SUMMARY
Our understanding of the evolution, formation, and pathological disruption of human brain circuits is impeded by a lack of comprehensive data on the developing brain transcriptome. Thus, we have undertaken whole-genome, exon-level expression analysis of thirteen regions from left and right sides of the mid-fetal human brain, finding 76% of genes to be expressed, and 44% of these to be differentially regulated. These data reveal a large number of specific gene expression and alternative splicing patterns, as well as co-expression networks, associated with distinct regions and neurodevelopmental processes. Of particular relevance to cognitive specializations, we have characterized the transcriptional landscapes of prefrontal cortex and perisylvian speech and language areas, which exhibit a population-level global expression symmetry. Finally, we show that differentially expressed genes are more frequently associated with human-specific evolution of putative cis-regulatory elements. Altogether, these data provide a wealth of novel biological insights into the complex transcriptional and molecular underpinnings of human brain development and evolution.
INTRODUCTION
The human brain is an immensely complex organ composed of billions of precisely interconnected neurons. The increase in both size and complexity of the brain, and in particular of the prefrontal cortex (PFC), defines us as a species more than any other evolutionary event (Kostovic, 1990; Hill and Walsh, 2005; Kaas and Preuss, 2007; Bystron et al., 2008). The development of human brain circuitry depends on the diversity and precise spatiotemporal regulation of its transcriptome. Thus, it has long been postulated that changes in the transcriptional regulation of key developmentally expressed genes contributed significantly to the evolution of human brain uniqueness (King and Wilson, 1975; Carroll, 2005; Khaitovich et al., 2006; Sikela, 2006; Vallender et al., 2008; Varki et al., 2008). Such changes are thought to have led to the creation of new combinatorial expression patterns from a relatively limited set of genes, and ultimately to the formation of distinct neuronal circuits that fostered the emergence of higher cognitive skills.
One essential mechanism for increasing the spatiotemporal complexity of the transcriptome is alternative splicing (AS), which generates multiple mRNA transcripts from a single gene. It has been estimated that 70% or more of human multi-exon genes are alternatively spliced (Johnson et al., 2003), and that the majority of splicing events are regulated in a tissue-specific manner (Clark et al., 2007). Moreover, in the adult human, the brain expresses more alternatively spliced transcripts than any other tissue (Yeo et al., 2004), and the importance of specific AS programs to a number of neurodevelopmental and neurological processes has been recognized (Licatalosi and Darnell, 2006; Coutinho-Mansfield et al., 2007). Perhaps most intriguingly, AS has also been implicated as a significant source of evolutionary diversity between human and chimpanzee (Calarco et al., 2007). Nevertheless, the extent and spatial specificity of AS within the developing human brain have not yet been evaluated.
The evolution of the human brain not only provided us with remarkable cognitive and motor abilities, but may also have increased our susceptibility to a spectrum of neurological and psychiatric disorders. Substantial evidence suggests that the symptoms of many human brain disorders are dramatically influenced by pre-existing regional molecular profiles and neuronal circuitry (Morrison and Hof, 1997). For example, schizophrenia and autism are linked to dysfunction of specific cortical circuits, in particular those of the PFC (Levitt, 2005). This suggests that such disorders are defined in part during development by differential gene expression determining regional differences in neuronal circuits.
Despite these motivations, technological and practical limitations have until now precluded a spatially comprehensive transcriptome survey of the human brain during the prenatal period, when key gene expression differences responsible for the unique features of human brain development and evolution are prominent. In this study, we used whole-genome exon microarrays to survey the transcriptome of the human left and right cerebellum, thalamus, striatum, hippocampus, and nine neocortical areas at mid-fetal gestation, a crucial developmental period for the formation of neuronal circuits and the appearance of structural asymmetry in perisylvian neocortical areas. Through multifaceted analysis of these data, we identified novel and potentially species-specific regional, intra-neocortical areal, and cell type-restricted patterns of differential gene expression, coordinated activity and AS associated with various neurodevelopmental processes. In addition, we discovered a disproportionate association of differentially expressed genes with conserved putative cis-regulatory elements displaying human-specific accelerated evolution. Altogether, these data represent a unique and crucial resource, and provide biologically relevant insights into the genetic and molecular mechanisms underlying the development, evolution, and increased disease susceptibility of the human brain.
RESULTS
Microdissection and Evaluation of Late Mid-Fetal Human Brain Specimens
The late mid-fetal stage is a crucial developmental period encompassing multiple cellular processes that are essential to the development of the human brain (Kostovic, 1990; Bystron et al., 2008; Fertuzinhos et al., 2009), and which have also likely undergone substantial evolutionary changes. The regions depicted schematically in Figure 1A, representing five major embryonic brain regions, were microdissected from left and right sides of four late mid-fetal human brains (18, 19, 21 and 23 weeks of gestation [wg]). Samples included cerebellum (CBL), thalamus (THM), striatum (STR), hippocampus (HIP), and nine areas of neocortex (NCTX) individually representing the major sensory and association cortices, with the exception of the somatosensory and motor cortices, which cannot be reliably distinguished at this fetal stage and were therefore combined into one “motor-somatosensory” (MS) sample. Furthermore, we sampled four distinct areas of prefrontal cortex (PFC): orbital (OPFC), dorsolateral (DLPFC), medial (MPFC), and ventrolateral (VLPFC). For complete details of tissue sources and specimens, see Table S1 in the Supplemental Data available with this article online.
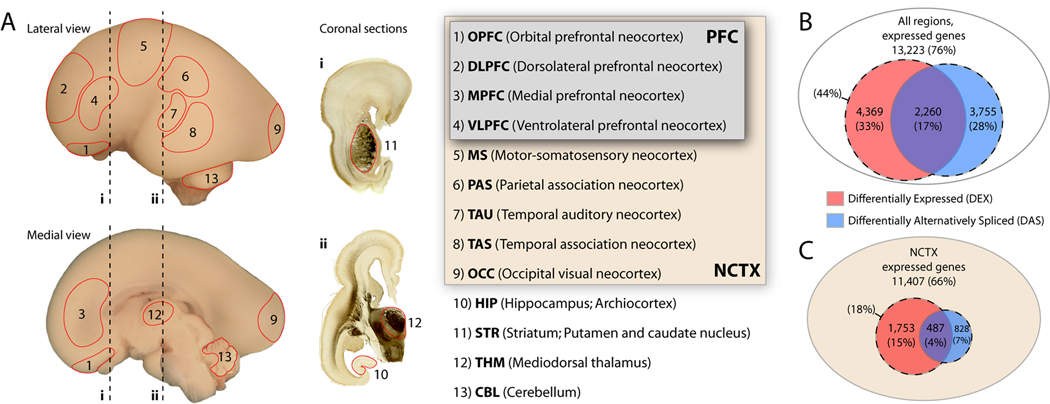
Figure 1. Regional and Neocortical Areal Gene Expression Profiling in the Mid-Fetal Human Brain.
(A) Human late mid-fetal brain illustrating locations of tissue samples microdissected from nine areas of neocortex (NCTX; including four areas of the prefrontal cortex [PFC]), hippocampus (HIP), striatum (STR), mediodorsal thalamus (THM), and cerebellum (CBL) from both sides of four mid-fetal brains. Dashed lines (i, ii) indicate levels of acetylcholine esterase-reacted coronal tissue sections.
(B) Comparison among NCTX, HIP, STR, THM, and CBL detected 76% of core genes expressed above background in at least one brain region. At a FDR of 10−5, 33% of these were DEX and 28% DAS between regions.
(C) Intra-NCTX analysis yielded fewer DEX and DAS genes, even at a relaxed threshold (FDR = 0.01).
Tissue remaining after microdissection was fixed and analyzed for possible neuropathological defects and the presence of all major neuronal and glial cell types present at this developmental age (Figure S1–Figure S2 and data not shown). Genomic analysis with Illumina BeadChip whole-genome genotyping assays confirmed the absence of large-scale genomic structural defects such as aneuploidy. Furthermore, copy number variant (CNV) predictions for all four brains fell within the range of variation found in a sample of 120 well-characterized HapMap individuals, and the majority of predictions corresponded to known CNVs (Figure S3 and Table S2). Thus, by multiple measures, these brains did not exhibit any obvious signs of neurodevelopmental or genetic pathology.
Spatially Comprehensive, Genome-Wide, Exon-Level Expression Profiling
We hybridized RNA isolated from the regions illustrated in Figure 1A to Affymetrix GeneChip Human Exon 1.0 ST Arrays ("Exon Arrays") to obtain independent genome-wide “whole transcript coverage” expression data from each of the four microdissected brains (Table S3). Probesets on the Exon Array are classified as Core, Extended, or Full according to the level of annotation of the source sequence(s); all analyses reported here utilized only the highest confidence “core” probesets, which are based on RefSeq and GenBank full-length mRNAs. Probesets are grouped into “transcript clusters” corresponding to all possible isoforms transcribed from a single locus or gene; therefore, for simplicity we refer to transcript clusters as genes throughout these results.
Hybridization of selected samples to both Affymetrix U133 and Exon microarrays revealed comparable differential expression results between the two platforms (R2 > 0.5; Figure S4). In addition, we performed principal component analysis (PCA) to assess the consistency of Exon Array data within and between brain regions, in comparison to the variability across subjects or hybridizations (Figure S5). PCA confirmed that brain region, rather than individual differences, contributed the majority of variance to the data. Together, these results supported the validity of the Exon Array for comprehensive expression profiling of the mid-fetal human brain.
Regional Differential Gene Expression and Alternative Splicing
We first analyzed gene expression and splicing differences across the five major embryonic brain divisions (NCTX, HIP, STR, THM, and CBL) represented in our samples. Out of 17,421 core transcripts, just over 76% (13,223) were present in at least one brain region, indicating that the great majority of human genes are significantly expressed in the developing brain (Figure 1B). Moreover, at a stringent false discovery rate (FDR) of 10−5, 4,369 genes, or about 33% of those present, were differentially expressed (DEX). In addition, 3,755 genes (28%) exhibited differential exon usage suggestive of region-dependent splicing (“differentially alternatively spliced,” DAS), for a combined 44% of expressed genes showing evidence of some form of differential regulation. A total of 2,260 genes (17%) fell into both categories (DEX and DAS). Of particular note are 1,495 genes, or 11% of those present, detected as DAS but not DEX, which represent a cohort of genes whose differential regulation across brain regions would not be detected by older 3’-biased array platforms. For our purposes, AS encompasses alternative promoter usage, polyadenylation site usage, and cassette exon splicing. Numbers of genes present, DEX, and DAS from all analyses are given in Table S4.
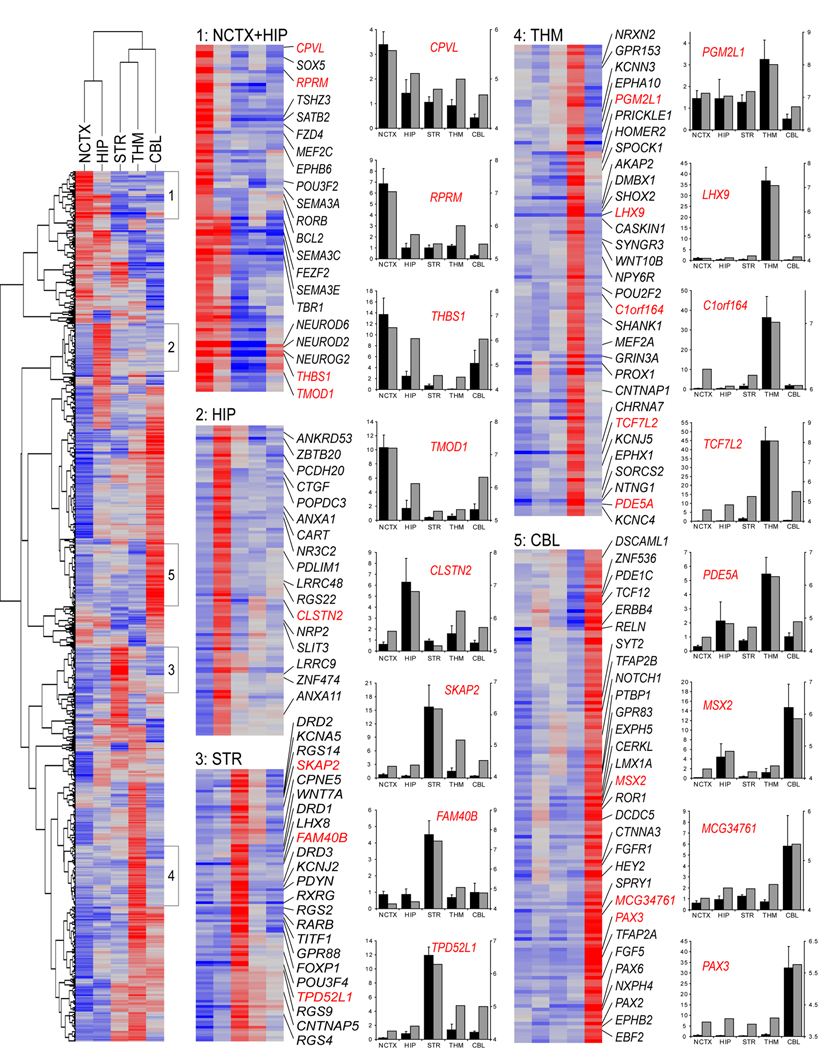
To identify candidates for further study, we selected DEX genes (FDR < 10−5) with a minimum two-fold difference in expression between any two brain regions and performed unsupervised hierarchical clustering on both genes and brain regions. We used the resulting heatmaps to identify groups of genes with the most specific or restricted expression patterns (Figure 2). In addition to large numbers of novel expression patterns, these clusters included transcription factors whose mouse orthologs are critical for the development of region-specific neuronal cell types, including FEZF2, SATB2,SOX5 and TBR1 in cortex, and TITF1 in STR (Sur and Rubenstein, 2005; Chen et al., 2005; Molyneaux et al., 2007; Leone et al., 2008; Britanova et al., 2008; Kwan et al., 2008). This validates our approach to identifying region-specific expression patterns in the developing human brain. Correlations and sizes of these and all subsequent clusters are given in Table S5; complete lists of genes in these clusters are given in Table S6.
Figure 2. Unsupervised Hierarchical Clustering and qRT-PCR Validation of Selected Genes Differentially Expressed Between Brain Regions.
Genes with FDR<10−5 and greater than twofold maximum expression difference were clustered using uncentered correlation and average linkage. Selected highly correlated (r>0.75) clusters of genes with the most restricted expression patterns, indicated in the heatmap at left, are shown in detail at right, with representative validated genes labeled in red. Red is higher expression, blue is lower expression. Complete gene lists for these clusters are given in Table S6. Bar graphs show qRT-PCR (black bars) next to Exon Array data (grey bars) for representative validated genes. Bar graphs represent fold-change relative to the average of all samples for qRT-PCR (black bars; mean±SEM) and median normalized log expression level for Exon Array (grey bars).
Consistent with their ontogenetic and phylogenetic closeness and similar cellular composition, NCTX and HIP were the only two brain regions with a positive correlation (r=0.13) across all of the genes clustered, as reflected in the 57 genes in cluster one enriched in both regions (Figure 2 and Table S5 and Table S6). CBL was the most distinct of the brain regions sampled, consistent with previous findings in the adult human brain (Khaitovich et al., 2004; Roth et al., 2006; see also Table S11) and reflecting differences in both its cytoarchitecture and possibly its developmental time course relative to forebrain structures.
These data provide a survey of the proportions of the human transcriptome with broad or specific expression in the late mid-fetal brain, as well as identifying a large number of regionally enriched or alternatively spliced genes not previously identified as such. In addition, the detection of many orthologs of important rodent neurodevelopmental genes suggests their human counterparts play evolutionarily conserved roles in establishing regional neuronal identity and connectivity.
Validation of Regional Differential Expression Data
We next sought to validate some of these novel expression patterns by additional methods. Genes were prioritized by expression level, fold change, functional classification, and existing rodent and monkey data. Particular emphasis was placed on previously unknown or uncharacterized genes, and on those that appeared to differ in their regional enrichment from available data in other species. Sixty-five of 68 genes validated by qRT-PCR (bar graphs in Figure 2; Table S6 and Table S9) correlated with Exon Array data (median r=0.95), giving us extremely high confidence in Exon Array gene-level analysis. We further examined over 20 candidate genes by in situ hybridization (ISH) or immunohistochemistry (IHC) in whole sagittal or coronal sections of more than ten additional mid-fetal human brains (Table S6, Figure S6, and data not shown; brain specimens listed in Table S1). These confirmation data provide further confidence in the Exon Array platform, as well as identifying an initial cohort of strong candidate genes for functional analyses.
Genetic Patterning of the Developing Human Neocortex
Next, we used a similar approach to search for areal differences in gene expression and splicing within the NCTX. Previous studies have found few differences between adult human neocortical areas (Khaitovich et al., 2004; Roth et al., 2006). We hypothesized that developing NCTX would show more genetic differences, both due to the possibility of an increased number of genes expressed during development and because of the neuronal differentiation and establishment of connectivity that occur during the late mid-fetal period. Comparing among nine areas of NCTX and using the same FDR threshold as the previous analysis (10−5), we identified 471 DEX (4.1%) and 496 DAS (4.3%) genes from the total of 11,407 expressed (Table S4). We hypothesized that important gene expression differences among neocortical areas might be smaller in magnitude than those between brain regions, and thus relaxed the FDR to 1% for exploratory cluster analysis, yielding 1,753 DEX (15.4%) and 828 DAS (7.3%) genes (Figure 1C). Clustering revealed a clear division between the four PFC areas and the four non-frontal lobe areas (PAS, TAU, TAS, OCC) (Figure S7). Interestingly, the MS area, which was dissected from the border region of the frontal and parietal lobes, was not highly correlated with either prefrontal or non-frontal areas. Rather, genes enriched in MS were roughly evenly divided into those correlated with PFC and those correlated with more posterior cortical areas (Figure S7), consistent with the mixed frontal/parietal nature of the tissue sample.
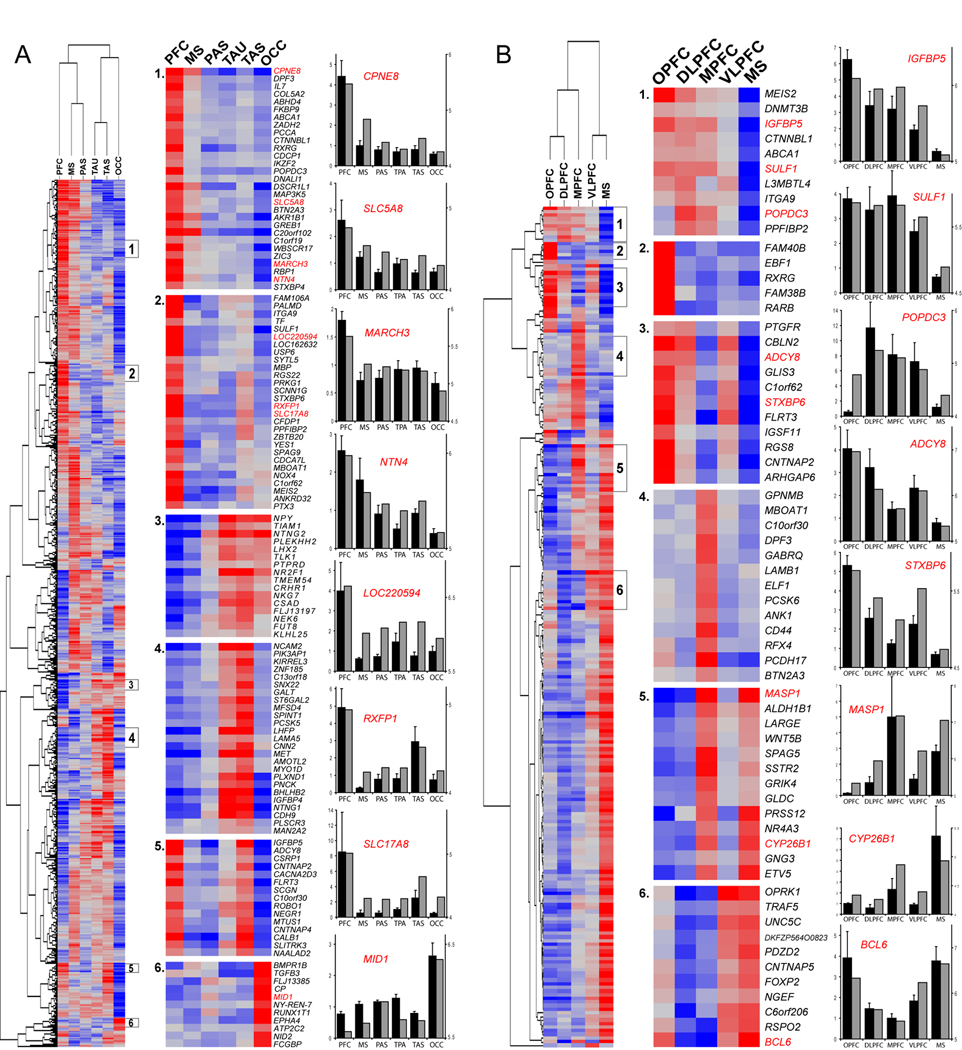
In order to investigate both PFC- and non-frontal-enriched genes in greater detail, we performed two additional, targeted intra-NCTX analyses. First, we grouped together the four PFC samples and compared them to the remaining NCTX areas. Hierarchical clustering of DEX genes (Figure 3A and Table S5 and Table S6) revealed, in addition to PFC- and non-frontal-enriched genes, more specific patterns of enrichment including TAU+TAS, PFC+TAS, and OCC. This analysis confirmed previous reports of enrichment of PCDH17 and CNTNAP2 (Figure 3B) in mid-fetal human frontal NCTX (Abrahams et al., 2007) and EPHA3 and EPHA7 (Table S9) in the fetal rhesus macaque monkey occipital and temporal NCTX, respectively (Sestan et al., 2001). However, the vast majority of our results revealed a complexity of expression patterns in the developing NCTX that has not been previously recognized in either humans (see Table S11), non-human primates, or rodents (see Table S12).
Figure 3. Unsupervised Hierarchical Clustering and qRT-PCR Validation of Selected Genes Differentially Expressed Between NCTX Areas.
(A) With PFC areas grouped, correlated clusters (r>0.8) of gene enrichment included: (1 and 2) PFC; (3) temporal (TAU+TAS) +OCC; (4) temporal lobe (TAS+TAU); (5) PFC+TAS; and (6) OCC.
(B) We separately analyzed the four PFC areas plus MS, and identified genes enriched within specific frontal lobe areas. Selected clusters include (1) pan-PFC, but not MS; (2) OPFC; (3) orbital and lateral PFC; (4) MPFC; (5) MPFC+MS; (6) VLPFC+MS. Red is higher expression, blue is lower expression. Bar graphs show qRT-PCR confirmation (black bars; mean±SEM) next to array data (grey bars) (axes as in Figure 2) with a median ANOVA p<0.0004 and correlation to array data r=0.9. Complete lists of genes in these clusters are given in Table S7–Table S8.
Areal Differences in the Transcriptome of the Developing Prefrontal Cortex
Next, we hypothesized that small but significant gene expression differences might exist among functionally distinct PFC areas during development. Thus, in our second targeted intra-NCTX analysis, we compared the four PFC areas and the MS sample that partially clustered with them (Figure 3B). This analysis yielded 233 DEX genes (2.1% of those present; FDR 1%; Table S4 and Table S8), representing the first evidence for genetic differences between functionally distinct PFC areas in human or non-human primate developing brain. We found the most specific gene enrichment in OPFC, followed by MPFC; other clusters defined various combinations of PFC areas and MS. Interestingly, VLPFC, which encompasses the prospective Broca’s speech area and its right hemisphere homolog, was more closely correlated to MS than to other prefrontal areas at this developmental stage (r=0.36), suggesting a molecular similarity to the neighboring orofacial motor cortex controlling muscles involved in speech production. Moreover, genes enriched in VLPFC+MS included FOXP2 (Figure 3B, cluster 6), haploinsufficiency of which causes a severe speech and language disorder associated with morphological abnormalities and functional underactivation of Broca's area (Lai et al., 2003; Liégeois et al., 2003). This differential expression of FOXP2 within the human frontal lobe has not been observed in previous studies (Lai et al., 2003; Teramitsu et al., 2004), suggesting that the late mid-fetal stage may be a critical time-point for the role of this gene in establishing speech-related cortical circuitry. In addition, genes that clustered with FOXP2 may be candidates for further investigation in speech and language disorders (e.g., CNTNAP5, BCL6, C6orf206).
Finally, this result prompted us to search more specifically for genes enriched in the perisylvian cortical region that encompasses future speech and language-related areas: VLPFC, MS, PAS, TAS, and TAU. An analysis contrasting these areas with the remaining NCTX samples identified many slightly but significantly enriched genes (Table S10). FOXP2 showed a modest 1.1-fold increase in perisylvian areas, while the greatest enrichment was found for TRPC7, TRPC4, and the unknown locus DKFZp547H025 (1.3- to 1.4-fold enrichment). In addition, NR4A2, which was 1.2-fold enriched in perisylvian areas, has previously been found to be enriched in mid-fetal temporal NCTX (Abrahams et al., 2007). These results suggest that despite being dispersed across the frontal, parietal, and temporal lobes, the combined perisylvian cortical areas may express a developmental genetic signature related to their common involvement in speech and language in humans. Altogether, these patterns of expression suggest genetic programs for the development of PFC and perisylvian areas involved in higher cognitive functions, and thus represent promising candidate genes for evolutionary and functional analyses.
Validation and Detailed Cellular Mapping of Intra-Neocortical Differentially Expressed Genes
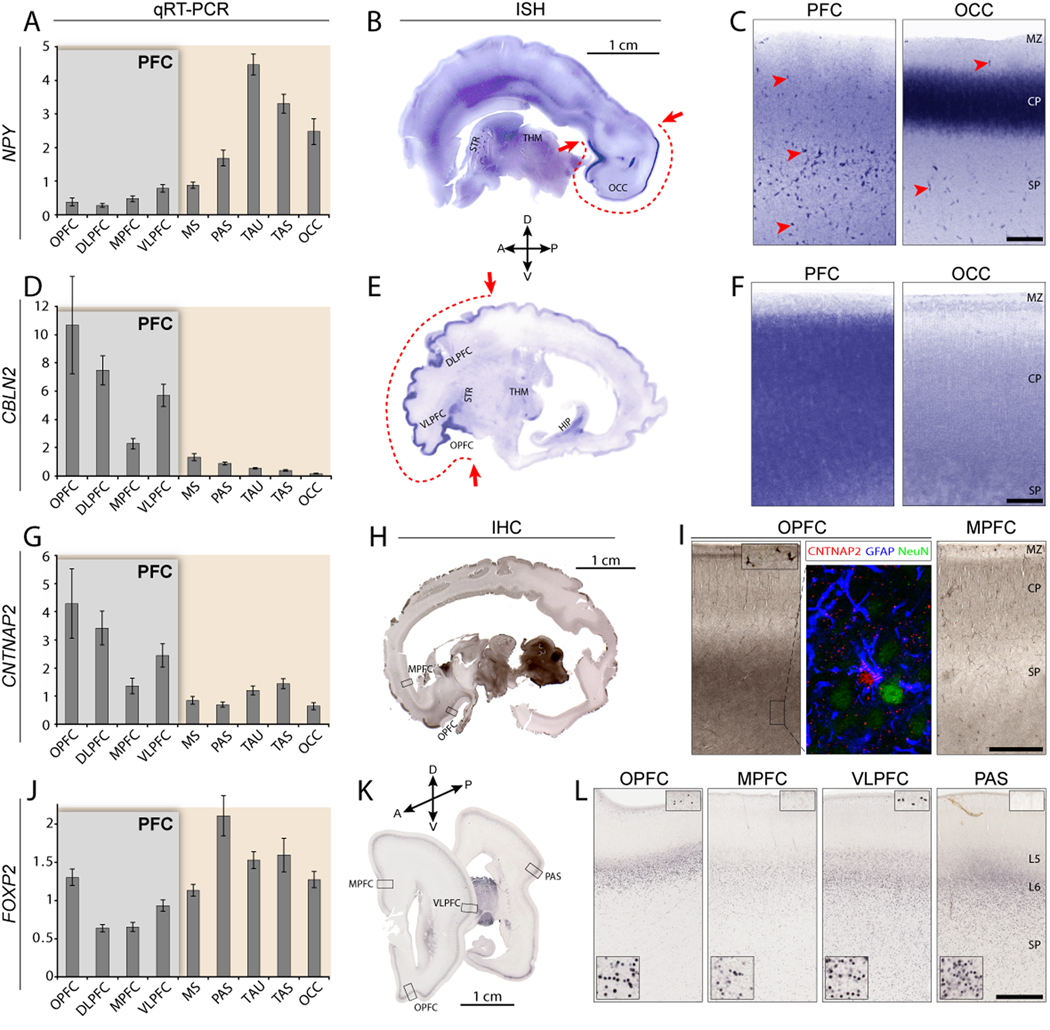
Genes identified as DEX within NCTX (Figure 3A) or PFC (Figure 3B) were chosen for further confirmation based on previous association with disorders such as autism, dyslexia, or speech and language impairments (e.g., CNTNAP2, ROBO1, FOXP2; Bakkaloglu et al., 2008; Hannula-Jouppi et al., 2005; Lai et al., 2003) or for apparent divergence from available rodent expression data (e.g., ANKRD32, CPNE8, POPDC3; see Table S12). We validated 75 intra-NCTX DEX genes by qRT-PCR (bar graphs in Figure 3; Table S7–Table S9), with a focus on PFC-enriched candidates, again finding a very high level of correlation between Exon Array and qRT-PCR results (median r > 0.9; median ANOVA p < 0.005). In addition, we confirmed 18 intra-NCTX DEX genes by ISH and/or IHC (Figure 4 and Table S9). These analyses revealed that despite the cellular heterogeneity of NCTX, Exon Array analysis was able to detect areal differences not only in genes exhibiting widespread expression within a neocortical area (Figure 4F) but also in genes whose expression differs in specific cell types, including astrocytes (Figure 4I), subplate neurons and marginal zone cells (Figure 4L), and cortical layer-specific neurons (Figure 4C). Thus, gene expression in various cell types contributes to neocortical areal molecular differences during development.
Figure 4. Confirmation and Cellular Mapping of Selected Neocortical Areal Expression Differences.
Confirmation of array results by qRT-PCR, and the detection of areal- and cell type-restricted expression patterns by ISH and IHC.
(A–C) NPY enrichment in non-frontal areas is confirmed by qRT-PCR, with highest expression in temporal (TAS, TAU) and occipital lobes (OCC). (B) ISH on a whole sagittal section of 24 wg human brain confirms NPY enrichment in OCC. NPY enrichment in temporal cortex is not visible in this very medial tissue section. (C) Higher magnification reveals specific enrichment in the middle of the occipital cortical plate (CP), and high expression in scattered cells throughout the subplate (SP) (red arrowheads).
(D–F) CBLN2 enrichment in OPFC and lateral PFC is confirmed by qRT-PCR (D) and ISH on a whole sagittal section of 24 wg brain (E). (F) Higher magnification reveals that CBLN2 is enriched throughout the prefrontal CP and SP, but absent from the marginal zone (MZ).
(G–I) CNTNAP2 is selectively enriched in OPFC and lateral PFC areas. (H–I) IHC reveals specific diffuse enrichment in orbitofrontal SP and high expression in scattered MZ cells. Triple-immunofluorescent staining (I, middle panel) reveals colocalization of CNTNAP2 with astrocytic marker GFAP but not neuronal marker NeuN, suggesting differential expression of CNTNAP2 in SP astrocytes.
(J–L) FOXP2 is differentially expressed within the frontal cortex, and enriched in perisylvian cortex. IHC in coronal 22 wg brain sections (K–L) suggests that these differences are accounted for by a combination of higher cellular expression levels in the CP, particularly in OPFC (lower insets), and greater numbers of FOXP2-immunopositive SP cells, especially in VLPFC and PAS. Interestingly, strongly FOXP2-positive cells were present in the MZ in VLPFC and OPFC, but were completely absent from the MZ in other areas (upper insets). Bar graphs are mean±SEM. Scale bars in C, F, I, L: 500 µm.
Global Left-Right Symmetry of Late Mid-Fetal Neocortical Gene Expression
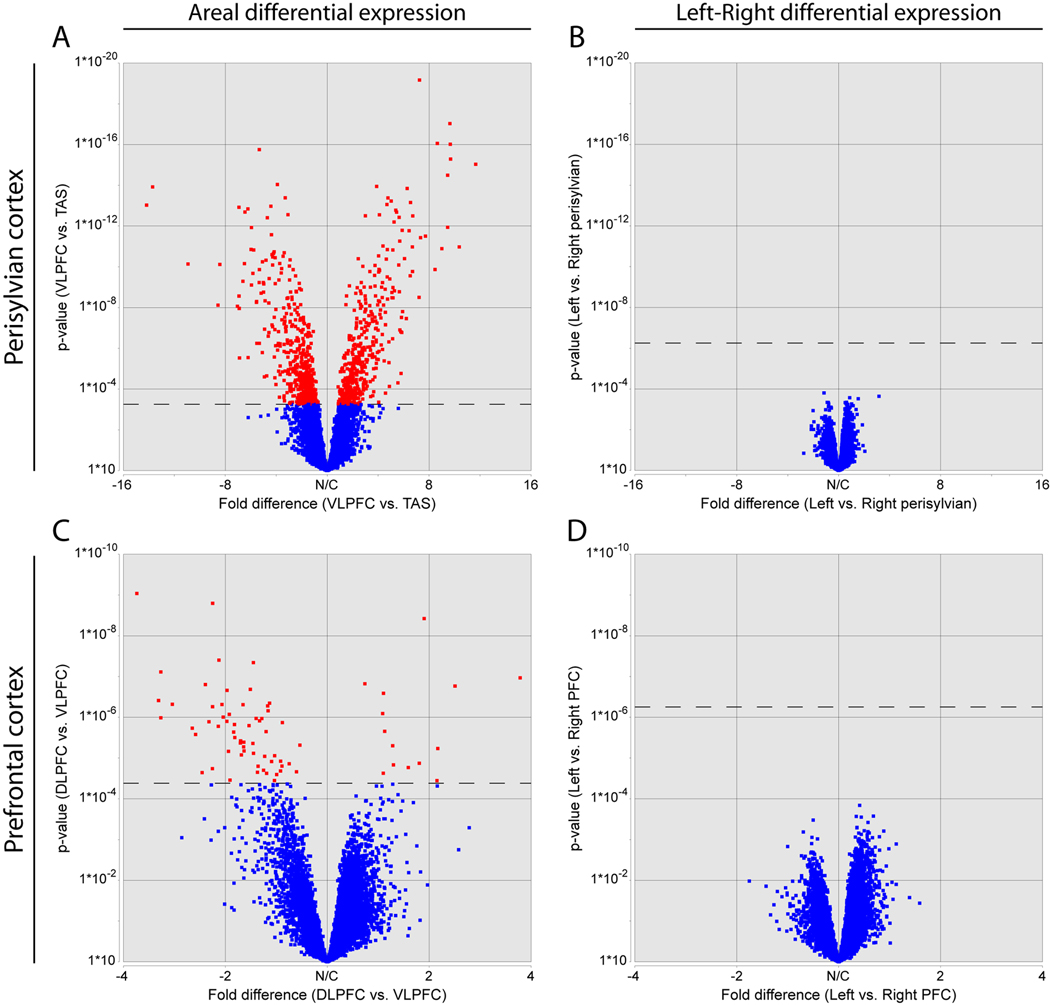
The human brain exhibits structural and functional left-right differences, a prominent example of which is the interhemispheric asymmetry of perisylvian NCTX underlying the functional lateralization in hand preference and speech and language processing (Galaburda et al., 1978). Subtle neocortical structural asymmetries first become evident during the late mid-fetal period analyzed in this study (Chi et al., 1977). Furthermore, a recent study by Sun et al. (2005) identified left-right differences in expression of LMO4 and other genes in the perisylvian regions of the human neocortex during the early mid-fetal period. To investigate whether such molecular correlates of cortical structural asymmetry persist into the late mid-fetal period, we compared gene expression and alternative exon usage between the left and right cortical hemispheres. Our Exon Array analysis did not detect any population-level hemispheric bias of gene expression or AS in individual neocortical areas or other analyzed brain regions. For example, we analyzed perisylvian areas (VLPFC, PAS, TAS, TAU) involved in speech and language processing, but were unable to detect significant differences either in perisylvian cortex as a whole (Figure 5B) or in individual areas (Figure S8). To further validate these findings, we performed qRT-PCR for over 120 genes displaying nonsignificant trends towards neocortical interhemispheric asymmetry on the Exon Arrays (data not shown). Only four of these genes showed a significant hemispheric bias (p<0.05; THBS1, SUMO2, CXorf1, and FAM5C), and all four of these results were driven by large but inconsistent asymmetry in specific neocortical areas, possibly reflecting inter-individual differences in these genes’ spatial or temporal regulation. For example, THBS1, a member of a gene family implicated in synaptogenesis (Christopherson et al., 2005) and human neocortical evolution (Cáceres et al., 2007), was ~2-fold enriched in the right VLPFC in two of the four brains tested. Overall, our results reveal a population-level global interhemispheric symmetry of gene expression in the late mid-fetal NCTX.
Figure 5. Population-level Global Left-Right Symmetry of Gene Expression in the Late Mid-Fetal Prefrontal and Perisylvian Neocortex.
Volcano plots depicting the range of fold-differences and uncorrected p-values in representative perisylvian (A–B) and prefrontal (C–D) areas of NCTX. A large number of genes show significant differences in expression between NCTX areas (A and C). In contrast, no genes showed significant differences between left and right hemispheres within areas (B and D). Dashed lines indicate a corrected p-value of 0.01 (step-up FDR).
Spatial Regulation of Alternative Splicing
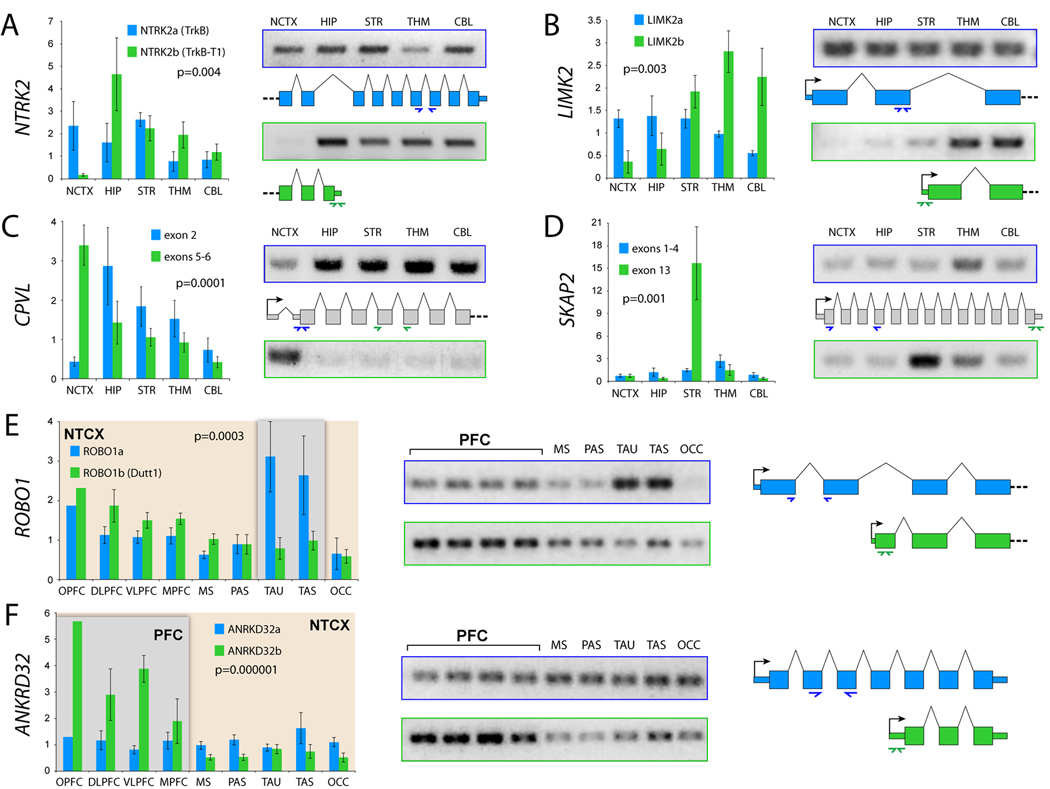
A major advantage of the Exon Array platform is the ability to probe individual exons within a transcript and thus test for tissue-specific expression of alternative isoforms. Analysis of probeset-level expression data identifies genes with a significant interaction between exon expression and tissue as candidates for DAS (Figure S9). From the 28% of genes significantly DAS across brain regions (Figure 1B), we selected a number of promising candidates, some with known alternative isoforms and some previously unknown to be alternatively spliced, and used exon- and isoform-specific qRT-PCR for validation (Figure 6). One of these, NTRK2, encodes multiple isoforms, including the full-length NTRK2a and the truncated NTRK2b (also known as TrkB-T1), which through distinct signaling pathways promote cortical neurogenesis or astrogliogenesis, respectively, in response to BDNF in mice (Cheng et al., 2007). We found that the truncated isoform, NTRK2b, is drastically and specifically downregulated in NCTX (Figure 6A), suggesting that the transition from neurogenesis to astrogliogenesis is delayed in the NCTX compared to other brain regions.
Figure 6. Validation of Selected Region-Specific Alternative Splicing Patterns.
Validation of differential AS across brain regions (A–D) or neocortical areas (E–F) by exon- or isoform-specific qRT-PCR.
(A) Confirmation of a specific reduction of truncated NTRK2b in NCTX, whereas full-length NTRK2a is more evenly expressed throughout the brain.
(B) The LIMK2b splice variant, which lacks the N-terminal LIM protein-binding domain, is predominantly enriched in THM and CBL.
(C–D) CPVL and SKAP2 are each predicted to encode a previously unknown short isoform as a result of alternative promoter usage. Exon 2 of CPVL is drastically reduced in NCTX, while exons 5–6 are expressed in a complementary pattern (C). A more than 10-fold enrichment in STR of SKAP2 exon 13 is consistent with expression of a novel variant composed of exons 8–3 (D).
(E) ROBO1 encodes two known isoforms, the full-length ROBO1a and the alternative short isoform ROBO1b. In the developing human NCTX, ROBO1a is highly enriched in temporal lobe, while ROBO1b is slightly enriched in PFC.
(F) ANKRD32 is a previously uncharacterized gene that appears to encode two splice isoforms, ANKRD32a, which is evenly expressed across the NCTX, and ANKRD32b, which is significantly enriched in PFC. Bar graphs are mean±SEM. P-values represent the interaction between brain region or NCTX area and splice isoform.
Another DAS gene, LIMK2, is one of two LIM kinases that regulate actin dynamics and may be involved in neurite morphogenesis (Endo et al., 2007). LIMK2 has three known isoforms, at least two of which, LIMK2a and LIMK2b, are regulated by distinct promoters, encode distinct proteins, and display expression differences between human brain and other tissues (Nomoto et al., 1999). We found that within mid-fetal brain, LIMK2b is specifically enriched in THM and CBL (Figure 6B). Although functional differences between these two splice variants have yet to be characterized, region-specific splicing within the developing brain suggests that they may play distinct roles in neuronal morphogenesis. Interestingly, Nomoto et al. (1999) found evidence of a role for RORA in transcriptional regulation of LIMK2b, but not LIMK2a. Our data also supported such a relationship, with RORA and LIMK2b showing parallel enrichment in THM and CBL (Figure S10), suggesting the potential for Exon Array data to help predict transcriptional regulatory relationships at the level of individual splice variants.
We also identified DAS candidates with previously unknown alternative splice variants. For example, Exon Array data indicated differential expression of novel variants of CPVL and SKAP2, two genes with unknown functions in brain development (Figure S9). Our qRT-PCR data confirmed a specific reduction of CPVL exon 2 in NCTX (Figure 6C), and a more than 10-fold enrichment of SKAP2 exon 13 in STR (Figure 6D). Together, Exon Array and qRT-PCR data predict novel short isoforms of CPVL and SKAP2 consisting of exons 4–13 and 8–13, respectively, of their full-length transcripts (Figure 6 and Figure S9).
Finally, we identified and confirmed several intra-NCTX DAS candidates. These included ROBO1 (Figure 6E), a key axon guidance gene whose mouse ortholog is required for the formation of major cortical axonal projections (Andrews et al., 2006). It encodes two main isoforms, ROBO1a and ROBO1b (Dutt1), recently shown to be differentially expressed in the embryonic mouse brain (Nural et al., 2007). We found that human ROBO1a is enriched in temporal lobe, while ROBO1b is enriched in PFC (Figure 6E), suggesting that ROBO1 AS may be involved in patterning human intracortical connectivity.
In addition, many intra-NCTX DAS genes have not been previously charaterized. For example, we found that PFC enrichment of ANKRD32 (Figure 3A, cluster 2) reflects differential expression of a novel short isoform, ANKRD32b, whereas full-length ANKRD32a is expressed at lower levels uniformly across the NCTX (Figure 6F). Altogether, our evidence for regional differential splicing and predictions of novel isoforms provide insights into specific functional mechanisms of human brain development and suggest a wealth of biological hypotheses for future work. More generally, our Exon Array AS data illuminate a new level of complexity in the transcriptome of the developing human brain.
Regulatory Evolution of Genes Expressed in the Developing Human Brain
Recently, the completed genome sequences of humans, chimpanzees, and other species have been leveraged to identify highly conserved noncoding sequences (CNSs) that often act as transcriptional cis-regulatory elements (Pennacchio et al., 2006). A subset of these elements appears to have undergone human-specific accelerated evolution (haCNSs) (Prabhakar et al., 2006). We tested whether DEX genes are disproportionately associated with these haCNSs. We first identified the list of 6,921 genes nearest to CNSs, 10% of which are near haCNSs. We found that out of 1,045 intra-NCTX DEX genes near CNSs, 181 (17%) were near haCNSs (Table S13a), representing a highly significant enrichment of accelerated evolution in regulatory regions (p=7.15×10−16; Table 1). In contrast, in a control set of 985 genes equally highly expressed in NCTX but with no differential expression across areas, only 8% were found near haCNSs, a significant decrease relative to the genome-wide rate (p=0.0056). Thus, for NCTX-expressed genes, those with differential or restricted expression were twice as likely to be associated with accelerated human evolution of cis-regulatory elements. Consistent with this effect, a disproportionate 20% of genes most significantly enriched in perisylvian areas involved in speech and language processing were associated with haCNSs, including FOXP2, PCDH9, LPHN2, and SORCS3 (Table S10). In order to determine whether this effect was specific to NCTX or was more generally related to fine spatial control of gene expression during human brain development, we tested the top regionally DEX genes, as well as another control list of highly expressed, non-DEX genes. Again, DEX genes were more than twice as likely to be associated with haCNSs as non-DEX genes (16% versus 7%; Table 1 and Table S13b). Finally, because many genes are also associated with chimpanzee-accelerated CNSs (caCNSs; Prabhakar et al., 2006), we tested whether this trend was preserved among genes near haCNSs but not caCNSs. We again found a significant, though lesser, enrichment of haCNSs near DEX genes (Table 1, right-hand columns). Together, these results suggest that accelerated evolution of putative cis-regulatory elements is a feature of a subset of genes with highly specific expression in the developing human brain.
Table 1. Accelerated Human Evolution of Putative cis-Regulatory Elements Near Differentially Expressed Genes.
Throughout the genome, 10% of genes nearest to conserved non-coding sequences (CNSs) are near those displaying evidence of accelerated evolution in the human lineage (haCNSs). Genes differentially expressed either within the NCTX or throughout the brain are significantly more likely to be near haCNSs (17% and 16%, respectively). In contrast, genes that are highly expressed without spatial specificity show a slight but significant decrease in association with haCNSs (~7–8%). This trend is preserved when excluding genes that are also near chimpanzee-accelerated elements (caCNSs). P-values represent significance by the hypergeometric distribution of the difference between the observed rate of haCNSs and the rate among all CNS-associated human gene (10%).
| Nearest to CNSs |
Nearest to haCNSs | Nearest to haCNSs but not caCNSs | |||||
|---|---|---|---|---|---|---|---|
| # | # | % | p-value | # | % | p-value | |
| All human genes | 6921 | 694 | 10.0% | 425 | 6.1% | ||
| Regionally DEX genes | 1263 | 203 | 16.1% | 2.5E-14 | 101 | 8.0% | 6.2E-04 |
| Highly expressed, not DEX | 1033 | 70 | 6.8% | 2.1E-05 | 51 | 4.9% | 0.012 |
| Intra-NCTX DEX genes | 1045 | 181 | 17.3% | 7.1E-16 | 85 | 8.1% | 9.8E-04 |
| Intra-NCTX high, not DEX | 985 | 81 | 8.2% | 0.006 | 51 | 5.2% | 0.023 |
Biological Themes of Differential Gene Expression
To characterize the major biological themes present in regional DEX genes, we assigned Gene Ontology, protein domain, pathway, and other annotations to the five clusters illustrated in Figure 2, and tested for significant enrichment of functionally related terms (Table S14). Not surprisingly, most of the enriched terms involved neural development or function. In the cortex (HIP+NCTX; cluster 1), transcriptional regulation-related annotations were both the most significant (top four terms, all p<0.004) and most numerous (79% of enriched genes). These included transcription factors known to be important in the development of cortical projection neurons such as FEZF2, SATB2, and SOX5, as well as functionally uncharacterized factors such as TSHZ3 and ZNF238. Another over-represented class of genes enriched in cortex were those involved in axon guidance (e.g., SEMA3A, SEMA3C, EPHB6, UNC5D), which may reflect the transition of afferent axons from the subplate to the deep cortical plate at this stage (Kostovic, 1990). In contrast, in STR (cluster 3), the most enriched annotation clusters involved synaptic transmission or specific receptor signaling pathways (e.g., dopamine receptor and GPCR signaling), consistent with the more developed and neurochemically diverse striatal circuits present at this stage (Sajin et al., 1992).
We next annotated the intra-NCTX DEX gene clusters highlighted in Figure 3 and identified similar themes, including signal transduction, transmembrane proteins, and neurotransmitter receptor activity (Table S14). Notably, there was again a highly significant enrichment of axon guidance-related genes (e.g., NTN4, ROBO1, EPHA4, PLXND1, NTNG1), consistent with the notion that differences in neuronal connectivity are critical for the definition of distinct functional areas. However, there was little overlap between the axon guidance genes uniformly enriched in NCTX and those DEX between NCTX areas, suggesting that while the former may be responsible for the general establishment of cortical afferent and efferent projections, the latter may be involved in more specific targeting of neocortical area-specific circuitry.
Network Organization of the Mid-Fetal Human Brain Transcriptome
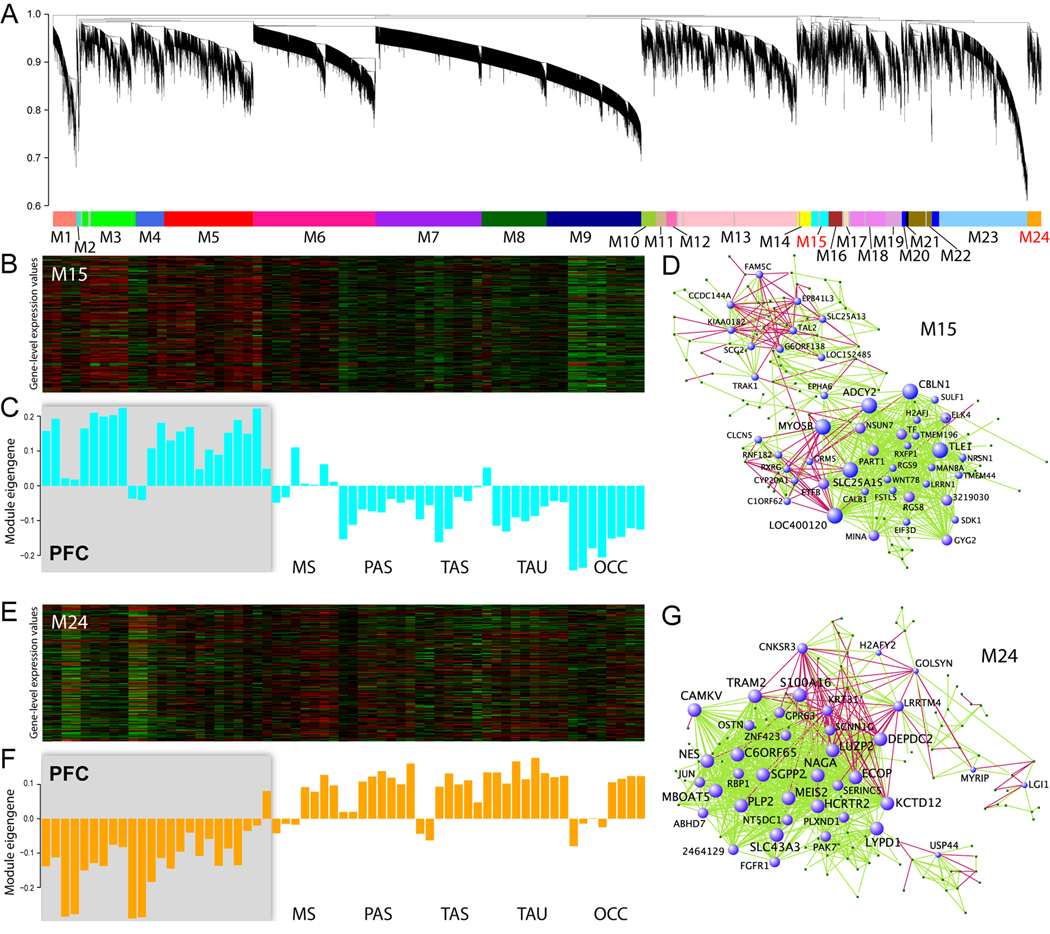
Finally, we sought to go beyond conventional differential gene expression analysis, which alone is not sufficient to convey all of the biological information embedded in large, high-dimensional data sets. We therefore performed weighted gene co-expression network analysis to identify groups of co-regulated genes, or “modules,” with similar patterns of connectivity (high topological overlap) (Oldham et al., 2008). We identified modules corresponding to both brain regions, e.g. NCTX and THM (modules M32 and M31; Figure S11 and Table S15–Table S16), and NCTX areas (modules M15 and M24; Figure 7 and Table S17–Table S18). These modules overlapped significantly with DEX genes, consistent with their regional identity. Notably, the two clearest examples of intra-NCTX modules, shown in Figure 7, replicate our earlier finding of a broad PFC-versus-non-frontal division of cortical gene expression profiles (see Figure S7).
Figure 7. Network Structure of Gene Co-Regulatory Relationships in Developing Human Neocortex.
Network analysis was performed to identify modules of co-regulated genes.
(A) Dendrogram showing clustering of genes based on topological overlap to identify modules of co-regulated genes in the NCTX. Modules were determined by dynamic tree cutting, numbered, and color-coded.
(B–C) Heatmap (B) and first principal component (C) of expression data for genes in module M15 (cyan) suggest identification of the module with PFC.
(D) The network structure of the PFC module illustrates which genes are the most interconnected. The PFC hub gene LOC400120 is an uncharacterized locus that was not DEX or DAS by conventional expression analysis.
(E–G) Module M24 (orange), in contrast, corresponds to non-PFC areas. Hubs in this network include known forebrain patterning genes MEIS2 and FGFR1, as well as genes previously unknown or uncharacterized in nervous tissue, such as TRAM2 and C6orf65.
Genes with the highest degree of within-module connectivity, termed “hub genes,” are expected to play important functional roles in the biology of the network. Thus, while traditional expression analysis may identify a list of genes, all of which are highly enriched in a given tissue, network analysis provides some insight into which of those genes may be more functionally relevant. In the NCTX module (Figure S11D), hub genes included ZIC2 and ZIC4, genes crucial for midline patterning of the dorsal forebrain (Aruga, 2004); LRRC7, a post-synaptic protein involved in dendritic morphology (Quitsch et al., 2005); and FOXG1, a transcription factor essential for neocortical specification in rodents (Sur and Rubenstein, 2005) and linked to microcephaly and mental retardation in humans (Shoichet et al., 2005). In the THM module (Figure S11G), the presence of hub gene TCF7L2, an important WNT signaling pathway transcription factor (Clevers, 2004), led us to identify 13 additional WNT pathway-annotated genes in the module (Table S16), suggesting a central role for WNT signaling in human THM development.
In the PFC network, hubs included PFC-enriched or -suppressed genes such as CBLN1, RGS8, and PART1 (Figure 7D and Table S17), as well as genes of unknown function, such as LOC400120 (C13orf36), that were not DEX or DAS in any of our conventional expression analyses. Non-PFC network hubs included MEIS2 and FGFR1 (Figure 7G), consistent with the roles of retinoic acid and FGF signaling, respectively, in mouse forebrain and neocortical patterning (LaMantia, 1999; Sur and Rubenstein, 2005; Rash and Grove, 2006; O'Leary et al., 2007).
Other modules exhibited no obvious relationship to a particular brain region, and may represent more distributed transcriptional networks important for mid-fetal human brain development. Notably, only two modules, M39 and M41, contained more DAS than DEX genes, suggesting that a common program of AS may contribute to these genes’ transcriptional co-regulation. This hypothesis was supported for at least one of these modules (M39; Figure S12A) by Gene Ontology analysis: the most significantly over-represented Biological Process annotations for this module included RNA splicing, mRNA processing, and related terms. Genes in this module included the neuronal-specific splicing factor PTBP2 (nPTB; Coutinho-Mansfield et al., 2007), as well as several HNRP, DEAD box, and other splicing factor family genes (Table S19). Although these GO terms also appeared in module M41 (Figure S12B; Table S20), this module was dominated by the more general annotations “RNA metabolic process” and “gene expression,” suggesting a broader transcriptional machinery network. Altogether, network analysis has identified coordinated regulation of gene activity and biological pathways involved in patterning and development of the mid-gestation human brain, elucidating an additional level of complexity in the functional organization of the fetal brain transcriptome. These networks contain mostly uncharacterized transcripts, as well as some key developmental and disease-relevant genes, and provide a rich source of new hypotheses about the functional and transcriptional relationships between genes involved in human brain development.
DISCUSSION
Our genome-wide exon-resolution analysis of the mid-fetal human brain transcriptome revealed complex spatial patterns of gene expression and alternative exon usage, as well as co-expression networks, the vast majority of which have not been previously described. We have found that approximately 76% of well-annotated human genes are expressed at this crucial neurodevelopmental stage. At a conservative false discovery rate of 10−5, 33% of these are DEX and 28% are DAS (Figure 1B and Table S4). The vast majority of these genes have not previously been studied, emphasizing how little is known about the transcriptome of the human fetal brain. Our exploration of these data through various analyses has generated a large number of specific and testable hypotheses with biological relevance to human brain evolution, development, and dysfunction.
Spatial and Functional Organization of the Human Mid-Fetal Brain Transcriptome
These analyses reveal novel developmental gene expression patterns corresponding to known anatomical and functional subdivisions of the human brain. While some spatial expression specificities may reflect underlying structural differences, others may reflect transient cellular developmental events, temporal neurogenic and maturational gradients, or spatially restricted signaling centers.
Our analysis has identified more DEX genes in the fetal human brain than have previously been reported in studies of adult human or embryonic mouse brain (Funatsu et al., 2004; Khaitovich et al., 2004; Roth et al., 2006; Kudo et al., 2007; Mühlfriedel et al., 2007). Multiple factors most likely contribute to this difference, including the increased sensitivity and genomic coverage of the Exon Array and other methodological differences. In addition, due to both the evolutionary differences of neocortical development between rodent and human (Preuss, 1995) as well as differences in developmental time-points surveyed, the larger number of DEX genes found in the present study may represent in large part a greater molecular diversity of human cortical areas and cell types. Furthermore, although methodological differences once again contribute, our finding of roughly two orders of magnitude more gene expression differences compared to the adult human NCTX suggests that prenatal differences in gene expression are more robust and complex than those present in the adult human brain.
Global Population-level Interhemispheric Genetic Symmetry of the Late Mid-fetal Neocortex
Structural left-right asymmetry is a prominent feature of the human NCTX and first appears during the late mid-fetal stage (Chi et al., 1977; Galaburda et al., 1977). However, we have found global population-level symmetry of gene expression during this period. Using the SAGE technique, Sun et al. (2005) reported significant asymmetric expression of the transcription factor LMO4 at 12 and 14 wg, but this difference was reduced at 16 to 17 wg and not detectable by 19 wg. Together, these data indicate that significant interhemispheric asymmetry of gene expression is likely a transient feature of the embryonic and early fetal NCTX, thus preceding by several weeks the structural asymmetry visible at late mid-gestation. If gene expression asymmetries are present in the late mid-fetal NCTX, they are likely limited to small differences in a few genes, and in a limited set of cell types.
Spatial Regulation of Alternative Splicing
Although the importance of AS in nervous system development has by now been well established, it has not previously been studied on a genome-wide scale in the developing human brain, nor has the prevalence of differential splicing between brain regions been comprehensively addressed. Our study has uncovered spatial patterns of enrichment of both known and novel splice variants. These data include, to our knowledge, the first evidence for intra-NCTX differential splicing, including enrichment of specific variants in the developing human PFC. One example is the axon guidance gene ROBO1, which is required for formation of major cortical connections in mouse (Andrews et al., 2006). Interestingly, a translocation breakpoint in the ROBO1 gene that has been associated with developmental dyslexia, a disorder linked to alterations in cortical circuits, results in loss of ROBO1a transcription while leaving ROBO1b unaffected (Hannula-Jouppi et al., 2005). In addition, recent work implicates differential splicing of mouse Robo3 in the midline crossing of spinal cord axons (Chen et al., 2008). Intriguingly, cortical midline (callosal) projections exhibit a rostrocaudal developmental gradient and prominent areal differences in the fetal rhesus macaque monkey (Dehay et al., 1988). Thus, although a role for AS in cortical axon guidance has not previously been identified, our finding of ROBO1 intra-NCTX DAS, together with several other lines of evidence, suggests that differential areal expression of axon guidance gene splice variants is likely an important mechanism of cortical circuit formation.
Our splicing analysis also identified 19 members of the protocadherin family of cell adhesion molecules (data not shown), consistent with previous reports of extensive splicing in this gene group (Wu and Maniatis, 1999). In contrast, neither DSCAM, famous for extensive AS in Drosophila (Schmucker et al., 2000), nor the related gene DSCAML1 appeared to be DAS in our analysis. We expect that the public availability of these Exon Array data will enable discovery and functional analysis of many more AS patterns and variants, generating a more complete picture of the transcriptional and posttranscriptional complexity of the developing human brain transcriptome.
Transcriptional Landscape of the Developing Prefrontal Cortex
We have identified more than 200 genes with putative expression differences within the mid-fetal human frontal lobe, many of which appear to be absent from or uniformly expressed in the developing mouse cortex (Table S12). These expression patterns may reflect species-specific differences in functionally specialized prefrontal areas, such as the complex social and emotional processing of the OPFC, and may also suggest new hypotheses regarding the genetic mechanisms controlling arealization of the human PFC. Many of these genes encode members of the same or related families of proteins, e.g., cerebellins; contactin associated proteins; and cadherins, suggesting a particular relevance of specific pathways or functions. In addition, several of the genes identified have been previously implicated in disorders that are thought to involve alterations of human PFC circuitry. These include CNTNAP2, which is not only related to language delay in autism, but is a target of the language-related transcriptional repressor FOXP2, and has a more general role as a susceptibility factor for specific language impairment (Vernes et al., 2008). Our data elaborate on these results, showing specific co-enrichment of CNTNAP2 and FOXP2 in OPFC and VLPFC. Thus, our study has uncovered complex spatial patterns of gene expression and AS that may reflect the underlying developmental, cellular, and species-specific differences between distinct PFC areas.
Implications for Clinical Research
Our data reveal previously unknown spatial expression patterns for many human disease-relevant genes. Furthermore, our data can help evaluate the results of genome-wide association or linkage studies by narrowing the focus to those genes that are specifically expressed or restricted to a relevant brain circuit during development. Finally, we contribute a rare resource in the form of whole-genome genotyping and expression data from the same individuals, enabling correlateion of copy number variation to expression levels across different regions of the developing human brain.
Implications for the Genetic Mechanisms of Human Brain Evolution
For more than a quarter century, the hypothesis has been advanced that variation in regulation of gene expression during development, rather than protein sequence, was the dominant factor in human phenotypic evolution (King and Wilson, 1975). In fact, a number of recent comparative studies on the evolution of coding sequences have shown that brain-enriched and -specific proteins have evolved more slowly than those enriched in other tissues (Duret and Mouchiroud, 2000), as well as more slowly and more rarely in humans than in other primates (Bakewell et al., 2007; Wang et al., 2007). Furthermore, consisitent with a critical role of regulatory changes in the evolution of uniquely human traits, a number of recent studies have identified signatures of positive selection or accelerated evolution in the human genome in non-coding sequences related to neural development or function (Pollard et al., 2006; Prabhakar et al., 2006; Haygood et al. 2007). Importantly, recent work has demonstrated that the human-specific substitutions in some of these regions can dramatically alter the spatial extent of their enhancer activity in transgenic mice (Prabhakar et al., 2008). However, the lack of spatially comprehensive transcriptome data from prenatal development, at which time crucial genetic and molecular processes direct the formation of neuronal circuits, has precluded systematic investigation of the relationship between the evolution of regulatory elements and spatial patterns of gene expression in the developing human brain. Our analysis finds that CNSs proximal to mid-fetal brain DEX genes, likely acting in many cases as cis-regulatory elements, show a disproportionate frequency of human-specific accelerated evolution (Table 1). Therefore, assuming an initially random distribution of tolerated mutations in CNSs, our results suggest that human-specific regulatory evolution at the level of CNSs has contributed to an increased spatial specificity of developmental brain expression in a subset of genes. This may provide a genetic mechanism for increased expression of human cortical genes (Preuss et al., 2004), mosaic changes in developmental and evolutionary trends confined to specific subsystems, or the emergence of novel phenotypic traits (Barton and Harvey, 2000; Rilling, 2001; Sherwood et al., 2008). Notably, many haCNS-associated DEX genes are enriched in PFC and perisylvian areas involved in higher cognitive functions (Table S13). Thus, this small subset of DEX genes represents candidates for involvement in critical aspects of human cognitive development and evolution. At the same time, the fact that a great majority of both DEX and non-DEX genes are not associated with haCNSs suggests general genetic and allometric constraints on the developmental trends and coordinated evolution of brain regions (Finlay and Darlington, 1995). These findings are a necessary step in a process that will require comparative analyses with expression data from multiple primate species and developmental time-points, in an effort to elucidate transcriptional mechanisms that led to the phenotypic specializations of human and non-human primate brains.
EXPERIMENTAL PROCEDURES
Human Brain Specimens and Tissue Processing
This study was carried out using post-mortem human brain specimens collected from the Human Fetal Tissue Repository at the Albert Einstein College of Medicine (AECOM). Dissected tissue was fresh-frozen in Trizol for RNA and DNA extraction, with a post-mortem interval of less than 1 hour. Remaining tissue was fixed and frozen, and sections were analyzed for neuropathological or developmental defects. Details of specimens, tissue processing, microdissection, and neuropathological assessment are given in the Supplemental Experimental Procedures and Table S1. These studies were approved by the Human Investigation Committees of AECOM and Yale University.
RNA Isolation, Processing, and Microarray Hybridization
Total RNA was extracted using TRIzol (Invitrogen), followed by treatment with RNeasy Mini Kit (Qiagen) to exclude smaller RNAs. The quality of total RNA was evaluated by 2100 Bioanalyzer (Agilent) and RNA 6000 Nano Kit (Agilent) before being processed with the Affymetrix GeneChip Whole Transcript Sense Target Labeling Assay and hybridized to the Affymetrix Exon 1.0 ST Arrays following the recommended Affymetrix protocols. Hybridized arrays were scanned on an Affymetrix GeneChip Scanner 3000 and visually inspected for hybridization artifacts.
Exon Array Data Analysis
Exon Array data were preprocessed using standard RMA normalization, DABG, and probeset summarization methods in either Partek Genomics Suite (Partek, Inc.) or the Excel Array Analysis software (XRAY; Biotique Systems, Inc.). Principal component analysis, left-right hemisphere ANOVAs, and t-tests were performed in Partek using gene summary values for all core transcript clusters. Global DEX and DAS ANOVAs were performed in XRAY using default parameters. All ANOVAs included brain specimen and date of hybridization as co-factors, to eliminate batch effects and variations due to individual genetic differences. All p-values were corrected for multiple comparisons using the FDR step-down method. Unsupervised hierarchical clustering was performed in Cluster 3.0 (bonsai.ims.u-tokyo.ac.jp/~mdehoon/software/cluster), and heatmaps were generated using Java Treeview (jtreeview.sourceforge.net). For further details, see the Supplemental Experimental Procedures.
Analysis of Patterns of Cis-Regulatory Evolution and Gene Expression
Genomic coordinates of all ~110k CNSs identified by Prabhakar et al. (2006) were mapped to the human genome (hg18; NCBI build 36.1) on the UCSC Genome Browser (genome.ucsc.edu) and cross-referenced with the RefSeq Genes track using Galaxy (main.g2.bx.psu.edu) to identify the nearest human RefSeq gene to each CNS. After removing duplicates, this yielded 6921 genes, of which 694 were near CNSs reported as showing evidence of accelerated evolution in the human lineage (haCNSs). We then intersected these gene lists with lists of DEX and non-DEX genes, calculated the proportion of haCNSs for each condition, and assigned a p-value according to the hypergeometric distribution.
Gene Ontology Annotation Analysis
Annotation analysis was performed using the web-based DAVID software (david.abcc.ncifcrf.gov; Dennis et al., 2003). Intra-NCTX DEX gene clusters from Figure 3 were grouped together to control for the length of annotated gene lists and allow direct comparison with the annotation of Figure 2 clusters. See Supplemental Experimental Procedures for details.
Weighted Gene Co-expression Network Analysis
Network analysis was performed as previously described (Oldham et al., 2008). Annotated R code used for our network analysis is available at www.sestanlab.org/resources. General information on network analysis methodology, as well as WGCNA software, is available at www.genetics.ucla.edu/labs/horvath/CoexpressionNetwork. For further details, see the Supplemental Experimental Procedures.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Bradford Poulos for assistance with tissue acquisition, Aiping Lin for help with microarray platform comparisons, James Noonan for advice on analysis of haCNSs, Fuying Gao for statistical analysis, Donna Karolchik and Andy Pohl for assistance in creating tracks for the Genome Browser, and many colleagues for their help and comments. This work was supported by NIH grants NS054273, MH081896 (to N.S.), NS056276 (to M.W.S.), NS051869 (to S.M.M.), MH060233 (to D.H.G.), Kavli Foundation, NARSAD, and the James S. McDonnell Foundation Scholar Award (to N.S.).
Footnotes
This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Accession Numbers
Microarray data can be accessed through the NCBI Gene Expression Ominbus (accession GSE13344), or viewed as a track on the UCSC Human Genome Browser at genome.ucsc.edu.
SUPPLEMENTAL DATA
Supplemental Data include Supplemental Experimental Procedures, 12 figures, 20 tables, and Supplemental References, and can be found with this article online at www.cell.com.
REFERENCES
- Abrahams BS, Tentler D, Perederiy JV, Oldham MC, Coppola G, Geschwind DH. Genome-wide analyses of human perisylvian cerebral cortical patterning. Proc. Natl. Acad. Sci. USA. 2007;104:17849–17854. doi: 10.1073/pnas.0706128104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews W, Liapi A, Plachez C, Camurri L, Zhang J, Mori S, Murakami F, Parnavelas JG, Sundaresan V, Richards LJ. Robo1 regulates the development of major axon tracts and interneuron migration in the forebrain. Development. 2006;133:2243–2252. doi: 10.1242/dev.02379. [DOI] [PubMed] [Google Scholar]
- Aruga J. The role of Zic genes in neural development. Mol. Cell Neurosci. 2004;26:205–221. doi: 10.1016/j.mcn.2004.01.004. [DOI] [PubMed] [Google Scholar]
- Bakewell MA, Shi P, Zhang J. More genes underwent positive selection in chimpanzee evolution than in human evolution. Proc. Natl. Acad. Sci. USA. 2007;104:7489–7494. doi: 10.1073/pnas.0701705104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakkaloglu B, O'Roak BJ, Louvi A, Gupta AR, Abelson JF, Morgan TM, Chawarska K, Klin A, Ercan-Sencicek AG, Stillman AA, et al. Molecular cytogenetic analysis and resequencing of contactin associated protein-like 2 in autism spectrum disorders. Am. J. Hum. Genet. 2008;82:165–173. doi: 10.1016/j.ajhg.2007.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton RA, Harvey PH. Mosaic evolution of brain structure in mammals. Nature. 2000;405:1055–1058. doi: 10.1038/35016580. [DOI] [PubMed] [Google Scholar]
- Britanova O, de Juan Romero C, Cheung A, Kwan KY, Schwark M, Gyorgy A, Vogel T, Akopov S, Mitkovski M, Agoston D, et al. Satb2 is a postmitotic determinant for upper-layer neuron specification in the neocortex. Neuron. 2008;57:378–392. doi: 10.1016/j.neuron.2007.12.028. [DOI] [PubMed] [Google Scholar]
- Bystron I, Blakemore C, Rakic P. Development of the human cerebral cortex: Boulder Committee revisited. Nat. Rev. Neurosci. 2008;9:110–122. doi: 10.1038/nrn2252. [DOI] [PubMed] [Google Scholar]
- Cáceres M, Lachuer J, Zapala MA, Redmond JC, Kudo L, Geschwind DH, Lockhart DJ, Preuss TM, Barlow C. Elevated gene expression levels distinguish human from non-human primate brains. Proc. Natl. Acad. Sci. USA. 2003;100:13030–13035. doi: 10.1073/pnas.2135499100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calarco JA, Xing Y, Cáceres M, Calarco JP, Xiao X, Pan Q, Lee C, Preuss TM, Blencowe BJ. Global analysis of alternative splicing differences between humans and chimpanzees. Genes Dev. 2007;21:2963–2975. doi: 10.1101/gad.1606907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll SB. Evolution at two levels: On genes and form. PLoS Biology. 2005;3 doi: 10.1371/journal.pbio.0030245. e245 EP. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Rasin M, Kwan KY, Sestan N. Zfp312 is required for subcortical axonal projections and dendritic morphology of deep-layer pyramidal neurons of the cerebral cortex. Proc. Natl.Acad. Sci. USA. 2005;102:17792–17797. doi: 10.1073/pnas.0509032102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Gore BB, Long H, Ma L, Tessier-Lavigne M. Alternative splicing of the Robo3 axon guidance receptor governs the midline switch from attraction to repulsion. Neuron. 2008;58:325–332. doi: 10.1016/j.neuron.2008.02.016. [DOI] [PubMed] [Google Scholar]
- Cheng A, Coksaygan T, Tang H, Khatri R, Balice-Gordon RJ, Rao MS, Mattson MP. Truncated tyrosine kinase B brain-derived neurotrophic factor receptor directs cortical neural stem cells to a glial cell fate by a novel signaling mechanism. J. Neurochem. 2007;100:1515–1530. doi: 10.1111/j.1471-4159.2006.04337.x. [DOI] [PubMed] [Google Scholar]
- Chi JG, Dooling EC, Gilles FH. Gyral development of the human brain. Ann. Neurol. 1977;1:86–93. doi: 10.1002/ana.410010109. [DOI] [PubMed] [Google Scholar]
- Christopherson KS, Ullian EM, Stokes CC, Mullowney CE, Hell JW, Agah A, Lawler J, Mosher DF, Bornstein P, Barres BA. Thrombospondins are astrocyte-secreted proteins that promote CNS synaptogenesis. Cell. 2005;120:421–433. doi: 10.1016/j.cell.2004.12.020. [DOI] [PubMed] [Google Scholar]
- Clark A, Schweitzer C, Chen X, Staples K, Lu G, Wang H, Williams A, Blume E. Discovery of tissue-specific exons using comprehensive human exon microarrays. Genome Biol. 2007;8:R64. doi: 10.1186/gb-2007-8-4-r64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clevers H. Wnt breakers in colon cancer. Cancer Cell. 2004;5:5–6. doi: 10.1016/s1535-6108(03)00339-8. [DOI] [PubMed] [Google Scholar]
- Coutinho-Mansfield GC, Xue Y, Zhang Y, Fu X. PTB/nPTB switch: a post-transcriptional mechanism for programming neuronal differentiation. Genes Dev. 2007;21:l573–1577. doi: 10.1101/gad.1575607. [DOI] [PubMed] [Google Scholar]
- Dehay C, Kennedy H, Bullier J, Berland M. Absence of interhemispheric connections of area 17 during development in the monkey. Nature. 1988;331:348–350. doi: 10.1038/331348a0. [DOI] [PubMed] [Google Scholar]
- Duret L, Mouchiroud D. Determinants of substitution rates in mammalian genes: expression Pattern affects selection intensity but not mutation rate. Mol. Biol.Evol. 2000;17:68–70. doi: 10.1093/oxfordjournals.molbev.a026239. [DOI] [PubMed] [Google Scholar]
- Endo M, Ohashi K, Mizuno K. LIM kinase and slingshot are critical for neurite extension. J. Biol. Chem. 2007;282:13692–13702. doi: 10.1074/jbc.M610873200. [DOI] [PubMed] [Google Scholar]
- Fertuzinhos S, Krsnik Z, Kawasawa YI, Rasin M, Kwan KY, Chen J, Judas M, Hayashi M, Sestan N. Selective depletion of molecularly defined cortical interneurons in human holoprosencephaly with severe striatal hypoplasia. Cereb Cortex Advance online access. 2009 doi: 10.1093/cercor/bhp009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finlay BL, Darlington RB. Linked regularities in the development and evolution of mammalian brains. Science. 1995;268:1578–1584. doi: 10.1126/science.7777856. [DOI] [PubMed] [Google Scholar]
- Funatsu N, Inoue T, Nakamura S. Gene expression analysis of the late embryonic mouse cerebral cortex using DNA microarray: identification of several region- and layer-specific genes. Cereb.Cortex. 2004;14:1031–1044. doi: 10.1093/cercor/bhh063. [DOI] [PubMed] [Google Scholar]
- Galaburda AM, LeMay M, Kemper TL, Geschwind N. Right-left asymmetrics in the brain. Science. 1978;199:852–856. doi: 10.1126/science.341314. [DOI] [PubMed] [Google Scholar]
- Hannula-Jouppi K, Kaminen-Ahola N, Taipale M, Eklund R, Nopola-Hemmi J, Kääriäinen H, Kere J. The axon guidance receptor gene ROBO1 is a candidate gene for developmental dyslexia. PLoS Genet. 2005;1:e50. doi: 10.1371/journal.pgen.0010050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haygood R, Fedrigo O, Hanson B, Yokoyama K, Wray GA. Promoter regions of many neural- and nutrition-related genes have experienced positive selection during human evolution. Nat Genet. 2007;39:1140–1144. doi: 10.1038/ng2104. [DOI] [PubMed] [Google Scholar]
- Hill RS, Walsh CA. Molecular insights into human brain evolution. Nature. 2005;437:64–67. doi: 10.1038/nature04103. [DOI] [PubMed] [Google Scholar]
- Johnson JM, Castle J, Garrett-Engele P, Kan Z, Loerch PM, Armour CD, Santos R, Schadt EE, Stoughton R, Shoemaker DD. Genome-wide survey of human alternative pre-mRNA splicing with exon junction microarrays. Science. 2003;302:2141–2144. doi: 10.1126/science.1090100. [DOI] [PubMed] [Google Scholar]
- Kaas JH, Preuss TM, et al., editors. The Evolution of Primate Nervous Systems. Vol 4. Oxford: Elsevier; 2007. Evolution of Nervous Sytems. [Google Scholar]
- Khaitovich P, Enard W, Lachmann M, PääPbo S. Evolution of primate gene expression. Nat. Rev. Genet. 2006;7:693–702. doi: 10.1038/nrg1940. [DOI] [PubMed] [Google Scholar]
- Khaitovich P, Muetzel B, She X, Lachmann M, Hellmann I, Dietzsch J, Steigele S, Do HH, Weiss G, Enard W, et al. Regional patterns of gene expression in human and chimpanzee brains. Genome Res. 2004;14:1462–1473. doi: 10.1101/gr.2538704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King MC, Wilson AC. Evolution at two levels in humans and chimpanzees. Science. 1975;188:107–116. doi: 10.1126/science.1090005. [DOI] [PubMed] [Google Scholar]
- Kostovic I. Structural and histochemical reorganization of the human prefrontal cortex during perinatal and postnatal life. Prog. Brain Res. 1990;85:223–239. doi: 10.1016/s0079-6123(08)62682-5. discussion 239–240. [DOI] [PubMed] [Google Scholar]
- Kudo LC, Karsten SL, Chen J, Levitt P, Geschwind DH. Genetic analysis of anterior posterior expression gradients in the developing mammalian forebrain. Cereb. Cortex. 2007;17:2108–2122. doi: 10.1093/cercor/bhl118. [DOI] [PubMed] [Google Scholar]
- Kwan KY, Lam MMS, Krsnik Z, Kawasawa YI, Lefebvre V, Sestan N. SOX5 postmitotically regulates migration, postmigratory differentiation, and projections of subplate and deep-layer neocortical neurons. Proc. Natl. Acad. Sci. USA. 2008;105:16021–16026. doi: 10.1073/pnas.0806791105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai CS, Gerrelli D, Monaco AP, Fisher SE, Copp AJ. FOXP2 expression during brain development coincides with adult sites of pathology in a severe speech and language disorder. Brain. 2003;126:2455–2462. doi: 10.1093/brain/awg247. [DOI] [PubMed] [Google Scholar]
- LaMantia AS. Forebrain induction, retinoic acid, and vulnerability to schizophrenia: insights from molecular and genetic analysis in developing mice. Biol. Psychiatry. 1999;46:19–30. doi: 10.1016/s0006-3223(99)00002-5. [DOI] [PubMed] [Google Scholar]
- Leone DP, Srinivasan K, Chen B, Alcamo E, McConnell SK. The determination of projection neuron identity in the developing cerebral cortex. Curr. Opin. Neurobiol. 2008;18:28–35. doi: 10.1016/j.conb.2008.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levitt P. Developmental neurobiology and clinical disorders: lost in translation? Neuron. 2005;46:407–412. doi: 10.1016/j.neuron.2005.04.015. [DOI] [PubMed] [Google Scholar]
- Licatalosi DD, Darnell RB. Splicing regulation in neurologic disease. Neuron. 2006;52:93–101. doi: 10.1016/j.neuron.2006.09.017. [DOI] [PubMed] [Google Scholar]
- Liégeois F, Baldeweg T, Connelly A, Gadian DG, Mishkin M, Vargha-Khadem F. Language fMRI abnormalities associated with FOXP2 gene mutation. Nat. Neurosci. 2003;6:1230–1237. doi: 10.1038/nn1138. [DOI] [PubMed] [Google Scholar]
- Molyneaux BJ, Arlotta P, Menezes JRL, Macklis JD. Neuronal subtype specification in the cerebral cortex. Nat. Rev. Neurosci. 2007;8:427–437. doi: 10.1038/nrn2151. [DOI] [PubMed] [Google Scholar]
- Morrison JH, Hof PR. Life and death of neurons in the aging brain. Science. 1997;278:412–419. doi: 10.1126/science.278.5337.412. [DOI] [PubMed] [Google Scholar]
- Mühlfriedel S, Kirsch F, Gruss P, Chowdhury K, Stoykova A. Novel genes differentially expressed in cortical regions during late neurogenesis. Eur. J. Neurosci. 2007;26:33–50. doi: 10.1111/j.1460-9568.2007.05639.x. [DOI] [PubMed] [Google Scholar]
- Nomoto S, Tatematsu Y, Takahashi T, Osada H. Cloning and characterization of the alternative promoter regions of the human LIMK2 gene responsible for alternative transcripts with tissue-specific expression. Gene. 1999;236:259–271. doi: 10.1016/s0378-1119(99)00280-2. [DOI] [PubMed] [Google Scholar]
- Nural HF, Todd Farmer W, Mastick GS. The Slit receptor Robo1 is predominantly expressed via the Dutt1 alternative promoter in pioneer neurons in the embryonic mouse brain and spinal cord. Gene Expr. Patterns. 2007;7:837–845. doi: 10.1016/j.modgep.2007.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldham MC, Konopka G, Iwamoto K, Langfelder P, Kato T, Horvath S, Geschwind DH. Functional organization of the transcriptome in human brain. Nat. Neurosci. 2008;11:1271–1282. doi: 10.1038/nn.2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Leary DD, Chou SJ, Sahara S. Area patterning of the mammalian cortex. Neuron. 2007;56:252–269. doi: 10.1016/j.neuron.2007.10.010. [DOI] [PubMed] [Google Scholar]
- Pennacchio LA, Ahituv N, Moses AM, Prabhakar S, Nobrega MA, Shoukry M, Minovitsky S, Dubchak I, Holt A, Lewis KD, et al. In vivo enhancer analysis of human conserved non-coding sequences. Nature. 2006;444:499–502. doi: 10.1038/nature05295. [DOI] [PubMed] [Google Scholar]
- Pollard KS, Salama SR, Lambert N, Lambot MA, Coppens S, Pedersen JS, Katzman S, King B, Onodera C, Siepel A, et al. An RNA gene expressed during cortical development evolved rapidly in humans. Nature. 2006;443:167–172. doi: 10.1038/nature05113. [DOI] [PubMed] [Google Scholar]
- Prabhakar S, Noonan JP, Paabo S, Rubin EM. Accelerated evolution of conserved noncoding sequences in humans. Science. 2006;314:786. doi: 10.1126/science.1130738. [DOI] [PubMed] [Google Scholar]
- Prabhakar S, Visel A, Akiyama JA, Shoukry M, Lewis KD, Holt A, Plajzer-Frick I, Morrison H, Fitzpatrick DR, Afzal V, et al. Human-specific gain of function in a developmental enhancer. Science. 2008;321:1346–1350. doi: 10.1126/science.1159974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preuss TM. Do rats have prefrontal cortex? The Rose-Woolsey-Akert program reconsidered. J.Cogn. Neurosci. 1995;7:1–24. doi: 10.1162/jocn.1995.7.1.1. [DOI] [PubMed] [Google Scholar]
- Preuss TM, Cáceres M, Oldham MC, Geschwind DH. Human brain evolution: insights from microarrays. Nat. Rev. Genet. 2004;5:850–860. doi: 10.1038/nrg1469. [DOI] [PubMed] [Google Scholar]
- Quitsch A, Berhorster K, Liew CW, Richter D, Kreienkamp H. Postsynaptic shank antagonizes dendrite branching induced by the leucine-rich repeat protein Densin-180. J. Neurosci. 2005;25:479–487. doi: 10.1523/JNEUROSCI.2699-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rash BG, Grove EA. Area and layer patterning in the developing cerebral cortex. Curr.Opin. Neurobiol. 2006;16:25–34. doi: 10.1016/j.conb.2006.01.004. [DOI] [PubMed] [Google Scholar]
- Rilling JK. Allometric departures for the human brain provide insights into hominid brain evolution. Behav. Brain Sci. 2001;24:292–293. [Google Scholar]
- Roth RB, Hevezi P, Lee J, Willhite D, Lechner SM, Foster AC, Zlotnik A. Gene expression analyses reveal molecular relationships among 20 regions of the human CNS. Neurogenetics. 2006;7:67–80. doi: 10.1007/s10048-006-0032-6. [DOI] [PubMed] [Google Scholar]
- Sajin B, Sestan N, Dmitrovic B. Compartmentalization of NADPH-diaphorase staining in the developing human striatum. Neurosci Lett. 1992;140:117–120. doi: 10.1016/0304-3940(92)90695-4. [DOI] [PubMed] [Google Scholar]
- Schmucker D, Clemens JC, Shu H, Worby CA, Xiao J, Muda M, Dixon JE, Zipursky SL. Drosophila Dscam is an axon guidance receptor exhibiting extraordinary molecular diversity. Cell. 2000;101:671–684. doi: 10.1016/s0092-8674(00)80878-8. [DOI] [PubMed] [Google Scholar]
- Sestan N, Rakic P, Donoghue MJ. Independent parcellation of the embryonic visual cortex and thalamus revealed by combinatorial Eph/ephrin gene expression. Curr. Biol. 2001;11:39–43. doi: 10.1016/s0960-9822(00)00043-9. [DOI] [PubMed] [Google Scholar]
- Sherwood CC, Subiaul F, Zawidzki TW. A natural history of the human mind: tracing evolutionary changes in brain and cognition. J. Anat. 2008;212:426–454. doi: 10.1111/j.1469-7580.2008.00868.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoichet SA, Kunde SA, Viertel P, Schell-Apacik C, von Voss H, Tommerup N, Ropers HH, Kalscheuer VM. Haploinsufficiency of novel FOXG1B variants in a patient with severe mental retardation, brain malformations and microcephaly. Hum. Genet. 2005;117:536–544. doi: 10.1007/s00439-005-1310-3. [DOI] [PubMed] [Google Scholar]
- Sikela JM. The jewels of our genome: the search for the genomic changes underlying the evolutionarily unique capacities of the human brain. PLoS Genet. 2006;2:e80. doi: 10.1371/journal.pgen.0020080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun T, Patoine C, Abu-Khalil A, Visvader J, Sum E, Cherry TJ, Orkin SH, Geschwind DH, Walsh CA. Early asymmetry of gene transcription in embryonic human left and right cerebral cortex. Science. 2005;308:1794–1798. doi: 10.1126/science.1110324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sur M, Rubenstein JL. Patterning and plasticity of the cerebral cortex. Science. 2005;310:805–810. doi: 10.1126/science.1112070. [DOI] [PubMed] [Google Scholar]
- Teramitsu I, Kudo LC, London SE, Geschwind DH, White SA. Parallel FoxP1 and FoxP2 expression in songbird and human brain predicts functional interaction. J. Neurosci. 2004;24:3152–3163. doi: 10.1523/JNEUROSCI.5589-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallender EJ, Mekel-Bobrov N, Lahn BT. Genetic basis of human brain evolution. Trends Neurosci. 2008;31:637–644. doi: 10.1016/j.tins.2008.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varki A, Geschwind DH, Eichler EE. Explaining human uniqueness: genome interactions with environment, behaviour and culture. Nat. Rev. Genet. 2008;9:749–763. doi: 10.1038/nrg2428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernes SC, Newbury DF, Abrahams BS, Winchester L, Nicod J, Groszer M, Alarcón M, Oliver PL, Davies KE, Geschwind DH, et al. A functional genetic link between distinct developmental language disorders. N Engl J Med. 2008;359:2337–2345. doi: 10.1056/NEJMoa0802828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang HY, Chien HC, Osada N, Hashimoto K, Sugano S, Gojobori T, Chou CK, Tsai SF, Wu CI, Shen CK. Rate of evolution in brain-expressed genes in humans and other primates. PLoS Biology. 2007;5:e13. doi: 10.1371/journal.pbio.0050013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Q, Maniatis T. A striking organization of a large family of human neural Cadherin-like cell adhesion genes. Cell. 1999;97:779–790. doi: 10.1016/s0092-8674(00)80789-8. [DOI] [PubMed] [Google Scholar]
- Yeo G, Holste D, Kreiman G, Burge C. Variation in alternative splicing across human tissues. Genome Biol. 2004;5:R74. doi: 10.1186/gb-2004-5-10-r74. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.