Abstract
Lipoproteins are centrally important in lipid transport, fuel metabolism, and cardiovascular disease. The prototypic lipoprotein has an outer shell of amphipathic lipids and proteins that solubilizes a hydrophobic lipid core. Lipoprotein-associated proteins have classically been viewed as structural elements and factors important in lipid metabolism. Recent mass spectrometric analyses reveal that the protein cargo of lipoproteins is much more diverse than previously appreciated, raising the possibility that lipoproteins play previously unsuspected roles in host defense mechanisms and inflammation. They further suggest that lipoprotein-associated proteins can identify humans at increased risk of cardiovascular disease. Here, we summarize recent developments in lipoproteomics, the proteomic analysis of lipoproteins. We also discuss the promises and challenges this powerful analytical strategy offers for expanding our understanding of the biology and structures of lipoproteins.
Keywords: cardiovascular disease, lipoprotein metabolism, innate immunity, inflammation, macrophages
Lipoproteins play a central role in extracellular lipid transport in species ranging from insects to mammals (1–3). As couriers of lipids, they transport triglycerides derived from the diet to peripheral tissues for energy metabolism. They also are important for the bidirectional movement of cholesterol between the liver and peripheral tissues. However, lipoproteins are implicated in pathways distinct from lipid metabolism. For example, inflammation markedly alters lipoprotein metabolism (4), and recent studies implicate lipoproteins as important mediators of the immune response and host defense mechanisms (5, 6).
Lipoproteins have attracted wide interest clinically because they are major risk factors for cardiovascular disease (CVD) in humans. For example, high levels of LDL cholesterol are a major risk factor for myocardial infarction and sudden death (7). In contrast, low levels of HDL cholesterol strongly associate with an increased risk of CVD (8), whereas high levels associate with longevity (9). LDL and HDL are the major carriers of cholesterol in plasma and serum, accounting for the strong relationship between cholesterol levels and CVD.
Apolipoprotein (apo)B-100 is the major protein structural component of VLDL and LDL, whereas apoA-I serves that function in HDL. Chylomicrons contain apoB-48, a truncated form of apoB-100. Clinical studies suggest that levels of apoB-100 and apoA-I may be better predictors of CVD risk than LDL and HDL cholesterol levels (10). Other protein components of LDL and HDL play important roles in lipid metabolism, energy homeostasis, and inflammation.
Most studies of lipoproteins have focused on single proteins. Proteomics, a global approach to understanding protein expression, regulation, and function, transcends analysis of individual components. One of its major tools is mass spectrometry (MS) (11, 12), which can detect and quantify hundreds or even thousands of proteins in one sample. MS measures molecular mass and therefore can also detect and characterize posttranslational modifications of proteins (13). Its ability to identify disease-related biomarkers is also a powerful advantage (14).
Recent mass spectrometric studies have revealed that lipoproteins carry a diverse array of previously unsuspected proteins. For example, shotgun proteomics has implicated regulation of the complement pathway and proteolysis in HDL’s cardioprotective effects (15). This review highlights lipoproteomics, using MS to study lipoprotein-associated proteins and their biology. We first provide brief overviews of lipoprotein physiology and MS. Then we summarize lipoproteomic studies, focusing on recent insights they have provided into lipoprotein function and biology. We also discuss the limitations of our current approaches as well as important technical and conceptual issues that should be addressed in future studies.
APOLIPOPROTEINS ARE OF CENTRAL IMPORTANCE IN LIPOPROTEIN BIOLOGY
Lipoproteins are classically considered to be spherical particles containing a hydrophobic core of neutral lipid (cholesteryl ester and triglycerides) and a surface rich in amphipathic proteins, phospholipids, and free cholesterol (1–3, 16). These surface components solubilize the hydrophobic lipid core. In mammals, including humans, there are two major classes of lipoproteins: those containing apoB (chylomicrons, LDL, VLDL) and those containing apoA-I (HDL). The metabolism of each lipoprotein class is dictated by its major structural protein (16). For instance, apoB-100 is required for binding LDL to the LDL receptor, which plays the essential role in uptake of LDL from the circulation by peripheral cells and the liver (1). In contrast, the truncated form of apoB (apoB-48) present in chylomicrons lacks the LDL binding domain. Thus, remnant particles derived from chylomicrons by lipolysis of core lipids are cleared from the liver by pathways distinct from that of the LDL receptor.
Enterocytes, which line the small intestine, assemble chylomicrons from triglycerides, phospholipids, and cholesterol derived from the diet and then secrete the nascent lipoprotein into the blood (Fig. 1). Chylomicrons are the largest lipoproteins in humans (16). Together with VLDL, which is synthesized by the liver, these triglyceride-rich lipoproteins undergo lipolysis in muscle and adipose tissue to provide fatty acids for energy metabolism. ApoB-48 and apoB-100 serve as structural supports for chylomicrons and VLDL, respectively, permitting the formation of a large, hydrophobic core (17). ApoC-II, another protein carried by triglyceride-rich lipoproteins that contain apoB, is a cofactor for lipoprotein lipase, which hydrolyzes triglycerides in the particles to free fatty acids (18). Deficiency of either protein results in profound disturbances in lipid metabolism, demonstrating the pivotal role apolipoproteins play in lipoprotein physiology.
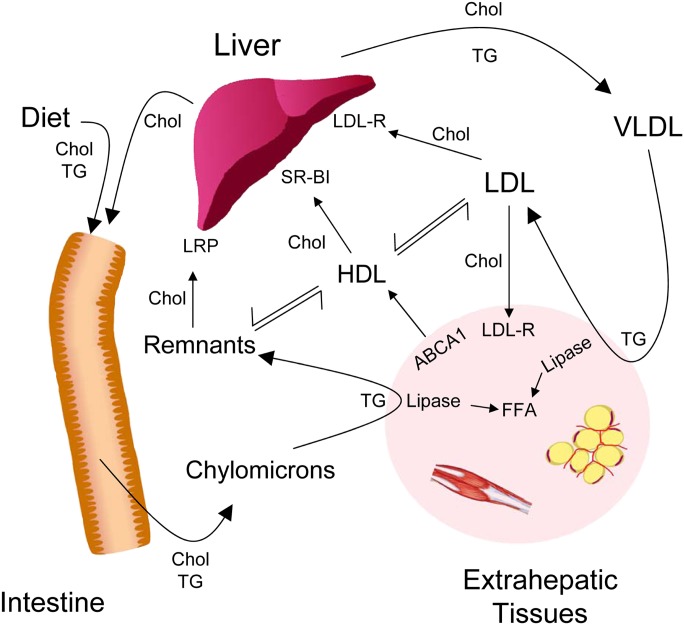
Fig. 1.
Overview of lipoprotein metabolism. Chylomicrons deliver triglycerides derived from the intestine into blood. Following triglyceride lipolysis to free fatty acids (FFAs) by peripheral tissues, chylomicron remnant particles are cleared by the liver via LDL receptor (LDL-R) related protein (LRP). The liver uses endogenously synthesized triglyceride and cholesterol as well as lipids derived from chylomicron remnants to synthesize VLDL. Triglyceride-rich VLDL is converted by lipolysis to intermediate density lipoprotein (IDL; not shown) and then cholesterol-rich LDL. Peripheral tissues and liver take up LDL-derived cholesterol via the LDL receptor. HDL accepts cholesterol from peripheral tissue for transport back to the liver. The protein components of lipoproteins play key roles as structural elements, enzymes, enzyme cofactors, and ligands for cell surface receptors. For example, apoB is a ligand for the LDL receptor whereas apoA-I interacts with the scavenger receptor SR-BI. Recent mass spectrometric studies demonstrate that lipoproteins carry a diverse array of lower abundance proteins, raising the possibility that lipoproteins play previously unsuspected role in vascular disease and inflammation.
In contrast to the triglyceride-rich chylomicrons and VLDL particles, the major lipids in HDL and LDL are cholesterol and cholesteryl esters (3). LDL is largely derived from circulating VLDL by the action of lipases on triglycerides (18). ApoB-100 provides structural support for VLDL and LDL (17). It also is the ligand for the LDL receptor on liver and peripheral tissue (1, 19). Modified forms of apoB-containing particles, such as oxidized LDL, promote the conversion of artery wall macrophages into cholesteryl ester-laden foam cells, the hallmark of the atherosclerotic lesion (20).
ApoA-I functions as the scaffold for HDL assembly (21). Unlike apoB, apoA-I is associated with very little lipid when it is secreted from hepatocytes and enterocytes. In this poorly lipidated state, it serves as the ligand for ABCA1, which exports phospholipid and cholesterol into nascent HDL particles, promoting their maturation (22, 23). HDL is protein-rich compared with other lipoproteins and contains many other apolipoproteins. Its apolipoproteins, in concert with other proteins like phospholipid transfer protein (PLTP) and cholesteryl ester transfer protein (CETP), collaborate with apoA-I to orchestrate the exchange of lipids between lipoprotein particles (24). ApoA-I also has a central role in removing cholesterol from macrophage foam cells, making a key contribution to the cardioprotective effects of HDL (25).
Apolipoproteins are often classified as nonexchangeable and exchangeable and each class has distinct structural features. ApoB exhibits both amphipathic α-helical and β-pleated sheet structure (26). Each chylomicron, VLDL, or LDL particle contains 1 mol of apoB, which is not exchanged between different particles. In contrast, apoA-I exists largely as an amphipathic α-helical protein (21). Each circulating HDL particle contains 2–4 mol of apoA-I that can move between lipoprotein particles. A wide variety of other apolipoproteins (apoCs, apoE, etc.) are found on lipoproteins containing apoB and apoA-I, though in smaller amounts than in the major structural proteins.
Lipoproteins also contain low-abundance proteins that generally do not exhibit the typical structural features of apolipoproteins. Certain of these, such as PLTP, CETP, and LCAT, have important roles in lipid metabolism. However, over the past decade, there has been an explosion of interest in minor protein components of lipoproteins that have functions distinct from lipid metabolism. For example, paraoxonase and clusterin, which are cotransported with HDL in plasma, have been proposed to have antioxidant and anti-inflammatory properties (4, 5, 27, 28). Importantly, levels of these proteins are markedly altered in humans with CVD and in mice that are susceptible to atherosclerosis (27, 28). Loss of anti-inflammatory and antioxidant proteins, perhaps in concert with gain of proinflammatory proteins, may thus be another key contributor to the antiatherogenic effects of HDL.
LIPOPROTEOMICS: MASS SPECTROMETRIC ANALYSIS OF LIPOPROTEINS
There are three general approaches to identifying proteins in complex mixtures (reviewed in Refs. 29–31). The first separates proteins based on isoelectric point (pI) and molecular size, using two-dimensional (2D) gel electrophoresis (32). Protein spots are visualized by staining and extracted from the gel. Following proteolytic digestion, peptides are analyzed with MS and/or MS/MS. This approach can be useful for quantifying proteomes of limited complexity that contain proteins that are relatively abundant. 2D gel electrophoresis is also a powerful strategy for identifying different isoforms or posttranslational modifications of the same protein. However, this analytical strategy also suffers from many limitations; for example: i) it can be difficult to reproducibly analyze multiple samples; ii) extraction and analysis of multiple protein spots by MS is laborious; iii) individual gel spots generally contain more than one protein, which complicates protein quantification; and iv) large, hydrophobic proteins, such as apoB, are unable to enter gels (33–39), making it difficult to quantify the contributions of individual proteins to the overall composition of lipoprotein particles.
The second approach uses matrix assisted laser desorption ionization-time of flight-MS (MALDI-TOF-MS) to identify intact proteins (40–46). Because the mass of an intact protein offers limited diagnostic information, this method is generally limited to identifying known proteins. It also is best applied to the detection of relatively small proteins (typically less than 35 kDa). Protein quantification by MALDI-TOF-MS can also be problematic, and this approach also suffers from a limited dynamic range. Thus, MADLI-TOF-MS is typically limited to detecting relatively abundant proteins that are already known to exist in lipoproteins (such as apoC, apoA, and apoE and their isoforms).
The third approach, termed shotgun proteomics (47), uses liquid chromatography (LC) in concert with electrospray ionization and tandem MS analysis (LC-ESI-MS/MS) to identify peptides. The peptide digest is separated by LC, introduced into the gas phase by ESI, and analyzed by tandem MS (15, 48, 49). To identify the proteins in the original mixture, the MS/MS spectrum collected for each peptide is searched against a database of all the theoretical spectra for a digest of the relevant genome. This method is powerful and can detect more than a thousand proteins and their posttranslational modifications in a single sample. It also is readily applied to the analysis of large numbers of samples, which is critical for clinical and translational studies. Data-base searching can be time-consuming and the analyses expensive, however (47), and protein quantification by this method is generally semi-quantitative.
Identification of proteins by MS/MS generally involves the analysis of peptide digests (29–31). Proteins that associate with lipids contain sizable hydrophobic portions that are stabilized by strong secondary structural features and these regions can resist complete proteolytic digestion (50, 51). In addition, the lipids in lipoproteins make protein digestion and mass spectrometric analysis challenging. However, recent technical advances have addressed many of these issues (reviewed in Ref. 51).
It is important to note that the method used to isolate lipoproteins significantly affects the protein content of the resulting particles (52). Most proteomic studies have relied on ultracentrifugation, which is convenient because particle classes are defined by density and ultracentrifugation is widely used for clinical studies. However, the typical isolation involves high concentrations of KBr, a potent chaotropic agent, which can dissociate proteins from lipoproteins (53, 54). Substituting D2O/sucrose for KBr may overcome these limitations (39).
Methods involving size exclusion chromatography can enrich for specific size classes of lipoprotein, but can result in contamination by nonlipoprotein proteins, especially when serum or plasma is fractionated. Immunoaffinity chromatography has potential advantages because it should more readily preserve lipoprotein-associated proteins (15, 36), but it classifies lipoproteins on the basis of a specific protein component (e.g., apoA-I or apoB).
EXPANDING THE LIPOPROTEOME
The proteins that have been identified to date by proteomics analysis of lipoproteins are listed in Table 1. Significantly more proteins have been detected in human HDL than in LDL or VLDL, however, only HDL has been analyzed by shotgun proteomics (15, 48, 49). More complete proteomes for LDL and VLDL might be defined by similar analyses. At this time, no proteomic studies of chylomicrons have been reported.
TABLE 1.
Proteins detected in lipoproteins by mass spectrometryaa
| Protein | VLDL | LDL | HDL |
|---|---|---|---|
| Actin | 1b | ||
| Albumin | 1b | 3e | 5,6,12 |
| Alpha-1-acid glycoprotein | 5,6 | ||
| Alpha-1-antitrypsin inhibitor | 1b | 4 | 5,6,7,9,12 |
| Alpha-1B-glycoprotein | 6,7 | ||
| Alpha-1-microglobulin/bikunin | 6 | ||
| Alpha-2-antiplasmin | 6 | ||
| Alpha-2HS-glycoprotein | 1b | 5,6 | |
| Alpha-2-macroglobulin | 3d | 5 | |
| Alpha-amylase (salivary) | 9 | ||
| Angiotensinogen | 6 | ||
| Apolipoprotein(a) | |||
| ApoA-I | 1 | 3e,4 | 5,6,7,8,9,10,12 |
| ApoA-II | 5,6,8,9,10,12 | ||
| ApoA-IV | 1 | 3c | 6,7,8,9,10,12 |
| ApoB | 1 | 3d | 6,12 |
| ApoC-I | 5,6,9,12 | ||
| ApoC-II | 3c,4 | 6,8,9,12 | |
| ApoC-III | 1 | 3c,4 | 5,6,7,8,9,10,12 |
| ApoC-IV | 1 | 6,12 | |
| ApoD | 5,6,8,10,12 | ||
| ApoE | 1 | 3e,4 | 5,6,8,9,10,12 |
| ApoF | 5,6,12 | ||
| ApoH | 6 | ||
| ApoJ (Clusterin) | 3c,4 | 5,6,8,12 | |
| ApoL-I | 1 | 6,7,8,9,12 | |
| ApoM | 1 | 3c | 6,8,9,12 |
| C4b binding protein | 10 | ||
| Calgranulin A | 3c | ||
| CETP | 6,10 | ||
| Complement C3 | 6,8 | ||
| Complement C4 | 6 | ||
| Complement C9 | 6 | ||
| Fibrinogen | 1b | 3d | 6,7,10,12 |
| Fibronectin | 3d | ||
| Haptoglobin-related protein | 5,6,12 | ||
| Haptopglobin | 3d | 8,10 | |
| Hemoglobin | 11 | ||
| Hemopexin | 1b | 6 | |
| Immunoglobulin Mu | 1b | 3d | |
| Inter-α-trypsin inhibitor chain H4 | 6,7 | ||
| Kininogen | 6 | ||
| LCAT | 6,10 | ||
| LPS binding protein | 7 | ||
| Lysozyme C | 3c | ||
| Paraoxonase 1 | 5,6,8,12 | ||
| Paraoxonase 3 | 6,12 | ||
| Platelet activating factor-acetyl hydrolase | 12 | ||
| Platelet basic protein | 12 | ||
| PLTP | 6,12 | ||
| Prenylcysteine lyase | 1 | 6 | |
| Prothrombin | 12 | ||
| Retinol binding protein | 6 | ||
| SAA1/2 | 6,9,12 | ||
| SAA4 | 3c,4 | 5,6,9,12 | |
| SerpinF1 | 6 | ||
| Transferrin | 5,6,8 | ||
| Transthyreitin | 6,7,8,12 | ||
| Vitamin D binding globulin | 1b | 5,6 | |
| Vitronectin | 6 |
Studies include: 1. Mancone, 2007 (37); 2. Bondarenko, 1999 (40); 3. Karlsson, 2005 (34); 4. Stahlman, 2008 (39); 5. Heller, 2005 (44); 6. Vaisar, 2007 (15); 7. Hortin, 2006 (69); 8. Rezaee, 2006 (38); 9. Karlsson, 2005 (33); 10. Kunitake, 1994 (36); 11. Watanabe, 2007 (46); 12. Davidson, 2009 (64).
Proteins detected in ultracentrifuge-purified VLDL and described by the authors as contaminants.
Proteins detected in ultracentrifuge-purified LDL and not in LDL prepared by size exclusion chromatography.
Proteins detected in LDL prepared by size exclusion chromatography and not in ultracentrifuge-purified LDL.
Proteins detected in LDL prepared by size exclusion chromatography and ultracentrifugation.
The most important message from proteomic analysis is simple: each lipoprotein class contains many proteins involved in processes distinct from lipid metabolism. For example, LDL contains lysozyme C, which can kill invading pathogens (34). This observation may be physiologically relevant, because apoB itself was recently shown to modulate the virulence of Staphylococcus aureus in vivo (55).
The HDL lipoproteome
HDL is anti-inflammatory (27), which may contribute to its atheroprotective capacity. Potential mechanisms include stimulation of nitric oxide secretion by endothelial cells (56), inhibition of neutrophil degranulation (57), regulation of the formation of the complement membrane attack complex (58, 59), and reduction of lipid peroxides (60).
We used LC-ESI-MS/MS to investigate the possibility that HDL carries proteins that might play a previously unsuspected role in its anti-inflammatory properties (15). Shotgun proteomics identified 48 proteins in HDL and HDL3 (the dense fraction of HDL) isolated by ultracentrifugation from healthy controls and/or CVD subjects. This analytical approach provides a comprehensive view of the HDL proteome because we identified 22 of 23 HDL proteins with established roles in lipid metabolism.
We used Gene Ontology (GO) Consortium analysis to associate the complex array of HDL proteins to biological processes (15). Unexpectedly, there were more acute-phase-response proteins, whose plasma concentrations are altered markedly by acute inflammation, than proteins implicated in lipid metabolism. We also identified 13 proteins not previously known to reside in HDL, including complement factors C9 and C4A/C4B as well as the complement regulatory protein vitronectin. Pioneering studies of HDL protein complexes that kill protozoa (61, 62) led to the proposal that HDL serves as a platform for the assembly of proteins. Our detection of a series of proteins involved in complement activation is consistent with this hypothesis.
Proteolysis of structural proteins in atherosclerotic lesions is thought to play a critical role in plaque rupture, the major cause of myocardial infarction and sudden death in subjects with CVD (16). It is therefore of interest that we found a family of proteins in HDL that contain proteinase inhibitor domains (15). Serine protease inhibitors, termed serpins, are key regulators of numerous biological pathways involved in coagulation, matrix remodeling, and inflammation. These observations raise the possibility that HDL is cardioprotective, in part, by inhibiting proteolysis and plaque rupture.
Complement activation increases tissue damage in animal models of acute myocardial infarction. It is noteworthy that we identified multiple complement regulatory proteins in HDL, which may limit injury to cardiac cells and prevent activation of the coagulant cascade.
HDL subspecies contain distinct protein populations that associate with antioxidant effects
LDL oxidation is implicated in the unregulated accumulation of cholesterol by macrophages, a key event in atherogenesis (63). HDL has been proposed to inhibit LDL oxidation in vivo, which may contribute significantly to its cardioprotective effects (27). Systemic inflammation has been proposed to convert HDL to a dysfunctional form that loses these antiatherogenic effects. However, the underlying factors that render HDL dysfunctional remain poorly understood.
Proteins such as paraoxonase (PON) that are cotransported with HDL in plasma have been proposed to have antioxidant properties (27). To begin to investigate the role of specific subspecies in the antiatherogenic effects of HDL, Davidson et al. (64) used LC-ESI-MS/MS to define the protein composition of five HDL fractions isolated by isopycnic density gradient ultracentrifugation. In parallel studies, they evaluated the ability of the different HDL subspecies to inhibit LDL oxidation in vitro. Importantly, levels of three specific proteins (apoL, PON1, and PON3) were highly enriched in the dense subfractions of HDL. Moreover, the ability of HDL to inhibit LDL oxidation strongly associated with the HDL content of those proteins. These observations support the proposal that HDL is composed of distinct subpopulations of particles that have discreet biological properties. Understanding the role of specific proteins and HDL subspecies may lead to new diagnostic and therapeutic approaches to atherosclerosis.
The LDL/VLDL lipoproteome
We used GO analysis to associate the proteins detected in VDLD and LDL (Table 1) with biological process. When compared with the whole human genome using the database for annotation, visualization, and integrated discovery (DAVID) resource (65, 66), the only category of proteins in VLDL that was significantly enriched associated with lipid transport (p<1 × 10−12). LDL was enriched in proteins associated with both lipid transport (p<1 × 10−9) and the inflammatory response (p<1 × 10−4).
In contrast, our analysis revealed that the HDL proteome (15) was enriched in proteins associated with lipid transport (p<1 × 10−21), the inflammatory response (p<1 × 10−17), complement activation (p<1 × 10−6), the acute phase response (p<1 × 10−11), and protease inhibition (p<1 × 10−11). Thus, the available data suggest that the HDL proteome is much more diverse than that of other lipoproteins.
Mechanistic implications of the HDL and apoB-associated lipoproteomes
The large number of proteins carried by lipoproteins supports the concept of significant heterogeneity in biological functions among lipoprotein classes. Consistent with their well-established roles in energy and cholesterol metabolism, the apoB-associated lipoproteome appears to be strongly linked to lipid transport. In contrast, HDL carries a much broader array of proteins, including those associated with inflammation, complement activation, and protease inhibition, raising the possibility that this class of lipoprotein particles plays an important role in host defense mechanisms and the acute-phase response.
The relatively low abundance of most of the proteins detected in lipoproteins, together with the known size of the lipoprotein classes on size-exclusion chromatography, indicates that each lipoprotein particle carries, at most, a small number of proteins. One possibility is that lipoproteins can mix and match proteins to form multimolecular complexes with precise biological functions. A seminal example is the trypanosome lytic factor, a heteromeric complex of apoA-I, apoL-I, and haptoglobin-related protein, which is a minor component of HDL (61, 62).
Peptidomics
In parallel with proteomics, peptidomics attempts to identify all of the peptides in a complex mixture. This may be particularly relevant to HDL because peptides derived from two HDL apolipoproteins, apoA-I and apoJ (clusterin), are atheroprotective in hypercholesterolemic animal models (67, 68). Peptides from a number of different proteins have been detected by MS/MS analysis of HDL, including apoA-I, apoA-IV, and fibrinogen (69). The presence of fibrinogen supports the proposal that dysfunctional HDL might promote thrombosis, the leading cause of sudden death and myocardial infarction in humans suffering from CVD.
Protein isoforms
Proteomics has also identified new isoforms of many apolipoproteins (34, 35, 37, 40, 41). Whereas some studies cataloged the novel protein forms, others went farther by identifying the primary structures of the new proteins. For example, one study demonstrated that a significant portion of apoA-II lacks the C-terminal glutamine (41). Another detected an apoE isoform in the HDL3 subfraction but not in the HDL2 subfraction (34). It will be of great interest to explore the physiological ramifications of these apolipoprotein isoforms.
Posttranslational modifications of proteins
Lipoprotein oxidation is implicated in atherogenesis (63). It is noteworthy that HDL isolated from humans with established CVD contains much higher levels of two oxidized amino acids, chlorotyrosine and nitrotyrosine, than does HDL of apparently healthy control subjects (70–72). Chlorotyrosine is a specific oxidation product of the heme protein myeloperoxidase (73) and macrophages in human atherosclerotic lesions express high levels of that enzyme (74). When specific tyrosine and methionine residues in apoA-I, the major protein in HDL, are oxidized by myeloperoxidase, it loses its ability to remove cholesterol from cells by the ABCA1 pathway (70, 75). This pathway may be important in generating dysfunctional HDL because the interaction of apoA-I with ABCA1 is critical in protecting macrophages from cholesterol accumulation in mouse models of hypercholesterolemia and accelerated atherosclerosis (25).
These observations suggest that the inflamed atherosclerotic lesion is one potential location where oxidative damage of proteins could be clinically important. In future studies, it will be of great interest to use targeted MS/MS to determine if site-specific modifications of apoA-I characteristic of myeloperoxidase can be detected in HDL isolated from humans with established CVD.
TOWARD QUANTITATIVE AND TRANSLATIONAL LIPOPROTEOMICS
In addition to identifying the proteins present in complicated mixtures of proteins, proteomics can be used to estimate the relative abundance of proteins (reviewed in Refs. 29, 31, 76–78). In the case of 2D gels, the intensity of protein spots detected with certain dyes is proportional to the protein concentration in the sample. The intensity of each spot can be normalized using spiked proteins, an endogenous protein whose concentration is constant from sample to sample, or with chemical tags (79). However, this approach relies on the assumption that all proteins in the mixture enter the gel, and that each protein spot consists of a single protein species (80). Neither of these assumptions is true for LDL and VLDL, which contain apoB as their major structural protein. In particular, apoB is problematic because of its large size and hydrophobicity, which causes the protein to precipitate out of solution during isoelectric focusing. It is also difficult to standardize 2D gel electrophoresis to yield reproducible results with large numbers of samples.
MALDI-TOF-MS can provide relative quantification of intact proteins, particularly if the protein mixture is not complex and there are large differences in relative abundance. This approach was used to demonstrate that apoC-III is highly enriched in the small dense LDL of subjects with type 2 diabetes or carotid atherosclerosis when compared with the small dense LDL of normal control subjects (42). In the same study, apoE was significantly depleted from the same fraction. Both proteins have important biological roles and the changes in concentration in this fraction could turn out to have important clinical implications.
A variety of label-free methods have been developed for quantifying relative protein abundance by MS, including spectral counting, summing all the MS/MS spectra observed for peptides derived from a single protein in an LC-ESI-MS/MS analysis (81, 82). Because proteins that are more abundant in a sample have a higher probability of being identified during data-dependent MS/MS scanning, the number of tandem spectra acquired for individual peptides reflects protein abundance. Pioneering studies demonstrated that spectral counting quantified exogenously added proteins in yeast cell lysates over a dynamic range of two orders of magnitude (81). Quantitative changes in protein expression in cultured cells and biological fluids, as assessed by immunoblotting, correlated strongly with spectral counting (83, 84). Collectively, these observations provide strong evidence that spectral counting is a sampling statistic that can assess relative protein abundance in biological material.
We used spectral counting to demonstrate that HDL3 isolated from the plasma of subjects with established CVD had significantly higher concentrations of apoE, complement C3, apoC-IV, PON-1, and apoA-IV than HDL3 from normal controls (15). To confirm that this approach can estimate protein abundance semi-quantitatively, we measured apoE levels immunochemically in HDL3 isolated from a different set of 64 subjects: 32 with established CVD and 32 age-matched controls. Levels of apoE were significantly higher in HDL3 isolated from the CVD subjects. In striking contrast, the two groups had similar levels of apoA-I and apoA-II. Importantly, relative protein abundance, as assessed by MS and biochemical assays, was similar, supporting the proposal that spectral counting can assess relative protein abundance in lipoproteins.
We recently used MS to test the hypothesis that aggressive lipid therapy with atorvastatin and niacin modifies the HDL proteome in humans with established CVD (48). To quantify changes in protein abundance, we employed two complementary methods, spectral counting and extracted ion chromatograms. Both methods are label-free (i.e., do not require introduction of a mass tag into the proteins), which makes them well-suited for analyzing HDL obtained in clinical studies. For extracted ion chromatograms, the ion current for a given peptide and charge state are extracted from the full scan mass spectrum and used to construct a chromatogram. The area under the curve then provides a quantitative measure of relative peptide abundance.
We used spectral counting as an initial screen to identify proteins that appeared to be differentially expressed and then used extracted ion chromatograms to quantify the relative abundance of these proteins in HDL3 isolated from CVD subjects before and during treatment with atorvastatin and niacin (48). This approach offers two important advantages. First, extracted ion chromatograms estimate protein ratios more accurately than spectral counting (81). Second, it is possible to compare the extracted ion chromatogram ratios of multiple peptides detected from the same protein (48), which should increase confidence in the results.
Spectral counting, together with extracted ion chromatograms, identified four HDL3 proteins whose relative abundance appeared to change as a result of treatment: apoE, apoJ, apoF, and PLTP (48). Levels of apoE fell, whereas levels of apoJ, apoF, and PLTP rose. We used two approaches to confirm that our mass spectrometric techniques accurately assessed changes in the relative abundance of proteins in HDL. First, when we analyzed HDL3 isolated from a different set of 13 subjects, we observed a strong linear correlation between apoE levels as assessed by extracted ion chromatograms and biochemically. Second, we used an immunochemical approach to confirm that combination therapy with niacin and statin reduced levels of apoE in HDL3 in an independent group of different subjects. Our observations indicate that when newly diagnosed CVD subjects are treated with combination therapy, the HDL3 proteome is remodeled to more closely resemble that of HDL3 of healthy control subjects (48).
CHALLENGES AND FUTURE DIRECTIONS
The elaborate cast of proteins in lipoprotein particles begs the question of how the proteins get there in the first place. For example, interactions between apoB and other lipoprotein-associated proteins during assembly of VLDL in the liver have not been detected (85, 86). Moreover, apoA-I is apparently secreted from the liver devoid of other proteins and lipid (87). Lipoprotein particles thus seem to acquire most of their nonstructural proteins while coursing the lymphatics, bloodstream, and tissues. Apolipoproteins are generally assumed to associate with lipoproteins via hydrophobic interactions, but studies of the trypanosome lytic factor strongly suggest that protein-protein interactions are also involved (61, 62). It will be of great interest to determine the molecular basis for assembling the protein cargo of lipoproteins.
Abundant blood proteins have been detected in isolated lipoproteins, raising concern about possible contamination (15, 33, 34, 36, 37). The best-studied of these putative contaminant proteins, albumin, plays a role in lipid metabolism via its fatty acid-binding sites (88). One group of investigators isolated albumin using seven different biochemical and immunochemical techniques (89). ApoA-I was readily detected in every preparation, suggesting that it normally interacts with albumin in blood. Other investigators found a relatively constant molar ratio of albumin to apoA-I in lipoproteins ranging from the density of HDL to that of VLDL (44). These observations raise the possibility that albumin interacts with apoA-I, and perhaps other lipoproteins, and that these interactions play previously unsuspected roles in lipoprotein function in vivo. In future studies, it will clearly be important to use complementary biochemical and functional studies to determine whether minor protein components of lipoproteins that are abundant in blood truly associate with the lipoproteins.
The availability of biomarkers that could reveal the presence of CVD in time to ward off a heart attack or stroke would have an enormous impact on health-care costs and public health, given that CVD is the leading cause of death in the industrialized world. However, the ability to identify subjects at increased risk for heart disease is limited. Our recent studies suggest that the HDL proteome may distinguish patients with CVD from healthy people (15). Moreover, we have shown that lipid lowering therapy remodels the HDL3 proteome in subjects with established CVD, yielding a protein composition that resembles that in apparently healthy subjects (48). These studies raise the possibility that quantifying the lipoproteome could provide insights into the efficacy of lipid therapy and help identify agents with cardioprotective actions.
Footnotes
Abbreviations:
- apo
- apolipoprotein
- CVD
- cardiovascular disease
- CETP
- cholesteryl ester transfer protein
- GO
- Gene Ontology
- LC-ESI-MS/MS
- liquid chromatography-electrospray ionization-tandem mass spectrometry
- MALDI-TOF-MS
- matrix assisted laser desorption ionization-time of flight-MS
- PLTP
- phospholipid transfer protein
- PON
- paraoxonase
- 2D
- two-dimensional
REFERENCES
- 1.Brown M. S., Goldstein J. L. 1986. A receptor-mediated pathway for cholesterol homeostasis. Science. 232: 34–47. [DOI] [PubMed] [Google Scholar]
- 2.Liu H., Ryan R. O. 1991. Role of lipid transfer particle in transformation of lipophorin in insect oocytes. Biochim. Biophys. Acta. 1085: 112–118. [DOI] [PubMed] [Google Scholar]
- 3.Rader D. J. 2006. Molecular regulation of HDL metabolism and function: implications for novel therapies. J. Clin. Invest. 116: 3090–3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Khovidhunkit W., Kim M. S., Memon R. A., Shigenaga J. K., Moser A. H., Feingold K. R., Grunfeld C. 2004. Effects of infection and inflammation on lipid and lipoprotein metabolism: mechanisms and consequences to the host. J. Lipid Res. 45: 1169–1196. [DOI] [PubMed] [Google Scholar]
- 5.Chait A., Han C. Y., Oram J. F., Heinecke J. W. 2005. Thematic review series: the immune system and atherogenesis. Lipoprotein-associated inflammatory proteins: markers or mediators of cardiovascular disease? J. Lipid Res. 46: 389–403. [DOI] [PubMed] [Google Scholar]
- 6.Getz G. S. 2005. Thematic review series: the immune system and atherogenesis. Bridging the innate and adaptive immune systems. J. Lipid Res. 46: 619–622. [DOI] [PubMed] [Google Scholar]
- 7.Anderson K. M., Odell P. M., Wilson P. W., Kannel W. B. 1991. Cardiovascular disease risk profiles. Am. Heart J. 121: 293–298. [DOI] [PubMed] [Google Scholar]
- 8.Gordon D. J., Rifkind B. M. 1989. High-density lipoprotein–the clinical implications of recent studies. N. Engl. J. Med. 321: 1311–1316. [DOI] [PubMed] [Google Scholar]
- 9.Barzilai N., Atzmon G., Schechter C., Schaefer E. J., Cupples A. L., Lipton R., Cheng S., Shuldiner A. R. 2003. Unique lipoprotein phenotype and genotype associated with exceptional longevity. JAMA. 290: 2030–2040. [DOI] [PubMed] [Google Scholar]
- 10.Walldius G., Jungner I., Holme I., Aastveit A. H., Kolar W., Steiner E. 2001. High apolipoprotein B, low apolipoprotein A-I, and improvement in the prediction of fatal myocardial infarction (AMORIS study): a prospective study. Lancet. 358: 2026–2033. [DOI] [PubMed] [Google Scholar]
- 11.Aebersold R., Mann M. 2003. Mass spectrometry-based proteomics. Nature. 422: 198–207. [DOI] [PubMed] [Google Scholar]
- 12.Cravatt B. F., Simon G. M., Yates J. R., 3rd. 2007. The biological impact of mass-spectrometry-based proteomics. Nature. 450: 991–1000. [DOI] [PubMed] [Google Scholar]
- 13.Cantin G. T., Yates J. R., 3rd. 2004. Strategies for shotgun identification of post-translational modifications by mass spectrometry. J. Chromatogr. A. 1053: 7–14. [DOI] [PubMed] [Google Scholar]
- 14.Heinecke J. W. 2009. A “new” thematic series: mass spectrometry-based proteomics of lipid biology. J. Lipid Res. 50: 777–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vaisar T., Pennathur S., Green P. S., Gharib S. A., Hoofnagle A. N., Cheung M. C., Byun J., Vuletic S., Kassim S., Singh P., et al. 2007. Shotgun proteomics implicates protease inhibition and complement activation in the antiinflammatory properties of HDL. J. Clin. Invest. 117: 746–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rader D. J., Hobbs H. H. 2008. Disorders of Lipoprotein Metabolism. Harrison's Principles of Internal Medicine. Fauci A. S., Kasper D. L., Longo D. L., Braunwald E., Hauser S. L., Jameson J. L., Loscalzo J., McGraw-Hill, New York: 2416–2429. [Google Scholar]
- 17.Schonfeld G. 2003. Familial hypobetalipoproteinemia: a review. J. Lipid Res. 44: 878–883. [DOI] [PubMed] [Google Scholar]
- 18.Fojo S. S., Brewer H. B. 1992. Hypertriglyceridemia due to genetic-defects in lipoprotein-lipase and apolipoprotein-C-II. J. Intern. Med. 231: 669–677. [DOI] [PubMed] [Google Scholar]
- 19.Brown M. S., Goldstein J. L. 1985. The receptor model for transport of cholesterol in plasma. Ann. N. Y. Acad. Sci. 454: 178–182. [DOI] [PubMed] [Google Scholar]
- 20.Gaut J. P., Heinecke J. W. 2001. Mechanisms for oxidizing low-density lipoprotein. Insights from patterns of oxidation products in the artery wall and from mouse models of atherosclerosis. Trends Cardiovasc. Med. 11: 103–112. [DOI] [PubMed] [Google Scholar]
- 21.Davidson W. S., Thompson T. B. 2007. The structure of apolipoprotein A-I in high density lipoproteins. J. Biol. Chem. 282: 22249–22253. [DOI] [PubMed] [Google Scholar]
- 22.Oram J. F., Vaughan A. M. 2006. ATP-binding cassette cholesterol transporters and cardiovascular disease. Circ. Res. 99: 1031–1043. [DOI] [PubMed] [Google Scholar]
- 23.Tall A. R., Yvan-Charvet L., Terasaka N., Pagler T., Wang N. 2008. HDL, ABC transporters, and cholesterol efflux: implications for the treatment of atherosclerosis. Cell Metab. 7: 365–375. [DOI] [PubMed] [Google Scholar]
- 24.Bruce C., Chouinard R. A., Jr., Tall A. R. 1998. Plasma lipid transfer proteins, high-density lipoproteins, and reverse cholesterol transport. Annu. Rev. Nutr. 18: 297–330. [DOI] [PubMed] [Google Scholar]
- 25.Oram J. F., Heinecke J. W. 2005. ATP-binding cassette transporter A1: a cell cholesterol exporter that protects against cardiovascular disease. Physiol. Rev. 85: 1343–1372. [DOI] [PubMed] [Google Scholar]
- 26.Segrest J. P., Jones M. K., De Loof H., Dashti N. 2001. Structure of apolipoprotein B-100 in low density lipoproteins. J. Lipid Res. 42: 1346–1367. [PubMed] [Google Scholar]
- 27.Barter P. J., Nicholls S., Rye K. A., Anantharamaiah G. M., Navab M., Fogelman A. M. 2004. Antiinflammatory properties of HDL. Circ. Res. 95: 764–772. [DOI] [PubMed] [Google Scholar]
- 28.Kontush A., Chapman M. J. 2006. Functionally defective high-density lipoprotein: a new therapeutic target at the crossroads of dyslipidemia, inflammation, and atherosclerosis. Pharmacol. Rev. 58: 342–374. [DOI] [PubMed] [Google Scholar]
- 29.Gorg A., Weiss W., Dunn M. J. 2004. Current two-dimensional electrophoresis technology for proteomics. Proteomics. 4: 3665–3685. [DOI] [PubMed] [Google Scholar]
- 30.Johnson R. S., Davis M. T., Taylor J. A., Patterson S. D. 2005. Informatics for protein identification by mass spectrometry. Methods. 35: 223–236. [DOI] [PubMed] [Google Scholar]
- 31.Kiehntopf M., Siegmund R., Deufel T. 2007. Use of SELDI-TOF mass spectrometry for identification of new biomarkers: potential and limitations. Clin. Chem. Lab. Med. 45: 1435–1449. [DOI] [PubMed] [Google Scholar]
- 32.O'Farrell P. H. 1975. High resolution two-dimensional electrophoresis of proteins. J. Biol. Chem. 250: 4007–4021. [PMC free article] [PubMed] [Google Scholar]
- 33.Karlsson H., Leanderson P., Tagesson C., Lindahl M. 2005. Lipoproteomics II: mapping of proteins in high-density lipoprotein using two-dimensional gel electrophoresis and mass spectrometry. Proteomics. 5: 1431–1445. [DOI] [PubMed] [Google Scholar]
- 34.Karlsson H., Leanderson P., Tagesson C., Lindahl M. 2005. Lipoproteomics I: mapping of proteins in low-density lipoprotein using two-dimensional gel electrophoresis and mass spectrometry. Proteomics. 5: 551–565. [DOI] [PubMed] [Google Scholar]
- 35.Karlsson H., Lindqvist H., Tagesson C., Lindahl M. 2006. Characterization of apolipoprotein M isoforms in low-density lipoprotein. J. Proteome Res. 5: 2685–2690. [DOI] [PubMed] [Google Scholar]
- 36.Kunitake S. T., Carilli C. T., Lau K., Protter A. A., Naya-Vigne J., Kane J. P. 1994. Identification of proteins associated with apolipoprotein A-I-containing lipoproteins purified by selected-affinity immunosorption. Biochemistry. 33: 1988–1993. [DOI] [PubMed] [Google Scholar]
- 37.Mancone C., Amicone L., Fimia G. M., Bravo E., Piacentini M., Tripodi M., Alonzi T. 2007. Proteomic analysis of human very low-density lipoprotein by two-dimensional gel electrophoresis and MALDI-TOF/TOF. Proteomics. 7: 143–154. [DOI] [PubMed] [Google Scholar]
- 38.Rezaee F., Casetta B., Levels J. H. M., Speijer D., Meijers J. C. M. 2006. Proteomic analysis of high-density lipoprotein. Proteomics. 6: 721–730. [DOI] [PubMed] [Google Scholar]
- 39.Stahlman M., Davidsson P., Kanmert I., Rosengren B., Boren J., Fagerberg B., Camejo G. 2008. Proteomics and lipids of lipoproteins isolated at low salt concentrations in D2O/sucrose or in KBr. J. Lipid Res. 49: 481–490. [DOI] [PubMed] [Google Scholar]
- 40.Bondarenko P. V., Cockrill S. L., Watkins L. K., Cruzado I. D., Macfarlane R. D. 1999. Mass spectral study of polymorphism of the apolipoproteins of very low density lipoprotein. J. Lipid Res. 40: 543–555. [PubMed] [Google Scholar]
- 41.Bondarenko R., Farwig Z. N., McNeal C. J., Macfarlane R. D. 2002. MALDI- and ESI-MS of the HDL apolipoproteins; new isoforms of apoA-I, II. Int. J. Mass Spectrom. 219: 671–680. [Google Scholar]
- 42.Davidsson P., Hulthe J., Fagerberg B., Olsson B. M., Hallberg C., Dahllof B., Camejo G. 2005. A proteomic study of the apolipoproteins in LDL subclasses in patients with the metabolic syndrome and type 2 diabetes. J. Lipid Res. 46: 1999–2006. [DOI] [PubMed] [Google Scholar]
- 43.Farwig Z. N., Campbell A. V., Macfarlane R. D. 2003. Analysis of high-density lipoprotein apolipoproteins recovered from specific immobilized pH gradient gel pI domains by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Anal. Chem. 75: 3823–3830. [DOI] [PubMed] [Google Scholar]
- 44.Heller M., Stalder D., Schlappritzi E., Hayn G., Matter U., Haeberli A. 2005. Mass spectrometry-based analytical tools for the molecular protein characterization of human plasma lipoproteins. Proteomics. 5: 2619–2630. [DOI] [PubMed] [Google Scholar]
- 45.Levels J. H. M., Bleijlevens B., Rezaee F., Aerts J., Meijers J. C. M. 2007. SELDI-TOF mass spectrometry of high-density lipoprotein. Proteome Sci. 5: 15–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Watanabe J., Chou K. J., Liao J. C., Miao Y., Meng H. H., Ge H., Grijalva V., Hama S., Kozak K., Buga G., et al. 2007. Differential association of hemoglobin with proinflammatory high density lipoproteins in atherogenic/hyperlipidemic mice. A novel biomarker of atherosclerosis. J. Biol. Chem. 282: 23698–23707. [DOI] [PubMed] [Google Scholar]
- 47.Washburn M. P., Wolters D., Yates J. R., 3rd. 2001. Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat. Biotechnol. 19: 242–247. [DOI] [PubMed] [Google Scholar]
- 48.Green P. S., Vaisar T., Pennathur S., Kulstad J. J., Moore A. B., Marcovina S., Brunzell J., Knopp R. H., Zhao X. Q., Heinecke J. W. 2008. Combined statin and niacin therapy remodels the high-density lipoprotein proteome. Circulation. 118: 1259–1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Heller M., Schlappritzi E., Stalder D., Nuoffer J. M., Haeberli A. 2007. Compositional protein analysis of high density lipoproteins in hypercholesterolemia by shotgun LC-MS/MS and probabilistic peptide scoring. Mol. Cell. Proteomics. 6: 1059–1072. [DOI] [PubMed] [Google Scholar]
- 50.MacCoss M. J., Yates J. R., 3rd. 2001. Proteomics: analytical tools and techniques. Curr. Opin. Clin. Nutr. Metab. Care. 4: 369–375. [DOI] [PubMed] [Google Scholar]
- 51.Vaisar T. 2009. Proteomic analysis of lipid-protein complexes. J. Lipid Res. 50: 781–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Scanu A. M., Edelstein C. 2008. HDL: bridging past and present with a look at the future. FASEB J. 22: 4044–4054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Curry M. D., Alaupovic P., Suenram C. 1976. Determination of apolipoprotein A and its constitutive A-I and A-II polypeptides by separate electroimmunoassays. Clin. Chem. 22: 315–322. [PubMed] [Google Scholar]
- 54.Kunitake S. T., Kane J. P. 1982. Factors affecting the integrity of high density lipoproteins in the ultracentrifuge. J. Lipid Res. 23: 936–940. [PubMed] [Google Scholar]
- 55.Peterson M. M., Mack J. L., Hall P. R., Alsup A. A., Alexander S. M., Sully E. K., Sawires Y. S., Cheung A. L., Otto M., Gresham H. D. 2008. Apolipoprotein B is an innate barrier against invasive Staphylococcus aureus infection. Cell Host Microbe. 4: 555–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Terasaka N., Yu S., Yvan-Charvet L., Wang N., Mzhavia N., Langlois R., Pagler T., Li R., Welch C. L., Goldberg I. J., et al. 2008. ABCG1 and HDL protect against endothelial dysfunction in mice fed a high-cholesterol diet. J. Clin. Invest. 118: 3701–3713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Blackburn W. D., Jr., Dohlman J. G., Venkatachalapathi Y. V., Pillion D. J., Koopman W. J., Segrest J. P., Anantharamaiah G. M. 1991. Apolipoprotein A-I decreases neutrophil degranulation and superoxide production. J. Lipid Res. 32: 1911–1918. [PubMed] [Google Scholar]
- 58.Calero M., Tokuda T., Rostagno A., Kumar A., Zlokovic B., Frangione B., Ghiso J. 1999. Functional and structural properties of lipid-associated apolipoprotein J (clusterin). Biochem. J. 344: 375–383. [PMC free article] [PubMed] [Google Scholar]
- 59.Rosenfeld S. I., Packman C. H., Leddy J. P. 1983. Inhibition of the lytic action of cell-bound terminal complement components by human high density lipoproteins and apoproteins. J. Clin. Invest. 71: 795–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Garner B., Waldeck A. R., Witting P. K., Rye K. A., Stocker R. 1998. Oxidation of high density lipoproteins. II. Evidence for direct reduction of lipid hydroperoxides by methionine residues of apolipoproteins AI and AII. J. Biol. Chem. 273: 6088–6095. [DOI] [PubMed] [Google Scholar]
- 61.Shiflett A. M., Bishop J. R., Pahwa A., Hajduk S. L. 2005. Human high density lipoproteins are platforms for the assembly of multi-component innate immune complexes. J. Biol. Chem. 280: 32578–32585. [DOI] [PubMed] [Google Scholar]
- 62.Vanhamme L., Paturiaux-Hanocq F., Poelvoorde P., Nolan D. P., Lins L., Van Den Abbeele J., Pays A., Tebabi P., Van Xong H., Jacquet A., et al. 2003. Apolipoprotein L–I is the trypanosome lytic factor of human serum. Nature. 422: 83–87. [DOI] [PubMed] [Google Scholar]
- 63.Heinecke J. W. 1997. Mechanisms of oxidative damage of low density lipoprotein in human atherosclerosis. Curr. Opin. Lipidol. 8: 268–274. [DOI] [PubMed] [Google Scholar]
- 64.Davidson W. S., Silva R. A., Chantepie S., Lagor W. R., Chapman M. J., Kontush A. 2009. Proteomic analysis of defined HDL subpopulations reveals particle-specific protein clusters: relevance to antioxidative function. Arterioscler. Thromb. Vasc. Biol. 29: 870–876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dennis G., Jr., Sherman B. T., Hosack D. A., Yang J., Gao W., Lane H. C., Lempicki R. A. 2003. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 4: P3. [PubMed] [Google Scholar]
- 66.Huang da W., Sherman B. T., Lempicki R. A. 2009. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protocols. 4: 44–57. [DOI] [PubMed] [Google Scholar]
- 67.Navab M., Anantharamaiah G. M., Reddy S. T., Van Lenten B. J., Wagner A. C., Hama S., Hough G., Bachini E., Garber D. W., Mishra V. K., et al. 2005. An oral apoJ peptide renders HDL antiinflammatory in mice and monkeys and dramatically reduces atherosclerosis in apolipoprotein E-null mice. Arterioscler. Thromb. Vasc. Biol. 25: 1932–1937. [DOI] [PubMed] [Google Scholar]
- 68.Navab M., Ruchala P., Waring A. J., Lehrer R. I., Hama S., Hough G., Palgunachari M. N., Anantharamaiah G. M., Fogelman A. M. 2009. A novel method for oral delivery of apolipoprotein mimetic peptides synthesized from all L-amino acids. J. Lipid Res. 50: 1538–1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hortin G. L., Shen R. F., Martin B. M., Remaley A. T. 2006. Diverse range of small peptides associated with high-density lipoprotein. Biochem. Biophys. Res. Commun. 340: 909–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bergt C., Pennathur S., Fu X., Byun J., O’Brien K., McDonald T. O., Singh P., Anantharamaiah G. M., Chait A., Brunzell J., et al. 2004. The myeloperoxidase product hypochlorous acid oxidizes HDL in the human artery wall and impairs ABCA1-dependent cholesterol transport. Proc. Natl. Acad. Sci. USA. 101: 13032–13037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pennathur S., Bergt C., Shao B., Byun J., Kassim S. Y., Singh P., Green P. S., McDonald T. O., Brunzell J., Chait A., et al. 2004. Human atherosclerotic intima and blood of patients with established coronary artery disease contain high density lipoprotein damaged by reactive nitrogen species. J. Biol. Chem. 279: 42977–42983. [DOI] [PubMed] [Google Scholar]
- 72.Zheng L., Nukuna B., Brennan M. L., Sun M., Goormastic M., Settle M., Schmitt D., Fu X., Thomson L., Fox P. L., et al. 2004. Apolipoprotein A-I is a selective target for myeloperoxidase-catalyzed oxidation and functional impairment in subjects with cardiovascular disease. J. Clin. Invest. 114: 529–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gaut J. P., Yeh G. C., Tran H. D., Byun J., Henderson J. P., Richter G. M., Brennan M. L., Lusis A. J., Belaaouaj A., Hotchkiss R. S., et al. 2001. Neutrophils employ the myeloperoxidase system to generate antimicrobial brominating and chlorinating oxidants during sepsis. Proc. Natl. Acad. Sci. USA. 98: 11961–11966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Daugherty A., Dunn J. L., Rateri D. L., Heinecke J. W. 1994. Myeloperoxidase, a catalyst for lipoprotein oxidation, is expressed in human atherosclerotic lesions. J. Clin. Invest. 94: 437–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Shao B., Oda M. N., Bergt C., Fu X., Green P. S., Brot N., Oram J. F., Heinecke J. W. 2006. Myeloperoxidase impairs ABCA1-dependent cholesterol efflux through methionine oxidation and site-specific tyrosine chlorination of apolipoprotein A-I. J. Biol. Chem. 281: 9001–9004. [DOI] [PubMed] [Google Scholar]
- 76.Bantscheff M., Schirle M., Sweetman G., Rick J., Kuster B. 2007. Quantitative mass spectrometry in proteomics: a critical review. Anal. Bioanal. Chem. 389: 1017–1031. [DOI] [PubMed] [Google Scholar]
- 77.Mueller L. N., Brusniak M. Y., Mani D. R., Aebersold R. 2008. An assessment of software solutions for the analysis of mass spectrometry based quantitative proteomics data. J. Proteome Res. 7: 51–61. [DOI] [PubMed] [Google Scholar]
- 78.Wang M., You J., Bemis K. G., Tegeler T. J., Brown D. P. 2008. Label-free mass spectrometry-based protein quantification technologies in proteomic analysis. Brief. Funct. Genomic Proteomic. 7: 329–339. [DOI] [PubMed] [Google Scholar]
- 79.Wheelock A. M., Morin D., Bartosiewicz M., Buckpitt A. R. 2006. Use of a fluorescent internal protein standard to achieve quantitative two-dimensional gel electrophoresis. Proteomics. 6: 1385–1398. [DOI] [PubMed] [Google Scholar]
- 80.Gygi S. P., Aebersold R. 1999. Absolute quantitation of 2-D protein spots. Methods Mol. Biol. 112: 417–421. [DOI] [PubMed] [Google Scholar]
- 81.Liu H., Sadygov R. G., Yates J. R., 3rd. 2004. A model for random sampling and estimation of relative protein abundance in shotgun proteomics. Anal. Chem. 76: 4193–4201. [DOI] [PubMed] [Google Scholar]
- 82.Old W. M., Meyer-Arendt K., Aveline-Wolf L., Pierce K. G., Mendoza A., Sevinsky J. R., Resing K. A., Ahn N. G. 2005. Comparison of label-free methods for quantifying human proteins by shotgun proteomics. Mol. Cell. Proteomics. 4: 1487–1502. [DOI] [PubMed] [Google Scholar]
- 83.Fu X., Gharib S. A., Green P. S., Aitken M. L., Frazer D. A., Park D. R., Vaisar T., Heinecke J. W. 2008. Spectral index for assessment of differential protein expression in shotgun proteomics. J. Proteome Res. 7: 845–854. [DOI] [PubMed] [Google Scholar]
- 84.Lu P., Vogel C., Wang R., Yao X., Marcotte E. M. 2007. Absolute protein expression profiling estimates the relative contributions of transcriptional and translational regulation. Nat. Biotechnol. 25: 117–124. [DOI] [PubMed] [Google Scholar]
- 85.Rashid K. A., Hevi S., Chen Y., Le Caherec F., Chuck S. L. 2002. A proteomic approach identifies proteins in hepatocytes that bind nascent apolipoprotein B. J. Biol. Chem. 277: 22010–22017. [DOI] [PubMed] [Google Scholar]
- 86.Wang H., Gilham D., Lehner R. 2007. Proteomic and lipid characterization of apolipoprotein B-free luminal lipid droplets from mouse liver microsomes: implications for very low density lipoprotein assembly. J. Biol. Chem. 282: 33218–33226. [DOI] [PubMed] [Google Scholar]
- 87.Chau P., Nakamura Y., Fielding C. J., Fielding P. E. 2006. Mechanism of prebeta-HDL formation and activation. Biochemistry. 45: 3981–3987. [DOI] [PubMed] [Google Scholar]
- 88.Spector A. A., John K., Fletcher J. E. 1969. Binding of long-chain fatty acids to bovine serum albumin. J. Lipid Res. 10: 56–67. [PubMed] [Google Scholar]
- 89.Gundry R. L., Fu Q., Jelinek C. A., Van Eyk J. E., Cotter R. J. 2007. Investigation of an albumin-enriched fraction of human serum and its albuminome. Proteomics Clin. Appl. 1: 73–88. [DOI] [PMC free article] [PubMed] [Google Scholar]