Abstract
The voltage dependent anion channel (VDAC) is an essential protein in the eukaryotic outer mitochondrial membrane, providing the pore for substrate diffusion. Three high-resolution structures of the isoform 1 of VDAC in detergent micelles and bicelles have recently been published, using solution NMR and X-ray crystallography. They resolve longstanding discussions about the membrane topology of VDAC and provide the first eukaryotic β-barrel membrane protein structure. The structure contains a surprising feature that had not been observed in an integral membrane protein before: A parallel β-strand pairing and thus an odd number of strands. The studies also give a structural and functional basis for the voltage gating mechanism of VDAC and its modulation by NADH, however they do not fully explain these functions yet. With the de novo structure of VDAC-1, as well as those of half a dozen other proteins, the number of integral membrane protein structures solved by solution NMR has doubled in the past two years. Numerous further structural and functional studies on many different membrane proteins show that solution NMR has become an important tool for membrane protein molecular biology.
Introduction
Mitochondria are complex organelles that fulfill multiple functions in eukaryotic cells. Their key task is the production of cellular energy, but they are also involved in other metabolic pathways, apoptosis, cellular differentiation and in the control of the cell cycle [1]. As a result from their probable bacterial ancestry, mitochondria are enveloped by a double membrane [2]. The energy production occurs near and within the inner mitochondrial membrane, and the outer membrane is thus a high-traffic zone of the eukaryotic cell [3]. Low-energy metabolites, mainly ADP, have to go into the mitochondria and high-energy metabolites, mainly ATP, have to migrate out again. The protein that provides membrane passage to these molecules, as well as to ions, small proteins, cofactors and other small compounds is the voltage-dependent anion channel (VDAC), also called the mitochondrial porin [4,5,6,7,8].
VDAC had been discovered in 1975 and immediately drew attention, since it stood out from other observed channels due to its voltage-gating properties [9]. Due to its central function and its interesting properties it has been extensively studied for decades. Numerous structural models were generated, but essentially the β-strand topology remained unclear [6,10,11,12]. Last year, 33 years after the discovery of VDAC, three long-term efforts to determine the three-dimensional structure of its isoform 1 at atomic resolution were finalized within a few weeks time [""13, ""14, ""15]. Remarkably, the technical approaches used for these three structure determinations were different from each other. One structure was solved by NMR alone [""13], one by X-ray crystallography alone [""15] and one using a hybrid method of both techniques [""14].
Here, we want to compare the three VDAC-1 structures and describe structural and functional aspects of VDAC-1 that can now be concluded on the basis of its high-resolution three-dimensional structure and associated data. We discuss the role of solution NMR in the structure determination of VDAC-1, as well as in other recent NMR membrane protein structure determinations and assess the use of protein refolding and detergent deuteration in these studies.
The β-barrel of VDAC-1
The basic structural feature of VDAC-1 is its large β-barrel. The topology of the barrel, i.e. the number of strands and their orientation relative to each other, is precisely the same in all three structures (Figure 1; compare also Figures 1 in [""13, ""14, ""15]). The amino acid sequence of VDAC is highly conserved from yeast to human and it is thus generally assumed that the overall fold of VDAC and its isoforms is the same in all eukaryotes [8]. Among the three determined structures, one is the mouse VDAC-1 [""15], and the other two are human VDAC-1 [""13, ""14]. These two forms are different by just four amino acid replacements, i.e. mouse VDAC-1 = human VDAC-1(T55N, M129V, A160S, I227V). It is now clear that these four changes are conservative or in loops and do not cause significant structural differences. Further, in each of the three structural studies, VDAC-1 was initially refolded from a denatured state into the detergent LDAO. For the crystallization in one study, VDAC-1 was then transferred into DMPC/CHAPSO bicelles [""15]. The VDAC-1 barrel structure is the same in both environments and thus not affected by the different biophysical properties of these two membrane mimics. VDAC-1 is currently the only known eukaryotic β-barrel membrane protein structure, and it is also the only β-barrel membrane protein featuring an odd number of strands. All other β-barrel membrane proteins exhibit exclusively antiparallel β-strand pairings and thus an even number of β-strands (Figure 2a). Intrinsically, antiparallel β-sheets are energetically slightly more favorable than parallel β-sheets by about 0.2 kcal/mol per residue pair (0.1 kcal/mol per formed hydrogen bond) [16] and this may be one reason for this bias, but likely, properties of the insufficiently understood folding mechanisms also play a role [17,18].
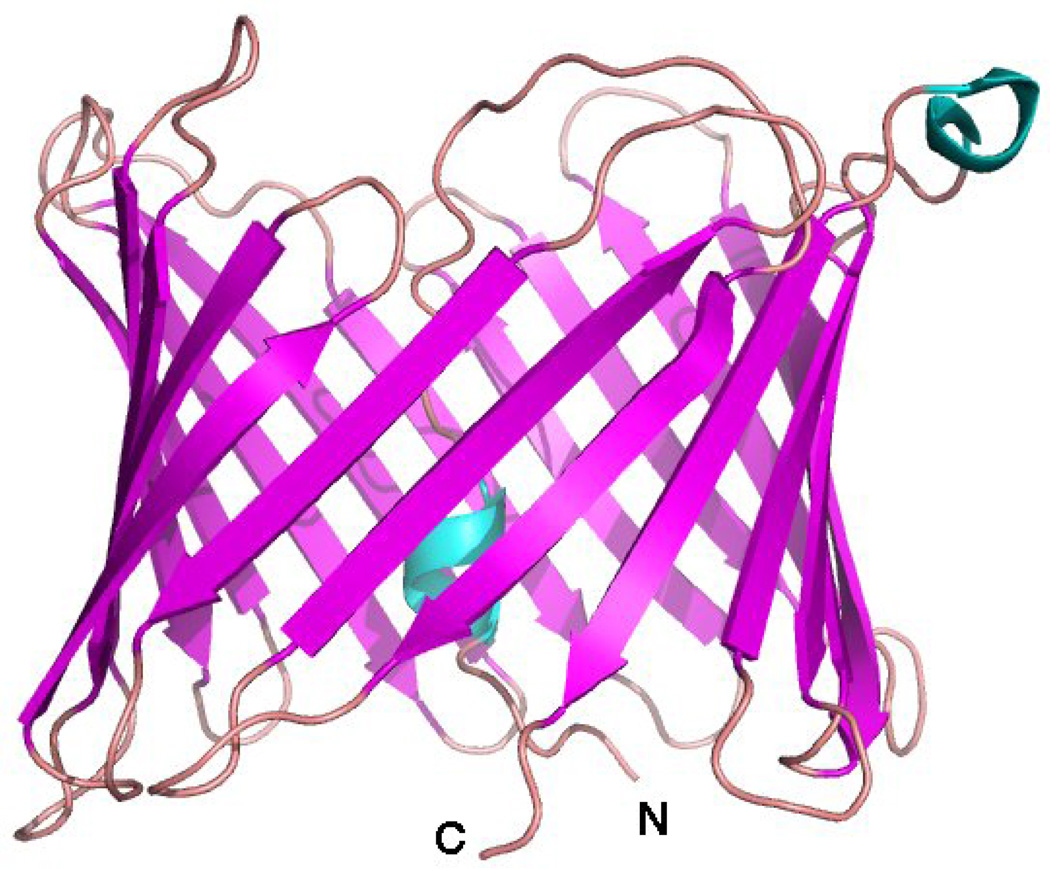
Figure 1.
NMR solution structure of VDAC-1 in LDAO micelles (PDB: 2K4T) [""13]. β-sheets are colored magenta, loops salmon, and helical secondary structure blue. N- and C-terminus of the 283-residue polypeptide are marked.
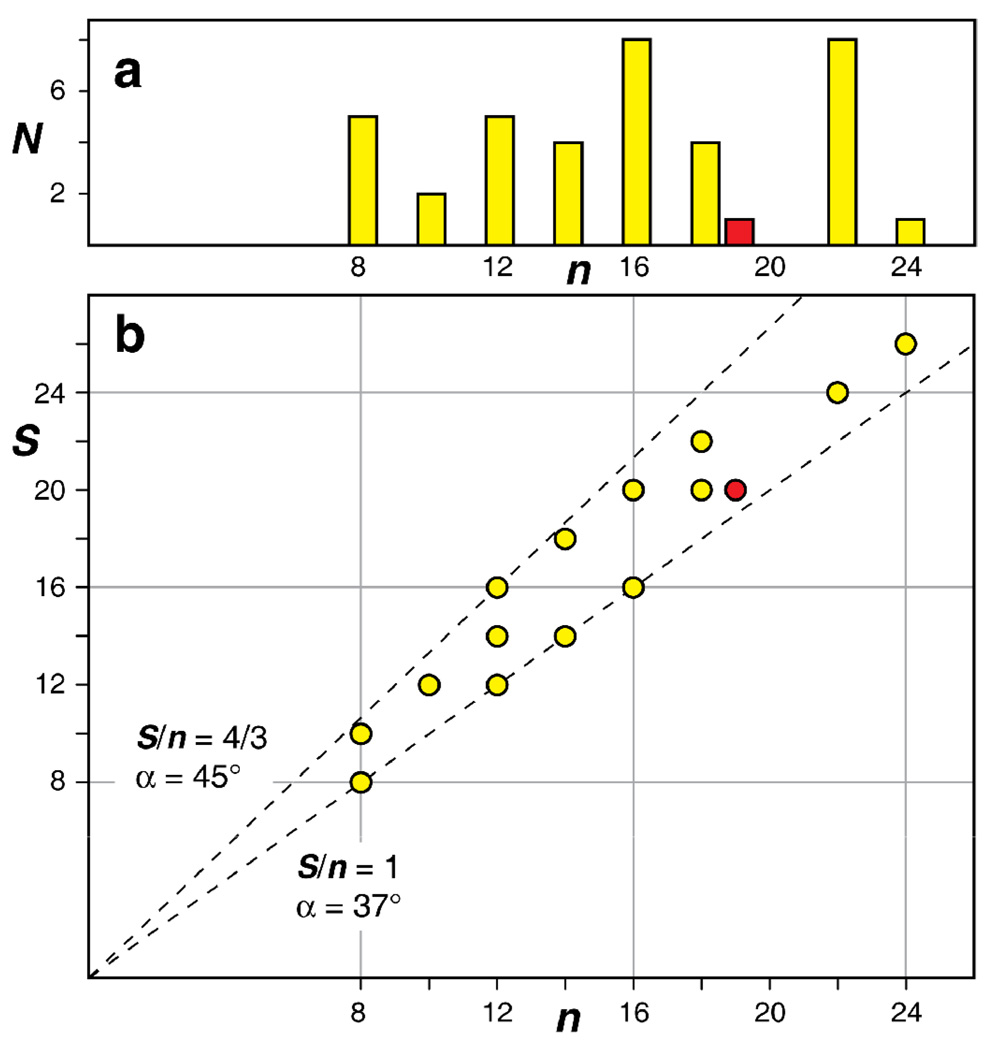
Figure 2.
Statistical analysis of all known β-barrel membrane protein structures [25]. (a) Histogram of the number of known unique β-barrel membrane proteins, N, vs. the number of β-strands, n. Yellow are prokaryotic proteins, red is the eukaryotic VDAC-1. (b) Combinations of shear numbers, S, and strand numbers, n that have been observed at least once are marked by circles in the same color coding as in panel a. The dashed lines mark the S/n ratios of 1 and 4/3. The corresponding angles between the membrane normal and the β-strands are indicated [19]. β-barrels resulting from a multimerization were not included in the analysis.
Just two numbers are able to define the structure of a β-barrel to a large extent [19]: The number of strands, n, and the shear number, S. In its simplest form for regular β-barrels without bulges, the shear number can be defined as follows [20]. In a cylindrical barrel the pairs of α-carbon atoms in adjacent strands, which have their side chains pointed to the same side of the sheet (outside or inside), lie on a helical trace across the surface of the barrel. Following this trace once around the cylinder one arrives back at the first strand a certain number of residues away from the starting point. This number is the shear number. To preserve the orientation of the hydrophobic outside residues, the shear number in membrane protein β-barrels is always even. VDAC-1, with 19 strands, has a shear number of 20. All other β-barrel membrane proteins have a shear number in the range n and n+4, (Figure 2b). The ratio of shear number and strand number S/n is linked via the equation tan α = ¾ S/n to the tilt angle α of the β-strands with respect to the membrane normal, where the factor ¾ results from the β-sheet geometry [19]. Interestingly, all currently known β-barrel membrane protein structures (except those resulting from a multimerization) lie within S/n = 1 and S/n = 4/3 and the tilt angle α thus in the narrow range between 37° and 45° (Figure 2b).
The flexible N-terminus of VDAC-1
A second aspect for the comparison of the three VDAC-1 structures concerns the residues 1–23, the only residues that are not part of the β-barrel or the loops and turns. Out of these residues, only the α-helical segment of residues 6–10 could be securely assigned by NMR so far. This segment has a close contact to the barrel wall. Structural constraints establishing this contact are based on many NOESY cross peaks between the methyl groups of Leucine 10, Valine 143 and Leucine 150, as well as between these moieties and several amide groups of the helix and the barrel wall [""13]. Intriguingly, Val 143 and Leu 150 are also the only hydrophobic side chains in the barrel wall pointing to the barrel interior. The close proximity of these three aliphatic residues was subsequently confirmed in the crystal structure of mouse VDAC-1 (Figure 3). Residues 11–20 appear well structured in the two published crystal structures but in different conformations [""14, ""15]. It is thus possible that the polypeptide changes its conformation between these two and potentially further structures. The crystal structures were obtained at cryogenic temperatures and even if the transitions occur also in the crystal, single conformations may be frozen out or fitted to the electron density. These residues were so far not assigned by NMR and the conformational exchange could be a reason for this. Depending on the exchange rate, conformational exchange can lead to multiple resonance lines or to line broadening and reduced signal intensity, hindering the resonance assignment [21]. A dynamic behavior of the segment 11–20 would additionally be supported by the observation that it has the highly flexible pivot sequence GYGFG on one end (residues 21–25) and a break in the helix (residue Gly11) at the other end in one of the crystal structures [""15].
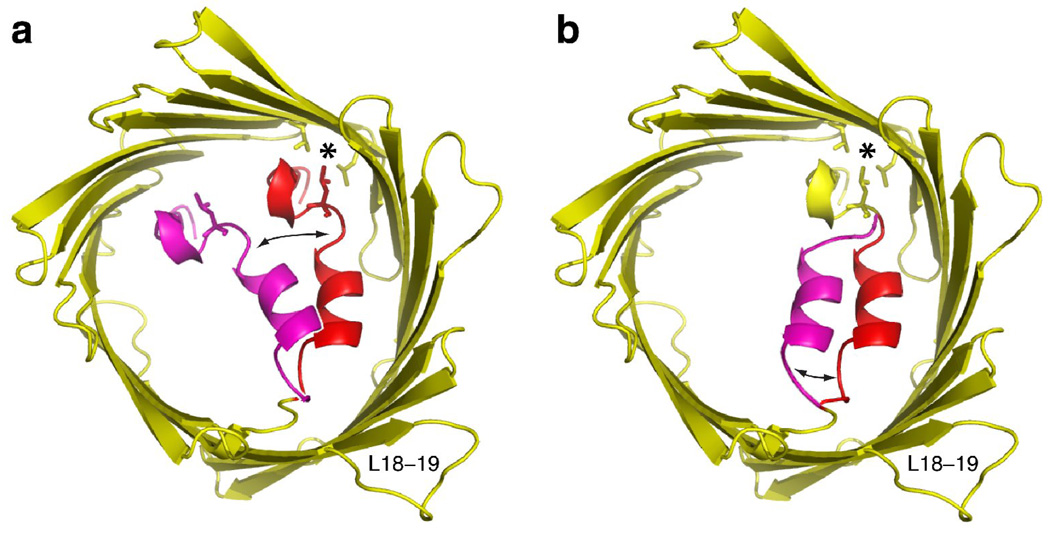
Figure 3.
Gating models for VDAC-1. Shown in yellow and red is the 2.3 Å crystal structure of VDAC-1 (PDB: 3EMN) [""15]. The side chains of residues Leu 10, Val 143 and Leu 150 are shown in stick representation and their hydrophobic contact point is marked with an asterisk. In each model, a certain amino acid segment, which is marked red, undergoes a conformational exchange to a fictive, magenta conformation, indicated by black arrows. (a) Model proposed by Ujwal et al. [""15]. Residues 1–20 undergo conformational exchange, disrupting the hydrophobic contact. (b) Alternative gating model proposed here. Only residues 11–20 undergo conformational exchange, preserving the hydrophobic contact.
Models for VDAC gating
In lipid bilayers, recombinant as well as native VDAC exhibits voltage gating [""13,22] and it is been well established that the N-terminal part of VDAC has a central function in this process [22,23]. VDAC adopts an “open” state at 0 mV and a “closed” state at higher voltages. In the open state, VDAC has a conductivity of about 4 nS and in the closed state, 40–60% thereof. The conductivity in the closed state is however mainly due to small cations, since VDAC is impermeable for ATP and other anionic molecules in the closed state [8]. With an energy difference of 10 kJ/mole between open and closed state, the closed state is populated also in the absence of voltage to about 2.3% and the closed state might thus also be present in the micelle-bound form at room temperature.
In context with the 2.3 Å crystal structure, it has been suggested that upon voltage gating, the whole N-terminal part, residues 1–20, moves to decrease the effective pore diameter [""15] (Figure 3a). Based on the above considerations in the mobility of the N-terminal parts, an alternative model is suggested here (Figure 3b). Only residues 11–20 might participate in the gating and the hydrophobic contact of residues 6–10 at the barrel wall would then remain preserved. Note however that at this point we do not have experimental evidence that unambiguously links either of these two models to the voltage gating process.
The role of NMR for structure determination of VDAC and other membrane proteins
As indicated, one of the three recent VDAC-1 structures was solved with NMR only [""13], a second one with a combined NMR / X-ray approach to 4.0 Å resolution [""14] and the third structure with X-ray, but to 2.3 Å resolution [""15]. Whereas the pure NMR and pure X-ray approaches used respective state-of-the-art techniques of their fields, the mixed structure applied an innovative technique, BUSTER-TNT [24] which combines NMR and diffraction data. In this particular case, according to the authors, their NMR and the X-ray data sets alone were not sufficient to solve the structure [""14]. However, as shown by the NMR structure, contemporary solution NMR techniques alone are sufficient to solve the structure of such a large β-barrel. Including VDAC-1, nine de novo structures of integral membrane proteins with more than two transmembrane segments have now been solved by solution NMR (Table 1, Table 2) [25]. Five of them have been determined within the last two years. The arsenal of solution NMR methods to study this class of proteins is large [26,27], and the technical approaches thus vary between the projects. Here, we want to focus on two technical features that have a substantial impact on the ability to determine membrane protein structures with NMR: Protein refolding and deuteration of the detergent micelle.
Table 1.
NMR structures of integral β-barrel membrane proteins solved by solution NMR.a
| protein | organis m |
number of residues |
number of β- strands |
detergent | deuterated detergent |
refolding | reference |
|---|---|---|---|---|---|---|---|
| OmpX | E. coli | 148 | 8 | DHPC | no | yes | [41] |
| PagP | E. coli | 164 | 8 | DPC, β-OG |
yes, yes |
yes | [42] |
| OmpA | E. coli | 177 | 8 | DPC | yes | yes | [43] |
| OmpA |
K. pneumo niae |
210 | 8 | DHPC | yes | yes | [44] |
| OmpG | E. coli | 280 | 14 | DPC | yes | yes | ["45] |
| VDAC-1 |
H. sapiens |
283 | 19 | LDAO | yes | yes | [""13] |
Abbreviations: DHPC, di-hexanoyl-phosphatidylcholine; DPC, dodecyl-phosphocholine; β-OG, β-octylglucopyranoside; LDAO, lauryldimethylamineoxide
Table 2.
NMR structures of integral α-helical membrane proteins with more than 2 transmembrane helices solved by solution NMR.a
| protein | organism | number of residues |
number of TM α- helices |
detergent | deuterated detergent |
refolding | |
|---|---|---|---|---|---|---|---|
| M2 |
Influenza virus |
42 | 4 × 1 | DHPC | yes | yes | ["46] |
| DsbB | E. coli | 176 | 4 | DPC | yes | no | ["47] |
| Phospholamban |
H. sapiens |
52 | 5 × 1 | DPC | yes | yes | ["48] |
Abbreviations: DHPC, di-hexanoyl-phosphatidylcholine; DPC, dodecyl-phosphocholine; TM, transmembrane
The role of protein refolding
Refolding from a denatured state may not be feasible for all membrane proteins and the search for a suitable refolding protocol may be tedious, but if this strategy is successful, the benefits are large: (i) Very high yield. For example, in the case of VDAC-1, typical yields are 40 mg of pure protein from 1 L of E. coli cell culture in minimal medium. (ii) Efficient purification to a high degree. We estimate the purity of VDAC-1 after His-tag purification in the denatured state and refolding to be above 98% as judged from SDS-PAGE (iii) High reproducibility. Due to the high purity, identical sample conditions and thus NMR spectra can be obtained. (iv) Efficient perdeuteration, and selective isotope labeling. Due to the high yields, cost-effective sample preparation is possible (v) Complete back exchange of amide protons. Since the perdeuterated protein produced by growing E. coli in D2O goes through a denatured state, all amide moieties can readily be protonated and are observable for assignments and structure determination.
As a consequence of these advantages, all solution NMR structures of integral membrane proteins, except one, were obtained by refolding from a denatured state (Table 1, Table 2).
Impact of deuterated detergent
For large particles with long rotational correlation times, perdeuteration of the protein is a prerequirement for good NMR spectra. Otherwise, the resonance lines are broadened due to strong dipole–dipole couplings and the overall spectral sensitivity is substantially reduced. For membrane proteins, the detergent micelle is in close contact with the protein and a deuteration of the detergent does thus also have an impact on the spectral quality. Our experience with VDAC shows that the quality of the [15N,1H]-TROSY correlation spectra is virtually independent of the degree of detergent deuteration. Also the TROSY-HNCA spectra could be recorded with good quality in either deuterated or protonated LDAO. In the 15N-resolved [1H,1H]-NOESY spectra, a sensitivity decrease of about 10–30% was observed when going from deuterated to protonated detergent, but the strongest cross peaks, including many interstrand amide–amide NOEs were still observable in protonated detergent. A strong difference between protonated and deuterated detergent was however observed for spectra of the Isoleucine, Leucine and Valine methyl groups. NOESY-type spectra of ILV labeled sample in protonated detergent did not produce useful spectra (T. Malia, personal communication), but the use of deuterated detergent substantially improved this situation and enabled recording of 3D- and 4D-resolved NOESY spectra. This trend is generally observed for membrane proteins and structural studies are thus found to use deuterated detergent whenever possible (Table 1 and Table 2).
Ligand binding and protein–protein interactions
The ability to characterize protein–ligand and protein–protein interaction sites structurally is a particular strength of NMR in solution [28,29]. The metabolite β-NADH, as well as the anti-apoptotic protein Bcl-xL are known to modulate the VDAC voltage gating process [30,31]. We could determine the NADH binding site on VDAC-1, which is located at one end of strands 17 and 18, near the extrusion of the N-terminus and could also map the site of the interaction with Bcl-xL, using data reported in our previous publication ["32].
Many further solution NMR studies of membrane protein structure, function and dynamics have revealed significant biological information. These include, to mention only a few, studies revealing mechanistic features of ion selectivity and gating in the KcsA channel [33,34], showing the domain interactions that may be related to the mechanism for the Kv channel inactivation [35], revealing how phosphorylation regulates the CFTR chlorine channel [36], solving the structure of the signaling domains of FecA to understand signaling within the FecA transporter [37], mapping protein–protein interactions to show how HasA delivers the heme to the membrane protein HasR [38], using methyl-NMR to propose a model for the interaction of signal recognition particle with its receptor [39], characterizing the backbone dynamics of sensory rhodopsin pSRII [40].
Conclusion
In the past two years, solution NMR was used in five de novo structure determinations of integral membrane proteins, including the first eukaryotic β-barrel membrane protein structure, VDAC-1. Furthermore, solution NMR has made substantial impact by a large number of other structural and functional studies of membrane proteins. These developments show that solution NMR has become an important tool for membrane protein molecular biology and let expect further significant contributions in the future.
Acknowledgements
The research described here was supported by NIH (grants GM075875, GM47467). S.H. was supported in part by the Swiss National Science Foundation. We thank Marco Colombini for critical discussions.
Footnotes
This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Scheffler IE. Mitochondria. New York: Wiley; 2007. [Google Scholar]
- 2.Margulis L. Origin of Eukaryotic Cells. New Haven, CT: Yale University Press; 1970. [Google Scholar]
- 3.Dolezal P, Likic V, Tachezy J, Lithgow T. Evolution of the molecular machines for protein import into mitochondria. Science. 2006;313:314–318. doi: 10.1126/science.1127895. [DOI] [PubMed] [Google Scholar]
- 4.Mannella CA. Structure of the mitochondrial outer membrane channel derived from electron microscopy of 2D crystals. J Bioenerg Biomembr. 1989;21:427–437. doi: 10.1007/BF00762515. [DOI] [PubMed] [Google Scholar]
- 5.Blachly-Dyson E, Forte M. VDAC channels. IUBMB Life. 2001;52:113–118. doi: 10.1080/15216540152845902. [DOI] [PubMed] [Google Scholar]
- 6.Casadio R, Jacoboni I, Messina A, De Pinto V. A 3D model of the voltage-dependent anion channel (VDAC) FEBS Lett. 2002;520:1–7. doi: 10.1016/s0014-5793(02)02758-8. [DOI] [PubMed] [Google Scholar]
- 7.Shoshan-Barmatz V, Gincel D. The voltage-dependent anion channel: characterization, modulation, and role in mitochondrial function in cell life and death. Cell Biochem Biophys. 2003;39:279–292. doi: 10.1385/CBB:39:3:279. [DOI] [PubMed] [Google Scholar]
- 8.Colombini M. VDAC: The channel at the interface between mitochondria and the cytosol. Mol Cell Biochem. 2004;256:107–115. doi: 10.1023/b:mcbi.0000009862.17396.8d. [DOI] [PubMed] [Google Scholar]
- 9.Schein SJ, Colombini M, Finkelstein A. Reconstitution in planar lipid bilayers of a voltage-dependent anion-selective channel obtained from Paramecium mitochondria. J Membr Biol. 1976;30:99–120. doi: 10.1007/BF01869662. [DOI] [PubMed] [Google Scholar]
- 10.Forte M, Guy HR, Mannella CA. Molecular genetics of the VDAC ion channel: structural model and sequence analysis. J Bioenerg Biomembr. 1987;19:341–350. doi: 10.1007/BF00768537. [DOI] [PubMed] [Google Scholar]
- 11.Song J, Midson C, Blachly-Dyson E, Forte M, Colombini M. The topology of VDAC as probed by biotin modification. J Biol Chem. 1998;273:24406–24413. doi: 10.1074/jbc.273.38.24406. [DOI] [PubMed] [Google Scholar]
- 12.Engelhardt H, Meins T, Poynor M, Adams V, Nussberger S, Welte W, Zeth K. High-level expression, refolding and probing the natural fold of the human voltage-dependent anion channel isoforms I and II. J Membr Biol. 2007;216:93–105. doi: 10.1007/s00232-007-9038-8. [DOI] [PubMed] [Google Scholar]
- ""13.Hiller S, Garces RG, Malia TJ, Orekhov VY, Colombini M, Wagner G. Solution structure of the integral human membrane protein VDAC-1 in detergent micelles. Science. 2008;321:1206–1210. doi: 10.1126/science.1161302.This paper shows the three-dimensional structure of human VDAC-1, solved by solution NMR in LDAO micelles. The binding site for NADH is discovered and the previously detected Bcl-xL interaction site is mapped on the VDAC structure.
- ""14.Bayrhuber M, Meins T, Habeck M, Becker S, Giller K, Villinger S, Vonrhein C, Griesinger C, Zweckstetter M, Zeth K. Structure of the human voltage-dependent anion channel. Proc Natl Acad Sci USA. 2008;105:15370–15375. doi: 10.1073/pnas.0808115105.The structure of human VDAC-1 in LDAO micelles is solved using a combination of NMR and X-ray data. The N-terminal α-helix adopts a previously undescribed conformation.
- ""15.Ujwal R, Cascio D, Colletier JP, Faham S, Zhang J, Toro L, Ping P, Abramson J. The crystal structure of mouse VDAC1 at 2.3 Å resolution reveals mechanistic insights into metabolite gating. Proc Natl Acad Sci USA. 2008;105:17742–17747. doi: 10.1073/pnas.0809634105.The three-dimensional structure of recombinant mouse VDAC-1 in bicelles is solved by X-ray diffraction. The structure shows a new conformation of the N-terminal α-helix.
- 16.Kobayashi K, Granja JR, Ghadiri MR. β-Sheet peptide architecture: Measuring the relative stability of parallel vs. antiparallel β-sheets. Angew Chem Int Ed. 1995;34:95–98. [Google Scholar]
- 17.Kleinschmidt JH. Membrane protein folding on the example of outer membrane protein A of Escherichia coli. Cell Mol Life Sci. 2003;60:1547–1558. doi: 10.1007/s00018-003-3170-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Neupert W, Herrmann JM. Translocation of proteins into mitochondria. Annu Rev Biochem. 2007;76:723–749. doi: 10.1146/annurev.biochem.76.052705.163409. [DOI] [PubMed] [Google Scholar]
- 19.Schulz GE. The structure of bacterial outer membrane proteins. Biochim Biophys Acta. 2002;1565:308–317. doi: 10.1016/s0005-2736(02)00577-1. [DOI] [PubMed] [Google Scholar]
- 20.McLachlan AD. Gene duplications in the structural evolution of chymotrypsin. J Mol Biol. 1979;128:49–79. doi: 10.1016/0022-2836(79)90308-5. [DOI] [PubMed] [Google Scholar]
- 21.Bain AD. Chemical exchange in NMR. Prog Nucl Magn Reson Spectrosc. 2003;43:63–103. [Google Scholar]
- 22.Koppel DA, Kinnally KW, Masters P, Forte M, Blachly-Dyson E, Mannella CA. Bacterial expression and characterization of the mitochondrial outer membrane channel. Effects of n-terminal modifications. J Biol Chem. 1998;273:13794–13800. doi: 10.1074/jbc.273.22.13794. [DOI] [PubMed] [Google Scholar]
- 23.De Pinto V, Tomasello F, Messina A, Guarino F, Benz R, La Mendola D, Magri A, Milardi D, Pappalardo G. Determination of the conformation of the human VDAC1 N-terminal peptide, a protein moiety essential for the functional properties of the pore. ChemBioChem. 2007;8:744–756. doi: 10.1002/cbic.200700009. [DOI] [PubMed] [Google Scholar]
- 24.Blanc E, Roversi P, Vonrhein C, Flensburg C, Lea SM, Bricogne G. Refinement of severely incomplete structures with maximum likelihood in BUSTER-TNT. Acta Crystallogr D Biol Crystallogr. 2004;60:2210–2221. doi: 10.1107/S0907444904016427. [DOI] [PubMed] [Google Scholar]
- 25.Membrane proteins of known structure on World Wide. Web URL: http://blanco.biomol.uci.edu/Membrane_Proteins_xtal.html.
- 26.Krueger-Koplin RD, Sorgen PL, Krueger-Koplin ST, Rivera-Torres AO, Cahill SM, Hicks DB, Grinius L, Krulwich TA, Girvin ME. An evaluation of detergents for NMR structural studies of membrane proteins. J Biomol NMR. 2004;28:43–57. doi: 10.1023/B:JNMR.0000012875.80898.8f. [DOI] [PubMed] [Google Scholar]
- 27.Tamm LK, Liang BY. NMR of membrane proteins in solution. Prog Nucl Magn Reson Spectrosc. 2006;48:201–210. [Google Scholar]
- 28.Marintchev A, Wagner G. Translation initiation: structures, mechanisms and evolution. Q Rev Biophys. 2004;37:197–284. doi: 10.1017/S0033583505004026. [DOI] [PubMed] [Google Scholar]
- 29.Reibarkh M, Malia TJ, Hopkins BT, Wagner G. Identification of individual protein-ligand NOEs in the limit of intermediate exchange. J Biomol NMR. 2006;36:1–11. doi: 10.1007/s10858-006-9028-7. [DOI] [PubMed] [Google Scholar]
- 30.Zizi M, Forte M, Blachly-Dyson E, Colombini M. NADH regulates the gating of VDAC, the mitochondrial outer membrane channel. J Biol Chem. 1994;269:1614–1616. [PubMed] [Google Scholar]
- 31.Vander Heiden MG, Li XX, Gottleib E, Hill RB, Thompson CB, Colombini M. Bcl-xL promotes the open configuration of the voltage-dependent anion channel and metabolite passage through the outer mitochondrial membrane. J Biol Chem. 2001;276:19414–19419. doi: 10.1074/jbc.M101590200. [DOI] [PubMed] [Google Scholar]
- "32.Malia TJ, Wagner G. NMR structural investigation of the mitochondrial outer membrane protein VDAC and its interaction with antiapoptotic Bcl-xL. Biochemistry. 2007;46:514–525. doi: 10.1021/bi061577h.This paper characterizes the interaction between the anti-apoptotic protein Bcl-xL and VDAC-1. It gives a refolding protocol and shows the first solution spectra for VDAC-1.
- 33.Chill JH, Louis JM, Miller C, Bax A. NMR study of the tetrameric KcsA potassium channel in detergent micelles. Protein Sci. 2006;15:684–698. doi: 10.1110/ps.051954706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Baker KA, Tzitzilonis C, Kwiatkowski W, Choe S, Riek R. Conformational dynamics of the KcsA potassium channel governs gating properties. Nat Struct Mol Biol. 2007;14:1089–1095. doi: 10.1038/nsmb1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Baker KA, Hilty C, Peti W, Prince A, Pfaffinger PJ, Wider G, Wüthrich K, Choe S. NMR-derived dynamic aspects of N-type inactivation of a Kv channel suggest a transient interaction with the T1 domain. Biochemistry. 2006;45:1663–1672. doi: 10.1021/bi0516430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Baker JM, Hudson RP, Kanelis V, Choy WY, Thibodeau PH, Thomas PJ, Forman-Kay JD. CFTR regulatory region interacts with NBD1 predominantly via multiple transient helices. Nat Struct Mol Biol. 2007;14:738–745. doi: 10.1038/nsmb1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ferguson AD, Amezcua CA, Halabi NM, Chelliah Y, Rosen MK, Ranganathan R, Deisenhofer J. Signal transduction pathway of TonB-dependent transporters. Proc Natl Acad Sci USA. 2007;104:513–518. doi: 10.1073/pnas.0609887104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Caillet-Saguy C, Piccioli M, Turano P, Izadi-Pruneyre N, Delepierre M, Bertini I, Lecroisey A. Mapping the interaction between the hemophore HasA and its outer membrane receptor HasR using CRINEPT-TROSY NMR spectroscopy. J Am Chem Soc. 2009;131:1736–1744. doi: 10.1021/ja804783x. [DOI] [PubMed] [Google Scholar]
- 39.Neher SB, Bradshaw N, Floor SN, Gross JD, Walter P. SRP RNA controls a conformational switch regulating the SRP-SRP receptor interaction. Nat Struct Mol Biol. 2008;15:916–923. doi: 10.1038/nsmb.1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gautier A, Kirkpatrick JP, Nietlispach D. Solution-state NMR spectroscopy of a seven-helix transmembrane protein receptor: backbone assignment, secondary structure, and dynamics. Angew Chem Int Ed. 2008;47:7297–7300. doi: 10.1002/anie.200802783. [DOI] [PubMed] [Google Scholar]
- 41.Fernández C, Wüthrich K. NMR solution structure determination of membrane proteins reconstituted in detergent micelles. FEBS Lett. 2003;555:144–150. doi: 10.1016/s0014-5793(03)01155-4. [DOI] [PubMed] [Google Scholar]
- 42.Hwang PM, Choy WY, Lo EI, Chen L, Forman-Kay JD, Raetz CRH, Prive GG, Bishop RE, Kay LE. Solution structure and dynamics of the outer membrane enzyme PagP by NMR. Proc Natl Acad Sci USA. 2002;99:13560–13565. doi: 10.1073/pnas.212344499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Arora A, Abildgaard F, Bushweller JH, Tamm LK. Structure of outer membrane protein A transmembrane domain by NMR spectroscopy. Nat Struct Biol. 2001;8:334–338. doi: 10.1038/86214. [DOI] [PubMed] [Google Scholar]
- 44.Renault M, Saurel O, Czaplicki J, Demange P, Gervais V, Löhr F, Reat V, Piotto M, Milon A. Solution state NMR structure and dynamics of KpOmpA, a 210 residue transmembrane domain possessing a high potential for immunological applications. J Mol Biol. 2009;385:117–130. doi: 10.1016/j.jmb.2008.10.021. [DOI] [PubMed] [Google Scholar]
- " 45.Liang B, Tamm LK. Structure of outer membrane protein G by solution NMR spectroscopy. Proc Natl Acad Sci USA. 2007;104:16140–16145. doi: 10.1073/pnas.0705466104.The NMR structure of the integral membrane protein OmpG from E. coli is presented, a 14-stranded β-barrel. The extracelular loops are more flexibility and the strands are thus shorter than in the X-ray structure. This work establishes a significant advancement for membrane protein NMR in terms of molecular size.
- "46.Schnell JR, Chou JJ. Structure and mechanism of the M2 proton channel of influenza A virus. Nature. 2008;451:591–595. doi: 10.1038/nature06531.The structure of the tetrameric channel M2 explains the mechanism for channel gating. Intriguingly, the binding site of the drug adamantine is on the channel outside in the lipid phase, as defined by intermolecular NOEs.
- "47.Zhou Y, Cierpicki T, Jimenez RH, Lukasik SM, Ellena JF, Cafiso DS, Kadokura H, Beckwith J, Bushweller JH. NMR solution structure of the integral membrane enzyme DsbB: functional insights into DsbB-catalyzed disulfide bond formation. Mol Cell. 2008;31:896–908. doi: 10.1016/j.molcel.2008.08.028.The authors present the structure of the E. coli enzyme DsbB by NMR and elucidate its functional mechanism. The technical aspects of this work, including protein isolation from the cell membrane and advanced NMR methods, are outstanding.
- " 48.Oxenoid K, Chou JJ. The structure of phospholamban pentamer reveals a channel-like architecture in membranes. Proc Natl Acad Sci USA. 2005;102:10870–10875. doi: 10.1073/pnas.0504920102.This work presents the structure of human phospholamban, a channel for calcium flux across the membrane, by solution NMR. The technical aspects of this work, in particular the experiments for intermolecular NOE detection in the homopentamer, are excellent.