Abstract
Bile acids are physiological detergents that generate bile flow and facilitate intestinal absorption and transport of lipids, nutrients, and vitamins. Bile acids also are signaling molecules and inflammatory agents that rapidly activate nuclear receptors and cell signaling pathways that regulate lipid, glucose, and energy metabolism. The enterohepatic circulation of bile acids exerts important physiological functions not only in feedback inhibition of bile acid synthesis but also in control of whole-body lipid homeostasis. In the liver, bile acids activate a nuclear receptor, farnesoid X receptor (FXR), that induces an atypical nuclear receptor small heterodimer partner, which subsequently inhibits nuclear receptors, liver-related homolog-1, and hepatocyte nuclear factor 4α and results in inhibiting transcription of the critical regulatory gene in bile acid synthesis, cholesterol 7α-hydroxylase (CYP7A1). In the intestine, FXR induces an intestinal hormone, fibroblast growth factor 15 (FGF15; or FGF19 in human), which activates hepatic FGF receptor 4 (FGFR4) signaling to inhibit bile acid synthesis. However, the mechanism by which FXR/FGF19/FGFR4 signaling inhibits CYP7A1 remains unknown. Bile acids are able to induce FGF19 in human hepatocytes, and the FGF19 autocrine pathway may exist in the human livers. Bile acids and bile acid receptors are therapeutic targets for development of drugs for treatment of cholestatic liver diseases, fatty liver diseases, diabetes, obesity, and metabolic syndrome.
Keywords: cholesterol 7α-hydroylase, nuclear receptors, farnesoid X receptor, fibroblast growth factor 19, cell signaling, lipid metabolism, cholesterol 7α-hydroxylase, drug therapy, cholestasis, liver diseases
Since the last special review of cholesterol 7α-hydroxylase (CYP7A1) published in the Journal of Lipid Research in 1977 (1), there has been remarkable progress on the molecular mechanisms of regulation of bile acid synthesis. The cloning of the key regulatory gene CYP7A1 about 20 years ago (2–4), followed by the identification of the bile acid-activated receptor farnesoid X receptor (FXR, NR1H4) 10 years later (5–7), has generated high interest in bile acid research. New functions of bile acids in metabolic regulation have been unraveled. It is now well recognized that bile acids are important signaling molecules that coordinately regulate a network of metabolic pathways, including lipid, glucose, drug, and energy metabolism (reviewed in Refs. 8–15).
The enterohepatic circulation of bile acids serves as an important physiological route not only for recycling of bile acids and absorption of nutrients but also for regulation of whole-body lipid metabolism. However, the mechanism underlying this remarkably efficient and complex physiological process has only recently been unraveled. This review will provide an update on the current understanding of the molecular mechanism of regulation of bile acid synthesis, with a focus on the most critical regulatory gene in the pathway, CYP7A1.
It should be emphasized that the bile acid pool in mice consists mostly of hydrophilic bile acids, muricholic acids, and cholic acid and is very different from the hydrophobic bile acid pool consisting predominantly chenodeoxycholic acid (CDCA), cholic acid (CA), and deoxycholic acid (DCA) in humans. Hydrophobic, but not hydrophilic, bile acids are efficacious endogenous ligands of the nuclear receptors FXR (NR1H4), pregnane X receptor (PXR; NR1I2), and vitamin D receptor (VDR; NR1I1) that play critical roles in the regulation of bile acid synthesis and metabolism. Therefore, results from studying bile acid synthesis in the mouse models may not be extrapolated to humans without verification in suitable human models. This review will focus on the regulation of bile acid synthesis in human livers and will address the species differences in regulation.
BILE ACIDS ARE VERSATILE SIGNALING MOLECULES
Bile acids are derived from cholesterol. Bile acid synthesis is the predominant metabolic pathway for catabolism of cholesterol in humans. Hydroxylation and modification of cholesterol to bile acids converts a hydrophobic membrane constituent to amphipathic molecules that can serve as powerful physiological detergents for absorption and transport of nutrients, fats, and vitamins but also as the versatile signaling molecules that are specific ligands for activation of nuclear and membrane receptors. Both free and conjugated bile acids bind to the ligand-binding domain of FXR, which forms a heterodimer with retinoid X receptor and binds to the inverted repeat of AGGTCA-like sequence with one nucleotide spacing (IR1) located in the promoters of the FXR target genes to stimulate gene transcription (5–7). FXR plays a central role in the regulation of bile acid synthesis, excretion, and transport (16, 17) as well as lipid, glucose, and energy metabolism (10, 12, 13, 18, 19). The hydrophobic bile acid CDCA is the most efficacious endogenous FXR ligand, whereas hydrophilic bile acids, such as ursodeoxycholic acid and muricholic acids, do not activate FXR. Bile acids also bind and activate PXR (20) and VDR (21). These two receptors play important roles in detoxification of bile acids, drugs, and xenobiotics (20, 22, 23).
Bile acids have been shown to modulate cellular signaling pathways, including calcium mobilization, cyclic AMP synthesis, and protein kinase C activation (9). It has been reported that bile acids activate the protein kinase C/Janus N-termina kinase pathway (24). Bile acids stimulate secretion of pro-inflammatory cytokines, tumor necrosis factor α (TNFα), and interleuken-1β (IL-1β) from Kupffer cells (resident macrophages in hepatocytes) that activate TNF receptor signaling and the mitogen-activated protein kinase (MAPK)/JNK pathway (25, 26). Conjugated bile acids induce mitochondrial reactive oxidizing species, which activates the epidermal growth factor receptor and Raf-1/MEK/ERK signaling pathway (27, 28). Conjugated bile acids activate the ERK and PI3K/AKT pathways via a pertussis toxin-sensitive mechanism involving Gαi protein-coupled receptor (29, 30). DCA activates the FAS receptor and the JNK pathway by induction of acidic sphingomyelinase-generated ceramide in rat primary hepatocytes (31). Bile acids also stimulate insulin receptor signaling (32). In brown adipose tissue, bile acids activate TGR5, a Gαi protein-coupled receptor (33, 34). TGR5 stimulates production of cAMP, which induces iodothyrone deiodinase (D2) and production of thyroid hormone T3, leading to stimulation of energy metabolism and improving glucose tolerance and insulin sensitivity (10, 12, 19). TGR5 is not expressed in hepatocytes but has been localized in the sinusoid endothelial cells (35). In the enteroendocrine cells, TGR5 stimulates glucagon-like peptide 1 (36), which has antidiabetic activity.
NUCLEAR RECEPTORS REGULATION OF BILE ACID SYNTHESIS
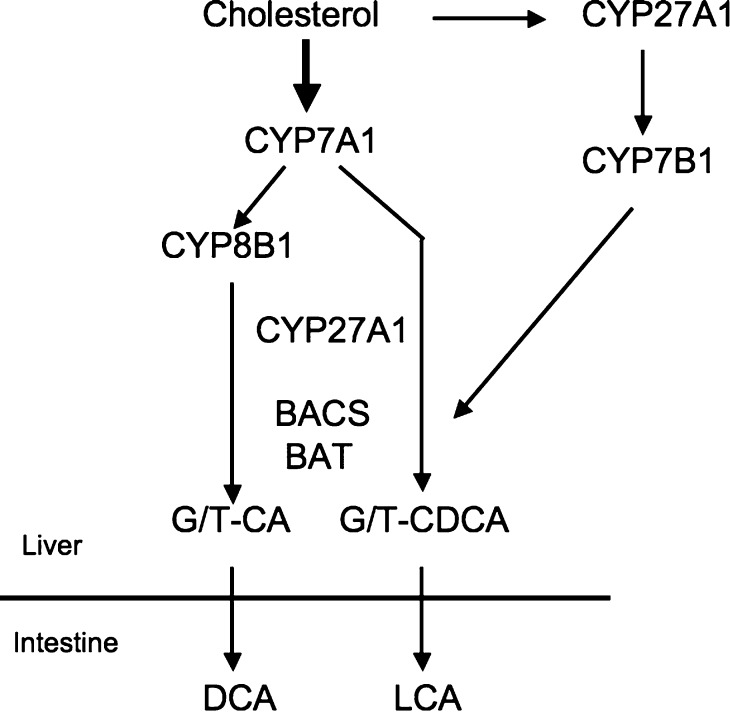
The liver is the only organ that has all 14 enzymes required for de novo synthesis of two primary bile acids in humans, CA (3α, 7α, 12α-trihydroxy-cholanoic acid) and CDCA (3α, 7α-dihydroxy-cholanoic acid) (Fig. 1) (37). The classic bile acid biosynthetic pathway is initiated by CYP7A1 (38). Sterol 12α-hydroxylase (CYP8B1) is required for synthesis of CA. Mitochondrial sterol 27 hydroxylase (CYP27A1) catalyzes sterol side chain oxidation, after which cleavage of a three-carbon unit in the peroxisomes leads to formation of a C24 bile acid. An alternative (acidic) pathway is initiated by CYP27A1, which in addition to the liver is expressed in macrophages and most other tissues, and may contribute significantly to total bile acid synthesis. Other minor pathways initiated by 25-hydroxylase in the liver and 24-hydroxylase in the brain also may contribute to bile acid synthesis. A nonspecific 7α-hydroxylase (CYP7B1) expressed in all tissues is involved in the generation of oxidized metabolites (oxysterols), which may be transported to the liver and converted to CDCA. Most bile acids are conjugated to glycine or taurine to decrease toxicity and increase solubility for secretion into bile. Bile acid:CoA synthase (BACS) and bile acid:amino acid transferase (BAT) are involved in amino acid conjugation of bile acids. In the intestine, glyco- and tauro-conjugated CA and CDCA are deconjugated, and 7α-dehydroxylase activity in bacteria flora removes a 7α-hydroxy group to form secondary bile acids DCA (3α, 12-dihydroxy) and lithocholic acid (LCA; 3α-monohydroxy), respectively. CA, CDCA, and DCA are reabsorbed in the intestine and transported back to the liver to inhibit bile acid synthesis. Most of the LCA is excreted in feces. A small amount of LCA circulated to the liver is sulfo-conjugated at the 3-hydroxy position by sulfotransferase (SULT2A1) and rapidly secreted into bile. Sulfation is the major pathway for detoxification of extremely hydrophobic bile acids in humans (39). Details of bile acid chemistry, biology, physiology, and synthesis have been reviewed recently (40, 41).
Fig. 1.
Bile acid synthesis. Cholesterol is converted to two primary bile acids in human liver, CA and CDCA. Key regulated enzymes, CYP7A1, CYP8B1, CYP27A1, and CYP7B1, in the pathways are indicated. CYP7A1 initiates the classic (neutral) bile acid biosynthetic pathway in the liver. CYP27A1 initiates the alternative (acidic) pathway in the liver and macrophages. CA and CDCA are conjugated to glycine (G) and taurine (T). BACS and BAT are two key enzymes involved in amino conjugation of bile acids. In the intestine, conjugated CA and CDCA are deconjugated and then dehydroxylated at the 7α-position to the secondary bile acids DCA and LCA, respectively.
Regulation of the rate-limiting enzyme in bile acid biosynthetic pathway CYP7A1 has been studied extensively. The CYP7A1 mRNA transcripts in the 3′-untranslated region are unusually long (3) and have a very short half-life of about 30 min (42, 43). It has been reported that bile acids reduce CYP7A1 mRNA stability via the bile acid response elements located in the 3′-untranslated region (43, 44). Numerous studies have demonstrated that bile acids, steroid hormones, inflammatory cytokines, insulin, and growth factors inhibit CYP7A1 transcription through the 5′-upstream region of the promoter (45–50).
Analysis of the proximal promoter of the rat Cyp7a1 identified two regions (footprints) that are putative binding sites for nuclear receptors (51), which are ligand-activated transcription factors that play important roles in embryogenesis, development, and metabolism (16). The sequence located at −73 to −55 of the rat CYP7A1 promoter is highly conserved and was identified as a putative bile acid response element (BARE-I) that might be involved in conferring bile acid inhibition. This sequence contains a DR4 (direct repeat spaced by four nucleotides) motif in all species except the human, which binds liver X receptor (LXRα or NR1H3), an oxysterol-activated nuclear receptor. The CYP7A1 is the first LXRα target gene identified (52, 53). This has been confirmed by the finding that when fed a high cholesterol diet, bile acid synthesis increases in wild-type mice but not in Lxrα null mice, which accumulate high levels of cholesterol in the liver (54). In contrast, the human CYP7A1 promoter does not bind LXRα and is not induced by LXRα due to alteration of the DR4 motif in the BARE-I sequence (55). This has been confirmed by the finding that transgenic mice carrying a human CYP7A1 do not respond to a high cholesterol diet and that the transgene is not induced and bile acid synthesis is not stimulated in these mice (56, 57). Another bile acid response element (BARE-II) is located in a region from −149 to −118 of the rat Cyp7a1 promoter, which has an 18-nucleotide sequence that is completely conserved in many species (58). This sequence contains a DR1 motif, which binds hepatocyte nuclear factor 4α (HNF4α; NR2A1). HNF4α transactivates CYP7A1 promoter activity by interacting with a coactivator, peroxisome proliferator-activated receptor γ coactivator 1α (PGC-1α). Mutation of the DR1 sequence drastically reduced basal CYP7A1 promoter activity and its response to bile acid inhibition (45).
Several earlier studies report that bile acid pool size increases in diabetic rats and insulin inhibits CYP7A1 and CYP8B1 activities (reviewed in Ref. 38). Recent studies show that insulin-regulated transcription factor FoxO1 binds to an insulin response sequence in the rat Cyp7a1 promoter and induces rat Cyp7a1 transcription (47). Insulin signaling phosphorylates FoxO1, which is excluded from the nucleus, and results in inhibiting rat CYP7A1. However, the FoxO1 binding site is not present in the human CYP7A1 promoter, and FoxO1 functions as a repressor that inhibits HNF4α and PGC-1α activation of the human CYP7A1 (47). In this study, insulin at the physiological concentrations rapidly stimulates CYP7A1 expression by inhibiting FoxO1, while high concentrations of insulin found in the insulin resistance state activate steroid response element binding protein-1c, which inhibits CYP7A1 expression by interacting with HNF4α. On the other hand, glucagon and cAMP strongly inhibit CYP7A1 expression via activation of PKA, which phosphorylates HNF4α and abolishes HNF4α DNA-binding activity, resulting in inhibition of CYP7A1 expression in human hepatocytes (48). This is in contrast to other studies in mice that cAMP and fasting induce Cyp7a1 expression, which parallels the induction of PGC-1α and phosphoenolpyruvate carboxykinase (59, 60). These investigators suggest that Cyp7a1 expression and bile acid synthesis is upregulated during fasting and that there is a coordinated regulation of bile acid synthesis and gluconeogenesis by the fasting-to-fed cycle. Biosynthetic pathways should be inactivated during fasting to conserve energy, except glucogeneogenesis, which provides energy during starvation. The induction of Cyp7a1 in fasting response in mice is in contrast to the observation in human patients that the serum 7α-hydroxy-4-cholesten-3-one (C4), a serum marker for bile acid synthesis and CYP7A1 enzyme activity, is reduced during the fasting, increased during the postprandial state, gradually decreased during the postabsorptive state (61). In addition, bile acid synthesis increases in the morning regardless of food intake (62).
FXR plays a critical role in the regulation of bile acid synthesis and homeostasis (63). However, FXR inhibits CYP7A1 (64), CYP8B1 (65), and CYP27A1 (66) transcription by complicated mechanisms described later. A recent study identified an FXR-binding site in the human but not mouse CYP8B1 promoter, suggesting a species-specific regulation of bile acid synthesis by FXR (67). The transcriptional repressor GPS2 differentially regulates CYP7A1 and CYP8B1 transcription by interacting with small heterodimer partner (SHP), liver-related homolog-1 [LRH-1; or human α-fetoprotein transcription factor (FTF), NR5A2], FXR, and HNF4α bound to these two promoters. In the CYP7A1 and CYP8B1 promoters, the HNF4α binding site overlaps with a binding site for LRH-1. The promoter of the human CYP27A1 also has an HNF4α binding site (66). FXR stimulates bile acid conjugation by inducing the genes encoding BACS and BAT, which also are induced by HNF4α (68). Thus, FXR and HNF4α may coordinately regulate bile acid synthesis and conjugation.
It has been reported that peroxisome proliferator activated receptor α (PPARα) plays a role in the regulation of bile acid synthesis (69). Bile acids induce PPARα transcription via induction of FXR (70). PPARα inhibits human CYP7A1 transcription by inhibiting HNF4α transactivation activity (71). PPARα also regulates CYP27A1 expression in macrophages (72). Activation of PPARα has been shown to increase unconjugated bile acids by induction of peroxisomal bile acid thioesterase (73), indicating that PPARα may play a role in balancing the amount of conjugated and free bile acids.
Orphan nuclear receptor Rev-erbα plays a role in the regulation of bile acid synthesis (74). Rev-erbα is a clock gene involved in the control of circadian rhythmicity (74–77). The activity of Rev-erbα is activated by heme (78) and functions as a heme sensor and transcriptional repressor that regulates lipid metabolism and adipogenesis (79, 80). Rev-erbα may coordinate energy metabolism and circadian rhythm during feeding and starvation (81). It has been well documented that CYP7A1 and CYP8B1 expression exhibits a pronounced diurnal rhythm in both a 12 h light:12 h dark cycle and constant darkness (82–85). In humans, bile acid synthesis exhibits a diurnal rhythm with two peaks around 3 and 9 PM. CYP7A1 expression is induced by diurnal regulated D-site binding protein (86–89) and Rev-erbα (90) but repressed by the clock genes DEC2 and E4BP4 (90, 91). These authors conclude that the circadian rhythm of Cyp7a1 is regulated directly by D-site binding protein, DEC2, and Rev-erbα/β. However, in Rev-erbα knockout mice, Cyp7a1 expression is induced and bile acid synthesis is stimulated, but the diurnal rhythm of Cyp7a1 remains (74). These authors suggest that Rev-erbα does not regulate the diurnal rhythm of CYP7A1 but induces CYP7A1 by inhibiting SHP and E4BP4 expression (74). In contrast, Rev-erbα does not regulate the circadian rhythm of CYP8B1.
FXR REGULATION OF ENTEROHEPATIC CIRCULATION OF BILE ACIDS
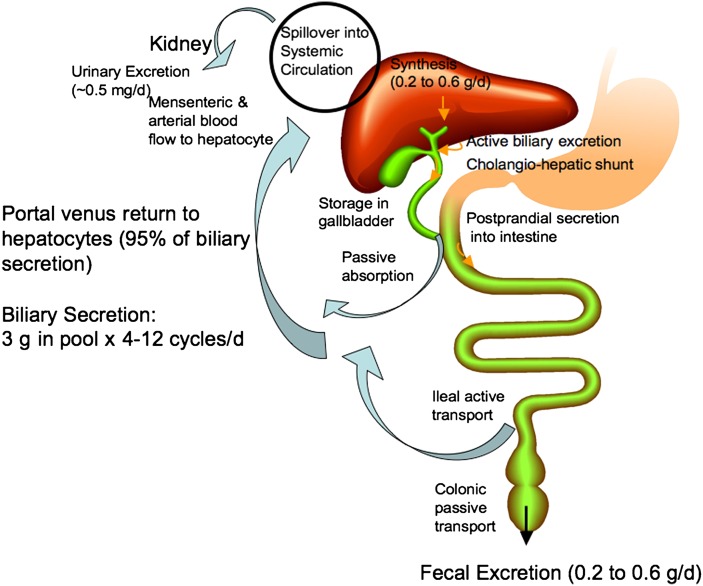
In humans, about 0.2–0.6 g of bile acids are synthesized daily in human liver. The enterohepatic circulation of bile acids is illustrated in Fig. 2. Conjugated bile acids are secreted into bile by canalicular bile salt export pump (BSEP; ABCB11) and stored in the gallbladder. Bile acids are spilled over into sinusoid blood when concentrations increase in the hepatocytes. Bile acids in blood circulation are reabsorbed when passing through the renal tubules in the kidney and are circulated back to the liver through systemic circulation. Some bile acids secreted in the bile duct are reabsorbed in the cholangiocytes and recycled back to hepatoytes (the cholangiohepatic shunt). After each meal, gallbladder contraction empties bile acids into the intestinal tract. When passing through the intestinal tract, some bile acids are reabsorbed in the upper intestine by passive diffusion, but most bile acids (95%) are reabsorbed in the ileum by apical sodium-dependent bile acid transporter (ASBT) located in the brush border membrane. Bile acids are transdiffused across the enterocyte to the basolateral membrane where the organic solute transporter α and β heterdimer (OSTα/OSTβ) effluxes bile acids into portal blood circulation, transported to the sinusoid, and taken up by Na+-dependent taurocholate cotransport peptide (NTCP) into hepatocytes. In the colon, DCA is reabsorbed and recycled with CA and CDCA to the liver. A bile acid pool of about 3 g is recycled 4–12 times a day. Bile acids lost in the feces (0.2–0.6 g/day) are replenished by de novo synthesis in the liver to main a constant bile acid pool.
Fig. 2.
Enterohepatic circulation of bile acids. In humans, about 0.2–0.6 g (averaging 0.5 g) bile acids are synthesized daily in human liver. Conjugated bile acids are secreted into bile and stored in the gallbladder. Some bile acids are spilled over into sinusoid blood and reabsorbed when passing through the renal tubules in the kidney and circulated back to the liver through mensenteric and arterial blood flow. Some bile acids secreted in the bile duct are reabsorbed in the cholangiocytes and recycled back to hepatocytes (cholangiohepatic shunt). After each meal, gallbladder contraction empties bile acids into the intestinal tract. When passing through the intestinal tract, some bile acids are reabsorbed in the upper intestine by passive diffusion, but most bile acids (95%) are reabsorbed in the ileum. Bile acids are transdiffused across the enterocyte to the basolateral membrane and excreted into portal blood circulation back to the sinusoid of hepatocytes. In the colon, DCA is reabsorbed by passive transport and recycled with CA and CDCA to the liver. A bile acid pool of about 3 g is recycled 4–12 times a day. Bile acids lost in the feces (0.2–0.6 g/day) are replenished by de novo synthesis in the liver to maintain a constant bile acid pool.
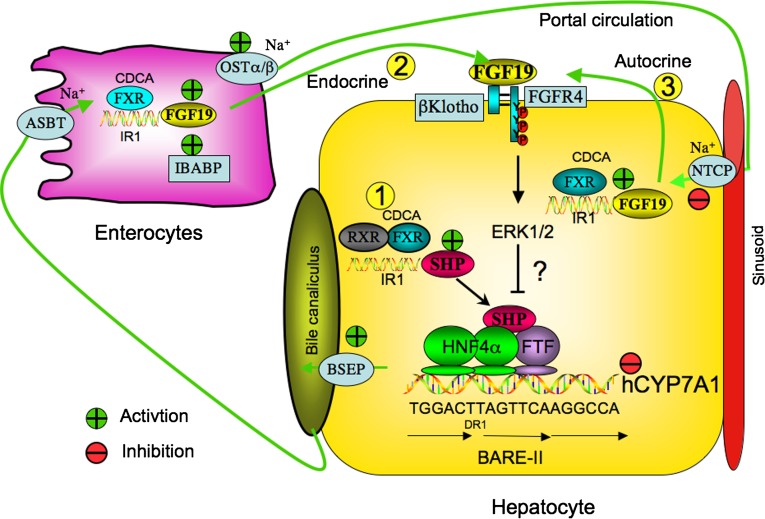
FXR plays a key role in control of enterohepatic circulation of bile acids (Fig. 3). FXR induces the expression of BSEP in the canalicular membrane, which is the driving force for bile formation. An FXR binding site in the human BSEP promoter has been identified (92). Mutation of BSEP causes progressive familial intrahepatic cholestasis type 2 (93, 94). In the intestine, bile acids are reabsorbed into the enterocytes by ASBT in the brush border membrane. Genetic knockout of Asbt in mice interrupts enterohepatic circulation of bile acids and causes bile acid malabsorption (95). FXR induces ASBT expression in mice and inhibits ASBT expression in rabbit but does not affect ASBT in humans (96). FXR induces the expression of ileal bile acid binding protein (IBABP), which is the first target gene of FXR identified in the gastrointestine system (97). The physiological function of IBABP is not clear. It may bind bile acids and reduce intracellular bile acid concentrations in the ileum. Bile acids are excreted from enterocytes via OSTα/OSTβ located in the basolateral (sinusoid) membrane (98, 99). The bile acid pool is reduced in Ostα/Ostβ −/− mice (100, 101). This is a result of increased fibroblast growth factor 15 (FGF15) expression in mouse intestine and reduced Cyp7a1 expression in mouse liver (see below). OST appears to be the major bile acid efflux transporter in the intestine. OSTα/OSTβ also has been localized in the sinusoid membrane of hepatocytes (100). The mouse Ostα/Ostβ promoters have both FXR and LRH-1 binding sites, indicating that OSTα/OSTβ expresson may be induced by FXR and inhibited by SHP/LRH-1 (102). FXR induces the hepatic OSTα/OSTβ expression in patients with primary biliary cirrhosis and in bile duct-ligated rats and mice (103). The FXR induction of OSTα/OSTβ may be an adaptive response to cholestasis to excrete bile acids from hepatocytes. Bile acids are transported back to hepatocytes via portal blood circulation, taken up by NTCP in the sinusoidal membrane. FXR inhibits Ntcp transcription by SHP-dependent inhibition of retinoid X receptor/RAR induction of Ntcp (104). Thus, FXR plays a critical role in the coordination of bile acid synthesis, biliary bile acid secretion, intestinal bile acid reabsorption and secretion, and bile acid uptake into hepatocytes. Deficiency in FXR regulation of enterohepatic circulation of bile acids may lead to cholestasis (8, 93, 105, 106).
Fig. 3.
Mechanisms of FXR regulation of enterohepatic circulation of bile acid. Bile acids synthesized in the liver are excreted into bile via BSEP and stored in the gallbladder. After each meal, bile acids are excreted into the intestinal tract. In the ileum, bile acids are reabsorbed by ASBT in the brush border membrane. Bile acids activate FXR to induce IBABP in enterocytes. OSTα/β transporter in the basolateral membrane effluxes bile acids to portal circulation to hepatocytes where they are taken up by NTCP. In the liver, bile acids activate FXR, which induces SHP expression. SHP then inhibits LRH-1 (or human FTF) and HNF4α transactivation of CYP7A1 (FXR/SHP pathway 1). In the endocrine pathway, intestinal bile acids activate FXR, which induces FGF19 expression. FGF19 may be transported to the liver to activate a liver-specific receptor tyrosine kinase FGFR4 (FXR/FGF19/FGFR4 pathway 2). In the autocrine pathway (pathway 3), cholestatic bile acids may activate FXR and FGF19/FGFR4 signaling, which activates the MAPK/ERK1/2 pathway to inhibit CYP7A1 transcription. It is not clear how the FGF19/ERK1/2 pathway downregulates CYP7A1 transcription. The endocrine pathway may be a physiological pathway for bile acid inhibition of bile acid synthesis, while the autocrine pathway may be an adaptive response to protect liver from cholestatic injury. BARE-II contains 18 bp sequence of overlapping HNF4α and FTF (α-fetoprotein transcription factor, a human homolog of mouse LRH-1) binding site, which is completely conserved in all species.
MECHANISMS OF FXR INHIBITION OF CYP7A1
Bile acid synthesis is feedback inhibited by bile acids returning to the liver via enterohepatic circulation to inhibit CYP7A1 (37). Currently, there are two FXR-dependent mechanisms for bile acid inhibition of CYP7A1 gene transcription (Fig. 3). In the liver, FXR induces SHP to inhibit CYP7A1. In the intestine, FXR induces FGF19 to activate liver FGF receptor 4 (FGFR4) signaling to inhibit CYP7A1.
The FXR/SHP pathway
The FXR binding site is not present in the CYP7A1 promoter (67). Three laboratories independently report a cascade mechanism of FXR inhibition of CYP7A1 (6, 107, 108). They show that FXR induces an atypical orphan nuclear receptor, SHP, that has no DNA-binding domain and is a common transcriptional repressor of nuclear receptors. SHP then inhibits the transactivating activity of LRH-1 and results in inhibiting CYP7A1 transcription (6, 107, 108). SHP also interacts with HNF4α to block HNF4α interaction with PGC-1α and results in inhibiting CYP7A1 and CYP8B1 transcription (109, 110). The LRH-1 and HNF4α binding sites overlap in the CYP7A1 and CYP8B1 promoter (109, 110). LRH-1 is a weak transcription factor and may compete with HNF4α for binding to the BARE and results in inhibiting CYP7A1 and CYP8B1 transcription.
The molecular mechanisms of SHP inhibition of gene expression have been studied in transgenic mice overexpressing SHP in hepatocytes (111). SHP transgenic mice have fat accumulation. Global gene expression profiling combined with chromatin immunoprecipitation assay reveals that SHP affects the genes involved in bile acid synthesis (CYP7A1, CYP8B1, and CYP7B1), conjugation (BAT), and transport (BSEP, NTCP, and MDR2) as expected. Interestingly SHP transgenic mice express lower levels of SR-B1 and CYP51b but higher levels of ABCA1, PPARγ, steroid response element binding protein-1c, CD36 (fatty acid transporter), fatty acid synthase, acyl-CoA carboxylase-1, and stearoyl-CoA desaturase-1 involved in fatty acid metabolism. These data suggest that SHP may play a role in downregulation of lipogenesis and protecting against steatosis. SHP alters chromatin configuration in SHP downregulated genes. Interestingly, SHP associates with unmodified and methylated-histone 3-Lys9 (H3K9) but not acetylated-H3K9. Furthermore, SHP interacts with histone deacetylase-1 (HDAC-1) and a histone methytransferase G9a (112), suggesting that SHP repression of gene transcription by multiple steps involving histone deacetylation, followed by H3K9 methylation, and stable association of SHP to chromatin. Another study reports that SHP interacts with a repressor, mSin3A, and recruits chromatin remodeling complex Swi/Snf/Brm to CYP7A1 chromatin containing the bile acid response elements (113). A follow-up study shows that SHP recruits HDACs and interacts with G9a to enhance SHP inhibition of CYP7A1 (114). It has been shown that bile acids stimulate translocation of HDACs to the nucleus to assemble a repressor complex consisting of HDAC7, SHP, and a common nuclear receptor corepressor silencing mediator of retinoid and thyroid receptor to inhibit CYP7A1 (115). Another nuclear receptor corepressor GPS2 interacts with SHP, LRH-1, HNF4α, and FXR and differentially regulates CYP7A1 and CYP8B1 (67). It is not clear how bile acids induce or recruit these repressor complexes to inhibit CYP7A1 transcription.
Two recent studies of liver conditional knockout of Lrh-1 in mice show that Cyp8b1 expression is abolished and CA is eliminated; however, basal Cyp7a1 expression is not affected and an FXR agonist GW4064 repressed Cyp7a1 expression (116, 117). These results suggest that LRH-1 may not be involved in FXR inhibition of CYP7A1 but is required for FXR inhibition of CYP8B1 gene transcription. This is consistent with the earlier finding that knockout of the Shp gene in mice did not prevent bile acid inhibition of CYP7A1 mRNA expression (118, 119). This raises the doubt that the FXR/SHP/LRH-1 pathway is involved in bile acid inhibition of CYP7A1 under normal physiological conditions. It should be noted that bile acid induction of SHP is very rapid and transient. It is likely that SHP is transiently induced by bile acids or inflammatory cytokines in acute phase response to liver injury and cholestasis and during liver regeneration (120) to protect liver against toxicity of bile acids and other metabolites (50, 121).
The FXR/FGF19/FGFR4 pathway
Holt et al. (122) first identified the fibroblast growth factor 19 (FGF19) as an FXR target gene by microarray analysis of human primary hepatocytes treated with an FXR agonist GW4064. They report that FGF19 activates receptor tyrosine kinase FGFR4 signaling to inhibit CYP7A1 mRNA expression levels in hepatocytes (122). A subsequent study has demonstrated that FXR induces FGF15, a mouse ortholog of FGF19 in mouse intestine, and the expression of FGF15 mRNA is inversely correlated to CYP7a1 mRNA expression levels in mouse liver (123). In Fgfr4 −/− or Fgf 15−/− mice, FGF15 and GW4064 do not affect Cyp7a1 mRNA expression levels, supporting the role of FGF15/FGFR4 signaling in mediating bile acid inhibition of Cyp7a1 expression. These investigators proposed that FGF15 might function as an enterohepatic signal to regulate bile acid synthesis (Fig. 3). Furthermore, GW4064 represses Cyp7a1 in liver-specific Fxr knockout mice but not in intestine-specific Fxr knockout mice, unequivocally confirming that the intestinal FXR but not liver FXR is required for bile acid inhibition of Cyp7a1 gene expression (124). These data also suggest that CYP8B1 is preferentially regulated by the liver FXR/SHP pathway and not by the FXR/FGF19/FGFR4 pathway. It appears that FGF19 is the bile acid-induced intestinal factor that is secreted in the ileum to inhibit bile acid synthesis in hepatocytes as proposed by Pandak et al. in 1995 (125).
FGF19 activation of FGFR4 signaling requires β-Klotho, a membrane-bound glycosidase coexpressed with FGFR4 in hepatocytes (Fig. 3). Genetic ablation of the β-Klotho gene in mice increases bile acid synthesis and secretion and Cyp7a1 expression (126). However, Cyp8b1 expression was not altered in the β-Klotho knockout mice, suggesting differential regulation of Cyp7a1 and Cyp8b1 by β-Klotho. Similar to the β-Klotho knockout mice, the Fgfr4-deficient mice have the same phenotypes of increased bile acid synthesis (127). Overexpression of a constitutively active human FGFR4 represses Cyp7a1 expression and decreases bile acid pool in wild-type mice (128). All these findings support the critical role of FGFR4 signaling in mediating bile acid feedback regulation. FGF19 has been detected in human patient sera. Interestingly, serum FGF19 levels exhibit diurnal variation that peaked 90–120 min after a postprandial rise in serum bile acids and serum 7α-hydroxy-4-cholesten-3-one (C4) levels (129). Feeding of CDCA increased FGF19 expression, whereas a bile acid-binding resin cholestyramine reduced FGF19 in humans. These results support that FGF19 plays an important role in inhibiting bile acid synthesis in humans. It is likely that the FXR/FGF19/FGFR4 pathway is the physiological mechanism for bile acid feedback regulation of bile acid synthesis.
THE AUTOCRINE FUNCTIONS OF FGF19
FGF15 has not been identified in mouse sera and livers, and bile acids do not induce FGF15 expression in mouse liver. Song et al. (50) reported recently that bile acids are able to induce FGF19 in primary human hepatocytes. This study shows that CDCA and GW4064 rapidly induce FGF19 mRNA expression, FGF19 protein secretion, and tyrosine phosphorylation of FGFR4 but inhibit CYP7A1 mRNA expression in primary human hepatocytes, suggesting that liver-produced FGF19 is secreted from hepatocytes to activate FGFR4 signaling in hepatocytes by an autocrine or paracrine mechanism. Furthermore, knockdown of SHP expression by small interfering RNA does not affect FGF19 inhibition of CYP7A1 mRNA expression in primary human hepatocytes, suggesting that SHP may not be required in FGF19 signaling (50). It is also noted that induction of FGF19 is sustained for at least 24 h, but induction of SHP mRNA by CDCA and GW4064 is transient, reaching the maximum in 1–3 h and reducing to the basal levels after 6 h of treatment (50). All these data show a lack of correlation between SHP and CYP7A1 expression levels in FGF19 signaling, in contrast to the inverse relationship between SHP and CYP7A1 expression levels that supports the FXR/SHP pathway. The study by Song et al. (50) demonstrates that FGF19/FGFR4 signaling activates and phosphorylates mainly the MAPK/ERK1/2 signaling pathway in human primary hepatocytes (Fig. 3). However, the downstream factor(s) of the FGF19 pathway that inhibits CYP7A1 gene transcription remains unknown.
Schaap et al. (130) report a study of FGF19 expression in patients with extrahepatic cholestasis. These investigators observed a 6- and 8-fold higher plasma FGF19 in cholestatic patients than noncholestatic patients and postcholestatic patients receiving a biliary stent to drain bile acids, respectively. FGF19 mRNA could be detected in the majority of liver specimens with a wide range of expression levels and was 31- to 374-fold higher in the cholestatic group than in the drained and noncholestatic group, respectively. CYP7A1 mRNA expression levels were 37- and 9.8-fold lower in the cholestatic group than in the control and drained groups, respectively. These investigators reasoned that FGF19 should be decreased in extrahepatic cholestasis patients, if FGF19 is only produced in the intestine. They suggest that bile acids accumulated in cholestatic liver could induce FGF19 expression as an adaptive response to cholestatic liver injury.
FXR-INDEPENDENT BILE ACID INHIBITION OF CYP7A1
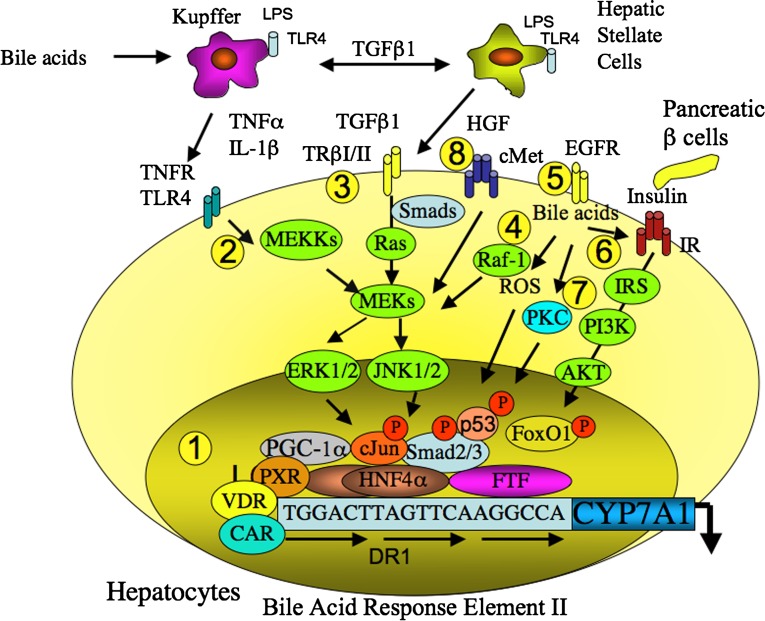
There are several FXR-independent mechanisms for bile acid inhibition of CYP7A1 (Fig. 4). The secondary bile acid LCA is a ligand of PXR and VDR. These two receptors bind to the BARE-I sequence in the human CYP7A1 promoter and inhibit CYP7A1 promoter activity (131, 132). PXR interacts with HNF4α and blocks HNF4α recruitment of PGC-1α to CYP7A1 chromatin and results in inhibiting CYP7A1 transcription. Guggulsterone, an FXR antagonist and PXR agonist, inhibits CYP7A1 transcription by activating PXR (133). Interestingly, PXR induces CYP27A1 expression in intestinal cells but not in liver cells, suggesting that PXR may play a role in the regulation of ABCA1 efflux of cholesterol from intestine cells to synthesize HDL for reverse transport of cholesterol to the liver (134). It has been reported that the constitutive androstane receptor (CAR) binds to the DR1 motif and competes with HNF4α for coactivators PGC-1α and glucocorticoid receptor interacting protein-1 and results in inhibiting CYP7A1 transcription (135). The LCA-activated VDR inhibits CYP7A1 gene transcription by several mechanisms; VDR may interact with HNF4α and blocks HNF4α interaction with PGC-1α; VDR and HNF4α competes for binding to the DR1 motif; VDR competes with HNF4α for coactivators; and VDR recruits corepressors to the CYP7A1 promoter (136).
Fig. 4.
FXR-independent and bile acid-activated cell signaling pathways in regulation of CYP7A1 transcription. 1) LCA activates PXR, CAR (indirectly), and VDR, which inhibit CYP7A1 transcription by interacting with HNF4α and blocking PGC-1α activation of HNF4α. 2) Bile acids stimulate inflammatory cytokines TNFα and IL-1β in Kupffer cells, which activate the TNF receptor and the MAPK/JNK pathways to inhibit CYP7A1 transcription. 3) TGFβ1 secreted from the HSC cells activates TGFβ1 receptor and the MAPK/Smad pathway. Smad3 interacts with HNF4α and recruitment of HDAC and mSin3A to inhibit CYP7A1 transcription. 4) Bile acids and TGFβ1 induce reactive oxidizing species (ROS) and activate p53, which interacts with HNF4α and inhibits HNF4α transactivation of CYP7A1. 5) Bile acids also activate epidermal growth factor receptor (EGFR) and the Raf-1/MEK/ERK signaling pathway to inhibit CYP7A1. 6) Bile acids enhance the insulin receptor signaling to phosphorylate and activate insulin receptor substrate, PI3K and AKT, which phosphorylates FoxO1 and inhibits CYP7A1. 7) Bile acids also activate protein kinase C, which phosphorylates cJun to inhibit CYP7A1. 8) During liver injury and regeneration, HGF secreted from HSC cells activates the HGF receptor cMet and the MAPK pathways to inhibit CYP7A1. All these signaling pathways may converge to regulate chromatin structure by the epigenetic mechanism.
During the acute phase response, lipopolysaccharide activates the Toll-like receptor 4 to induce and release TNFα and IL-1β from the Kupffer cells. Bile acids also induce TNFα and IL-1β, which may cross the sinusoid membrane to activate the TNF receptor and the MAPK/JNK pathway to inhibit CYP7A1 transcription (25). JNK phosphorylates cJun and HNF4α and results in inhibiting CYP7A1 and CYP8B1 transcription and bile acid synthesis (47, 136, 137). TGFβ-1 secreted from Kupffer cells activates Toll-like receptor 4 in hepatic stellate cells (HSCs) and secretion of TGFβ-1, which activates its receptor TRβII and the SMAD signaling pathway in hepatocytes. Bile acids stimulate TGFβ-1 expression in hepatocytes by induction of thrombospondin-1, which activates latent TGFβ-1 in HSC leading to the activation of HSC (138). SMAD3 enters the nucleus and recruits HDACs and mSin3A to inhibit HNF4α activation of CYP7A1 transcription (139). TGFβ1 and bile acids activate the Ras/MAPK/JNK pathway, induce reactive oxidative species, and phosphorylate a tumor suppressor p53 (140, 141). It has been reported that p53 interacts with HNF4α and inhibits HNF4α activity (142) and may result in inhibiting CYP7A1. In contrast, TGFβ1 stimulates the rat CYP7A1 promoter, which contains the binding sites for SMAD, FoxO1, and HNF4α. These three transcription factors synergistically induce rat CYP7A1 transcription (143). This may explain the paradoxical induction of CYP7A1 transcription in bile duct-ligated rats (144). Bile acids also activate epidermal growth factor receptor and the Raf-1/MEK/ERK signaling pathway to inhibit CYP7A1 transcription (27). Bile acids enhance the insulin receptor signaling in rat primary hepatocytes (32). Insulin signaling phosphorylates and activates the insulin receptor leading to activation of insulin receptor substrates, PI3K and AKT, which phosphorylates FoxO1 and inhibits CYP7A1 transcription (47). Bile acid-activated protein kinase C phosphorylates cJun to inhibit CYP7A1 transcription (24). During acute phase response to liver injury and regeneration, hepatocyte growth factor (HGF) released from HSCs activates the HGF receptor cMet and MAPK pathways to inhibit CYP7A1 transcription and bile acid synthesis (49). All these cell signaling pathways may play critical roles in protection against bile acid toxicity during liver injury and cholestasis and may converge to regulate CYP7A1 chromatin structure by epigenetic mechanism.
CONCLUSIONS AND FUTURE PERSPECTIVES
Bile acid research in the last two decades has contributed significantly to our understanding of the mechanisms of bile acid synthesis and pathogenesis of liver diseases and metabolic syndrome. The CYP7A1 mRNA expression has become a biomarker for studying lipid metabolism in animal models of fatty liver disease, diabetes, obesity, and cholestasis. The mechanism underlying FXR/FGF19/FGFR4 signaling in inhibition of CYP7A1 transcription and bile acid synthesis remains to be elucidated. The genetically modified mouse models are widely used for studying bile acid synthesis and regulation. However, distinct species differences in bile acid composition, synthesis, and regulation between humans and mice exist. It is imperative that a suitable human model system be developed to verify results from animal studies.
Bile acids and bile acid-activated receptors FXR, PXR, CAR, and VDR are therapeutic targets for development of drugs for treatment of gallstone, fatty liver disease, cholestatic liver disease, obesity, diabetes, and metabolic syndrome (12, 145, 146). A bile acid derivative, 6α-ethyl-CDCA, has a protective effect on cholestasis (147) and is in the second phase of clinical trials for primary biliary cirrhosis. Another bile acid derivative, 6α-ethyl, 23(S)-methyl-CDCA, is a selective TGR5 agonist and has therapeutic potential for the treatment of obesity, diabetes, and metabolic syndrome (148). Fatty acid-bile acid conjugates are in clinical trials for treatment of nonalcoholic fatty liver disease and nonalcoholic steatohepatitis, gallstone disease, and cardiovascular diseases (149–152). It is anticipated that basic research in bile acid metabolism will be translated into clinical diagnosis and treatment of liver and metabolic diseases in the near future.
Footnotes
Abbreviations:
- ASBT
- apical sodium-dependent bile acid transporter
- BACS
- bile acid:CoA synthase
- BARE
- bile acid response element
- BAT
- bile acid:amino acid transferase
- BSEP
- bile salt export pump
- CA
- cholic acid
- CAR
- constitutive androstane receptor
- CDCA
- chenodeoxycholic acid
- CYP7A1
- cholesterol 7α-hydroylase
- CYP8B1
- sterol 12α-hydroxylase
- CYP27A1
- sterol 27 hydroxylase
- DCA
- deoxycholic acid
- FGF15
- fibroblast growth factor 15
- FGF19
- fibroblast growth factor 19
- FGFR4
- FGF receptor 4
- FTF
- α-fetoprotein transcription factor
- FXR
- farnesoid X receptor
- H3K9
- histone 3-Lys9
- HDAC
- histone deacetylase
- HGF
- hepatocyte growth factor
- HNF4α
- hepatocyte nuclear factor 4α
- HSC
- hepatic stellate cell
- IBABP
- ileal bile acid binding protein
- IL-1β
- interleuken-1β
- LCA
- lithocholic acid
- LXR
- liver orphan receptor
- LRH-1
- liver-related homolog-1
- MAPK
- mitogen-activated protein kinase
- NTCP
- Na+-dependent taurocholate cotransport peptide
- OST
- organic solute transporter
- PXR
- pregnane X receptor
- PPARα
- peroxisome proliferator activated receptor α
- PGC-1α
- peroxisome proliferator-activated receptor γ coactivator 1α
- SHP
- small heterodimer partner
- TNFα
- tumor necrosis factor α
- VDR
- vitamin D receptor
This work was supported by National Institutes of Health Grants DK-44442 and DK-58379. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the National Institutes of Health.
REFERENCES
- 1.Myant N. B., Mitropoulos K. A. 1977. Cholesterol 7α-hydroxylase. J. Lipid Res. 18: 135–153. [PubMed] [Google Scholar]
- 2.Noshiro M., Nishimoto M., Morohashi K., Okuda K. 1989. Molecular cloning of cDNA for cholesterol 7α-hydroxylase from rat liver microsomes. Nucleotide sequence and expression. FEBS Lett. 257: 97–100. [DOI] [PubMed] [Google Scholar]
- 3.Li Y. C., Wang D. P., Chiang J. Y. 1990. Regulation of cholesterol 7α-hydroxylase in the liver. Cloning, sequencing, and regulation of cholesterol 7α-hydroxylase mRNA. J. Biol. Chem. 265: 12012–12019. [PubMed] [Google Scholar]
- 4.Jelinek D. F., Andersson S., Slaughter C. A., Russell D. W. 1990. Cloning and regulation of cholesterol 7α-hydroxylase, the rate-limiting enzyme in bile acid biosynthesis. J. Biol. Chem. 265: 8190–8197. [PMC free article] [PubMed] [Google Scholar]
- 5.Makishima M., Okamoto A. Y., Repa J. J., Tu H., Learned R. M., Luk A., Hull M. V., Lustig K. D., Mangelsdorf D. J., Shan B. 1999. Identification of a nuclear receptor for bile acids. Science. 284: 1362–1365. [DOI] [PubMed] [Google Scholar]
- 6.Wang H., Chen J., Hollister K., Sowers L. C., Forman B. M. 1999. Endogenous bile acids are ligands for the nuclear receptor FXR/BAR. Mol. Cell. 3: 543–553. [DOI] [PubMed] [Google Scholar]
- 7.Parks D. J., Blanchard S. G., Bledsoe R. K., Chandra G., Consler T. G., Kliewer S. A., Stimmel J. B., Willson T. M., Zavacki A. M., Moore D. D., et al. 1999. Bile acids: natural ligands for an orphan nuclear receptor. Science. 284: 1365–1368. [DOI] [PubMed] [Google Scholar]
- 8.Zollner G., Marschall H. U., Wagner M., Trauner M. 2006. Role of nuclear receptors in the adaptive response to bile acids and cholestasis: pathogenetic and therapeutic considerations. Mol. Pharm. 3: 231–251. [DOI] [PubMed] [Google Scholar]
- 9.Nguyen A., Bouscarel B. 2008. Bile acids and signal transduction: role in glucose homeostasis. Cell. Signal. 20: 2180–2197. [DOI] [PubMed] [Google Scholar]
- 10.Houten S. M., Watanabe M., Auwerx J. 2006. Endocrine functions of bile acids. EMBO J. 25: 1419–1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Keitel V., Kubitz R., Haussinger D. 2008. Endocrine and paracrine role of bile acids. World J. Gastroenterol. 14: 5620–5629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thomas C., Pellicciari R., Pruzanski M., Auwerx J., Schoonjans K. 2008. Targeting bile-acid signalling for metabolic diseases. Nat. Rev. Drug Discov. 7: 678–693. [DOI] [PubMed] [Google Scholar]
- 13.Lefebvre P., Cariou B., Lien F., Kuipers F., Staels B. 2009. Role of bile acids and bile acid receptors in metabolic regulation. Physiol. Rev. 89: 147–191. [DOI] [PubMed] [Google Scholar]
- 14.Eloranta J. J., Kullak-Ublick G. A. 2008. The role of FXR in disorders of bile acid homeostasis. Physiology (Bethesda). 23: 286–295. [DOI] [PubMed] [Google Scholar]
- 15.Modica S., Murzilli S., Salvatore L., Schmidt D. R., Moschetta A. 2008. Nuclear bile acid receptor FXR protects against intestinal tumorigenesis. Cancer Res. 68: 9589–9594. [DOI] [PubMed] [Google Scholar]
- 16.Chiang J. Y. 2002. Bile acid regulation of gene expression: roles of nuclear hormone receptors. Endocr. Rev. 23: 443–463. [DOI] [PubMed] [Google Scholar]
- 17.Chiang J. Y. 2004. Regulation of bile acid synthesis: pathways, nuclear receptors, and mechanisms. J. Hepatol. 40: 539–551. [DOI] [PubMed] [Google Scholar]
- 18.Chiang J. Y. 2005. Nuclear receptor regulation of lipid metabolism: potential therapeutics for dyslipidemia, diabetes, and chronic heart and liver diseases. Curr. Opin. Investig. Drugs. 6: 994–1001. [PubMed] [Google Scholar]
- 19.Watanabe M., Houten S. M., Mataki C., Christoffolete M. A., Kim B. W., Sato H., Messaddeq N., Harney J. W., Ezaki O., Kodama T., et al. 2006. Bile acids induce energy expenditure by promoting intracellular thyroid hormone activation. Nature. 439: 484–489. [DOI] [PubMed] [Google Scholar]
- 20.Staudinger J. L., Goodwin B., Jones S. A., Hawkins-Brown D., MacKenzie K. I., LaTour A., Liu Y., Klaassen C. D., Brown K. K., Reinhard J., et al. 2001. The nuclear receptor PXR is a lithocholic acid sensor that protects against liver toxicity. Proc. Natl. Acad. Sci. USA. 98: 3369–3374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Makishima M., Lu T. T., Xie W., Whitfield G. K., Domoto H., Evans R. M., Haussler M. R., Mangelsdorf D. J. 2002. Vitamin D receptor as an intestinal bile acid sensor. Science. 296: 1313–1316. [DOI] [PubMed] [Google Scholar]
- 22.Xie W., Radominska-Pandya A., Shi Y., Simon C. M., Nelson M. C., Ong E. S., Waxman D. J., Evans R. M. 2001. An essential role for nuclear receptors SXR/PXR in detoxification of cholestatic bile acids. Proc. Natl. Acad. Sci. USA. 98: 3375–3380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sonoda J., Xie W., Rosenfeld J. M., Barwick J. L., Guzelian P. S., Evans R. M. 2002. Regulation of a xenobiotic sulfonation cascade by nuclear pregnane X receptor (PXR). Proc. Natl. Acad. Sci. USA. 99: 13801–13806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stravitz R. T., Vlahcevic Z. R., Gurley E. C., Hylemons P. B. 1995. Repression of cholesterol 7a-hydroxylase transcription by bile acids is mediated through protein kinase C in primary cultures of rat hepatocytes. J. Lipid Res. 36: 1359–1368. [PubMed] [Google Scholar]
- 25.Miyake J. H., Wang S. L., Davis R. A. 2000. Bile acid induction of cytokine expression by macrophages correlates with repression of hepatic cholesterol 7alpha-hydroxylase. J. Biol. Chem. 275: 21805–21808. [DOI] [PubMed] [Google Scholar]
- 26.Gutierrez A., Ratliff E. P., Andres A. M., Huang X., McKeehan W. L., Davis R. A. 2006. Bile acids decrease hepatic paraoxonase 1 expression and plasma high-density lipoprotein levels via FXR-mediated signaling of FGFR4. Arterioscler. Thromb. Vasc. Biol. 26: 301–306. [DOI] [PubMed] [Google Scholar]
- 27.Rao Y. P., Studer E. J., Stravitz R. T., Gupta S., Qiao L., Dent P., Hylemon P. B. 2002. Activation of the Raf-1/MEK/ERK cascade by bile acids occurs via the epidermal growth factor receptor in primary rat hepatocytes. Hepatology. 35: 307–314. [DOI] [PubMed] [Google Scholar]
- 28.Fang Y., Han S. I., Mitchell C., Gupta S., Studer E., Grant S., Hylemon P. B., Dent P. 2004. Bile acids induce mitochondrial ROS, which promote activation of receptor tyrosine kinases and signaling pathways in rat hepatocytes. Hepatology. 40: 961–971. [DOI] [PubMed] [Google Scholar]
- 29.Dent P., Fang Y., Gupta S., Studer E., Mitchell C., Spiegel S., Hylemon P. B. 2005. Conjugated bile acids promote ERK1/2 and AKT activation via a pertussis toxin-sensitive mechanism in murine and human hepatocytes. Hepatology. 42: 1291–1299. [DOI] [PubMed] [Google Scholar]
- 30.Fang Y., Studer E., Mitchell C., Grant S., Pandak W. M., Hylemon P. B., Dent P. 2007. Conjugated bile acids regulate hepatocyte glycogen synthase activity in vitro and in vivo via Galphai signaling. Mol. Pharmacol. 71: 1122–1128. [DOI] [PubMed] [Google Scholar]
- 31.Gupta S., Natarajan R., Payne S. G., Studer E. J., Spiegel S., Dent P., Hylemon P. B. 2004. Deoxycholic acid activates the c-Jun N-terminal kinase pathway via FAS receptor activation in primary hepatocytes: role of acidic sphingomyelinase-mediated ceramide generation in FAS receptor activation. J. Biol. Chem. 279: 5821–5828. [DOI] [PubMed] [Google Scholar]
- 32.Han S. I., Studer E., Gupta S., Fang Y., Qiao L., Li W., Grant S., Hylemon P. B., Dent P. 2004. Bile acids enhance the activity of the insulin receptor and glycogen synthase in primary rodent hepatocytes. Hepatology. 39: 456–463. [DOI] [PubMed] [Google Scholar]
- 33.Kawamata Y., Fujii R., Hosoya M., Harada M., Yoshida H., Miwa M., Fukusumi S., Habata Y., Itoh T., Shintani Y., et al. 2003. A G protein-coupled receptor responsive to bile acids. J. Biol. Chem. 278: 9435–9440. [DOI] [PubMed] [Google Scholar]
- 34.Maruyama T., Miyamoto Y., Nakamura T., Tamai Y., Okada H., Sugiyama E., Itadani H., Tanaka K. 2002. Identification of membrane-type receptor for bile acids (M-BAR). Biochem. Biophys. Res. Commun. 298: 714–719. [DOI] [PubMed] [Google Scholar]
- 35.Keitel V., Reinehr R., Gatsios P., Rupprecht C., Gorg B., Selbach O., Haussinger D., Kubitz R. 2007. The G-protein coupled bile salt receptor TGR5 is expressed in liver sinusoidal endothelial cells. Hepatology. 45: 695–704. [DOI] [PubMed] [Google Scholar]
- 36.Katsuma S., Hirasawa A., Tsujimoto G. 2005. Bile acids promote glucagon-like peptide-1 secretion through TGR5 in a murine enteroendocrine cell line STC-1. Biochem. Biophys. Res. Commun. 329: 386–390. [DOI] [PubMed] [Google Scholar]
- 37.Russell D. W. 2003. The enzymes, regulation, and genetics of bile acid synthesis. Annu. Rev. Biochem. 72: 137–174. [DOI] [PubMed] [Google Scholar]
- 38.Chiang J. Y. 1998. Regulation of bile acid synthesis. Front. Biosci. 3: d176–d193. [DOI] [PubMed] [Google Scholar]
- 39.Hofmann A. F. 2004. Detoxification of lithocholic acid, a toxic bile acid: relevance to drug hepatotoxicity. Drug Metab. Rev. 36: 703–722. [DOI] [PubMed] [Google Scholar]
- 40.Hofmann A. F., Hagey L. R. 2008. Bile acids: chemistry, pathochemistry, biology, pathobiology, and therapeutics. Cell. Mol. Life Sci. 65: 2461–2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Monte M. J., Marin J. J., Antelo A., Vazquez-Tato J. 2009. Bile acids: chemistry, physiology, and pathophysiology. World J. Gastroenterol. 15: 804–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pandak W. M., Stravitz R. T., Lucas V., Heuman D. M., Chiang J. Y. 1996. Hep G2 cells: a model for studies on regulation of human cholesterol 7α-hydroxylase at the molecular level. Am. J. Physiol. 270: G401–G410. [DOI] [PubMed] [Google Scholar]
- 43.Baker D. M., Wang S. L., Bell D. J., Drevon C. A., Davis R. A. 2000. One or more labile proteins regulate the stability of chimeric mRNAs containing the 3′-untranslated region of cholesterol 7α-hydroxylase mRNA. J. Biol. Chem. 275: 19985–19991. [DOI] [PubMed] [Google Scholar]
- 44.Agellon L. B., Cheema S. K. 1997. The 3′-untranslated region of the mouse cholesterol 7α-hydroxylase mRNA contains elements responsive to post-transcriptional regulation by bile acids. Biochem. J. 328: 393–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Crestani M., Sadeghpour A., Stroup D., Gali G., Chiang J. Y. L. 1998. Transcriptional activation of the cholesterol 7α-hydroxylase gene (CYP7A) by nuclear hormone receptors. J. Lipid Res. 39: 2192–2200. [PubMed] [Google Scholar]
- 46.Li T., Jahan A., Chiang J. Y. 2006. Bile acids and cytokines inhibit the human cholesterol 7α-hydroxylase gene via the JNK/c-jun pathway in human liver cells. Hepatology. 43: 1202–1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li T., Kong X., Owsley E., Ellis E., Strom S., Chiang J. Y. 2006. Insulin regulation of cholesterol 7α-hydroxylase expression in human hepatocytes: roles of forkhead box O1 and sterol regulatory element-binding protein 1c. J. Biol. Chem. 281: 28745–28754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Song K. H., Chiang J. Y. 2006. Glucagon and cAMP inhibit cholesterol 7alpha-hydroxylase (CYP7a1) gene expression in human hepatocytes: discordant regulation of bile acid synthesis and gluconeogenesis. Hepatology. 43: 117–125. [DOI] [PubMed] [Google Scholar]
- 49.Song K. H., Ellis E., Strom S., Chiang J. Y. 2007. Hepatocyte growth factor signaling pathway inhibits cholesterol 7α-hydroxylase and bile acid synthesis in human hepatocytes. Hepatology. 46: 1993–2002. [DOI] [PubMed] [Google Scholar]
- 50.Song K. H., Li T., Owsley E., Strom S., Chiang J. Y. 2009. Bile acids activate fibroblast growth factor 19 signaling in human hepatocytes to inhibit cholesterol 7alpha-hydroxylase gene expression. Hepatology. 49: 297–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chiang J. Y. L., Stroup D. 1994. Identification and characterization of a putative bile acid responsive element in cholesterol 7α-hydroxylase gene promoter. J. Biol. Chem. 269: 17502–17507. [PubMed] [Google Scholar]
- 52.Janowski B. A., Willy P. J., Devi T. R., Falck J. R., Mangelsdorf D. J. 1996. An oxysterol signalling pathway mediated by the nuclear receptor LXRα. Nature. 383: 728–731. [DOI] [PubMed] [Google Scholar]
- 53.Lehmann J. M., Kliewer S. A., Moore L. B., Smith-Oliver T. A., Oliver B. B., Su J-L., Sundseth S. S., Winegar D. A., Blanchard D. E., Spencer T. A., et al. 1997. Activation of the nuclear receptor LXR by oxysterols defines a new hormone response pathway. J. Biol. Chem. 272: 3137–3140. [DOI] [PubMed] [Google Scholar]
- 54.Peet D. J., Turley S. D., Ma W., Janowski B. A., Lobaccaro J. M., Hammer R. E., Mangelsdorf D. J. 1998. Cholesterol and bile acid metabolism are impaired in mice lacking the nuclear oxysterol receptor LXR alpha. Cell. 93: 693–704. [DOI] [PubMed] [Google Scholar]
- 55.Chiang J. Y., Kimmel R., Stroup D. 2001. Regulation of cholesterol 7α-hydroxylase gene (CYP7A1) transcription by the liver orphan receptor (LXRα). Gene. 262: 257–265. [DOI] [PubMed] [Google Scholar]
- 56.Agellon L. B., Drover V. A., Cheema S. K., Gbaguidi G. F., Walsh A. 2002. Dietary cholesterol fails to stimulate the human cholesterol 7α-hydroxylase gene (CYP7A1) in transgenic mice. J. Biol. Chem. 277: 20131–20134. [DOI] [PubMed] [Google Scholar]
- 57.Chen J. Y., Levy-Wilson B., Goodart S., Cooper A. D. 2002. Mice expressing the human CYP7A1 gene in the mouse CYP7A1 knock-out background lack induction of CYP7A1 expression by cholesterol feeding and have increased hypercholesterolemia when fed a high fat diet. J. Biol. Chem. 277: 42588–42595. [DOI] [PubMed] [Google Scholar]
- 58.Stroup D., Crestani M., Chiang J. Y. 1997. Identification of a bile acid response element in the cholesterol 7α-hydroxylase gene CYP7A. Am. J. Physiol. 273: G508–G517. [DOI] [PubMed] [Google Scholar]
- 59.De Fabiani E., Mitro N., Gilardi F., Caruso D., Galli G., Crestani M. 2003. Coordinated control of cholesterol catabolism to bile acids and of gluconeogenesis via a novel mechanism of transcription regulation linked to the fasted-to-fed cycle. J. Biol. Chem. 278: 39124–39132. [DOI] [PubMed] [Google Scholar]
- 60.Shin D. J., Campos J. A., Gil G., Osborne T. F. 2003. PGC-1α activates CYP7A1 and bile acid biosynthesis. J. Biol. Chem. 278: 50047–50052. [DOI] [PubMed] [Google Scholar]
- 61.Lundasen T., Liao W., Angelin B., Rudling M. 2003. Leptin induces the hepatic high density lipoprotein receptor scavenger receptor B type I (SR-BI) but not cholesterol 7α-hydroxylase (Cyp7a1) in leptin-deficient (ob/ob) mice. J. Biol. Chem. 278: 43224–43228. [DOI] [PubMed] [Google Scholar]
- 62.Galman C., Angelin B., Rudling M. 2005. Bile acid synthesis in humans has a rapid diurnal variation that is asynchronous with cholesterol synthesis. Gastroenterology. 129: 1445–1453. [DOI] [PubMed] [Google Scholar]
- 63.Rizzo G., Renga B., Mencarelli A., Pellicciari R., Fiorucci S. 2005. Role of FXR in regulating bile acid homeostasis and relevance for human diseases. Curr. Drug Targets Immune Endocr. Metabol. Disord. 5: 289–303. [DOI] [PubMed] [Google Scholar]
- 64.Chiang J. Y. L., Kimmel R., Weinberger C., Stroup D. 2000. FXR responds to bile acids and represses cholesterol 7α-hydroxylase gene (CYP7A1) transcription. J. Biol. Chem. 275: 10918–10924. [DOI] [PubMed] [Google Scholar]
- 65.Yang Y., Zhang M., Eggertsen G., Chiang J. Y. 2002. On the mechanism of bile acid inhibition of rat sterol 12α- hydroxylase gene (CYP8B1) transcription: roles of alpha-fetoprotein transcription factor and hepatocyte nuclear factor 4alpha. Biochim. Biophys. Acta. 1583: 63–73. [DOI] [PubMed] [Google Scholar]
- 66.Chen W., Chiang J. Y. 2003. Regulation of human sterol 27-hydroxylase gene (CYP27A1) by bile acids and hepatocyte nuclear factor 4alpha (HNF4alpha). Gene. 313: 71–82. [DOI] [PubMed] [Google Scholar]
- 67.Sanyal S., Bavner A., Haroniti A., Nilsson L. M., Lundasen T., Rehnmark S., Witt M. R., Einarsson C., Talianidis I., Gustafsson J. A., et al. 2007. Involvement of corepressor complex subunit GPS2 in transcriptional pathways governing human bile acid biosynthesis. Proc. Natl. Acad. Sci. USA. 104: 15665–15670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Inoue Y., Yu A. M., Inoue J., Gonzalez F. J. 2004. Hepatocyte nuclear factor 4alpha is a central regulator of bile acid conjugation. J. Biol. Chem. 279: 2480–2489. [DOI] [PubMed] [Google Scholar]
- 69.Hunt M. C., Yang Y. Z., Eggertsen G., Carneheim C. M., Gafvels M., Einarsson C., Alexson S. E. 2000. The peroxisome proliferator-activated receptor α (PPARα) regulates bile acid biosynthesis. J. Biol. Chem. 275: 28947–28953. [DOI] [PubMed] [Google Scholar]
- 70.Pineda Torra I., Claudel T., Duval C., Kosykh V., Fruchart J. C., Staels B. 2003. Bile acids induce the expression of the human peroxisome proliferator-activated receptor alpha gene via activation of the farnesoid X receptor. Mol. Endocrinol. 17: 259–272. [DOI] [PubMed] [Google Scholar]
- 71.Marrapodi M., Chiang J. Y. 2000. Peroxisome proliferator-activated receptor alpha (PPARα) and agonist inhibit cholesterol 7α-hydroxylase gene (CYP7A1) transcription. J. Lipid Res. 41: 514–520. [PubMed] [Google Scholar]
- 72.Quinn C. M., Jessup W., Wong J., Kritharides L., Brown A. J. 2005. Expression and regulation of sterol 27-hydroxylase (CYP27A1) in human macrophages: a role for RXR and PPARgamma ligands. Biochem. J. 385: 823–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Solaas K., Kase B. F., Pham V., Bamberg K., Hunt M. C., Alexson S. E. 2004. Differential regulation of cytosolic and peroxisomal bile acid amidation by PPAR alpha activation favors the formation of unconjugated bile acids. J. Lipid Res. 45: 1051–1060. [DOI] [PubMed] [Google Scholar]
- 74.Duez H., van der Veen J. N., Duhem C., Pourcet B., Touvier T., Fontaine C., Derudas B., Bauge E., Havinga R., Bloks V. W., et al. 2008. Regulation of bile acid synthesis by the nuclear receptor Rev-erbα. Gastroenterology. 135: 689–698. [DOI] [PubMed] [Google Scholar]
- 75.Preitner N., Damiola F., Lopez-Molina L., Zakany J., Duboule D., Albrecht U., Schibler U. 2002. The orphan nuclear receptor REV-ERBalpha controls circadian transcription within the positive limb of the mammalian circadian oscillator. Cell. 110: 251–260. [DOI] [PubMed] [Google Scholar]
- 76.Yin L., Lazar M. A. 2005. The orphan nuclear receptor Rev-erbalpha recruits the N-CoR/histone deacetylase 3 corepressor to regulate the circadian Bmal1 gene. Mol. Endocrinol. 19: 1452–1459. [DOI] [PubMed] [Google Scholar]
- 77.Yin L., Wang J., Klein P. S., Lazar M. A. 2006. Nuclear receptor Rev-erbalpha is a critical lithium-sensitive component of the circadian clock. Science. 311: 1002–1005. [DOI] [PubMed] [Google Scholar]
- 78.Raghuram S., Stayrook K. R., Huang P., Rogers P. M., Nosie A. K., McClure D. B., Burris L. L., Khorasanizadeh S., Burris T. P., Rastinejad F. 2007. Identification of heme as the ligand for the orphan nuclear receptors REV-ERBalpha and REV-ERBbeta. Nat. Struct. Mol. Biol. 14: 1207–1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Burris T. P. 2008. Nuclear hormone receptors for heme: REV-ERBα and REV-ERBbeta are ligand-regulated components of the mammalian clock. Mol. Endocrinol. 22: 1509–1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wang J., Lazar M. A. 2008. Bifunctional role of Rev-erbalpha in adipocyte differentiation. Mol. Cell. Biol. 28: 2213–2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yin L., Wu N., Curtin J. C., Qatanani M., Szwergold N. R., Reid R. A., Waitt G. M., Parks D. J., Pearce K. H., Wisely G. B., et al. 2007. Rev-erbα, a heme sensor that coordinates metabolic and circadian pathways. Science. 318: 1786–1789. [DOI] [PubMed] [Google Scholar]
- 82.Kawamoto T., Noshiro M., Furukawa M., Honda K. K., Nakashima A., Ueshima T., Usui E., Katsura Y., Fujimoto K., Honma S., et al. 2006. Effects of fasting and re-feeding on the expression of Dec1, Per1, and other clock-related genes. J Biochem. 140: 401–408. [DOI] [PubMed] [Google Scholar]
- 83.Noshiro M., Okuda K. 1990. Molecular cloning and sequence analysis of cDNA encoding human cholesterol 7α-hydroxylase. FEBS Lett. 268: 137–140. [DOI] [PubMed] [Google Scholar]
- 84.Noshiro M., Nishimoto M., Okuda K. 1990. Rat liver cholesterol 7α-hydroxylase. Pretranslational regulation for circadian rhythm. J. Biol. Chem. 265: 10036–10041. [PubMed] [Google Scholar]
- 85.Chiang J. Y., Miller W. F., Lin G. M. 1990. Regulation of cholesterol 7 alpha-hydroxylase in the liver. Purification of cholesterol 7α-hydroxylase and the immunochemical evidence for the induction of cholesterol 7α-hydroxylase by cholestyramine and circadian rhythm. J. Biol. Chem. 265: 3889–3897. [PubMed] [Google Scholar]
- 86.Mueller C. R., Maire P., Shibler U. 1990. DBP, a liver enriched transcriptional activator is expressed late in ontogeny and its tissue specificity is determined post-transcriptionally. Cell. 61: 279–291. [DOI] [PubMed] [Google Scholar]
- 87.Wuarin J., Schibler U. 1990. Expression of the liver enriched transcription activator protein DBP follows a stringent circadian rhythm. Cell. 63: 1257–1266. [DOI] [PubMed] [Google Scholar]
- 88.Lavery D. J., Schibler U. 1993. Circadian transcription of the cholesterol 7α-hydroxylase gene may involve the liver-enriched bZIP protein DBP. Genes Dev. 7: 1871–1884. [DOI] [PubMed] [Google Scholar]
- 89.Yamada M., Nagatomo J., Setoguchi Y., Kuroki N., Higashi S., Setoguchi T. 2000. Circadian rhythms of sterol 12alpha-hydroxylase, cholesterol 7α- hydroxylase and DBP involved in rat cholesterol catabolism. Biol. Chem. 381: 1149–1153. [DOI] [PubMed] [Google Scholar]
- 90.Noshiro M., Usui E., Kawamoto T., Kubo H., Fujimoto K., Furukawa M., Honma S., Makishima M., Honma K., Kato Y. 2007. Multiple mechanisms regulate circadian expression of the gene for cholesterol 7α-hydroxylase (Cyp7a), a key enzyme in hepatic bile acid biosynthesis. J. Biol. Rhythms. 22: 299–311. [DOI] [PubMed] [Google Scholar]
- 91.Noshiro M., Kawamoto T., Furukawa M., Fujimoto K., Yoshida Y., Sasabe E., Tsutsumi S., Hamada T., Honma S., Honma K., et al. 2004. Rhythmic expression of DEC1 and DEC2 in peripheral tissues: DEC2 is a potent suppressor for hepatic cytochrome P450s opposing DBP. Genes Cells. 9: 317–329. [DOI] [PubMed] [Google Scholar]
- 92.Ananthanarayanan M., Balasubramanian N. V., Makishima M., Mangelsdorf D. J., Suchy F. J. 2001. Human bile salt export pump (BSEP) promoter is transactivated by the farnesoid X receptor/bile acid receptor (FXR/BAR). J. Biol. Chem. 276: 28857–28865. [DOI] [PubMed] [Google Scholar]
- 93.Jansen P. L., Sturm E. 2003. Genetic cholestasis, causes and consequences for hepatobiliary transport. Liver Int. 23: 315–322. [DOI] [PubMed] [Google Scholar]
- 94.Carlton V. E., Pawlikowska L., Bull L. N. 2004. Molecular basis of intrahepatic cholestasis. Ann. Med. 36: 606–617. [DOI] [PubMed] [Google Scholar]
- 95.Dawson P. A., Haywood J., Craddock A. L., Wilson M., Tietjen M., Kluckman K., Maeda N., Parks J. S. 2003. Targeted deletion of the ileal bile acid transporter eliminates enterohepatic cycling of bile acids in mice. J. Biol. Chem. 278: 33920–33927. [DOI] [PubMed] [Google Scholar]
- 96.Li H., Chen F., Shang Q., Pan L., Shneider B. L., Chiang J. Y., Forman B. M., Ananthanarayanan M., Tint G. S., Salen G., et al. 2005. FXR-activating ligands inhibit rabbit ASBT expression via FXR-SHP-FTF cascade. Am. J. Physiol. Gastrointest. Liver Physiol. 288: G60–G66. [DOI] [PubMed] [Google Scholar]
- 97.Tu H., Okamoto A. Y., Shan B. 2000. FXR, a bile acid receptor and biological sensor. Trends Cardiovasc. Med. 10: 30–35. [DOI] [PubMed] [Google Scholar]
- 98.Dawson P. A., Hubbert M., Haywood J., Craddock A. L., Zerangue N., Christian W. V., Ballatori N. 2005. The heteromeric organic solute transporter α-β, Ostα-Ostß, is an ileal basolateral bile acid transporter. J. Biol. Chem. 280: 6960–6968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ballatori N., Christian W. V., Lee J. Y., Dawson P. A., Soroka C. J., Boyer J. L., Madejczyk M. S., Li N. 2005. OSTα-OSTβ: a major basolateral bile acid and steroid transporter in human intestinal, renal, and biliary epithelia. Hepatology. 42: 1270–1279. [DOI] [PubMed] [Google Scholar]
- 100.Ballatori N., Fang F., Christian W. V., Li N., Hammond C. L. 2008. Ostα-Ostβ is required for bile acid and conjugated steroid disposition in the intestine, kidney, and liver. Am. J. Physiol. Gastrointest. Liver Physiol. 295: G179–G186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Rao A., Haywood J., Craddock A. L., Belinsky M. G., Kruh G. D., Dawson P. A. 2008. The organic solute transporter α-β, Ostα-Ostβ, is essential for intestinal bile acid transport and homeostasis. Proc. Natl. Acad. Sci. USA. 105: 3891–3896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Frankenberg T., Rao A., Chen F., Haywood J., Shneider B. L., Dawson P. A. 2006. Regulation of the mouse organic solute transporter α-β, Ostα-Ostβ, by bile acids. Am. J. Physiol. Gastrointest. Liver Physiol. 290: G912–G922. [DOI] [PubMed] [Google Scholar]
- 103.Boyer J. L., Trauner M., Mennone A., Soroka C. J., Cai S. Y., Moustafa T., Zollner G., Lee J. Y., Ballatori N. 2006. Upregulation of a basolateral FXR-dependent bile acid efflux transporter OSTα-OSTβ in cholestasis in humans and rodents. Am. J. Physiol. Gastrointest. Liver Physiol. 290: G1124–G1130. [DOI] [PubMed] [Google Scholar]
- 104.Denson L. A., Sturm E., Echevarria W., Zimmerman T. L., Makishima M., Mangelsdorf D. J., Karpen S. J. 2001. The orphan nuclear receptor, shp, mediates bile acid-induced inhibition of the rat bile acid transporter, ntcp. Gastroenterology. 121: 140–147. [DOI] [PubMed] [Google Scholar]
- 105.Kullak-Ublick G. A., Meier P. J. 2000. Mechanisms of cholestasis. Clin. Liver Dis. 4: 357–385. [DOI] [PubMed] [Google Scholar]
- 106.Trauner M., Meier P. J., Boyer J. L. 1998. Molecular pathogenesis of cholestasis. N. Engl. J. Med. 339: 1217–1227. [DOI] [PubMed] [Google Scholar]
- 107.Goodwin B., Jones S. A., Price R. R., Watson M. A., McKee D. D., Moore L. B., Galardi C., Wilson J. G., Lewis M. C., Roth M. E., et al. 2000. A regulatory cascade of the nuclear receptors FXR, SHP-1, and LRH-1 represses bile acid biosynthesis. Mol. Cell. 6: 517–526. [DOI] [PubMed] [Google Scholar]
- 108.Lu T. T., Makishima M., Repa J. J., Schoonjans K., Kerr T. A., Auwerx J., Mangelsdorf D. J. 2000. Molecular basis for feedback regulation of bile acid synthesis by nuclear receptors. Mol. Cell. 6: 507–515. [DOI] [PubMed] [Google Scholar]
- 109.Zhang M., Chiang J. Y. 2001. Transcriptional regulation of the human sterol 12α-hydroxylase gene (CYP8B1): roles of hepatocyte nuclear factor 4α (HNF4α) in mediating bile acid repression. J. Biol. Chem. 276: 41690–41699. [DOI] [PubMed] [Google Scholar]
- 110.Del Castillo-Olivares A., Campos J. A., Pandak W. M., Gil G. 2004. Role of FTF/LRH-1 on bile acid biosynthesis. A known nuclear receptor activator that can act as a suppressor of bile acid biosynthesis. J. Biol. Chem. 279: 16813–16821. [DOI] [PubMed] [Google Scholar]
- 111.Boulias K., Katrakili N., Bamberg K., Underhill P., Greenfield A., Talianidis I. 2005. Regulation of hepatic metabolic pathways by the orphan nuclear receptor SHP. EMBO J. 24: 2624–2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Boulias K., Talianidis I. 2004. Functional role of G9a-induced histone methylation in small heterodimer partner-mediated transcriptional repression. Nucleic Acids Res. 32: 6096–6103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Kemper J. K., Kim H., Miao J., Bhalla S., Bae Y. 2004. Role of an mSin3A-Swi/Snf chromatin remodeling complex in the feedback repression of bile acid biosynthesis by SHP. Mol. Cell. Biol. 24: 7707–7719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Fang S., Miao J., Xiang L., Ponugoti B., Treuter E., Kemper J. K. 2007. Coordinated recruitment of histone methyltransferase G9a and other chromatin modifying enzymes in SHP-mediated regulation of hepatic bile acid metabolism. Mol. Cell. Biol. 27: 1407–1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Mitro N., Godio C., De Fabiani E., Scotti E., Galmozzi A., Gilardi F., Caruso D., Chacon A. B., Crestani M. 2007. Insights in the regulation of cholesterol 7α-hydroxylase gene reveal a target for modulating bile acid synthesis. Hepatology. 46: 885–897. [DOI] [PubMed] [Google Scholar]
- 116.Mataki C., Magnier B. C., Houten S. M., Annicotte J. S., Argmann C., Thomas C., Overmars H., Kulik W., Metzger D., Auwerx J., et al. 2007. Compromised intestinal lipid absorption in mice with a liver-specific deficiency of liver receptor homolog 1. Mol. Cell. Biol. 27: 8330–8339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Lee Y. K., Schmidt D. R., Cummins C. L., Choi M., Peng L., Zhang Y., Goodwin B., Hammer R. E., Mangelsdorf D. J., Kliewer S. A. 2008. Liver receptor homolog-1 regulates bile acid homeostasis but is not essential for feedback regulation of bile acid synthesis. Mol. Endocrinol. 22: 1345–1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Kerr T. A., Saeki S., Schneider M., Schaefer K., Berdy S., Redder T., Shan B., Russell D. W., Schwarz M. 2002. Loss of nuclear receptor SHP impairs but does not eliminate negative feedback regulation of bile acid synthesis. Dev. Cell. 2: 713–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Wang L., Lee Y. K., Bundman D., Han Y., Thevananther S., Kim C. S., Chua S. S., Wei P., Heyman R. A., Karin M., et al. 2002. Redundant pathways for negative feedback regulation of bile Acid production. Dev. Cell. 2: 721–731. [DOI] [PubMed] [Google Scholar]
- 120.Huang W., Ma K., Zhang J., Qatanani M., Cuvillier J., Liu J., Dong B., Huang X., Moore D. D. 2006. Nuclear receptor-dependent bile acid signaling is required for normal liver regeneration. Science. 312: 233–236. [DOI] [PubMed] [Google Scholar]
- 121.Zhang L., Huang X., Meng Z., Dong B., Shiah S., Moore D. D., Huang W. 2009. Significance and mechanism of CYP7a1 gene regulation during the acute phase of liver regeneration. Mol. Endocrinol. 23: 137–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Holt J. A., Luo G., Billin A. N., Bisi J., McNeill Y. Y., Kozarsky K. F., Donahee M., Wang D. Y., Mansfield T. A., Kliewer S. A., et al. 2003. Definition of a novel growth factor-dependent signal cascade for the suppression of bile acid biosynthesis. Genes Dev. 17: 1581–1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Inagaki T., Choi M., Moschetta A., Peng L., Cummins C. L., McDonald J. G., Luo G., Jones S. A., Goodwin B., Richardson J. A., et al. 2005. Fibroblast growth factor 15 functions as an enterohepatic signal to regulate bile acid homeostasis. Cell Metab. 2: 217–225. [DOI] [PubMed] [Google Scholar]
- 124.Kim I., Ahn S. H., Inagaki T., Choi M., Ito S., Guo G. L., Kliewer S. A., Gonzalez F. J. 2007. Differential regulation of bile acid homeostasis by the farnesoid X receptor in liver and intestine. J. Lipid Res. 48: 2664–2672. [DOI] [PubMed] [Google Scholar]
- 125.Pandak W. M., Heuman D. M., Hylemon P. B., Chiang J. Y., Vlahcevic Z. R. 1995. Failure of intravenous infusion of taurocholate to down-regulate cholesterol 7α-hydroxylase in rats with biliary fistulas. Gastroenterology. 108: 533–544. [DOI] [PubMed] [Google Scholar]
- 126.Ito S., Fujimori T., Furuya A., Satoh J., Nabeshima Y. 2005. Impaired negative feedback suppression of bile acid synthesis in mice lacking betaKlotho. J. Clin. Invest. 115: 2202–2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Yu C., Wang F., Kan M., Jin C., Jones R. B., Weinstein M., Deng C. X., McKeehan W. L. 2000. Elevated cholesterol metabolism and bile acid synthesis in mice lacking membrane tyrosine kinase receptor FGFR4. J. Biol. Chem. 275: 15482–15489. [DOI] [PubMed] [Google Scholar]
- 128.Yu C., Wang F., Jin C., Huang X., McKeehan W. L. 2005. Independent repression of bile acid synthesis and activation of c-Jun N-terminal kinase (JNK) by activated hepatocyte fibroblast growth factor receptor 4 (FGFR4) and bile acids. J. Biol. Chem. 280: 17707–17714. [DOI] [PubMed] [Google Scholar]
- 129.Lundasen T., Galman C., Angelin B., Rudling M. 2006. Circulating intestinal fibroblast growth factor 19 has a pronounced diurnal variation and modulates hepatic bile acid synthesis in man. J. Intern. Med. 260: 530–536. [DOI] [PubMed] [Google Scholar]
- 130.Schaap F. G., van der Gaag N. A., Gouma D. J., Jansen P. L. 2009. High expression of the bile salt-homeostatic hormone fibroblast growth factor 19 in the liver of patients with extrahepatic cholestasis. Hepatology. 49: 1228–1235. [DOI] [PubMed] [Google Scholar]
- 131.Li T., Chiang J. Y. 2004. Mechanism of rifampicin and pregnane X receptor (PXR) inhibition of human cholesterol 7α-hydroxylase gene (CYP7A1) transcription. Am. J. Physiol. Gastrointest. Liver Physiol. 288: G74–G84. [DOI] [PubMed] [Google Scholar]
- 132.Han S., Chiang J. Y. 2009. Mechanism of vitamin D receptor inhibition of cholesterol 7α-hydroxylase gene transcription in human hepatocytes. Drug Metab. Dispos. 37: 469–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Owsley E., Chiang J. Y. 2003. Guggulsterone antagonizes farnesoid X receptor induction of bile salt export pump but activates pregnane X receptor to inhibit cholesterol 7α-hydroxylase gene. Biochem. Biophys. Res. Commun. 304: 191–195. [DOI] [PubMed] [Google Scholar]
- 134.Li T., Chen W., Chiang J. Y. 2007. PXR induces CYP27A1 and regulates cholesterol metabolism in the intestine. J. Lipid Res. 48: 373–384. [DOI] [PubMed] [Google Scholar]
- 135.Miao J., Fang S., Bae Y., Kemper J. K. 2006. Functional inhibitory cross-talk between car and HNF-4 in hepatic lipid/glucose metabolism is mediated by competition for binding to the DR1 motif and to the common coactivators, GRIP-1 and PGC-1alpha. J. Biol. Chem. 281: 14537–14546. [DOI] [PubMed] [Google Scholar]
- 136.Gupta S., Stravitz R. T., Dent P., Hylemon P. B. 2001. Down-regulation of cholesterol 7α-hydroxylase (CYP7A1) gene expression by bile acids in primary rat hepatocytes is mediated by the c-Jun N-terminal kinase pathway. J. Biol. Chem. 276: 15816–15822. [DOI] [PubMed] [Google Scholar]
- 137.Jahan A., Chiang J. Y. 2005. Cytokine regulation of human sterol 12α-hydroxylase (CYP8B1) gene. Am. J. Physiol. Gastrointest. Liver Physiol. 288: G685–G695. [DOI] [PubMed] [Google Scholar]
- 138.Myung S. J., Yoon J. H., Gwak G. Y., Kim W., Yang J. I., Lee S. H., Jang J. J., Lee H. S. 2007. Bile acid-mediated thrombospondin-1 induction in hepatocytes leads to transforming growth factor-β-dependent hepatic stellate cell activation. Biochem. Biophys. Res. Commun. 353: 1091–1096. [DOI] [PubMed] [Google Scholar]
- 139.Li T., Chiang J. Y. 2007. A novel role of transforming growth factor beta1 in transcriptional repression of human cholesterol 7α-hydroxylase gene. Gastroenterology. 133: 1660–1669. [DOI] [PubMed] [Google Scholar]
- 140.Wilkinson D. S., Ogden S. K., Stratton S. A., Piechan J. L., Nguyen T. T., Smulian G. A., Barton M. C. 2005. A direct intersection between p53 and transforming growth factor beta pathways targets chromatin modification and transcription repression of the alpha-fetoprotein gene. Mol. Cell. Biol. 25: 1200–1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Cordenonsi M., Montagner M., Adorno M., Zacchigna L., Martello G., Mamidi A., Soligo S., Dupont S., Piccolo S. 2007. Integration of TGF-β and Ras/MAPK signaling through p53 phosphorylation. Science. 315: 840–843. [DOI] [PubMed] [Google Scholar]
- 142.Maeda Y., Seidel S. D., Wei G., Liu X., Sladek F. M. 2002. Repression of hepatocyte nuclear factor 4α tumor suppressor p53: involvement of the ligand-binding domain and histone deacetylase activity. Mol. Endocrinol. 16: 402–410. [DOI] [PubMed] [Google Scholar]
- 143.Li T., Ma H., Chiang J. Y. 2008. Transforming growth factor beta 1, tumor necrosis factor a, and insulin signaling crosstalk in regulation of the rat cholesterol 7α-hydroxylase gene expression. J. Lipid Res. 49: 1981–1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Dueland S., Reichen J., Everson G. T., Davis R. A. 1991. Regulation of cholesterol and bile acid homeostasis in bile-obstructed rats. Biochem. J. 280: 373–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Boyer J. L. 2005. Nuclear receptor ligands: rational and effective therapy for chronic cholestatic liver disease? Gastroenterology. 129: 735–740. [DOI] [PubMed] [Google Scholar]
- 146.Zollner G., Trauner M. 2009. Nuclear receptors as therapeutic targets in cholestatic liver diseases. Br. J. Pharmacol. 156: 7–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Fiorucci S., Clerici C., Antonelli E., Orlandi S., Goodwin B., Sadeghpour B., Sabatino G., Russo G., Castellani D., Willson T. M., et al. 2005. Protective effects of 6-ethyl chenodeoxycholic acid, a farnesoid X receptor (FXR) ligand, in estrogen induced cholestasis. J. Pharmacol. Exp. Ther. 313: 604–612. [DOI] [PubMed] [Google Scholar]
- 148.Pellicciari R., Sato H., Gioiello A., Costantino G., Macchiarulo A., Sadeghpour B. M., Giorgi G., Schoonjans K., Auwerx J. 2007. Nongenomic actions of bile acids. Synthesis and preliminary characterization of 23- and 6,23-alkyl-substituted bile acid derivatives as selective modulators for the G-protein coupled receptor TGR5. J. Med. Chem. 50: 4265–4268. [DOI] [PubMed] [Google Scholar]
- 149.Gilat T., Leikin-Frenkel A., Goldiner L., Laufer H., Halpern Z., Konikoff F. M. 2001. Arachidyl amido cholanoic acid (Aramchol) is a cholesterol solubilizer and prevents the formation of cholesterol gallstones in inbred mice. Lipids. 36: 1135–1140. [DOI] [PubMed] [Google Scholar]
- 150.Leikin-Frenkel A., Weinbroum A. A., Leikin-Gobbi D., Krupitzky L., Goldiner I., Shafat L., Gilat T., Konikoff F. M. 2004. Faecal sterol output is increased by arachidyl amido cholanoic acid (Aramchol) in rats. Biochem. Soc. Trans. 32: 131–133. [DOI] [PubMed] [Google Scholar]
- 151.Leikin-Frenkel A., Goldiner I., Leikin-Gobbi D., Rosenberg R., Bonen H., Litvak A., Bernheim J., Konikoff F. M., Gilat T. 2008. Treatment of preestablished diet-induced fatty liver by oral fatty acid-bile acid conjugates in rodents. Eur. J. Gastroenterol. Hepatol. 20: 1205–1213. [DOI] [PubMed] [Google Scholar]
- 152.Gilat T., Leikin-Frenkel A., Goldiner I., Halpern Z., Konikoff F. M. 2002. Dissolution of cholesterol gallstones in mice by the oral administration of a fatty acid bile acid conjugate. Hepatology. 35: 597–600. [DOI] [PubMed] [Google Scholar]