complex with cholesterol sulfate, which contains a sulfate in place of the 3β-hydroxyl group. Their data explained why hydrophilic substitutions on the isooctyl side chain prevent binding (Infante et al., 2008b). When cholesterol binds to NPC2, its isooctyl side chain is buried deep within a hydrophobic pocket. In contrast, the 3β-hydroxyl group is exposed on the surface. This exposure explains why a sulfate substitution (as in cholesterol sulfate) or a reversal of the orientation of the hydroxyl from 3β to 3α (as in epicholesterol) does not prevent binding. On the other hand, hydrophilic additions to the side chain, as in 25-HC, prevent burial of the side chain in the deep hydrophobic pocket.
NPC1 is much more complex than NPC2, and no high resolution structural information is available. NPC1 contains 13 predicted membrane-spanning helices and 3 large luminal domains (Davies and Ioannou, 2000). The first luminal domain is designated the N-terminal domain (NTD). It consists of ∼240 amino acids, which project into the lumen. This domain is hereafter designated NPC1(NTD). The other two large luminal domains are loops that span between transmembrane helices 2/3 and 8/9. We previously showed that full-length NPC1 binds [3H]cholesterol with nanomolar affinity (Infante et al., 2008a). Surprisingly, the binding site is localized to the soluble NTD (Infante et al., 2008b). In sharp contrast to NPC2, NPC1(NTD) binds not only cholesterol, but also its oxygenated derivatives, 25-HC and 27-HC. On the other hand, NPC1(NTD) does not bind sterols with modifications at the 3β-hydroxyl position such as cholesterol sulfate or epicholesterol. These data led to the suggestion that NPC1(NTD) binds cholesterol in an orientation opposite to that of NPC2 with the 3β-hydroxyl of cholesterol facing the interior of NPC1(NTD) and the isooctyl side chain exposed (Infante et al., 2008b).
In a subsequent study, we provided evidence that cholesterol could transfer between purified NPC1(NTD) and NPC2 in a bidirectional fashion (Infante et al., 2008c). When [3H]cholesterol was bound to NPC1(NTD) at 4°C, the dissociation rate into detergents was extremely slow, but the [3H]cholesterol transferred rapidly to NPC2. The transfer could also proceed in the opposite direction: NPC2 transferred its bound [3H]cholesterol to NPC1(NTD) two orders of magnitude faster than when [3H]cholesterol was delivered to NPC1(NTD) in detergent solution (Infante et al., 2008c). NPC2 can deliver bound cholesterol directly to liposomes even in the absence of NPC1 (Cheruku et al., 2006; Babalola et al., 2007; Xu et al., 2008; Infante et al., 2008c). However, NPC1(NTD) could not deliver its bound cholesterol to liposomes unless NPC2 was present as an intermediate carrier. Although these studies showed a unique ability of NPC2 to facilitate entry and exit of cholesterol from NPC1(NTD), they did not indicate which direction this transfer took within lysosomes, i.e., LDL to NPC2 to NPC1 vs. LDL to NPC1 to NPC2. Although the NPC2-to-NPC1 model seems more logical (Infante et al., 2008c; Subramanian and Balch, 2008; Schulze et al., 2009), direct data in support of this model are lacking.
In the current studies, we used x-ray crystallography to determine a high resolution structure of NPC1(NTD) in the apoprotein (apo) form and in complex with cholesterol or 25-HC. The protein contains a deep pocket that surrounds the sterol, burying the 3ß-hydroxyl group and the tetracyclic ring, but leaving the isooctyl side chain partially exposed. This orientation is opposite to the orientation of cholesterol bound to NPC2 in which the side chain is buried and the 3ß-hydroxyl exposed. Through mutational analysis, we identified two functional subdomains of NPC1(NTD) - one for sterol binding and the other for NPC2-mediated transfer. We use these data to advance a hypothetical working model for cholesterol transfer in which soluble NPC2 accepts cholesterol from LDL and carries it to the membrane-bound N-terminal domain of NPC1, which then inserts the cholesterol into the lysosomal membrane.
RESULTS
Crystal Structure of NPC1(NTD)
For structural studies, human NPC1(NTD) was produced in Hi-5 insect cells using a baculovirus vector. The soluble secreted protein was purified from the culture medium. Initial attempts at crystallization were hampered by heterogeneity attributable to the 5 potential N-linked glycosylation sites (4 of which are conserved in various species) and a proline-rich C-terminal region. After carrying out a systematic screen of mutations of these 4 conserved glycosylation sites in combination with C-terminal deletions, we obtained crystals from a construct that spanned residues 23-252 and harbored 3 mutations (N70Q, N122Q, N185Q) that removed 3 of the 4 conserved glycosylation sites. When assayed for its binding to [3H]cholesterol and [3H]25-HC, this protein exhibited binding kinetics similar to that of wild-type (WT) NPC1(NTD) produced in CHO-K1 cells (data not shown). Crystals were grown in the absence and presence of cholesterol or 25-HC (Figure 1A). The structure of NPC1(NTD) was determined by single wavelength anomalous dispersion (SAD) phasing (Table 1) and refined to 1.8 Å (apoprotein and cholesterol complex) and 1.6 Å (25-HC complex).
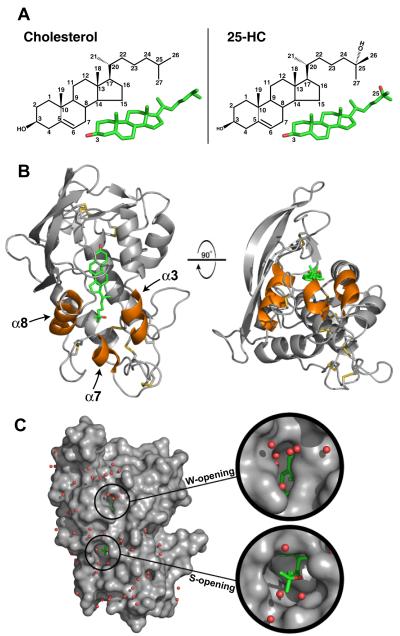
Figure 1. Structure of NPC1(NTD) Bound to 25-Hydroxycholesterol.
(A) Stick models of cholesterol (left) and 25-HC (right) with carbon positions numbered. Carbon and oxygen atoms are colored green and red, respectively.
(B) NPC1(NTD) is represented as a ribbon diagram in gray, and the disulfide bonds are shown in yellow. The positions of cholesterol and 25-HC are essentially identical. For simplicity, we show only the 25-HC molecule (colored in green). Helix3, helix7, and helix8 are colored orange.
(C) The surface of NPC1(NTD), colored in gray, reveals openings at either end of the bound sterol. The W-opening would allow passage of a single water molecule (red spheres), but not a sterol molecule. The S-opening would become large enough to allow entry or exit of a sterol if the opening were expanded slightly (see Figure 2D).
Table 1.
Data Collection and Refinement Statistics for NPC1(NTD)
| 25-HC | Cholesterol | Apo | SeMethionine | |
|---|---|---|---|---|
| Data Collection | ||||
| Space group | P64 | P64 | P64 | P64 |
| Cell dimensions (Å) | ||||
| a,b | 65.926 | 65.691 | 66.181 | 65.441 |
| c | 82.988 | 82.367 | 82.644 | 82.672 |
| Wavelength (Å) | 0.97940 | 0.97940 | 0.97940 | 0.97915 |
| Resolution (Å) (final shell) |
50 - 1.60 (1.63 - 1.60) |
50.0 - 1.80 (1.83 - 1.80) |
50 - 1.80 (1.83 - 1.80) |
50 - 2.00 (2.03 - 2.00) |
| Reflections | ||||
| Total | 113183 | 87312 | 85984 | 69646 |
| Unique | 26443 | 18504 | 18538 | 13189 |
| Completeness (%) | 97.8 (88.3) |
98.8 (94.0) |
98.1 (98.2) |
96.3 (78.0) |
| Rsym (%) | 3.1 | 5.9 | 8.0 | 7.6 |
| Refinement | ||||
| Rcryst (%) | 17.5 | 17.4 | 18.0 | |
| Rfree (%) | 20.3 | 20.7 | 22.1 | |
| rmsd | ||||
| Bond length (Å) | 0.012 | 0.012 | 0.012 | |
| Bond angle (°) | 1.476 | 1.477 | 1.374 | |
| Bave | ||||
| Protein (Å2) | 26.1 | 36.7 | 35.7 | |
| Ligand (Å2) | 22.9 | 43.6 | 54.6 | |
NPC1(NTD) is composed of 8 α-helices that are flanked by a 3-strand mixed β sheet (Figure 1B). NPC1(NTD) contains 18 cysteine residues, all of which form disulfide bonds (C25-C74, C31-C42, C63-C109, C75-C113, C97-C238, C100-C160, C177-C184, C227-C243, C240-C247). Electron density is present for 2 N-acetyl glucosamine molecules attached to Asn222 (Figure S1B), and they form hydrogen bonds with Glu110 and the carbonyl oxygen of Gly65 (Figure S1C). Electron density for an N-acetyl glucosamine is also present at a previously unanticipated site, Asn158 (sequence, NAC) (Figure S1A).
A comparison of the apoprotein (Figure 2A) and the sterol-bound forms (Figures 2B and 2C) reveals virtually no change in the structure of the protein upon sterol binding with a rmsd of 0.1 and 0.2 Å (apo vs. cholesterol and 25-HC, respectively) across all Cα atoms (Figure S2). For the three structures, electron density is visible for residues 23-247. Cholesterol and 25-HC are both clearly visible, but cholesterol is less well ordered than 25-HC (see B-factors in Table I).
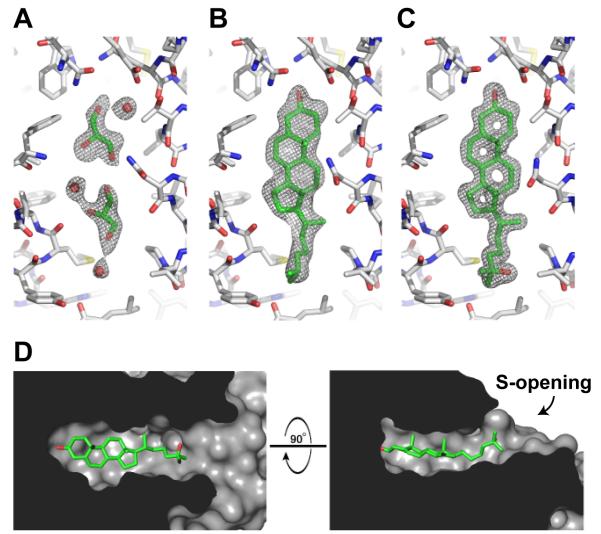
Figure 2. Sterol Binding Pocket.
(A-C) The sigmaA-weighted 2Fo-Fc electron density map contoured at 1σ within the sterol binding pocket is shown in gray mesh for the apoNPC1(NTD) (A), the cholesterol-bound (B), and 25-HC-bound (C) forms. The binding pocket is essentially identical in apoNPC1(NTD) and the sterol bound forms. The binding pocket of apoNPC1(NTD) is occupied by 2 glycerol and 3 water molecules. NPC1(NTD) is colored gray, bound ligands are colored green, and water molecules are shown as red spheres.
(D) The internal surface of the binding pocket is shown in gray. Bound 25-HC is shown in green.
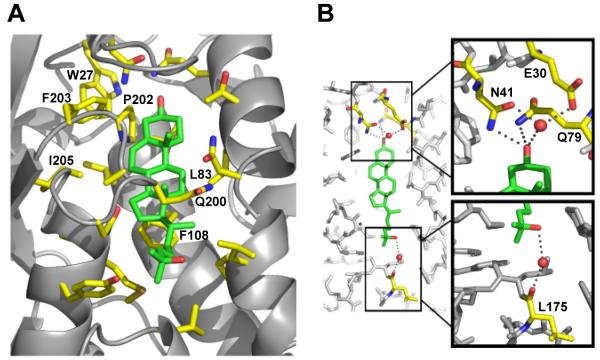
In the apo structure, the sterol binding pocket is occupied by 2 glycerol molecules and 3 water molecules (Figure 2A). The glycerol in the apo structure likely results from diffusion into the crystal during cryoprotection, as glycerol was not present during protein purification or crystallization. In the absence of sterols, the sterol binding pocket is likely lined with water molecules. At each end of the sterol binding pocket are openings toward solvent. One opening, located near the 3β-hydroxyl group of the sterol, is just large enough for a single water molecule to enter or exit (Figure 1C). We designate this region as the water opening (W-opening). The other opening, located at the end of the isooctyl side chain of the sterol molecule, is slightly larger, but not large enough to permit passage of the tetracyclic ring without a conformational change (Figures 1C and 2D). We designate this region as the sterol opening (S-opening). Cholesterol and 25-HC bind in a similar manner. The 25-hydroxyl group on 25-HC does not form a specific interaction with the protein, instead forming a water-mediated interaction with the main chain carbonyl of Leu175 (Figure 3B).
Figure 3. Expanded View of Sterol Binding Pocket.
(A) Residues that line the binding pocket are shown in yellow, and the sterol molecule is shown in green.
(B) Hydrogen bonds between residues of NPC1(NTD) and the hydroxyl groups of the sterol are denoted by gray dots. Residues involved in hydrogen bonds are colored yellow, and the sterol molecule is colored green. Nitrogen atoms are shown in blue, and oxygen atoms are shown in red. Water molecules are shown as red spheres.
Monomeric State of Soluble NPC1(NTD)
Based on size exclusion chromatography, soluble NPC1(NTD) was hypothesized to exist as a dimer (Infante et al., 2008b). Previous results with glycosylated proteins have shown anomalous migration behavior on size exclusion chromatography (Andrews, 1965). We used analytical ultracentrifugation sedimentation velocity (SV) analysis to determine the oligomerization state of NPC1(NTD) in solution. SV analysis of NPC1(NTD) produced in CHO-K1 cells with all 6 of the predicted or identified glycosylation sites intact showed a single species of 3.2 S corresponding to a molecular mass of 41.9 kDa (∼35% carbohydrate content by mass) (Figure S3). SV analysis of the protein produced in insect cells, with 3 of the glycosylation sites mutated, showed a single species of 2.2 S corresponding to a molecular mass of 30.3 kDa (∼15% carbohydrate content by mass) (Figure S3). Both results are consistent with NPC1(NTD) being a monomer in solution. Similar results were obtained from sedimentation equilibrium analysis (data not shown).
Sterol Binding Pocket
The sterol binding pocket of NPC1(NTD) is lined predominately with hydrophobic residues (Figure 3A), including Trp27, Leu83, Phe108, Pro202, Phe203, and Ile205, consistent with the hydrophobicity of cholesterol. The exceptions lie at the site of the 3β-hydroxyl group. Two polar residues, Asn41 and Gln79, form direct hydrogen bonds with the 3β-hydroxyl group (Figure 3B). In addition, Glu30 forms a water-mediated interaction with the 3β-hydroxyl, helping to stabilize binding of sterols and imposing stereospecificity. The residues in this region would not permit binding of the 3α-epimer of cholesterol (epicholesterol) or bulky additions at this position (Figure S4). These structural findings are consistent with our previous binding data showing that NPC1(NTD) does not bind epicholesterol or cholesterol sulfate (Infante et al., 2008b).
The binding pocket of NPC1(NTD) tightly encloses the four rings of the sterol. The pocket narrows around the C20 and C22 positions of the isooctyl side chain (Figure 2D) and begins to expand and open toward solvent after C23. The binding pocket is wider prior to C20, accommodating the larger size of the sterol tetracyclic ring. Our previous binding data showed that hydroxyl substitutions on the tetracyclic ring or on C20 diminish binding, while modifications on C24, C25, and C27 permit binding (Infante et al., 2008a; Infante et al., 2008b). These data are consistent with the size and shape of the binding pocket. The size and shape of the binding pocket restrict the entry and exit of sterols from NPC1(NTD) and would require expansion of the S-opening. Displacement of helix3, helix7, helix8 and/or the loop between helix8/strand7 would allow sterols to bind or dissociate (Figure 1B, orange).
Alanine Scan Mutagenesis to Identify Residues of NPC1(NTD) Required for Sterol Binding
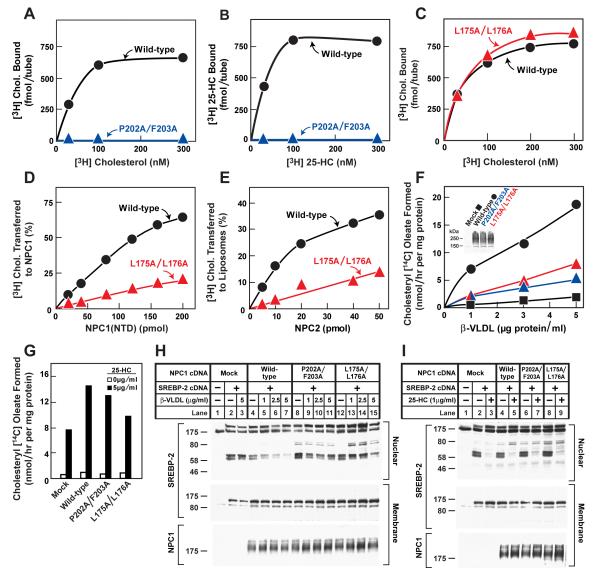
To determine key residues in NPC1(NTD) that are important for sterol binding, we performed an alanine scan mutagenesis across the protein (amino acids 23-264) and assayed each mutant for binding to [3H]cholesterol and [3H]25-HC (Figure S5). We generated a panel of 84 mutant plasmids in which 1, 2, or 3 contiguous amino acids were changed to alanines. This panel included all 242 residues in NPC1(NTD) except for the following: 4 conserved residues previously shown to have normal binding activity when replaced with alanine (Asn103, Gln117, Phe120, Tyr157) (Infante et al., 2008b); 1 conserved residue that when mutated to alanine (Gln79) showed reduced cholesterol binding (60% of normal) and virtually no 25-HC binding (Infante et al., 2008b); 6 naturally-occurring mutations that were shown previously to have normal binding activity (Gln92, Thr137, Pro166, Asn222, Asp242, Gly248) (Infante et al., 2008b); 18 cysteines; and the C-terminal 32 residues (amino acids 233-264), which when deleted do not decrease binding of [3H]cholesterol or [3H]25-HC (data not shown).
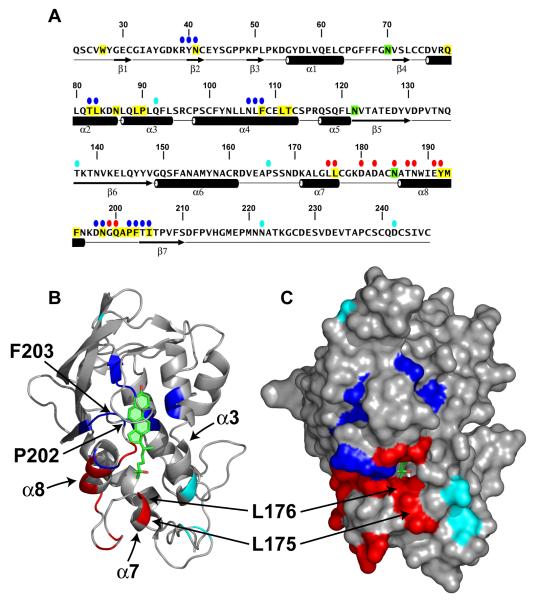
As described in Supplemental Experimental Procedures, we developed a rapid assay that allowed measurement of 3H-sterol binding to aliquots of concentrated media from CHO-K1 cells that had been transfected with plasmids encoding epitope-tagged WT or mutant versions of NPC1(NTD). The amount of secreted protein was determined by immunoblotting with an antibody directed against the FLAG epitope tag. Of the 84 mutants, 5 did not produce secreted protein (Y28-E30, L80/Q81, W189/I190, M193/F194, and V208/F209). Binding assays were performed by trapping the His-tagged proteins with bound 3H-sterol on nickel columns as described in Supplemental Experimental Procedures. Media from cells transfected with mock vector showed no binding of [3H]cholesterol or [3H]25-HC (<20 fmol/tube), whereas media from cells transfected with WT NPC1(NTD) showed 860-1500 fmol/tube and 780-1000 fmol/tube for [3H]cholesterol and [3H]25-HC, respectively (Figure S5B). Six of the 84 NPC1(NTD) mutant proteins showed binding that was less than 25% of WT (Figure S5B, blue). All of these mutations (R39-N41, T82/L83, N106-F108, D197/N198, P202/F203, and T204/I205) replace residues that map to the binding pocket (Figure 4, blue). These residues include the majority of the hydrophobic amino acids that line the binding pocket around the tetracyclic ring except for Trp27, whose replacement reduced [3H]cholesterol binding to a level slightly above the 25% threshold (Figure S5B). Of the two hydrophilic residues that form direct hydrogen bonds with the 3β-hydroxy group (Figure 3B), the replacement of Asn41 with alanine disrupted [3H]cholesterol binding by 90% (Figure S5B). The other hydrophilic residue (Gln79) was previously shown to produce a 40% reduction in cholesterol binding when replaced with alanine (Infante et al., 2008b). The only amino acid in the sterol binding domain that could not be evaluated in the alanine scan was Glu30; which forms a water-mediated bond with the 3ß-hydroxyl. This mutant protein was not secreted into the media. We identified 7 mutant NPC1(NTD) proteins that exhibited a preferential reduction of binding of [3H]25-HC as compared with [3H]cholesterol (Figure S5B, pink). All 7 of these mutations replaced amino acids that mapped either to the binding pocket (V26/W27 and G199/Q200) or to adjacent amino acids (F101/Y102, L144/Q145, Y146/Y147, T187/N188, and N195/K196) (Figure S7, pink).
Figure 4. Location of Functionally Important Residues in NPC1(NTD).
(A) Amino acid sequence of NPC1(NTD) with functionally important residues highlighted. Blue ovals denote residues that exhibit decreased binding of cholesterol and 25-HC by >75% when mutated to alanine. Red ovals denote residues that exhibit decreased transfer of cholesterol to liposomes by >70% when mutated to alanine. Cyan ovals denote naturally-occurring mutations in patients with NPC1 disease. Residues that line the binding pocket are shaded yellow. N-linked glycosylation sites that were eliminated are shaded green. The secondary structure of NPC1(NTD) is indicated below the sequence.
(B and C) Ribbon diagram (B) and surface representation (C) of NPC1(NTD), showing the positions of functionally important residues. Bound 25-HC is shown as a stick model in green. Color coding is the same as in (A). The location of the L175, L176, P202, and F203 residues are denoted by arrows.
To validate the major findings from the assays with unfractionated culture media, we purified the mutant protein with the least amount of cholesterol binding (P202A/F203A) and analyzed it in detail. As shown in Figures 5A and 5B, the purified protein showed no detectable binding of either [3H]cholesterol or [3H]25-HC.
Figure 5. Biochemical and Functional Analysis of Sterol Binding and Transfer Mutants.
(A-C) 3H-Sterol binding. Each reaction, in a final volume of 80 μl buffer C with 0.004% NP-40, contained 220 ng purified WT or mutant NPC1(NTD)-LVPRGS-His8-FLAG, 1 μg BSA, and indicated concentration of [3H]cholesterol (132 dpm/fmol) (A,C) or [3H]25-HC (165 dpm/fmol) (B). After incubation for 24 hr at 4°C, the amount of bound 3H-sterol was measured as described in Supplemental Experimental Procedures. Each value is the average of duplicate assays and represents total binding after subtraction of a blank value (10-70 fmol/tube). Mean variation for each of the duplicate assays in A, B, and C were 6.1%, 5.0%, and 5.0%, respectively.
(D) [3H]Cholesterol transfer from NPC2 to NPC1(NTD). Each reaction, in a final volume of 200 μl buffer D (pH 5.5) without detergent, contained ∼40 pmol of donor protein NPC2-FLAG complexed to [3H]cholesterol (830 fmol, 132 dpm/fmol) and increasing concentrations of purified WT or mutant NPC1(NTD)-LVPRGS-His8-FLAG acceptor protein. After incubation for 15 min at 4°C, the amount of [3H]cholesterol transferred to NPC1(NTD) was measured by Ni-NTA-agarose chromatography as described in the [3H]cholesterol transfer assay in Experimental Procedures. Each value is the average of duplicate assays and represents percentage of [3H]cholesterol transferred to NPC1(NTD). The 100% value for transfer from NPC2 was 830 fmol/tube. Mean variation for each of the duplicate assays for WT and mutant were 7.9% and 8.2%, respectively
(E) [3H]Cholesterol transfer from NPC1(NTD) to liposomes as a function of NPC2. Each reaction, in final a volume of 200 μl buffer D (pH 5.5) without detergent, contained ∼50 pmol of WT or L175A/L176A versions of NPC1(NTD)-LVPRGS-His8-FLAG, each complexed to [3H]cholesterol (950 and 660 fmol, respectively; 132 dpm/fmol); 20 μg PC liposomes labeled with Texas red dye; and increasing concentrations of NPC2-His10. After incubation for 10 min at 4°C, the amount of [3H]cholesterol transferred to liposomes was measured in the flow-through of the nickel column as described for the [3H]cholesterol transfer assay in Experimental Procedures. Each value is the average of duplicate assays and represents the percentage of [3H]cholesterol transferred to liposomes. Blank values in the absence of NPC2 (5-6% transfer) were subtracted. The 100% values for transfer from WT and L175A/176A versions of NPC1(NTD) were 950 and 660 fmol/tube, respectively. Mean variation for each of the duplicate assays for WT and mutant were 8.8% and 9.3%, respectively.
(F-I) Cholesterol esterification and SREBP-2 processing in mutant CHO cells lacking NPC1 function transfected with NPC1 cDNAs. Mutant CHO 4-4-19 cells were set up for experiments and transfected with 2 μg pcDNA3.1 or with WT or mutant versions of pCMV-NPC1-His8-FLAG (F,G); or co-transfected with 0.4 μg pcDNA3.1 or with WT or mutant versions of pTK-NPC1-His8-FLAG3 plus 3 μg pTK-HSV-BP2 (H,I) as described in Experimental Procedures. 24 hr after transfection, the medium was switched to medium A containing 5% newborn calf lipoprotein-deficient serum, 5 μM compactin, and 50 μM sodium mevalonate. After incubation for 24 hr, the medium was switched to the same medium containing 50 μM compactin and various concentrations of ß-VLDL (F,H) or 25-HC (G,I) as indicated.
(F,G) Cholesterol esterification. After incubation for 5 hr at 37°C, each cell monolayer was pulse-labeled for 1 hr with 0.2 mM sodium [14C]oleate (6301 dpm/pmol). The cells were then harvested for measurement of their content of cholesteryl [14C] oleate and [14C] triglycerides. Each value is the average of duplicate incubations. Mean variation for each of the duplicate incubations for WT, P202A/F203A, and L175A/L176A were 9.6%, 14.5%, and 4.3%, respectively. The rate of synthesis of [14C]triglycerides for mock, NPC1 WT, NPC1(P202A/F203A), and NPC1(L175A/L176A) transfected cells incubated with 5 μg/ml ß-VLDL was 340, 396, 352, and 365 nmol/hr per mg protein, respectively. The rate of synthesis of [14C]triglycerides incubated with 25-HC was 347, 496, 304, and 434 nmol/hr per mg protein, respectively.
(H,I) SREBP-2 processing. After incubation for 4 hr at 37°C, cells received a direct addition of 25 μg/ml of N-acetyl-leucinal-leucinal norleucinal. After 1 hr, triplicate dishes were harvested and pooled for preparation of nuclear extracts and 100,000 g membrane fractions, which were analyzed by immunoblotting for the indicated protein. The concentrations of antibodies were 0.2 and 4 μg/ml for SREBP-2 (anti-HSV) and NPC1 (anti-FLAG), respectively. All filters were exposed on x-ray film for 2-10 s.
(A-I) Similar results were obtained in 3 or more independent experiments.
Alanine Scan Mutagenesis to Identify Residues Required for NPC2-mediated Transfer
Previously, we showed that NPC2 catalyzes the bidirectional transfer of cholesterol between NPC1(NTD) and liposomes in vitro (Infante et al., 2008c). To test the amino acids in NPC1(NTD) that are important for this action, we performed a transfer assay with all of the alanine scan mutants except for the 8 mutants whose [3H]cholesterol binding activity was less than 50% of the WT. For this assay, we prepared stable complexes of [3H]cholesterol:NPC1(NTD) and purified them by immunoaffinity chromatography as described in Supplemental Experimental Procedures. We then measured the transfer of the bound [3H]cholesterol to liposomes in the presence and absence of purified NPC2. In the absence of NPC2, less than 8% of bound [3H]cholesterol from WT NPC1(NTD) was transferred to liposomes. In the presence of NPC2, WT NPC1(NTD) transferred an additional 54-90% of bound [3H]cholesterol in 3 experiments (Figure S6B, shaded region) with an average of 80%. Six mutant NPC1(NTD) proteins transferred less than 25% of their bound [3H]cholesterol to liposomes in an NPC2-dependent manner (Figures 4 and S6B, red). These mutations replaced residues L175/L176, D180/D182, N185, T187/N188, E191/Y192, and G199/Q200, all of which map to a surface of a subdomain of NPC1(NTD) spanning amino acids 162-200. This region includes helices 7 and 8 and the intervening loop (Figure 4, red). Of the 6 mutants in this region, the two most deficient in transfer are L175A/L176A and E191A/Y192A. Two of these 4 WT residues (L176 and Y192) are identical in 12 out of 12 mammalian species, and the other two (L175 and E191) are identical in 11 of the 12 species (Infante et al., 2008b).
To validate the findings from the above transfer assays done with culture media, we purified one of the mutants, L175A/L176A, that showed a marked decrease in the amount of NPC2-dependent [3H]cholesterol transfer from NPC1(NTD) to liposomes. When incubated with [3H]cholesterol for 24 hr at 4°C, a time necessary to achieve equilibrium binding in the absence of NPC2 (Infante et al., 2008c), the mutant protein bound cholesterol with similar affinity to that of the WT protein (Figure 5C). Moreover, its rates of association and dissociation of [3H]cholesterol at 4°C and 37°C were virtually identical to those of WT NPC1(NTD) (Figure S8). Equilibrium binding for both WT and mutant proteins was achieved within 30 min at 37°C, and dissociation of previously bound [3H]cholesterol for both proteins occurred rapidly at 37°C and extremely slowly at 4°C (Figure S8).
Despite its normal binding kinetics, the mutant protein showed a marked defect in NPC2-stimulated transfer of cholesterol to liposomes (Figure 5E). As compared to WT NPC1(NTD), the mutant protein required 5 times more NPC2 to transfer an equivalent amount of [3H]cholesterol to liposomes. To show that this transfer defect is the result of defective interaction with NPC2 and not with liposomes, we tested the ability of NPC2 to transfer its cholesterol to WT NPC1(NTD) and to this mutant. Within 15 min, NPC2 transferred 4-fold more [3H]cholesterol to WT NPC1(NTD) as compared to the L175A/L176A mutant (Figure 5D).
Cholesterol Binding and Transfer Mutants Fail to Restore Function to NPC1-deficient Cells
To assay the function of mutant NPC1 proteins, we measured the two reactions that occur when cholesterol or oxysterols reach the ER: 1) activation of acyl-CoA:cholesterol acyltransferase (ACAT), thereby increasing incorporation of radiolabeled fatty acids into cholesteryl esters (Pentchev et al., 1995; Liscum and Faust, 1987); and 2) inhibition of the proteolytic processing of sterol regulatory element-binding proteins (SREBPs), thereby inactivating transcription of genes for cholesterol synthesis and uptake (Brown and Goldstein, 1997; Infante et al., 2008a).
Figure 5F shows an experiment in which we transfected plasmids encoding full-length WT and mutant NPC1 into CHO 4-4-19 cells. These cells lack NPC1 function as a result of a point mutation (G660R) in transmembrane helix 3 (L. Liscum, personal communication; Dahl et al., 1992; Infante et al., 2008b). 48 hr after transfection, the cells were incubated for 5 hr with varying concentrations of β-VLDL, a cholesterol-rich lipoprotein that binds LDL receptors and delivers cholesterol to lysosomes. (Because of its high affinity for the LDL receptor, ß-VLDL is frequently used as a ligand in cell culture studies (van Driel et al., 1987)). After incubation with ß-VLDL, the cells then received [14C]oleate and the incorporation into cholesteryl [14C]oleate was measured. When transfected with a control (mock) plasmid, the CHO 4-4-19 cells showed little cholesteryl [14C]oleate formation. Expression of WT NPC1 allowed β-VLDL to stimulate cholesteryl oleate formation by 20-fold (Figure 5F). Full-length NPC1 harboring the P202A/F203A mutation in the NTD was markedly defective in restoring sensitivity to β-VLDL (Figure 5F, blue triangles). Similar defects were seen when full-length NPC1 contained the L175A/L176A mutation (red triangles). None of the mutant proteins interfered with the ability of 25-HC to stimulate cholesteryl ester formation, a response that is normal in the NPC1 mutant cells (Figure 5G).
Immunoblotting revealed that the mutant proteins were expressed at the same level as WT (Figure 5F, inset). Both mutant and WT proteins showed a diffuse band characteristic of fully glycosylated lysosomal proteins that have left the ER (Watari et al., 1999). Moreover, treatment of the mutant proteins with the glycosidase Endo H (which removes only high mannose chains) did not alter their migration pattern on SDS-PAGE, providing further evidence that the mutant proteins had folded properly and left the ER (data not shown). In the same experiment, Endo H treatment of a control protein containing a high mannose chain (Site-1 protease) (DeBose-Boyd et al., 1999) showed the expected increase in electrophoretic mobility.
To study the inhibition of SREBP processing, we employed a standard protocol in which CHO 4-4-19 cells were deprived of sterols and then incubated with β-VLDL for 5 hr after which membrane and nuclear extracts were subjected to immunoblotting to detect the full-length precursor and the processed nuclear form of SREBP-2. Mock-transfected CHO 4-4-19 cells failed to suppress SREBP-2 cleavage when incubated with β-VLDL (Figure 5H; lanes 2,3). Suppression was restored by transfection with a plasmid encoding WT NPC1 (lanes 4-7), but not with plasmids encoding the binding-defective mutant (lanes 8-11) or transfer-defective mutant (lanes 12-15). None of these plasmids affected suppression by 25-HC, which is normal in NPC1 deficient cells (Figure 5I).
DISCUSSION
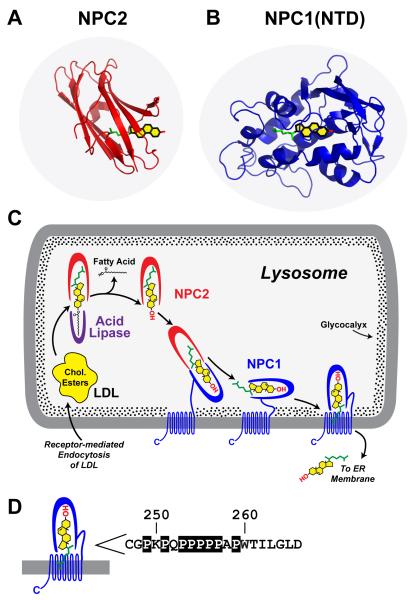
Considered together with previous studies of NPC1 and NPC2 (Xu et al., 2007; Infante et al., 2008a; Infante et al., 2008b; Infante et al., 2008c), the current studies permit us to formulate a hypothetical working model to explain the requirement for these two proteins in the egress of lipoprotein-derived cholesterol from lysosomes (see Figure 6C). The major consideration in formulating this model is the striking reversal in the orientation of cholesterol when it transfers between NPC2 and NPC1.
Figure 6. Model for Egress of Lipoprotein-derived Cholesterol from Lysosomes.
(A) Structure of NPC2 bound to cholesterol. Redrawn from Xu, et al. (2007).
(B) Structure of NPC1(NTD) bound to cholesterol.
(C) Proposed pathway for transfer of cholesterol from LDL or ß-VLDL to NPC2 to NPC1 to membranes. See Discussion for explanation of this working model.
(D) Sequence of amino acids 247-266 that link the NTD to the first transmembrane domain in NPC1 (Davies and Ioannou, 2000). Prolines in this sequence are boxed. These prolines are invariant in 12 vertebrate species (Infante et al., 2008b) except for P256, which is conserved in 8 of the 12 species.
As described previously by Xu et al. (2007), cholesterol binds to NPC2 with its 3β-hydroxyl group exposed to solvent (Figure 6A). The exposure of the 3ß-hydroxyl allows NPC2 to bind lipoprotein-derived cholesterol either before or immediately after the fatty acid on the hydroxyl group is removed by acid lipase. Even though NPC2 can deliver cholesterol directly to liposomes in vitro as mentioned in the Introduction, genetics reveals that NPC2 alone is not sufficient for cholesterol egress from lysosomes, and hence the requirement for NPC1 (Pentchev, 2004; Sleat et al., 2004). We hypothesize that the NPC1 requirement may be imposed to overcome either of two impediments: 1) cholesterol should enter the hydrophobic lysosomal membrane most readily when its hydrophobic isooctyl side chain leads the way. When cholesterol is bound to NPC2, the hydrophobic side chain is deeply buried in the protein and it cannot lead the way into the lysosomal membrane bilayer; and/or 2) the carbohydrate glycocalyx that lines the interior of the lysosomal membrane (Neiss, 1984) creates a diffusion barrier that would prevent NPC2 from interacting directly with the membrane (Kolter and Sandhoff, 2005; Schulze et al., 2009). Both of these impediments could be overcome if NPC2 transferred its cholesterol to NPC1(NTD). This would reverse the orientation of cholesterol so that its hydrophobic side chain could lead the way into the membrane, and it likely provides a mechanism for cholesterol to transit the glycocalyx.
The model of Figure 6C is supported by our previously published in vitro data showing that [3H]cholesterol binds to and dissociates from NPC1(NTD) extremely slowly and that both processes are accelerated in the presence of NPC2 (Infante et al., 2008c). As is apparent from the NPC1(NTD) crystal structure, entry or exit of cholesterol from NPC1(NTD) requires enlargement of the S-opening, which could be accomplished by displacement of the loop between helix8/strand7 and/or a shift in the position of helix3, helix7, or helix8 (Figures 1B, orange). Mutagenesis studies support the involvement of residues near the S-opening, but not the W-opening in the NPC2-facilitated transfer of cholesterol. Alanine substitutions at multiple sites in helix7, helix8, and the helix8/strand7 loop decreased the ability of NPC2 to open the NPC1(NTD) binding site as indicated by a reduced ability of NPC2 to facilitate the transfer of NPC1(NTD)-bound [3H]cholesterol to liposomes (Figures 4 and S6, red). This phenotype was not observed with mutations in helix3 or any other site in the protein. These data suggest that NPC2 interacts with NPC1(NTD) to elicit a rearrangement in helix7, helix8, and the intervening loop between helix8/strand7, thereby expanding the S-opening and allowing cholesterol to enter or exit from the binding pocket of NPC1(NTD). Helix3 may also contribute to the expansion of the S-opening. Comparison of the apo, cholesterol-bound, and 25-HC-bound structures shows that a slight displacement of helix3 is possible with the largest shift (∼1Å) occurring at Asn86/Leu87 (Figure S2). Exit through the S-opening is consistent with the idea that the hydrophobic side chain of cholesterol is the first part of the sterol to leave NPC1(NTD), thereby allowing it to insert into the lysosomal membrane.
The cell culture data of Figure 5 support the notion that transfer of cholesterol between NPC1 and NPC2 is required for exit of lipoprotein-derived cholesterol from lysosomes. When the L175A/L176A mutation was introduced into full-length NPC1, the protein could not restore egress of cholesterol from lysosomes to a normal degree as judged by a failure of normal cholesterol esterification (Figure 5F) and suppression of SREBP-2 cleavage (Figure 5H) in the presence of β-VLDL. These data provide strong support for the notion that cholesterol must be transferred between NPC2 and NPC1(NTD) in order to exit the lysosome. However, the result does not in itself determine that the transfer goes in the direction of NPC2-to-NPC1. In our model, this directionality is hypothesized in order to overcome the two impediments discussed above, i.e., the directionality of cholesterol insertion into the bilayer and the necessity to transit the glycocalyx.
The NTD is separated from the membrane domain of NPC1 by a linker composed of ∼20 amino acids, 8 of which are prolines (Figure 6D). It is likely that this sequence extends in such a way that the NTD domain can project through the glycocalyx, which has been measured at ∼8 nm (Neiss, 1984). This glycocalyx is shown as stipples in Figure 6C. In order to deliver cholesterol to the membrane, the NTD would have to move toward the membrane. Moreover, another protein would have to enlarge the S-opening on NPC1(NTD) in order for cholesterol to leave the NTD and enter the membrane. It is likely that these processes are mediated by the membrane domain of NPC1 which contains 13 transmembrane helices and two large loops that project into the lysosomal lumen. Five of the 13 transmembrane helices (no. 3-7) bear sequence homology to the sterol-sensing domains of Scap, a polytopic membrane protein in the SREBP pathway that has been shown to bind cholesterol with high affinity (Radhakrishnan et al., 2004). It seems likely that the sterol-sensing domain of NPC1 participates in cholesterol transfer. In Figure 6C, we show the NPC1(NTD) interacting with the membrane domain of the same protein. However, we have not ruled out the possibility that the NPC1(NTD) transfers the cholesterol directly to the membrane.
In transferring its bound cholesterol to the lysosomal membrane, the NTD of NPC1 could interact either with its own membrane domain (as shown in Figure 6C) in which case it might transfer the cholesterol to the putative sterol-sensing domain in transmembrane helices 3-7, or it could interact with the membrane domain of a neighboring NPC1 molecule. In this regard, Ohgami et al. (Ohgami et al., 2004) reported that they could crosslink photoactivated cholesterol to NPC1 when the photoactivated cholesterol was added to the culture medium of WT CHO cells. The reaction was reduced when the cells harbored mutant NPC1 with a point mutation in transmembrane helix3, which is part of the sterol-sensing domain. Once cholesterol is transferred to the membrane domain of NPC1, it could be flipped to the cytosolic leaflet from which it could then be picked up by cytosolic cholesterol-binding proteins. It seems likely that additional proteins would be required for the export process. Clues may come from study of patients with the NPC disease phenotype who do not have mutations in NPC1 or NPC2.
One caveat to the model of Figure 6C arises because we have thus far been unable to demonstrate a stable complex between NPC1(NTD) and NPC2. Therefore, the postulated interaction between these two proteins might be transient. We are currently exploring methods to capture such transient interactions using NMR, chemical crosslinkers, and fluorescence energy transfer methods.
Although the model of Figure 6 still remains to be proven, it offers a scheme that can be tested by further experiments using in vitro biochemistry and cell culture methodologies. It is hoped that testing of this model may lead to new insights into a fundamental biologic process - namely, how lipoprotein-derived cholesterol is transported out of the lysosomal compartment so that it can exert its structural and regulatory functions within the cell. Understanding this process hopefully will shed new light on a devastating disease.
EXPERIMENTAL PROCEDURES
Expression and Purification of NPC1(NTD) for Crystallization
Sf9 cells infected with His6-ENLYFQGA-NPC1(23-252)(N70Q/N122Q/N185Q) baculovirus (Wasilko and Lee, 2006) were used to infect Hi-5 cells at 1×106 cells/ml in Excel-405 medium. After incubation for 96 hr at 27°C, cells were pelleted by centrifugation, and the medium was concentrated by tangential flow filtration and exchanged into buffer A. The concentrated medium from 6 liters of culture was applied to a 10 ml Ni-NTA column equilibrated in buffer A, washed sequentially with buffer A containing 5 mM and then 25 mM imidazole, and eluted with buffer A containing 250 mM imidazole. Fractions containing NPC1(NTD) were pooled, concentrated, and buffer-exchanged into buffer B, after which 0.9 mg/ml of His-tagged TEV protease was added, and the mixture was gently rocked overnight at 21°C. Following removal of the His-tag from His6-ENLYFQGA-NPC1(23-252)(N70Q/N122Q/N185Q), the mixture was applied to a 10 ml Ni-NTA column, and the flow-through was directly applied to a MonoQ column equilibrated in buffer B. NPC1(NTD) was eluted using a linear gradient from 50 to 500 mM NaCl, and fractions containing NPC1(NTD) were pooled and concentrated. The concentrated NPC1(NTD) was then applied to a Superdex 200 gel filtration column equilibrated with buffer containing 10 mM Tris-chloride (pH 7.5), 50 mM NaCl, and 0.01% NaN3. Fractions containing NPC1(NTD) were pooled and concentrated to 30 mg/ml prior to crystallization.
Selenomethionine-containing NPC1(NTD) was generated in TNi-pro cells (Expression Systems) adapted to grow in ESF-921 methionine-free medium. At 16 and 40 hr after baculoviral infection with His6-ENLYFQGA-NPC1(23-252)(N70Q/N122Q/N185Q), the medium was supplemented with 100 μg/ml L-selenomethionine. Selenomethionine-containing NPC1(NTD) was purified as described above for the unlabeled protein.
Crystallization and Structure Determination
Prior to crystallization, cholesterol or 25-HC, at a final concentration of 1 mM in 5% (v/v) ethanol, was incubated with NPC1(NTD) overnight at 4°C. Initial crystals were obtained using the Fluidigm TOPAZ system. Crystals of apoNPC1(NTD) or the cholesterol-bound and 25-HC-bound forms were produced by mixing protein at 30 mg/ml with an equal volume of reservoir solution containing 25% (w/v) polyethylene glycol 1500, 100 mM MES acid, and 30 mM glycine at pH 3.25 (final pH, ∼4.0). Crystals were transferred stepwise into reservoir solution containing 25% (v/v) glycerol and soaked overnight at 21°C. Crystals were flash-frozen in a - 160°C nitrogen stream. The crystals belong to space group P64. Diffraction data were collected at the Advanced Photon Source (Argonne, IL) beam line 19-ID and 19-BM and processed with HKL2000 (Otwinowski and Minor, 1997) and the CCP4 suite (Collaborative Computation Project, 1994). Four selenium sites were found from SAD data using ShelXD (Schneider and Sheldrick, 2002), and the resulting maps were improved by density modification using DM (Cowtan, 1994). The model was built with the program COOT (Emsley and Cowtan, 2004). Initial refinement was performed with CNS (Brunger et al., 1998), and the final cycles of refinement were performed with REFMAC (Winn et al., 2001). Figures were generated with PYMOL (www.pymol.org).
[3H]Cholesterol Transfer Assays for Purified NPC Proteins
The transfer assays, including methods for isolation of complexes of [3H]cholesterol bound to NPC1(NTD) or NPC2 and for preparation of liposomes, have been previously described (Infante et al., 2008c). For transfer assays of NPC1(NTD) to liposomes, incubation conditions are described in Figure 5E. After incubation, the 200-μl mixture was diluted with 750 μl of buffer C and loaded onto a 2-ml column packed with 0.3 ml of Ni-NTA-agarose beads preequilibrated with buffer C. Each column was washed with 1 ml of buffer C. The amount of [3H]cholesterol transferred to liposomes was quantified by scintillation counting of the combined flow-through and wash fractions. For transfer assays of NPC2 to NPC1(NTD), incubation conditions are described in Figure 5D. After incubation, each mixture was processed as above except that the column was washed with 3 ml of buffer C and eluted with 250 mM imidazole, after which the eluate was subjected to scintillation counting.
Assays for Cholesterol Esterification and SREBP-2 Processing in Cultured Cells
Mutant CHO 4-4-19 cells, defective in NPC1 function (Dahl et al., 1992), were transfected and tested for incorporation of [14C]oleate into cholesteryl [14C]oleate and proteolytic processing of SREBP-2 as previously described (Infante et al., 2008b). For assays of cholesterol esterification, cells were transfected with 2 μg pcDNA3.1 or pCMV-NPC1-His8-FLAG (wild-type or mutant versions). For assays of SREBP-2 processing, cells were co-transfected with 0.5 μg pcDNA3.1 or pTK-NPC1-His8-FLAG3 (wild-type or mutant versions) plus 3 μg pTK-HSV-BP2. Incubation conditions are described in the legend to Figure 5.
Supplementary Material
ACKNOWLEDGMENTS
We thank Dorothy Goddard, Lisa Henry, Bethany Cartwright, and Maya Palnitkar for excellent technical assistance; Lisa Beatty and Shomanike Head for invaluable help with tissue culture; Dr. Chad Brautigam for help with analytical ultracentrifugation; and Dr. Laura Liscum for the gift of CHO 4-4-19 cells. This work was supported by grants from the National Institutes of Health (HL20948), Welch Foundation (I-1185), and Perot Family Foundation. R.E.I. and M.L.W. are supported by Medical Scientist Training Program Grant 5T32 (GM08014). R.E.I. is also supported by the Ara Parseghian Medical Research Foundation. J.D. is an Investigator of the Howard Hughes Medical Institute. Crystallographic structures were derived from work performed at Argonne National Laboratory, Structural Biology Center at the Advanced Photon Source (operated by UChicago Argonne, LLC, for the U.S. Department of Energy, Office of Biological and Environmental Research under Contract DE-AC02-06CH11357).
Accession Numbers
The atomic coordinates have been deposited in the Protein Data Bank with accession codes 3GKH (apoNPC1(NTD)), 3GKI (cholesterol-bound), and 3GKJ (25-HC-bound).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Andrews P. The gel-filtration behaviour of proteins related to their molecular weights over a wide range. Biochem. J. 1965;96:595–606. doi: 10.1042/bj0960595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babalola JO, Wendeler M, Breiden B, Arenz C, Schwarzmann G, Locatelli-Hoops S, Sandhoff K. Development of an assay for the intermembrane transfer of cholesterol by Niemann-Pick C2 protein. Biol. Chem. 2007;388:617–626. doi: 10.1515/BC.2007.063. [DOI] [PubMed] [Google Scholar]
- Brown MS, Goldstein JL. A receptor-mediated pathway for cholesterol homeostasis. Science. 1986;232:34–47. doi: 10.1126/science.3513311. [DOI] [PubMed] [Google Scholar]
- Brown MS, Goldstein JL. The SREBP pathway: Regulation of cholesterol metabolism by proteolysis of a membrane-bound transcription factor. Cell. 1997;89:331–340. doi: 10.1016/s0092-8674(00)80213-5. [DOI] [PubMed] [Google Scholar]
- Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 1998;D54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- Carstea ED, Morris JA, Coleman, et al. Niemann-Pick C1 disease gene: Homology to mediators of cholesterol homeostasis. Science. 1997;277:228–231. doi: 10.1126/science.277.5323.228. [DOI] [PubMed] [Google Scholar]
- Cheruku SR, Xu Z, Dutia R, Lobel P, Storch J. Mechanism of cholesterol transfer from the Niemann-Pick type C2 protein to model membranes supports a role in lysosomal cholesterol transport. J. Biol. Chem. 2006;281:31594–31604. doi: 10.1074/jbc.M602765200. [DOI] [PubMed] [Google Scholar]
- Collaborative Computation Project, N.4 The CCP4 suite: Programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 1994;D50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- Cowtan K. Joint CCP4 and ESF-EACBM Newsletter on Protein Crystallography. 1994;31:34–38. [Google Scholar]
- Dahl NK, Reed KL, Daunais MS, Faust JR, Liscum L. Isolation and characterization of Chinese hamster ovary cells defective in the intracellular metabolism of low density lipoprotein-derived cholesterol. J. Biol. Chem. 1992;267:4889–4896. [PubMed] [Google Scholar]
- Davies JP, Ioannou YA. Topological analysis of Niemann-Pick C1 protein reveals that the membrane orientation of the putative sterol-sensing domain is identical to those of 3-hydroxy-3-methylglutaryl-CoA reductase and sterol regulatory element binding protein cleavage-activating protein. J. Biol. Chem. 2000;275:24367–24374. doi: 10.1074/jbc.M002184200. [DOI] [PubMed] [Google Scholar]
- DeBose-Boyd RA, Brown MS, Li W-P, Nohturfft A, Goldstein JL, Espenshade PJ. Transport-dependent proteolysis of SREBP: Relocation of Site-1 protease from Golgi to ER obviates the need for SREBP transport to Golgi. Cell. 1999;99:703–712. doi: 10.1016/s0092-8674(00)81668-2. [DOI] [PubMed] [Google Scholar]
- Demel RA, De Kruyff B. The function of sterols in membranes. Biochim. Biophys. Acta. 1976;457:109–132. doi: 10.1016/0304-4157(76)90008-3. [DOI] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;D60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Friedland N, Liou H-L, Lobel P, Stock AM. Structure of a cholesterol-binding protein deficient in Niemann-Pick type C2 disease. Proc. Natl. Acad. Sci. USA. 2003;100:2512–2517. doi: 10.1073/pnas.0437840100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein JL, Dana SE, Faust JR, Beaudet AL, Brown MS. Role of lysosomal acid lipase in the metabolism of plasma low density lipoprotein: Observations in cultured fibroblasts from a patient with cholesteryl ester storage disease. J. Biol. Chem. 1975;250:8487–8495. [PubMed] [Google Scholar]
- Goldstein JL, DeBose-Boyd RA, Brown MS. Protein sensors for membrane sterols. Cell. 2006;124:35–46. doi: 10.1016/j.cell.2005.12.022. [DOI] [PubMed] [Google Scholar]
- Infante RE, Abi-Mosleh L, Radhakrishnan A, Dale JD, Brown MS, Goldstein JL. Purified NPC1 protein: I. Binding of cholesterol and oxysterols to a 1278-amino acid membrane protein. J. Biol. Chem. 2008a;283:1052–1063. doi: 10.1074/jbc.M707943200. [DOI] [PubMed] [Google Scholar]
- Infante RE, Radhakrishnan A, Abi-Mosleh L, Kinch LN, Wang ML, Grishin NV, Goldstein JL, Brown MS. Purified NPC1 protein: II. Localization of sterol binding to a 240-amino acid soluble luminal loop. J. Biol. Chem. 2008b;283:1064–1075. doi: 10.1074/jbc.M707944200. [DOI] [PubMed] [Google Scholar]
- Infante RE, Wang ML, Radhakrishnan A, Kwon HJ, Brown MS, Goldstein JL. NPC2 facilitates bidirectional transfer of cholesterol between NPC1 and lipid bilayers, a step in cholesterol egress from lysosomes. Proc. Natl. Acad. Sci. USA. 2008c;105:15287–15292. doi: 10.1073/pnas.0807328105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko DC, Binkley J, Sidow A, Scott MP. The integrity of a cholesterol-binding pocket in Niemann-Pick C2 protein is necessary to control lysosome cholesterol levels. Proc. Natl. Acad. Sci. USA. 2003;100:2518–2525. doi: 10.1073/pnas.0530027100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolter T, Sandhoff K. Principles of lysosomal membrane digestion: stimulation of sphingolipid degradation by sphingo-lipid activator proteins and anionic lysosomal lipids. Annu. Rev. Cell Dev. Biol. 2005;21:81–103. doi: 10.1146/annurev.cellbio.21.122303.120013. [DOI] [PubMed] [Google Scholar]
- Liscum L, Faust JR. Low density lipoprotein (LDL)-mediated suppression of cholesterol synthesis and LDL uptake is defective in Niemann-Pick type C fibroblasts. J. Biol. Chem. 1987;262:17002–17008. [PubMed] [Google Scholar]
- Naureckiene S, Sleat DE, Lackland H, Fensom A, Vanier MT, Wattiaux R, Jadot M, Lobel P. Identification of HE1 as the second gene of Niemann-Pick C disease. Science. 2000;290:2298–2301. doi: 10.1126/science.290.5500.2298. [DOI] [PubMed] [Google Scholar]
- Neiss WF. A coat of glycoconjugates on the inner surface of the lysosomal membrane in the rat kidney. Histochemistry. 1984;80:603–608. [PubMed] [Google Scholar]
- Ohgami N, Ko DC, Thomas M, Scott MP, Chang CCY, Chang T-Y. Binding between the Niemann-Pick C1 protein and a photoactivatable cholesterol analog requires a functional sterol-sensing domain. Proc. Natl. Acad. Sci. USA. 2004;101:12473–12478. doi: 10.1073/pnas.0405255101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura N, Kiuchi S, Tamba M, Kashima T, Hiramoto S, Baba T, Dacheux F, Dacheux J-L, Sugita Y, Jin Y-Z. A porcine homolog of the major secretory protein of human epididymis, HE1, specifically binds cholesterol. Biochim. Biophys. Acta. 1999;1438:377–387. doi: 10.1016/s1388-1981(99)00070-0. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. Processing of x-ray diffraction data collected in oscillation mode. Meth. Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Pentchev PG. Niemann-Pick C research from mouse to gene. Biochim. Biophys. Acta. 2004;1685:3–7. doi: 10.1016/j.bbalip.2004.08.005. [DOI] [PubMed] [Google Scholar]
- Pentchev PG, Vanier MT, Suzuki K, Patterson MC. Niemann-Pick disease type C: A cellular cholesterol lipidosis. In: Scriver CR, Beaudet AL, Sly WS, Valle D, editors. The Metabolic and Molecular Basis of Inherited Disease. McGraw-Hill Inc.; New York: 1995. pp. 2625–2639. [Google Scholar]
- Radhakrishnan A, Sun L-P, Kwon HJ, Brown MS, Goldstein JL. Direct binding of cholesterol to the purified membrane region of SCAP: mechanism for a sterol-sensing domain. Mol. Cell. 2004;15:259–268. doi: 10.1016/j.molcel.2004.06.019. [DOI] [PubMed] [Google Scholar]
- Roth MG. Clathrin-mediated endocytosis before fluorescent proteins. Nat. Rev. Mol. Cell Biol. 2006;7:63–68. doi: 10.1038/nrm1783. [DOI] [PubMed] [Google Scholar]
- Schneider TR, Sheldrick GM. Substructure solution with SHELXD. Acta Crystallogr. D Biol. Crystallogr. 2002;D58:1772–1779. doi: 10.1107/s0907444902011678. [DOI] [PubMed] [Google Scholar]
- Schulze H, Kolter T, Sandhoff K. Principles of lysosomal membrane degradation. Cellular topology and biochemistry of lysosomal lipid degradation. Biochim. Biophys. Acta. 2009;1793:674–683. doi: 10.1016/j.bbamcr.2008.09.020. [DOI] [PubMed] [Google Scholar]
- Simons K, Ikonen E. How cells handle cholesterol. Science. 2000;290:1721–1726. doi: 10.1126/science.290.5497.1721. [DOI] [PubMed] [Google Scholar]
- Sleat DE, Wiseman JA, El-Banna M, Price SM, Verot L, Shen MM, Tint GS, Vanier MT, Walkley SU, Lobel P. Genetic evidence for nonredundant functional cooperativity between NPC1 and NPC2 in lipid transport. Proc. Natl. Acad. Sci. USA. 2004;101:5886–5891. doi: 10.1073/pnas.0308456101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian K, Balch WE. NPC1/NPC2 function as a tag team duo to mobilize cholesterol. Proc. Natl. Acad. Sci. USA. 2008;105:15223–15224. doi: 10.1073/pnas.0808256105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Driel IR, Goldstein JL, Südhof TC, Brown MS. First cysteine-rich repeat in ligand-binding domain of low density lipoprotein receptor binds Ca2+ and monoclonal antibodies, but not lipoproteins. J. Biol. Chem. 1987;262:17443–17449. [PubMed] [Google Scholar]
- Wasilko D, Lee S. TIPS: Titerless infected-cells preservation and scale-up. Bioprocessing J. 2006;5:29–32. [Google Scholar]
- Watari H, Blanchette-Mackie EJ, Dwyer NK, Glick JM, Patel S, Neufeld EB, Brady RO, Pentchev PG, Strauss JF., III Niemann-Pick C1 protein: Obligatory roles for N-terminal domains and lysosomal targeting in cholesterol mobilization. Proc. Natl. Acad. Sci. USA. 1999;96:805–810. doi: 10.1073/pnas.96.3.805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winn MD, Isupov MN, Murshudov GN. Use of TLS parameters to model anisotropic displacements in macromolecular refinement. Acta Crystallogr. D Biol. Crystallogr. 2001;D57:122–133. doi: 10.1107/s0907444900014736. [DOI] [PubMed] [Google Scholar]
- Xu S, Benoff B, Liou H-L, Lobel P, Stock AM. Structural basis of sterol binding by NPC2, a lysosomal protein deficient in Niemann-Pick type C2 disease. J. Biol. Chem. 2007;282:23525–23531. doi: 10.1074/jbc.M703848200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z, Farver W, Kodukula S, Storch J. Regulation of sterol transplant between membranes and NPC2. Biochemistry. 2008;47:11134–11143. doi: 10.1021/bi801328u. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.