Abstract
Activation of ligand-gated channels is initiated by the binding of small molecules at extracellular sites and culminates with the opening of a membrane-embedded pore. To investigate how perturbations at ligand-binding domains influence the gating reaction, we examined current traces recorded from individual NMDA receptors in the presence of several subunit-specific partial agonists. Here we show that low-efficacy agonists acting at either the GluN1 or the GluN2A subunit had very similar effects on the receptor’s activation reaction, possibly reflecting a high degree of coupling between the two subunit-types during gating. In addition, we demonstrate that partial agonists increased the height of all energy barriers encountered by NMDA receptors during activation. This result stands in sharp contrast to the localized effects observed for pentameric ligand-gated channels and may represent a novel mechanism by which partial agonists reduce receptor activity.
Keywords: ligand-gated ion channel, glutamate receptor, NMDA receptor, neurotransmitter receptor, partial agonist, gating, energy landscape, single-channel, gating kinetics, synaptic transmission, neurotransmitter
Introduction
The mechanism by which the binding of small molecules at extracellular sites of ligand-gated channels triggers a conformational change in the receptor’s membrane-embedded pore remains poorly characterized1. For ionotropic glutamate receptors, crystallographic studies suggested that upon binding, the ligand initiates large-scale motions within the agonist-binding domains, which in turn prompt further intra-subunit or inter-subunit rearrangements, and culminate with a widening of the channel pore2,3. For the fast-gating nicotinic acetylcholine receptors (rise time, RT ~1 ms), free energy considerations predict that the gating reaction proceeds as a potent wave of conformational change, which begins at the agonist-binding sites and peaks in the pore, but whose intermediate conformers are too ephemeral to be observed experimentally4. More recently, single-channel studies have invoked the existence of short-lived pre-open receptor states within the activation reactions of several fast-gating ligand-activated channels, including AMPA, glycine and acetylcholine receptors5–7. For the much slower-gating NMDA receptors (RT ~20 ms), several fully-liganded pre-open intermediates are observed routinely in single–channel records8,9. More recently, these intermediates have been integrated within comprehensive gating schemes, thus defining a most probable temporal sequence in which they become populated during activation10–16. Still, the physical locations and the precise nature of the changes that underlie these kinetically defined transitions remain unknown.
NMDA receptors assemble as dimers of heterodimers and are composed of two glycine-binding GluN1 subunits and two glutamate-binding GluN2 subunits17,18. The GluN2 subunits, of which four (A–D) have been characterized, endow functional receptors with characteristic kinetics and pharmacology19,20. NMDA receptors are exceptional among ligand-gated channels in that they become active only after all agonist-binding sites have been occupied21,22. Because of this dual agonist-requirement and due to their remarkably slow kinetics, NMDA receptors present a unique opportunity to measure the contributions of individual subunit types to kinetically resolvable gating transitions. Based on kinetic analyses of single GluN1/GluN2B channels activated with subunit-specific ligands, it was proposed that partial agonists are less effective at opening the pore because they slow down, in a subunit-specific manner distinct pre-opening transitions10. More recently, kinetic analyses of single pentameric receptors suggested that agonist efficacy affects only the first pre-opening step during gating but not the channel-opening step per se6,7. These intriguing findings, and the better understanding of NMDA receptor gating that has emerged in recent years, motivated us to characterize in further detail the mechanism of partial agonism at NMDA receptors.
We found that in contrast to the localized effects observed for partial agonists at pentameric ligand-gated channels6,7, partial agonists result in long-ranging changes within the NMDA receptor gating sequence; however, agonists of similar efficacy initiate activations that have identical free energy profiles, regardless of the subunit-type to which they bind. Based on these results, we suggest that the two subunit-types are tightly coupled during gating and that agonist efficacy affects all activation steps detected. In light of the contained changes reported recently for pentameric channels, such wide-spread effects may reflect a novel mechanism for partial agonism at ligand-gated receptors.
Results
Glycine-site low-efficacy agonists lengthen specifically two time-components
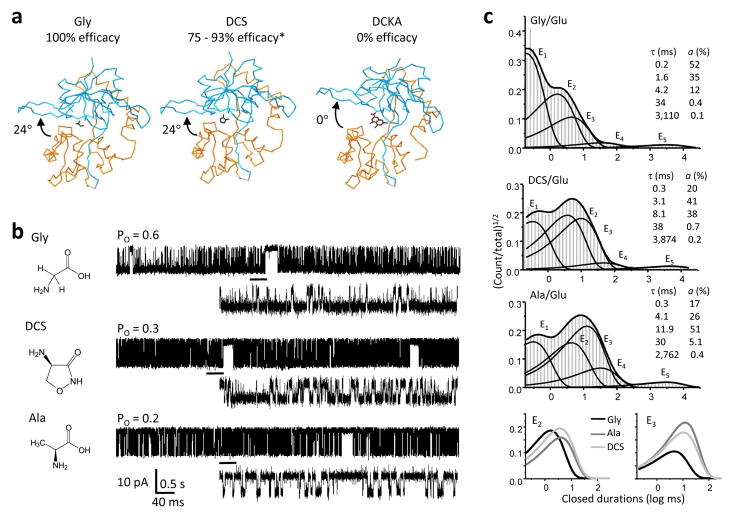
Comprehensive characterization of the NMDA receptor kinetics requires that long periods of activity be recorded to accumulate large numbers of openings and closures. This is because even when continuously exposed to maximally active concentrations of the physiological co-agonists glutamate and glycine, NMDA receptors reside in open conformations for only a fraction (<60%) of the observed time8,23. In addition, a quantitative description of the kinetic mechanism of NMDA receptors involves the estimation of at least ten rate constants that differ by four orders of magnitude10–13,16,24. For these reasons, we chose to examine steady-state currents from channels with relatively high intrinsic open probability: recombinant GluN1/GluN2A receptors in cell-attached membrane patches (Po = 0.4 – 0.8)11,15 exposed to saturating concentrations of agonists whose efficacies were at least 50% of those reported for glutamate or glycine. We focused first on the glycine-site ligands DCS (10 mM, 75 – 93%, n = 5)25,26 and ACPC (0.1 mM, 80%, n = 11)27 because high-resolution structures have been reported for the isolated agonist-binding domain of the GluN1 subunit in complex with these two ligands27,28. Importantly, these studies suggested that in contrast to the graded cleft-closure induced by partial agonists at AMPA-type glutamate receptor subunits, at NMDA-type subunits partial agonists may stabilize fully-closed cleft conformations, similar to those elicited by full-agonists, thus leaving unresolved the mechanism of partial agonism at these receptors (Figure 1a)27. We also recorded single-channel activity with saturating concentrations of the glycine-site partial agonists L-Alanine (Ala, 1 mM, 79%, n = 6)29 and ACBC (2 mM, 40 %, n = 11)27. We compared these records with those obtained with saturating concentrations of both glycine (0.1 mM) and glutamate (1 mM, n = 5).
Figure 1. NMDA receptor activities with GluN1 partial agonists.
a, Crystal structures of the GluN1 ligand-binding domain in complex with the full agonist Gly (PDB:1PB7), the partial agonist DCS (PDB:1PB9) and the antagonist DCKA (PDB: 1PBQ) suggested that full and partial agonists induce the same degree of cleft closure relative to the open structure stabilized by antagonist27. b, Chemical structure of the agonist used is shown to the left of a representative single-channel trace (10 s displayed filter at 1 kHz with openings down). Glutamate (1 mM) was included in all recordings alongside saturating concentrations of the indicated co-agonist. Underlined portion (500 ms) of each record is expanded below filtered at 12 kHz. c, Histograms of closed interval distributions for a record in each condition (events: Gly, 509,041; Ala, 317,593; DCS, 31,754) are overlaid with probability density function (thick lines) and exponential components E1 – E5 (thin lines) calculated from fits of the 5C3O state model to the entire sequence of events in the respective file. Insets show time constants and areas for the corresponding exponential components in the file shown. Bottom panel illustrates together the E2 and E3 components for the three agonists above. *Efficacy reported by refs. 26 and 25.
All the partial agonists studied elicited NMDA receptor currents of unitary amplitudes similar to those of full agonists (Figure 1b, Table 1). This observation is consistent with the view that partial agonists elicit sub-maximal macroscopic currents from NMDA receptors due entirely to a decrease in receptor open probability (Po) without changes in channel conductance, as is the case for the related AMPA-type channels10,30. For each agonist studied, the mean Po was consistent with the lower efficacy estimated previously from whole-cell current measurements (Table 1). This result suggested that the single-molecule behaviors captured in our data set were sufficient to reproduce the average behaviors estimated from ensemble measurements. We used the entire sequence of single-channel currents in each file to estimate mean closed (MCT) and mean open (MOT) durations for each of the receptors observed. The results showed that partial agonists elicited lower Po due to both longer closures and shorter openings (Table 1). This result is similar with the effects of partial agonists on both open and closed durations at pentameric ligand-gated receptors6. Although at GluN1/GluN2B receptors, no significant change in MOT was observed with partial agonists, it is likely that this reflected thee much shorter openings of GluN1/GluN2B isoforms, which may have hampered the detection of statistically-significant agonist-specific changes in this parameter10.
Table 1.
Effects of partial agonists on single GluN1/GluN2A channel currents
| Agonists GluN1/GluN2 | reported efficacy (%) | amplitude (pA) | Po * | MOT (ms) | MCT (ms) | n | period analyzed (min) | events analyzed |
|---|---|---|---|---|---|---|---|---|
| Glu/Gly | 100 | 9.1 ± 0.6 | 0.64 ± 0.06 | 11.2 ± 1.1 | 6.4 ± 1.7 | 5 | 190 | 1,256,063 |
| Glu/DCS | 75b, 93c | 9.2 ± 0.4 | 0.45 ± 0.11 | 8.4 ± 1.4 | 12 ± 3 | 5 | 109 | 438,502 |
| Glu/Ala | 79d | 9.5 ± 0.4 | 0.27 ± 0.04 | 5.3 ± 0.6 | 14 ± 2 | 6 | 165 | 941,880 |
| Glu/ACPC | 80a | 10.0 ± 0.3 | 0.27 ± 0.04 | 5.0 ± 0.8 | 12 ± 2 | 11 | 187 | 1,810,577 |
| Glu/ACBC | 40a | 10.0 ± 0.2 | 0.07 ± 0.02 | 3.8 ± 0.6 | 39 ± 12 | 5 | 62 | 99,860 |
| HCA/Gly | 86e | 10.0 ± 0.5 | 0.29 ± 0.06 | 4.7 ± 0.7 | 15 ± 3 | 10 | 287 | 1,791,508 |
| SYM/Gly | 72e | 10.0 ± 0.6 | 0.36 ± 0.09 | 5.7 ± 0.9 | 13 ± 3 | 5 | 98 | 554,762 |
| HQA/Gly | 75e | 10.2 ± 0.5 | 0.25 ± 0.04 | 5.8 ± 0.7 | 18 ± 2 | 5 | 217 | 997,465 |
| QA/Gly | 20f, 60g | 10.0 ± 0.6 | 0.13 ± 0.06 | 6.2 ± 1.0 | 218 ± 87 | 8 | 165 | 328,578 |
References are: a) Inanobe 2005, b) Sheinen 2001, c) Priestley 1995, d) Chen 2008, e) Erreger 2007, f ) Blanke 2008, g) de Cavalho 1996.
Open probability was calculated as the fractional occupancy of the open state when fitting the entire sequence of events to kinetic models having five closed and one open states (5C1O).
To probe in further detail the mechanism by which glycine-site partial agonists decreased receptor Po, we characterized for each record the closed and open interval distributions. For all files analyzed, 5 closed and 3 or 4 open components were necessary and sufficient to fully describe the data. This result is consistent with previous reports that within clusters of activity NMDA receptor channels visit with high frequencies three shorter closed components (E1 – E3), whereas the two longer closed components (E4, E5) terminate clusters and reflect infrequent visits into desensitized states10,31,32. Closed interval distributions with the overlaid probability density function and exponential components E1 – E5 calculated for glycine, DCS and Ala are illustrated for individual receptors in Figure 1c. For ACPC and ACBC, representative current traces and histograms, as well as a summary of changes in closed-time constants for all patches are shown in Supplementary Figure 1a–c.
Statistical analyses of these data showed that all partial agonists tested increased τE2 and τE3 by ~2-fold. In addition to these changes common to all GluN1 ligands tested, we also observed agonist-specific changes: τE1 was modestly increased with DCS and Ala; and τE4 was increased ~3-fold for ACBC (Supplementary Table 1). This result suggested that partial agonists increased all closed durations within clusters, a finding in contrast with previous measurements at GluN1/GluN2B receptors, where the glycine-site partial agonists DCS and HA-966 increased only one (τE2) of the three intra-cluster closed time-components10. Whereas it is possible that this discrepancy illustrates differences in gating between the two isoforms, it is more likely that statistical significance of differences in the other two components (τE1 and τE3) were obscured by the much higher variability and much lower open probability characteristic to GluN2B-containing receptors, particularly when observed in excised patches20. To determine whether the changes we observed are specific to the subunit-type to which the agonists-tested bind we recorded single-channel currents from one-channel patches using glutamate-site partial agonists.
Glutamate-site agonists lengthen the same two closed time-components
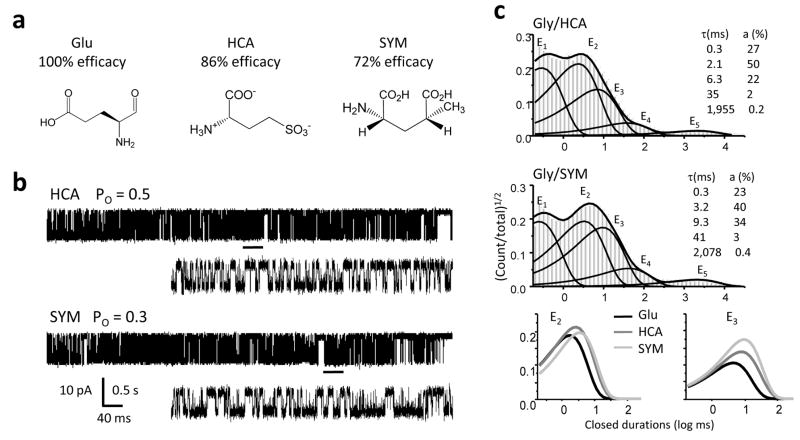
For the GluN2 family of subunits, only one high-resolution structure has been solved. It illustrates a back-to-back heterodimer of the GluN1 and GluN2A agonist-binding domains, both assuming cleft conformations fully closed around glycine and glutamate, respectively33. To probe the mechanism of partial agonism at the glutamate-site we recorded cell-attached single-channel currents in the presence of saturating glycine (0.1 mM) and the glutamate-site partial agonists L-homocysteate (HCA, 1 mM, 86%, n = 10)34, SYM2081 (SYM, 2 mM, 72%, n = 5)34, quinolinic acid (QA, 1 mM, 20 – 60%, n = 8)35,36 and homoquinolinic acid (HQA, 1 mM, 75%, n = 5)34 (Figure 2a). These agonists have macroscopic efficacies comparable to those of the glycine-site partial agonists we tested and thus, we reasoned that differences in magnitude would not confound possible differences in mechanism.
Figure 2. NMDA receptor activities with GluN2A partial agonists.
a, Chemical structures of the ligands used, with reported efficacies34.b, Representative single-channel traces (10 s, shown filtered at 1 kHz with openings down) obtained with glycine and the partial agonists indicated. Underlined portion (500 ms) of each record is shown below filtered at 12 kHz and higher resolution. c, Histograms of closed interval distributions for a record in each condition (events: HCA, 137,512; SYM, 226,688) superimposed with the probability density function (thick lines) and exponential components (E1 – E5, thin lines) estimated from fits of a 5C4O state model to the idealized data. Insets show time constants and areas for the corresponding exponential components in the file shown. Bottom panel singles out the E2 and E3 components of the distributions shown above.
Representative single-channel traces obtained with glutamate-site partial agonists (HCA and SYM, Figure 2b; HQA and QA, Supplementary Figure 2a) can be compared to the glutamate-elicited traces illustrated in Figure 1b. As with glycine-site partial agonists, single-channel current amplitudes were not affected and partial agonism at the glutamate site was fully explained by a reduction in channel Po (Table 1). Similarly, the observed lower Po was the combined result of both longer closures and shorter openings (Table 1). Interval distributions for all files analyzed revealed 5 closed and 3 or 4 open components. The closed interval distributions obtained with HCA and SYM were strikingly similar to those obtained with glycine-site partial agonists, having significantly longer time constants for the same two exponential components E2 and E3 (Figure 2c, Figure 3 and Supplementary Table 1). Representative traces and histograms, and a summary of closed-time changes for HQA and QA are illustrated in Supplementary Figure 2. QA resulted in single-channel traces with high patch-to-patch variability and prevented us from characterizing its kinetic mechanism with accuracy. However, for the other seven partial agonists investigated, regardless of subunit specificity, we identified as a common mechanism for the observed decrease in Po a statistically significant increase in τE2, and τE3.
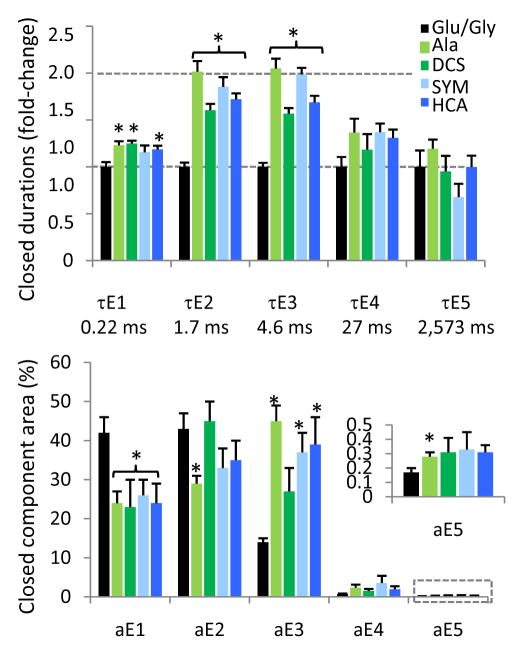
Figure 3. Partial agonists prolonged specifically the shorter, more frequent closures.
Top panel, Partial agonists caused a robust and significant increase in the mean duration of two closed time components, τE2 and τE3, relative to full agonists (black bars, mean values below); τE1 was also significantly, albeit modestly increased. Bottom panel, For all agonists tested, the majority of closures (aE1–aE3 >95%) were short (τE1–τE3 < 30 ms), whereas long closures (τE4–τE5) occurred with very low frequencies (aE4 <5%, aE5<0.5%), which were not changed by partial agonists. Bars represent means (sem): black, full agonists, Glu/Gly (n = 5); green, GluN1 ligands: Ala (light green, n = 6), DCS (dark green, n = 5); blue, GluN2A ligands: SYM (dark blue n = 5), HCA (light blue, n = 7). Color intensity illustrates partial agonists with higher (dark) and lower (light) efficacy at each site. Statistical significance of differences was assessed with the Student’s t-test (*, p < 0.05).
A previous study of GluN1/GluN2A receptors in excised patches identified τE3 as the only closed time-components increased by HQA37. Here, we also detected only a small change for τE2 (from 1.74 ± 0.08 ms, n = 5 for glutamate to 2.0 ± 0.1 ms, n = 5 for HQA). However, as with the other partial agonists tested, this increase was statistically significant (p < 0.03) (Supplementary Figure 2c and Supplementary Table 1). Based on these results we conclude that the binding of partial agonists at either the GluN1 or the GluN2A subunits have similar effects on NMDA receptor gating, at least at the level of closed time-components.
Low-efficacy agonists shorten NMDA receptor openings
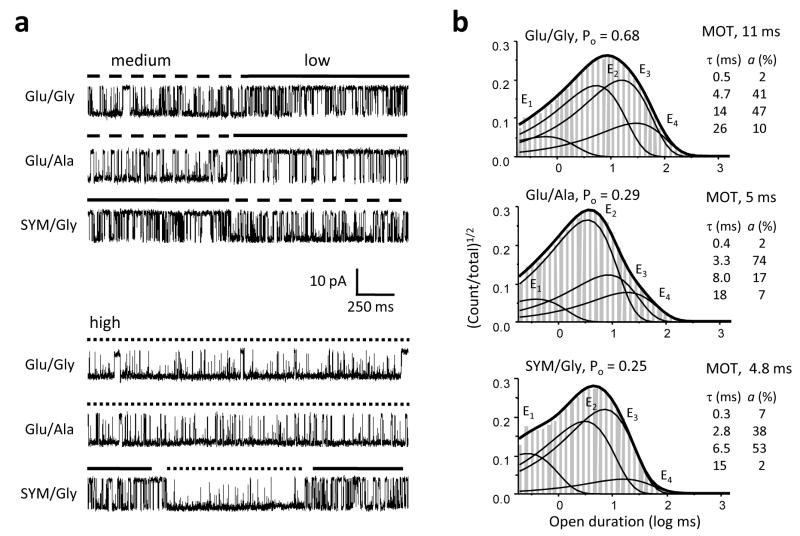
To investigate the mechanism underlying the observed decrease in channel MOT in the presence of partial agonists we examined in detail the open interval distributions for four of the eight partial agonists tested, DSC (n = 5), Ala (n = 6), HCA (n = 7) and SYM (n = 5), and compared these with those obtained from recordings with the full agonists glutamate and glycine (n = 5) (Supplementary Table 2). All records subjected to this analysis were best fit by state models with at least three (n = 18) and up to four (n = 10) open states, consistent with previous observations of modal gating11. This result and direct observations of shifts between periods with distinct open durations indicated that when activated with partial agonists, NMDA receptors retain modal behavior (Figure 4a). Thus a core gating mechanism composed of 5 closed and 2 open states is preserved, and the 4 open components identified in long-duration patches are the sum of two components in each mode: a fast component (τfast) present in all modes, and a second longer component, τlow, τmedium or τhigh, each characteristic of the low-, medium- or high-Po behavior, respectively.
Figure 4. Partial agonists decrease NMDA receptor open durations.
a, Single-channel current traces (2 s, displayed filtered at 1 kHz with downward openings) illustrate periods of activity recorded from patches containing just one active receptor, activated with the agonist combinations shown at left. Top panel illustrates switches between medium and low gating modes. Bottom panel illustrates periods of high-mode gating from the same records as in top panel, respectively. b, Histograms of open intervals for a record in each condition (events: Glu/Gly, 71,085; Ala/Glu, 283,797; SYM/Gly, 226,688) are overlaid with the probability density function (thick lines) and exponential components E1 – E4 (thin lines) calculated from the fits to the data from each patch with a 5C4O model. Insets show time constants and relative areas for the corresponding components. The fast component is present in all three modes whereas the three longer components correspond to the low-, medium- and high-mode, respectively.
By comparing open interval distributions obtained with full agonists with those obtained with the glycine-site (Ala) or the glutamate-site (SYM) partial agonists, we found that partial agonists at either site shortened significantly the duration of the longer time component in each mode (Figure 4b). By comparing the frequencies with which τlow, τmedium and τhigh occurred with full or partial agonists we observed that partial agonists also biased channels toward lower Po gating modes. However, due to the low number of modal shifts observed for each channel, this result was not statistically significant (Supplementary Table 2).
Partial agonists have wide raging effects on gating
As previously described, our results are consistent with a minimal gating mechanism for fully liganded NMDA receptors composed of 5 closed and 2 open states (5C2O) with superimposed, albeit infrequent, modal swings. Because we could not definitively resolve whether partial agonists lower MOT by only shortening openings in each mode or by a combined effect on open durations and a decreased frequency of higher Po modes, we chose to represent all observed openings with an aggregated open state (Figure 5a). This approximation has no consequence on the estimated rate constants for transitions between closed conformations because within each mode, the two coupled open states are distal to agonist-binding10–12,16,24,38. However, in this simplified model, the rate constants for transitions into and from the aggregated open state represented weighted averages of intra- and inter-mode transitions rather than microscopic opening rate constants. We used this operational model to identify the transitions sensitive to agonist efficacy.
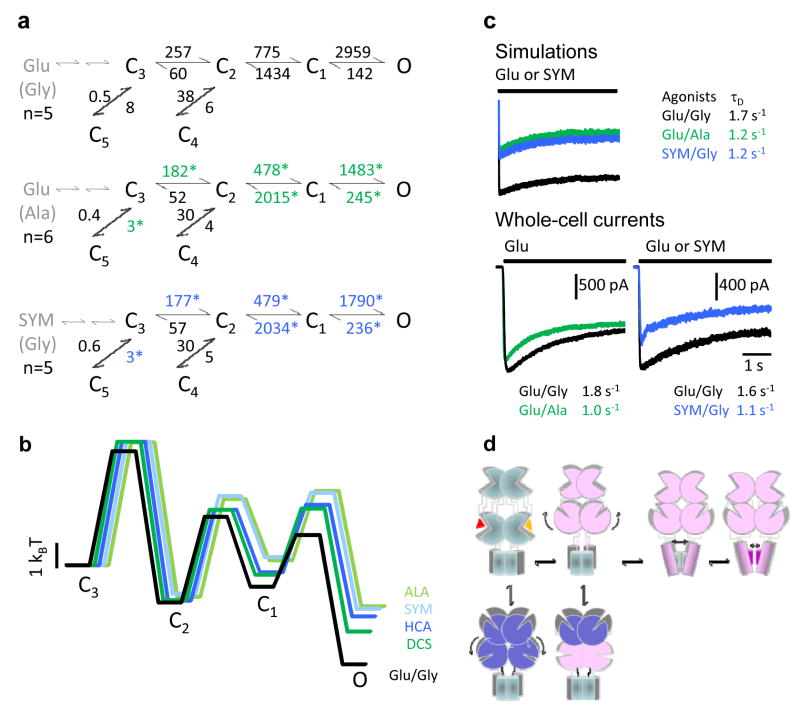
Figure 5. Kinetic mechanism of NMDA receptor activation by partial agonists.
a, State models with rate constants represented as averages of results from fits to individual records in each condition. Open states are represented as an aggregate O, for simplicity. Two sequential binding steps are represented in grey. Asterisks denote rates which were significantly different from Glu/Gly (p < 0.05). b, Calculated free-energy relationships were plotted with respect to C3. States C4 and C5, which represent desensitized conformations, were omitted from the energy diagram to emphasize the activation pathway. Traces were slightly shifted horizontally for clarity. c, Top, current traces simulated with the models and rate constants shown in panel a, and after appending two agonist-binding steps to state C3 (see Methods). Bottom, whole-cell voltage-clamp current traces in response to 5-s agonist-applications as indicated, in the continuous presence of the corresponding co-agonist. Overlaid traces represent averages of 5 interleaved sweeps recorded from the same cell. Time constants of fits with single exponential functions are given for the displayed traces. d, Cartoon representing rearrangements in quaternary structure possibly corresponding to the kinetic intermediates characterized in this study. The activation pathway is imagined to consist of: the aggregated closures of all ligand-binding domains (C3-C2), followed by movements in trans-membrane helices or/and the linker regions (C2-C1), and finally direct gating of the permeation pathway (C1-O) by fast oscillations of residues in the selectivity filter. Off-pathway intermediates result from non-productive closures of the ligand-binding domains, fostered perhaps by concurrent rearrangements in the N-terminal domains. This model takes into account our previous observation that glutamate dissociates with measurable rates from state C3 but not others (ref 24), and the current results that partial agonists affect to the same extent and in a symmetrical manner the C3-C5 and C3-C2 equilibria. It also incorporates current knowledge on the role of: the N-terminal domains in desensitization, the third transmembrane domain in activation and the selectivity filter in gating.
By comparing the rate constants obtained for a glycine-site agonist (Ala) and a glutamate-site agonist (SYM), we found that these partial agonists changed the same transitions (Figure 5a). Although small differences in gating were observed between agonists, in general partial agonists at either subunit-type decreased the activation rate constants C3→C2, C2→C1, and C1→O and increased the deactivation rate constants C1←O and C2←C1 (Supplementary Figure 3 and Supplementary Table 3). A pictorial representation of the calculated relative free-energy profiles for these reaction mechanisms illustrate that partial agonists at either the GluN1 or the GluN2A subunits resulted in activations that were energetically very similar with each other (Figure 5b). In addition, comparing these with the calculated activation profile elicited by full-agonists, it was readily apparent that partial agonists caused pervasive changes on the NMDA receptor activation reaction: they increased the height of all activation barriers and resulted in increased occupancies of closed pre-open states at the expense of open state occupancies.
As a common trend for the partial agonists investigated in this study, except for the C3←C2 deactivation step, which remained unchanged, all transitions located along the activation pathway were affected: those leading to open states were decreased and those leading away from open states were increased (Supplementary Table 3 and Supplementary Figure 3). In contrast, among off-pathway rate constants, only the C3→C5 rate constant was significantly decreased for all ligands tested. Functionally, the closed state C5 represents the principal desensitized state of NMDA receptors, and our observation that partial agonists decreased both the activation rate constants and the desensitization rate constant is in agreement with previously reported effects of partial agonists at homomeric GluA2 receptors5,39,40. These results are collectively consistent with the hypothesis that the closed state C3 represents fully liganded receptor conformations whose ligand-binding clefts are open, such that ligands can still dissociate and re-associate with measurable rates24. Further, our results are also in agreement with the premise that cleft-closure, with simultaneous ligand entrapment, can be productive when it leads to activation (C3→C2) or un-productive when it promotes desensitization (C3→C5)41,42.
Paradoxically, although the kinetic model clearly estimates that both the activation and the desensitization rate constants are decreased by partial agonists, it also predicted increased desensitization of macroscopic responses as a consequence of increased occupancy of state C3 from which receptors can desensitize (Figure 5c). To test this prediction we measured and compared the kinetics of macroscopic desensitization in the presence of saturating concentrations of partial agonists.
Partial agonists accelerate macroscopic desensitization
To examine how partial-agonists affect NMDA receptor macroscopic desensitization we recorded whole-cell current traces during 5-second applications of glutamate in the continuous presence of Ala, or applications of SYM in the continuous presence of glycine. Because measurements of macroscopic current decay can report on a range of mechanisms by which NMDA receptor responses are known to desensitize, we selected conditions where the time-dependent decline in current was expected to reflect mostly the accumulation of receptors in microscopic desensitized states (C5 and C4). To minimize contributions by a zinc-dependent mechanism, to which GluN1/GluN2A channels are particularly sensitive, or by a calcium-dependent mechanism brought about by disruption of endogenous calcium stores, we included metal chelators in both extra-cellular and intra-cellular solutions (EDTA and EGTA, respectively). In addition, knowing that channel open probabilities may decrease following cell break-in we anticipated differences in current amplitudes between the intact cell-attached preparation (represented by the model) and experimental currents. Keeping in mind these limitations, we examined the kinetics with which receptors desensitize in the continued presence of GluN1 or GluN2 partial agonists.
Current traces elicited with either Ala or SYM in the presence of the corresponding full co-agonist, declined significantly faster than interleaved controls, with time constants (Glu/Ala, 1.11 ± 0.02 s, n = 3, p < 0.001; SYM/Gly, 0.95 ± 0.05 s, n = 3, p < 0.002 versus Glu/Gly, 1.70 ± 0.05 s, n = 6) very similar to those predicted by the model (Glu/Ala,1.2 s; SYM/Gly, 1.2 s versus Glu/Gly, 1.7 s) (Figure 5c). Thus the experimental measurements reproduced the model’s surprising prediction that although partial agonists are less effective in promoting entry into the desensitized state, they elicit macroscopic currents which desensitize faster. The predicted mechanism is an increased occupancy of pre-open states (C3 and C2) from which desensitization can occur (C3→C5 and C2→C4): although the chance for a receptor to desensitize is smaller with partial agonists, more receptors have the chance to desensitize. Clearly, this result on macroscopic desensitization need not be a general effect of partial agonists, since this ensemble measurement is also influenced by changes in rate constants elsewhere in the system.
Discussion
Structural studies of the isolated ligand-binding domains of several ionotropic glutamate receptor subunits have advanced the hypothesis that agonist-induced conformational changes in the ligand-binding modules represent the initial large-scale rearrangement that impels subsequent gating or/and desensitization transitions. It is yet unclear how the sequence of structural changes postulated to take place within the ligand-binding domain of one subunit coordinate with movements in the remaining three subunits. One hypothesis, supported by functional data for GluA2 homomeric channels suggested that subunits can change conformation independently, such that subunits activate individually in random order, following unitary ligand-binding events43. However, this scenario may not apply to NMDA receptors given that these receptors require that all subunits be occupied with ligand before channels open22. Further, the results presented here show that perturbations in the ligand-binding domain of either subunit-type have similar effects on the receptor’s activation thus suggesting similar roles for the ligand-binding domains of GluN1 and GluN2 subunits and possibly a high degree of coupling between the two.
If such inter-subunit coupling exists, and given that most native glutamate receptors assemble as dimers of heterodimers, it may be that the kinetic transitions we observed for fully liganded receptors represent conformational changes within one heterodimer first (C3-C2), with subsequent rearrangements in the remaining heterodimer (C2-C1) with a final widening of the pore (C3-O). This mechanism would allow for both high-coupling between subunits within each heterodimer and relative independence of heterodimers. However, our results appear inconsistent with this hypothesis mostly because perturbations at either the GluN1 or GluN2 ligand binding-sites affected asymmetrically the C3-C2 and C2-C1 equilibria, with only the forward rates changed in the former and with both forward and reverse rates changed in the latter. This result would rather suggest that the structural changes represented by these two equilibria are of a distinct nature.
We thus suggest the alternative premise that the first kinetically resolvable transition during NMDA receptor gating (C3-C2), which we found to be sensitive to the efficacy of agonist at either subunit-type, may represent rearrangements within the ligand-binding domain of the entire receptor. In this scenario the ligand-binding domains of all four subunits have to switch into high-affinity conformations before the subsequent rearrangements necessary for gating or desensitization can occur with measurable probability (C2-C1). Structural information for regions other than the ligand-binding domains is currently lacking. However, functional data point strongly to gating-associated movements in locations additional to the hinged ligand-binding domains: conserved residues at the top of the third transmembrane domains and fast motions in the selectivity filters of trans-membrane pores have been shown to be important for gating. Thus, the pre-open intermediates postulated in the kinetic scheme may represent stepwise changes at locations positioned along the longitudinal axis of the protein: all four ligand-binding domains assuming the active conformation (C2) first, followed by movements in all four linker domains (C3), and with final widening of the pore (O) (Figure 5d). This sequence would allow perturbations at either ligand-binding domain to result in similar changes on the energetics of the entire gating reaction. It is also consistent with reports that the trajectory of the conformational wave that represents gating in the nicotinic acetylcholine receptor occurs along the longitudinal axis of the protein4.
We should note that the resolution of our recordings prevented us from detecting events shorter than 0.15 ms and that as with all kinetic studies, the conclusions derived here apply for the examined resolution and models. For these reasons, we cannot exclude the possibility that as was demonstrated for AChRs6,7, NMDA receptors also visit before opening, very brief states whose stability is agonist-independent. Nevertheless, the mechanism we propose, which does not include explicitly such brief pre-open intermediates, reproduces well that trajectory of the macroscopic response. Thus, it is likely that even if such states exist they contribute negligibly to the NMDA receptor physiological response.
In summary, we show that partial agonists specific for either GluN1 or GluN2 subunits have indistinguishable and wide ranging effects on the NMDA receptor activation reaction. The remarkably similar effects of perturbations at either NMDA receptor subunit-type suggest that the sequence of conformational changes that represents this receptor’s activation may involve structural rearrangements that cannot be traced to individual subunits or separate dimers. The pervasive influence of agonists on all gating equilibria differs markedly from the localized effects observed at pentameric receptors and may represent a novel mechanism by which partial agonists decrease the open probabilities of ligand-gated receptors.
Detailed Methods
Cells and transfections
HEK cells (ATCC CRL-1573), a gift from Dr. Auerbach (Buffalo, NY), were maintained in Dulbecco's Modified Eagle Medium (DMEM, Invitrogen, Grand Island, NY) with 10% Fetal Bovine Serum at 37°C in a 5% CO2 atmosphere and were passed when reaching 80 – 90 % confluence. Passages 22 – 40 were used for transfections.
Plasmids encoding rat GluN1–1a (U08261) and rat GluN2A (M91561) NMDA receptor subunits were gifts from Drs. Wenthold (Washington, DC) and Auerbach (Buffalo, NY), respectively. Open reading frames were sub-cloned into pcDNA3(+) under the control of the CMV promoter. Complete sequence of each insert was confirmed periodically by sequencing. Calcium precipitate-mediated transfections were done by incubating the cells for 2 hours with mixtures consisting of (in mM): 140 NaCl, 5 KCl, 0.75 Na2HPO4, 6 sucrose, 125 CaCl2, 25 HEPES/NaOH, pH 7.05, and 1 μg cDNA (NR1:NR2A: GFP = 1:1:1) per 35-mm dish. After removing the precipitate, the cells were grown 24–48 hrs in DMEM supplemented with 2 mM Mg2+.
Before each experiment, cells were washed, bathed in PBS and placed on the stage of an inverted microscope. Individual cells were selected visually, based on fluorescence intensity.
Electrophysiology
Single-channel currents were recorded with the cell-attached patch-clamp technique. The electrodes were filled with (extracellular) solutions containing (in mM): 150 NaCl, 2.5 KCl, 1 EDTA, 10 HEPBS, and agonists as indicated, adjusted to pH 8 (NaOH). Inward sodium currents were recorded after applying + 100 mV through the recording pipette. Currents were amplified and low-pass filtered at 10 kHz (Axopatch200B, 4-pole Bessel), sampled at 20 – 40 kHz (National Instruments card PCI-6229, M Series) and recorded into digital files with QUB acquisition software (www.qub.buffalo.edu).
Macroscopic currents were recorded with the whole-cell patch-clamp technique. Glass electrodes were filled with (intracellular) solutions containing (in mM): 135 CsF, 33 CsOH, 2 MgCl2, 1 CaCl2, 10 HEPES, and 10 BAPTA, adjusted to pH 7.4 (CsOH). After reaching the whole-cell configuration, the voltage was clamped at −70 mV and cells were superfused with extracellular solutions using a lightly pressurized perfusion system that exchanged solutions within 0.3 – 0.5 s (BPS-8, ALA Scientific Instruments). Extracellular solutions contained (in mM): 150 NaCl, 2.5 KCl, 0.01 EDTA and agonists as indicated, and were adjusted to pH 8 (NaOH). Peak and steady-state current amplitudes and desensitization time constants were estimated by fitting traces averaged for five or more sweeps with a mono-exponential function. Mean values obtained per each condition were compared with those obtained from the control.
Simulations
Macroscopic currents were simulated with the QuB software from 200 receptors (5 pA/each) as time-dependent accumulation of receptors in the open state. Glutamate-site agonist pulses were simulated as 5-second square steps from zero into saturation concentrations of agonist, as indicated for each. Prior to pulse application, all receptors occupied a resting unliganded state from which they could access state C3 of the indicated gating scheme (Figure 5a) by proceeding sequentially along two identical binding steps, each assumed to have the microscopic kinetics measured previously for glutamate: k+= 1.7x107 M−1s−1 and k- = 60 s−1 (ref24). The kinetics of the macroscopic traces were quantified as described above for experimental currents.
Analyses
Single-channel data selection and pre-processing
Each digital file was displayed with QUB software and inspected visually for simultaneous openings, signal-to-noise ratios, high-frequency artifacts and baseline drift. In the conditions tested, due to low receptor desensitization rate constants (< 10 s−1) and high Po within bursts (0.5 – 0.9), records consisted of long, almost solid bursts of activity separated by silent periods. As a result, records originating from patches with two or more active channels contained many superimposed bursts and were easy to detect and exclude from further processing and analyses. Although QUB software can handle files with two or more active channels (provided these have similar kinetics), such multi-channel records, become computationally expensive for minutes-long records. For this reason, we aimed to obtain multiple records with no apparent double openings for each condition tested. From this set, only records which required minimum processing were selected, mostly because it proved to be more time-consuming to ‘clean’ a noisy file than to record a new one, particularly for the long files necessary for this study (average was ~200,000 events/file).
Records with no apparent double openings were considered to originate from a single active receptor for the following reasons. First, in files where simultaneous openings were observed, the estimated time constant for the longest exponential component (E5) was substantially shorter, indicative of one channel opening while another was desensitized. Second, for files obtained with partial agonists where no simultaneous openings were observed, τ5 was of similar duration with that observed in one-channel patches obtained with full agonists. Thus, although the partial agonists used decreased Po substantially and made it more tenuous to equate the absence of double openings with the presence of only one channel, we are confident that the files selected for analyses originated from an individual receptor.
To correct errors due to occasional high frequency artifacts, periods of low signal-to-noise ratios, or baseline drift, the selected records were processed as follows. Brief (0.05 – 0.15 ms) current spikes were replaced with either closed- or open-level currents, matching the level of adjacent events, using the “erase” function. Longer portions (ms to min) with low signal-to-noise ratios were deleted and the remaining flanking traces were preserved as separate segments within the processed file, to indicate discontinuity in the record. Finally, baseline drifts were corrected by resetting the baseline to zero-current levels, as necessary.
Preprocessed data were idealized in QUB with the SKM algorithm after digitally low-pass filtering at 12 kHz with an imposed conservative dead time (0.15 ms, 3 – 6 samples) across all files.
Time constants and relative areas
We chose to perform all our single-channel measurements with saturating concentrations of agonists to minimize possible effects due to differences in the receptor’s affinity for the ligands tested. We assumed that at the high concentrations used, closures that reflect agonist dissociation are much shorter and more infrequent relative to closures that reflect fully liganded conformations. Based on this assumption, we attributed all the kinetic differences observed for the various ligands, to differences in ligand efficacy and not affinity.
Kinetic analyses of idealized records were performed with QUB software using the MIL algorithm. To determine the number of closed and open states required to best-fit the data in each digital file (one channel per file), we started to fit each file with a simple two-state model; to this we added closed and open states one-at-a-time, while monitoring the associated increase in log likelihood (LL) values. A minimal model was selected by arbitrarily imposing a threshold of 10 LL units per state added. Each of the files considered for statistical analyses had between 3x105 and 6x106 events. Values for the calculated time constants and relative area of closed and open components were calculated from the minimal models optimized as described above, and were reported as mean ± sem for each agonist pair along with the corresponding sample size (n). These parameters are independent of the arrangement of states within the model (see example in Supplementary Methods).
Rate constants
To estimate microscopic rate constants it is necessary to postulate a reaction mechanism, and thus presume the most likely path that receptors follow when transitioning from one state to another. This is necessary, most importantly, because each model reflects a particular physical reality but also because in some instances the optimized rate constant values may depend on the arrangement of transitions within the model. For NMDA receptors a kinetic model based on independently gating subunits was proposed. It postulates that after becoming fully liganded either the GluN2 (slow transition) or GluN1 (fast transition) can independently change conformation to become active (slow/fast or fast/slow) but must arrive to a pre-open conformation where both subunits have been activated before the pore opens and the receptor transitions into an open conformation10. We compared this model with a simple sequential model (Supplementary Methods). We found that a) the linear model returned consistently higher LL values despite having fewer parameters; and b) the rate constants estimated with the two models were very similar. For these reasons we used the simpler, linear model throughout our analyses. It was also useful that the linear model, having been derived entirely on statistical arguments, does not attribute any a priori physical significance for the postulated transitions. Estimated rate constants were reported as means ± sem, along with the respected sample size.
Statistics Changes in kinetics were evaluated using the two-tailed unpaired Student’s t-test between values obtained for a particular partial agonist and values for the same parameter obtained with the full agonists glutamate and glycine. Differences of means were considered significant for p < 0.05. We opted for individual t-test comparisons rather than multiple ANOVA testing because although we identified changes in kinetic parameters that were common to all conditions investigated the magnitudes of the differences were agonist-specific. This was expected because: a) the efficacies of the agonists used were not identical; and b) because in addition to changes that were common to all partial-agonists tested, we also observed agonist-specific changes. Since a multiple ANOVA analysis tests the null hypothesis by comparing individual sample means to the global mean for all conditions, it is not appropriate to assume that each tested parameter (time or rate constant) will have the same variance. For these reasons we chose the t-test to evaluate statistical significance, and aimed to minimize the possibility of reaching an erroneous conclusion by increasing the number of partial agonists tested at each site. Bar graphs (Figure 3, and Supplementary Figures 1c and 2c) were constructed by normalizing the mean value of the kinetic parameter illustrated to the mean value for the glutamate/glycine pair.
Energy landscapes (Figure 5b) were graphed by calculating and plotting changes in free energies relative to the free energies of the closed states C3, the first diliganded state to be populated during activation. Calculations were done with the rate constants illustrated in Figure 5a and Supplementary Table 3 and the relationship ΔG0 = -RT(lnKeq), where R is the molar gas constant, T is the absolute temperature, and Keq is the equilibrium constant of the transition considered, calculated as the ratio of the forward to reverse rate constants. Barrier heights were of arbitrary magnitude and were represented as E‡ = ΔGo + (10 − lnk+). Wells and peaks were arbitrarily positioned at equal distances along the reaction coordinate and the plots for each agonist pair were slightly shifted horizontally to optimize visibility.
Notations for NMDA receptor subunits are those currently recommended by IUPHAR18 (www.iuphar-db.org). Notations for kinetic components are those of Shelley and Magleby44.
Supplementary Material
Acknowledgments
We thank Jason Meyers and Andrei Popescu for obtaining some of the single-channel records. This work was supported by the NIH (GKP).
Abbreviations
- ACBC
1-Aminocyclobutane-1-carboxylic acid
- ACPC
1-Aminocyclopropane-1-carboxylic acid
- Ala
L-Alanine
- DSC
D-Cycloserine
- DCKA
5,7-Dichlorokynurenic acid
- GluN1
Glycine-binding NMDA receptor subunit (previously abbreviated NR1)
- GluN2
Glutamate-binding NMDA receptor subunit (previously abbreviated NR2)
- HCA
L-Homocysteic acid (L-2-amino-4-sulfobutyric acid)
- HQA
Homoquinolinic acid (pyridine-2-carboxy, 3-methylcarboxylic acid)
- NMDA
N-Methyl-D-aspartic acid
- QA
Quinolinic acid (pyridine-2, 3-dicarboxylic acid)
- SYM
SYM2081 (2S, 4R - 4-methylglutamic acid)
References
- 1.Colquhoun D. What have we learned from single ion channels? J Physiol. 2007;581(2):425–427. doi: 10.1113/jphysiol.2007.131656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Armstrong N, Sun Y, Chen GQ, Gouaux E. Structure of a glutamate-receptor ligand-binding core in complex with kainate. Nature. 1998;395(6705):913–917. doi: 10.1038/27692. [DOI] [PubMed] [Google Scholar]
- 3.Armstrong N, Gouaux E. Mechanisms for activation and antagonism of an AMPA-sensitive glutamate receptor: crystal structures of the GluR2 ligand binding core. Neuron. 2000;28(1):165–181. doi: 10.1016/s0896-6273(00)00094-5. [DOI] [PubMed] [Google Scholar]
- 4.Grosman C, Zhou M, Auerbach A. Mapping the conformational wave of acetylcholine receptor channel gating. Nature. 2000;403(6771):773–776. doi: 10.1038/35001586. [DOI] [PubMed] [Google Scholar]
- 5.Robert A, Armstrong N, Gouaux JE, Howe JR. AMPA receptor binding cleft mutations that alter affinity, efficacy, and recovery from desensitization. J Neurosci. 2005;25(15):3752–3762. doi: 10.1523/JNEUROSCI.0188-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lape R, Colquhoun D, Sivilotti LG. On the nature of partial agonism in the nicotinic receptor superfamily. Nature. 2008;454(7205):722–727. doi: 10.1038/nature07139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mukhtasimova N, Lee WY, Wang HL, Sine SM. Detection and trapping of intermediate states priming nicotinic receptor channel opening. Nature. 2009 doi: 10.1038/nature07923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Howe JR, Colquhoun D, Cull-Candy SG. On the kinetics of large-conductance glutamate-receptor ion channels in rat cerebellar granule neurons. Proc R Soc Lond B Biol Sci. 1988;233(1273):407–422. doi: 10.1098/rspb.1988.0030. [DOI] [PubMed] [Google Scholar]
- 9.Gibb AJ, Colquhoun D. Glutamate activation of a single NMDA receptor-channel produces a cluster of channel openings. Proc R Soc Lond B Biol Sci. 1991;243(1306):39–45. doi: 10.1098/rspb.1991.0007. [DOI] [PubMed] [Google Scholar]
- 10.Banke TG, Traynelis SF. Activation of NR1/NR2B NMDA receptors. Nat Neurosci. 2003;6(2):144–152. doi: 10.1038/nn1000. [DOI] [PubMed] [Google Scholar]
- 11.Popescu G, Auerbach A. Modal gating of NMDA receptors and the shape of their synaptic response. Nat Neurosci. 2003;6(5):476–483. doi: 10.1038/nn1044. [DOI] [PubMed] [Google Scholar]
- 12.Auerbach A, Zhou Y. Gating reaction mechanisms for NMDA receptor channels. J Neurosci. 2005;25(35):7914–7923. doi: 10.1523/JNEUROSCI.1471-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schorge S, Elenes S, Colquhoun D. Maximum likelihood fitting of single channel NMDA activity with a mechanism composed of independent dimers of subunits. J Physiol. 2005 doi: 10.1113/jphysiol.2005.095349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dravid SM, Prakash A, Traynelis SF. Activation of recombinant NR1/NR2C NMDA receptors. J Physiol. 2008;586(Pt 18):4425–4439. doi: 10.1113/jphysiol.2008.158634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Erreger K, Dravid SM, Banke TG, Wyllie DJ, Traynelis SF. Subunit-specific gating controls rat NR1/NR2A and NR1/NR2B NMDA channel kinetics and synaptic signalling profiles. J Physiol. 2005;563(Pt 2):345–358. doi: 10.1113/jphysiol.2004.080028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang W, Howe JR, Popescu GK. Distinct gating modes determine the biphasic relaxation of NMDA receptor currents. Nat Neurosci. 2008;11(12):1373–1375. doi: 10.1038/nn.2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cull-Candy S, Brickley S, Farrant M. NMDA receptor subunits: diversity, development and disease. Curr Opin Neurobiol. 2001;11(3):327–335. doi: 10.1016/s0959-4388(00)00215-4. [DOI] [PubMed] [Google Scholar]
- 18.Collingridge GL, Olsen RW, Peters J, Spedding M. A nomenclature for ligand-gated ion channels. Neuropharmacology. 2008 doi: 10.1016/j.neuropharm.2008.06.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Monyer H, et al. Heteromeric NMDA receptors: molecular and functional distinction of subtypes. Science. 1992;256(5060):1217–1221. doi: 10.1126/science.256.5060.1217. [DOI] [PubMed] [Google Scholar]
- 20.Vicini S, et al. Functional and pharmacological differences between recombinant N-methyl-D-aspartate receptors. J Neurophysiol. 1998;79(2):555–566. doi: 10.1152/jn.1998.79.2.555. [DOI] [PubMed] [Google Scholar]
- 21.Johnson JW, Ascher P. Glycine potentiates the NMDA response in cultured mouse brain neurons. Nature. 1987;325(6104):529–531. doi: 10.1038/325529a0. [DOI] [PubMed] [Google Scholar]
- 22.Kleckner NW, Dingledine R. Requirement for glycine in activation of NMDA-receptors expressed in Xenopus oocytes. Science. 1988;241(4867):835–837. doi: 10.1126/science.2841759. [DOI] [PubMed] [Google Scholar]
- 23.Jahr CE. High probability opening of NMDA receptor channels by L-glutamate. Science. 1992;255(5043):470–472. doi: 10.1126/science.1346477. [DOI] [PubMed] [Google Scholar]
- 24.Popescu G, Robert A, Howe JR, Auerbach A. Reaction mechanism determines NMDA receptor response to repetitive stimulation. Nature. 2004;430(7001):790–793. doi: 10.1038/nature02775. [DOI] [PubMed] [Google Scholar]
- 25.Priestley T, et al. Pharmacological properties of recombinant human N-methyl-D-aspartate receptors comprising NR1a/NR2A and NR1a/NR2B subunit assemblies expressed in permanently transfected mouse fibroblast cells. Mol Pharmacol. 1995;48(5):841–848. [PubMed] [Google Scholar]
- 26.Sheinin A, Shavit S, Benveniste M. Subunit specificity and mechanism of action of NMDA partial agonist D-cycloserine. Neuropharmacology. 2001;41(2):151–158. doi: 10.1016/s0028-3908(01)00073-9. [DOI] [PubMed] [Google Scholar]
- 27.Inanobe A, Furukawa H, Gouaux E. Mechanism of partial agonist action at the NR1 subunit of NMDA receptors. Neuron. 2005;47(1):71–84. doi: 10.1016/j.neuron.2005.05.022. [DOI] [PubMed] [Google Scholar]
- 28.Furukawa H, Gouaux E. Mechanisms of activation, inhibition and specificity: crystal structures of the NMDA receptor NR1 ligand-binding core. Embo J. 2003;22(12):2873–2885. doi: 10.1093/emboj/cdg303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen PE, et al. Modulation of glycine potency in rat recombinant NMDA receptors containing chimeric NR2A/2D subunits expressed in Xenopus laevis oocytes. J Physiol. 2008;586(1):227–245. doi: 10.1113/jphysiol.2007.143172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jin R, Banke TG, Mayer ML, Traynelis SF, Gouaux E. Structural basis for partial agonist action at ionotropic glutamate receptors. Nat Neurosci. 2003;6(8):803–810. doi: 10.1038/nn1091. [DOI] [PubMed] [Google Scholar]
- 31.Howe JR, Cull-Candy SG, Colquhoun D. Currents through single glutamate receptor channels in outside-out patches from rat cerebellar granule cells. J Physiol Lond. 1991;432:143–202. doi: 10.1113/jphysiol.1991.sp018381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gibb AJ, Colquhoun D. Activation of N-methyl-D-aspartate receptors by L-glutamate in cells dissociated from adult rat hippocampus. J Physiol (Lond) 1992;456:143–179. doi: 10.1113/jphysiol.1992.sp019331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Furukawa H, Singh SK, Mancusso R, Gouaux E. Subunit arrangement and function in NMDA receptors. Nature. 2005;438(7065):185. doi: 10.1038/nature04089. [DOI] [PubMed] [Google Scholar]
- 34.Erreger K, et al. Subunit-specific agonist activity at NR2A-, NR2B-, NR2C-, and NR2D-containing N-methyl-D-aspartate glutamate receptors. Mol Pharmacol. 2007;72(4):907–920. doi: 10.1124/mol.107.037333. [DOI] [PubMed] [Google Scholar]
- 35.de Carvalho LP, Bochet P, Rossier J. The endogenous agonist quinolinic acid and the non endogenous homoquinolinic acid discriminate between NMDAR2 receptor subunits. Neurochem Int. 1996;28(4):445–452. doi: 10.1016/0197-0186(95)00091-7. [DOI] [PubMed] [Google Scholar]
- 36.Blanke ML, VanDongen AM. The NR1 M3 domain mediates allosteric coupling in the N-methyl-D-aspartate receptor. Mol Pharmacol. 2008;74(2):454–465. doi: 10.1124/mol.107.044115. [DOI] [PubMed] [Google Scholar]
- 37.Erreger K, et al. Mechanism of partial agonism at NMDA receptors for a conformationally restricted glutamate analog. J Neurosci. 2005;25(34):7858–7866. doi: 10.1523/JNEUROSCI.1613-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schorge S, Colquhoun D. Studies of NMDA receptor function and stoichiometry with truncated and tandem subunits. J Neurosci. 2003;23(4):1151–1158. doi: 10.1523/JNEUROSCI.23-04-01151.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang W, Robert A, Vogensen S, Howe JR. The relationship between agonist potency and AMPA receptor kinetics. Biophys J. 2006 doi: 10.1529/biophysj.106.084426. biophysj.106.084426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang W, Cho Y, Lolis E, Howe JR. Structural and single-channel results indicate that the rates of ligand binding domain closing and opening directly impact AMPA receptor gating. J Neurosci. 2008;28(4):932–943. doi: 10.1523/JNEUROSCI.3309-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sun Y, et al. Mechanism of glutamate receptor desensitization. Nature. 2002;417(6886):245–253. doi: 10.1038/417245a. [DOI] [PubMed] [Google Scholar]
- 42.Howe JR. Desensitization at the interface. ACS Chem Biol. 2006;1(10):623–626. doi: 10.1021/cb6004382. [DOI] [PubMed] [Google Scholar]
- 43.Rosenmund C, Stern-Bach Y, Stevens CF. The tetrameric structure of a glutamate receptor channel. Science. 1998;280(5369):1596–1599. doi: 10.1126/science.280.5369.1596. [DOI] [PubMed] [Google Scholar]
- 44.Shelley C, Magleby KL. Linking exponential components to kinetic states in Markov models for single-channel gating. J Gen Physiol. 2008;132(2):295–312. doi: 10.1085/jgp.200810008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.