Abstract
Satellite cells purified from adult skeletal muscle can participate extensively in muscle regeneration and can also re-populate the satellite cell pool, suggesting that they have direct therapeutic potential for treating degenerative muscle diseases1,2. The paired-box transcription factor Pax7 is required for satellite cells to generate committed myogenic progenitors3. In this study we undertook a multi-level approach to define the role of Pax7 in satellite cell function. Using comparative microarray analysis, we identified several novel and strongly regulated targets; in particular, we identified Myf5 as a gene whose expression was regulated by Pax7. Using siRNA, fluorescence-activated cell sorting (FACS) and chromatin immunoprecipitation (ChIP) studies we confirmed that Myf5 is directly regulated by Pax7 in myoblasts derived from satellite cells. Tandem affinity purification (TAP) and mass spectrometry were used to purify Pax7 together with its co-factors. This revealed that Pax7 associates with the Wdr5–Ash2L–MLL2 histone methyltransferase (HMT) complex that directs methylation of histone H3 lysine 4 (H3K4, refs 4–10). Binding of the Pax7–HMT complex to Myf5 resulted in H3K4 tri-methylation of surrounding chromatin. Thus, Pax7 induces chromatin modifications that stimulate transcriptional activation of target genes to regulate entry into the myogenic developmental programme.
Satellite cells arise from a population of muscle progenitor cells that originate in the central domain of the dermomyotome. These progenitors express the paired-box transcription factors Pax3 and Pax7 (refs 11, 12), and although neither their emergence nor their maintenance requires Pax3 function13, recent studies have demonstrated that Pax7 is uniquely indispensable for these cells 14. In the absence of Pax7, satellite cells die and thus fail to re-populate their niche11,14,15. Pax7, is therefore essential for the formation and maintenance of a population of functional satellite cells. Analysis of the physiological functions of Pax7 has been hindered by relatively weak trans-activation properties resulting from cis-repression16. Consequently, the mechanisms by which Pax7 activates downstream target genes remain unclear. To address this problem, we used a comparative microarray approach to globally identify Pax7 myogenic targets and we propose that the regulation of those genes by Pax7 is intimately related to the protein complexes with which it interacts.
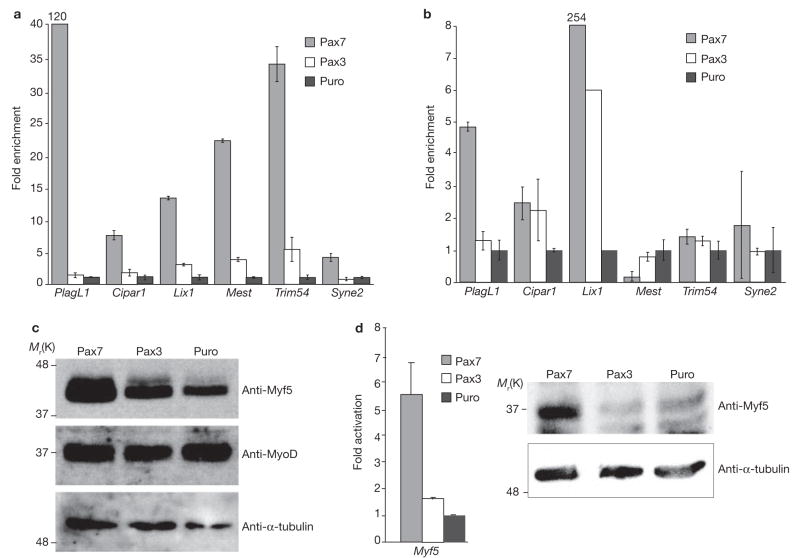
Pools of C2C12 myoblasts were transfected with retrovirus expressing either mouse Pax7–FLAG, or a control virus expressing only the puromycin resistance gene (Puro). As expected, persistent Pax7 expression resulted in the maintenance of a proliferative phenotype and inhibition of differentiation17 (Supplementary Information, S1). Total RNA was harvested to generate probes for hybridization to Affymetrix GeneChip microarrays; 43 genes were up-regulated by Pax7–FLAG, including several that exhibited striking changes in expression (Table 1). Real-time PCR analysis confirmed that the candidate transcripts were increased in response to Pax7 (Fig. 1a). Pax7 and Pax3 are very closely related proteins; however, Pax7 and Pax3 differed significantly in their abilities to regulate the target genes that were identified. When expressed in C2C12 myoblasts, the levels of each target were substantially increased by Pax7–FLAG whereas Pax3–FLAG had either no effect (PlagL1; Syne2) or only modest effects (Cipar1; Lix1; Mest; Trim54) (Fig. 1a). To further assess the specificity and responsiveness of these genes, 10T1/2 fibroblast cells were transfected with Pax7–FLAG, Pax3–FLAG or Puro control Despite the non-muscle environment of these cells, the levels of all the candidate genes tested, with the exception of Mest, were increased by Pax7 and they were more responsive to Pax7 than to Pax3 (Fig. 1b), confirming the observations made in C2C12 myoblasts. This is consistent with the phenotypic observations that Pax7 and Pax3 have divergent, non-redundant functions during both embryonic14 and post-natal15 myogenesis.
Table 1.
Pax7-induced increases in expression in C2C12 myoblasts
| Symbol | GID | Fold | Name | RLT-PCR |
|---|---|---|---|---|
| Plagl1 | NM_009538 | 385.0 | 1Pleiomorphic adenoma gene-like 1 | 135.8 |
| Lix1 | NM_025681 | 12.3 | Limb expression 1 homolog (chicken) | 34.8 |
| Syne2 | BF582734 | 11.3 | Synaptic nuclear envelope 2 | 2.2 |
| Cipar1 | AK008716 | 8.1 | Castration-induced prostatic apoptosis-related 1 | 166.6 |
| Trim54 | NM_021447 | 7.2 | Tripartite motif-containing 54 | 4.1 |
| Il13ra1 | NM_133990 | 5.4 | 2 Interleukin 13 receptor, alpha 1 | – |
| Mest | NM_008590 | 5.0 | Mesoderm specific transcript | 17.9 |
| Peg3 | NM_008817 | 4.6 | 2 Paternally expressed 3 | – |
| 3110001A13Rik | NM_025626 | 4.6 | 3 RIKEN cDNA 3110001A13 gene | – |
| Ahr | NM_013464 | 4.4 | Aryl-hydrocarbon receptor | – |
| Npnt | NM_033525 | 4.1 | 2 Nephronectin | – |
| Ltb4dh | NM_025968 | 4.1 | Leukotriene B4 12-hydroxydehydrogenase | – |
| Msln | NM_018857 | 4.1 | mesothelin | – |
| C1qtnf3 | NM_030888 | 3.8 | C1q and tumour necrosis factor related protein 3 | – |
| – | BB369191 | 3.7 | Sim. to mouse pentylenetetrazol-related mRNA | – |
| Pparg | NM_011146 | 3.4 | Peroxisome proliferator activated receptor gamma | – |
| Cd24a | NM_009846 | 3.1 | 2 CD24a antigen | – |
| Pkia | NM_008862 | 3.0 | 2 Protein kinase inhibitor, alpha | – |
| Nqo1 | NM_008706 | 2.9 | NAD(P)H dehydrogenase, quinone 1 | – |
| Marcks | NM_008538 | 2.6 | 3 Myristoylated alanine-rich protein kinase C substrate | – |
| Gch1 | NM_008102 | 2.6 | GTP cyclohydrolase 1 | – |
| Cdh11 | NM_009866 | 2.6 | Cadherin 11 | – |
| Myo1d | NM_177390 | 2.5 | Myosin ID | – |
| Cxcr4 | NM_009911 | 2.5 | Chemokine (C-X-C motif) receptor 4 | – |
| Crlf1 | NM_018827 | 2.5 | Cytokine receptor-like factor 1 | – |
| Sema3e | NM_011348 | 2.4 | Semaphorin 3E | – |
| Mdfic | NM_175088 | 2.4 | MyoD family inhibitor domain-containing | – |
| Ass1 | NM_007494 | 2.4 | Argininosuccinate synthetase 1 | – |
| Id3 | NM_008321 | 2.4 | Inhibitor of DNA binding 3 | – |
| Ppap2a2 | NM_008903 | 2.3 | 2 Phosphatidic acid phosphatase 2a isoform 2 | – |
| Myf5 | NM_008656 | 2.2 | Myogenic factor 5 | 3.0 |
| Cdh2 | NM_007664 | 2.2 | Cadherin 2 (N-cadherin) | – |
| Depdc6 | NM_145470 | 2.2 | DEP domain containing 6 | – |
| Prss23 | NM_029614 | 2.2 | 2 Protease, serine, 23 | – |
| Id2 | NM_010496 | 2.1 | Inhibitor of DNA binding 2 | – |
| Emb | NM_010330 | 2.1 | Embigin | – |
| Rnf128 | NM_023270 | 2.1 | 2 Ring finger protein 128 | – |
| Rnh1 | NM_145135 | 2.1 | Ribonuclease/angiogenin inhibitor 1 | – |
| Olfm1 | NM_019498 | 2.1 | Olfactomedin 1 | – |
| – | BG069607 | 2.1 | – | – |
| Tob1 | NM_009427 | 2.1 | Transducer of ErbB-2.1 | – |
| Atp11a | NM_015804 | 2.1 | ATPase, class VI, type 11A | – |
| 2210409B22Rik | BM207133 | 2.0 | RIKEN cDNA 2210409B22 gene | – |
Pax7–FLAG samples were stringently compared with Puro samples to derive sets of candidate Pax7-regulated genes. PlagL1 was not detected in Puro samples but was highly expressed in Pax7–FLAG samples. Others that were also considerably increased included Lix1 (12-fold); Syne2 (11-fold); Cipar1 (8-fold); Trim54 (7-fold); and Mest (5-fold).
Signal was very low and called Absent in control.
Mean fold change for two distinct probesets directed at the same transcript.
Mean fold change for 3 distinct probesets directed at the same transcript. Bold lines indicate those used for real-time PCR validation
Figure 1.
Candidate target genes are specifically activated by Pax7 in C2C12 myoblasts and are only weakly responsive to Pax3. (a) Pax7 strongly activated PlagL1, Cipar1, Lix1, Mest and Trim54 in C2C12 myoblasts whereas Pax3 was far less effective. Syne2 was only weakly increased by Pax7. Expression was normalized to GAPDH transcript levels and is shown relative to the empty-vector (Puro) controls. (b) PlagL1, and particularly Lix1, were both strongly induced by Pax7 in non-muscle 10T1/2 fibroblasts but were weakly induced or unresponsive to Pax3. Cipar, Trim54 and Syne2 transcripts are also increased by Pax7d in 10T1/2 cells. (c) Myf5 protein levels were also increased by Pax7–FLAG and Pax3–FLAG expression in C2C12 myoblasts whereas MyoD protein levels remained unaffected; α-tubulin was included as a control. (d) Real-time PCR and western blot analysis demonstrated that Myf5 RNA and protein, respectively, were increased in 10T1/2 cells expressing Pax7-FLAG (error bars are standard error (s.e.m.)). Full-length scans of western blot data can be found in Supplementary Information S4.
The most significant target identified in our microarray study was the gene for the myogenic regulatory factor Myf5, one of two well-established primary master regulators of skeletal muscle commitment (Myf5) and differentiation (MyoD) in satellite cells1,18,19. This suggests a direct link between Pax7 and myogenic commitment of adult myoblasts. Myf5 expression was increased by Pax7–FLAG, as shown by microarray analysis (2.2-fold; Table 1) and real-time PCR (~3-fold; data not shown). Importantly, changes in Myf5 protein levels were consistent with the observed increases in mRNA levels. Expression of Pax7–FLAG in C2C12 cells resulted in a dramatic increase in Myf5 protein compared with the Puro control (Fig. 1c). An exaggerated change in Myf5 protein versus mRNA levels has been reported previously in MyoD−/− myoblasts20. As shown for previously described targets, Pax3-induced levels of Myf5 protein were substantially lower than those induced by Pax7 in C2C12 myoblasts; in contrast, MyoD levels were unaffected by Pax 3 or Pax 7 (Fig. 1c). 10T1/2 fibroblasts are capable of myogenic differentiation in the presence of exogenous Myf5 or MyoD. Remarkably, Pax7 expression in 10T1/2 cells resulted in the induction of Myf5 at both RNA and protein levels (Fig. 1d), whereas Pax3 produced no detectable change. This indicates that Pax7 readily stimulates Myf5 transcription in both muscle and non-muscle cell lines and provides a molecular connection between Pax7 and myogenic commitment.
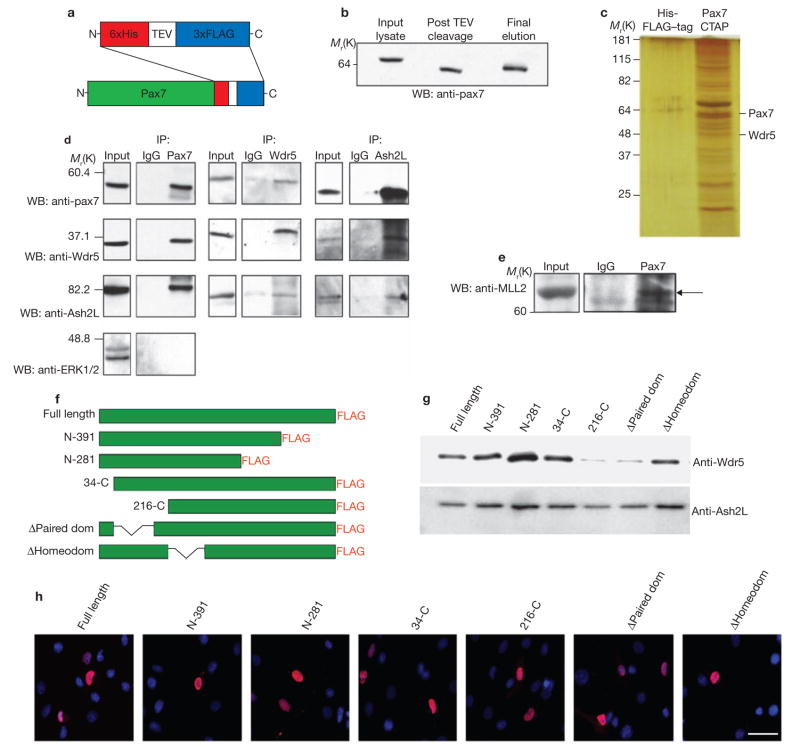
To test our hypothesis that Pax7 activates target genes by recruiting regulatory complexes to these loci, proteins interacting with Pax7 were purified using a 6x-histidine–TEV (tobacco etch virus)cleavage–3xFLAG tandem affinity purification (TAP) tag fused to the carboxy (C)-terminus of full-length mouse Pax7 (Fig. 2a). This construct (Pax7–CTAP), or the tag alone (HisFLAG–tag) control, was expressed in C2C12 myoblasts. The Pax7–CTAP protein migrated at the predicted size; showed no sign of degradation; was amenable to cleavage by TEV protease and was detected by western-blot analysis of the final eluate (Fig. 2b).
Figure 2.
Identification of Pax7-interacting co-factors. (a) A TAP tag, consisting of six histidine and three FLAG epitopes, separated by a TEV cleavage site, was fused to the C-terminus of Pax7 to create a Pax7–CTAP (His–TEV–FLAG) construct. A construct expressing only the tag (referred to as HisFLAG-tag) was used as a negative control. (b) High yield purification of the Pax7–CTAP protein as shown by detection of the fusion protein in the initial cell lysate, in the eluate following TEV cleavage, and in the final elution from Ni+-resin. (c,) TAP of Pax7–CTAP- compared with HisFLAG-tag-associated proteins from C2C12 cells. Protein matches were generated from multiple data sets and were identified following MALDI–TOF (analysed via Mascot). Pax7–CTAP-interacting proteins were compared with those identified in HisFLAG-tag purifications to unequivocally identify those that were Pax7-specific (versus those that represented contaminants). (d) Pax7 co-immunoprecipitated with Wdr5 and Ash2L, two conserved units of a HMT complex. As a negative control, the Pax7-immunoprecipitate was also probed with an antibody to a non-native-Pax7 interacting nuclear protein, ERK1/2. (e) Co-purification of Pax7 and MLL2 immunoprecipitated from primary myoblast nuclear extract. (f) Pax7-deletion constructs used to map Pax7 binding to the HMT complex. (g) Loss of the paired domain was observed to almost entirely abolish binding between Pax7 and Wdr5. (h) Pax7–FLAG truncations where observed to localize to the nucleus (DAPI-blue, Pax7–FLAG deletion constructs-red, scale bar = 20 μm). Full length scans of western blot data can be found in Supplementary Information S4.
We performed large-scale TAPs to identify interacting proteins via MALDI–TOF mass spectrometry of silver-stained SDS–PAGE bands (Fig. 2c). A complex mixture of largely uncharacterized, interacting proteins was identified, together with the target protein Pax7. Of particular interest, one of the Pax7-associated proteins identified was the WD40-domain-containing protein Wdr5. The observation that Pax7 was co-isolated with Wdr5 is intriguing given that Wdr5, the trithorax-group protein Ash2L, and RbBP5 are the three common components of histone methyltransferase (HMT) complexes that methylate histone H3 at lysine 4 (H3K4, refs 4–10, 21). Extensive biochemical and structural studies have revealed that Wdr5 is a crucial component of HMT because it associates directly with the amino acid tail of histone H3 (refs 4, 22–25), whereas Ash2L, RbBP5 and Wdr5 together create the core structural platform for subsequent protein complex assembly4. This suggests that Pax7 interacts with a core component of the complex, namely Wdr5, to recruit HMT complexes to target promoters for inscription of epigenetic modifications.
We used nuclear extracts from satellite cell-derived primary myoblasts to validate the Wdr5 interaction in an endogenous setting and to determine whether native Pax7 associates with the other core components of HMT. Using antibodies to the endogenous Pax7, Wdr5 and Ash2L proteins, reciprocal co-immunoprecipitation analysis revealed that antibodies reactive with Pax7 specifically co-precipitated Wdr5 and Ash2L (versus IgG controls). In addition, antibodies reactive with Wdr5 co-precipitated Pax7 and Ash2L, and antibodies reactive with Ash2L co-precipitated Pax7 and Wdr5 (Fig. 2d). These results support the assertion that the three proteins are found in the same complex. We also performed control western blots with α-Erk1/2, a nuclear protein present in abundance in many protein complexes. The failure to detect Erk1/2 in Pax7-immunoprecipitates demonstrated the specificity of the observed interactions. Together, these data confirm that in primary myoblasts Wdr5 and Ash2L interact with Pax7, and support an association of Pax7 with a HMT complex. To identify the methyltransferase responsible for this activity, we performed co-immunoprecipitation with Pax7 and probed for the members of the MLL family of HMTs. The MLL proteins are obvious candidates for this activity because of their known association with the Wdr5–Ash2L–RbBP5 core complex21. Indeed, Pax7 interacted with MLL2 (Fig. 2e) but not MLL1 (data not shown), indicating that MLL2 is a specific HMT recruited to this complex.
To further investigate these interactions we produced Pax7–FLAG constructs that contained amino (N)- and C-termini truncations, and specific deletions of the Pax7 paired-domain or homeodomain (Fig. 2f). Deletion of the paired-domain of Pax7, whether following truncation of a significant portion of the N-terminus or a more subtle deletion of only the paired-domain, resulted in near complete abolition of Wdr5 binding to Pax7 (a moderate decrease in binding was observed with Ash2L). In contrast, the effect of deleting the homeodomain appeared to be negligible (Fig. 2g). To ensure that these effects did not result from a failure of the constructs to enter the correct cellular compartment, we performed immunocytochemistry and noted that all of the Pax7–FLAG deletion constructs were localized to the nucleus (Fig. 2h). Given that the paired-domain is involved in the DNA-binding activity of Pax7, this result suggests that DNA binding may be required for the Pax7–HMT complex to form and/or remain bound.
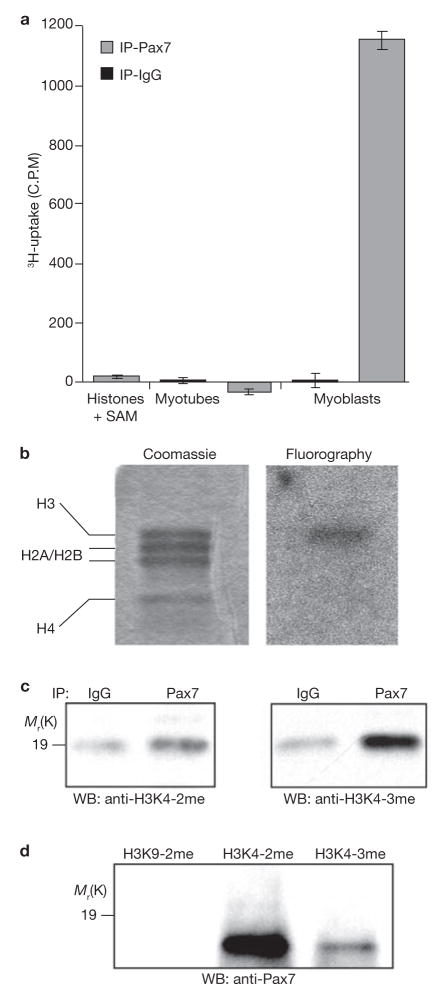
To directly confirm that the Pax7–Wdr5–Ash2L–MLL2 complex possesses methyltransferase activity, we incubated Pax7 immunocomplexes (from primary myoblast nuclear extracts) with core histones in the presence of the tritiated methyl-donor S-adenosyl-methionine (SAM). Histones incubated with the Pax7–immunoprecipitated complex showed an elevated level of tritium incorporation (>1,000 c.p.m.) compared with the IgG control, demonstrating that the Pax7–immunocomplex indeed possesses HMT activity. Histones incubated with immunoprecipitates from primary myotubes (which do not express Pax7) showed no incorporation of SAM above baseline levels of activity (a representative experiment is shown in Fig. 3a).
Figure 3.
Pax7 immunocomplex has HMT activity and associates with sites of H3K4 methylation. (a) Pax7 or IgG immunoprecipitates from differentiated myotubes (Pax7 negative) or primary myoblasts were incubated with core histones and 3H-SAM. Pax7 immunocomplexes from primary myoblasts were consistently observed to induce a markedly higher level of histone methylation than IgG controls (error bars represent s.e.m.). (b) Fluorography of histones incubated with Pax7 immunocomplexes from primary myoblasts indicated that the methyltransferase activity associated with the Pax7 immunocomplex were directed at histone H3. (c) Western blot analysis with antisera directed against dimethylated H3K4 (H3K42me) and trimethylated H3K4 (H3K43me) indicate that histones incubated with the Pax7 immunocomplexes show a markedly increased level of H3K43me, with a concurrent but lesser increase in H3K42me. (d) IP-western blot analysis revealed that Pax7 was highly associated with chromatin isolated from primary myoblasts using antisera reactive with H3K42me and H3K43me, but not at all with chromatin isolated with antisera reactive with dimethylated H3K9. Full-length scans of western blot data can be found in Supplementary Information S4.
Modifications of the histone tail significantly influence the nature of gene expression26. Methylation of H3K4 marks chromatin in a conformation permissive for transcription, while trimethylated H3K4 is restricted to the 53 promoter and coding regions and is considered to be a definitive marker of active genes27–30. To test the prediction that the Pax7-Wdr5–Ash2L–MLL2 complex specifically directs methylation of H3K4, we repeated the previous experiment and performed SDS–PAGE on the samples. Coomassie staining and fluorography indicated that methyltransferase activity is indeed directed against H3 (Fig. 3b). Using H3K4 modification-specific antibodies, we observed that the Pax7 complex mediated trimethylation and (to a markedly lesser extent) dimethylation of H3K4 (Fig. 3c). These modifications are indicative of active or transcriptionally permissive chromatin27. In contrast, western blots with antibodies specific for dimethylated H3K9 (a marker of repressed chromatin31) showed no increases over background levels in the presence of the Pax7 complex (data not shown).
These data demonstrate that Pax7 recruits a protein complex capable of methylating H3K4. As such, we predicted that Pax7 physically associates with sites of open chromatin that are methylated on H3K4. To test this, we performed immunoprecipitations using methylation-specific antibodies and probed western blots for the presence of Pax7. Pax7 was observed to associate strongly with regions of dimethylated H3K4, and, to a lesser extent, with trimethylated H3K4 (Fig. 3d). No association of Pax7 with dimethylated H3K9 was observed (Fig. 3d).
Intriguingly, certain structural data show that Wdr5 binds directly to the H3 tail with an affinity not affected by the state of lysine-4 methylation22,23, although other data indicate a preference for dimethylated H3K424,25. In neither case is the question addressed of how the HMT complex is recruited to specific target genes. This function must be conferred by the unique component(s) of the overall complex, such as a site-specific transcriptional activator, rather than any of the three core elements21,22,25. Our data indicate that Pax7 provides the missing selective function that targets HMT complexes to specific genes while remaining consistent with either model of Wdr5 function.
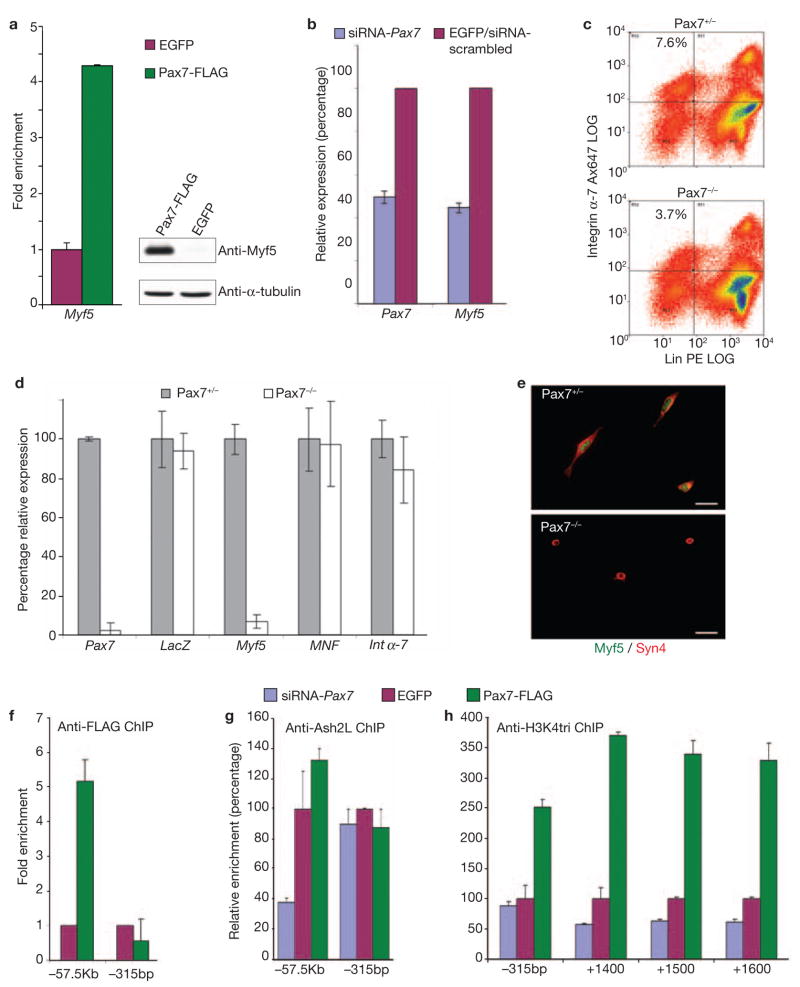
To directly examine the effect of Pax7 on myogenic commitment, we focused on the Pax7 regulation of Myf5 in satellite cells. We proposed that Myf5 would be regulated by Pax7 in satellite cells, would be bound by Pax7 and HMT–complex proteins and would exhibit an altered methylation status. Expression of Pax7–FLAG in satellite cell-derived myoblasts produced a large increase in Myf5 RNA expression, with an exaggerated change at the protein level (Fig. 4a), consistent with our observations in C2C12 myoblasts and those of other groups20. Accordingly, when we knocked down Pax7 expression by 50% using siRNA, Myf5 expression was reduced to approximately 40% of that observed in the scrambled siRNA control cells (Fig. 4b), demonstrating that Myf5 expression is responsive to Pax7 in satellite cell-derived myoblasts. To test this in vivo, we sorted satellite cells from Pax7-null mice. These mice possess reduced numbers of satellite cells (3.7% −/− versus 7.6% +/− Fig. 4c), and were previously shown to have a severely impaired ability to regenerate damaged muscle and maintain viable satellite cell populations3,11–13,15. However, we were able to obtain sufficient numbers of satellite cells from four-week old pups to examine the expression level of Myf5. Initially we examined the levels of LacZ, and two key markers of satellite cells, myocyte nuclear factor and integrin-α7. Expression was equivalent between the heterozygous and homozygous-null populations of cells, indicating that we were indeed sorting satellite cells from the mutant. Strikingly, Myf5 was absent from Pax7-null satellite cells (Fig. 4d). Immunocytochemistry of the sorted cells revealed that the Pax7−/− cells maintained a rounded, non-activated, non-myogenic phenotype (Fig. 4e). Thus, Pax7 is required for the expression of Myf5 and the commitment of satellite cells to the myogenic lineage.
Figure 4.
Pax7 regulates Myf5 expression directly in satellite cell-derived myoblasts. (a) Over-expression of Pax7 resulted in elevated levels of Myf5 RNA and protein. (b) siRNA knockdown of Pax7 in satellite cell-derived myoblasts results in concurrent knockdown of Myf5 expression. (c) FACS sorted satellite cells (Integrin α-7+; Lin: CD31−, CD45−, CD11−, Sca1−) from Pax7−/− mice show complete absence of Myf5 expression via both (d) RNA (n = 3 animals per group, P =: 2.9 × 10−5 for Pax7 and 1.7 × 10−5, for Myf5, * = P < 0.001), and (e) immunofluorescence (Syndecan 4-red, (Syn4) Myf5-green (Myf5), scale bar = 20 μm). Expression of LacZ was assayed as the Pax7 knockout was created via the insertion of a LacZ sequence into the Pax7 locus and as such all satellite cells should be expressing LacZ in place of Pax7. (f, g) ChIP demonstrated that Pax7 was bound directly to the −57.5 kb region of Myf5 in complex with Ash2L (f) and in a site-specific manner (g). The downstream −315 bp loci, which does not contain a binding site and hence did not show any enrichment, is shown for comparison purposes. (h) Antisera directed against H3K4 trimethylation ChIP demonstrated that, accordingly, the coding region of Myf5 showed a marked increase in activation as measured by trimethylation of H3K4 (error bars = s.e.m.). Full-length scans of western blot data can be found in Supplementary Information S4.
A Pax3-paired domain binding site (~57.5 kb upstream of the transcriptional start) that activates the Myf5 gene in primary myoblasts was recently defined32,33. Chromatin immunoprecipitations (performed from Pax7–FLAG and EGFP control satellite cell-derived myoblasts using α-FLAG) revealed that this site is bound by Pax7 (Fig. 4f). To test our model of Pax7 recruiting a HMT complex, we performed ChIP using anti?]-Ash2L antibodies from three satellite cell-derived myoblast populations: Pax7–FLAG, EGFP control, and siRNA–Pax7 knockdown cells. This revealed that, in the presence of high Pax7 (Pax7–FLAG), Ash2L levels on the −57.7 Myf5 promoter were increased by more than 125% compared with controls (EGFP); however, reduced Pax7 levels in myoblasts (siRNA–Pax7) decreased Ash2L binding to this region to 40% of that seen in the controls (Fig. 4g), demonstrating a Pax7-dependent recruitment of the HMT complex to this region.
In keeping with the hypothesis that the recruitment of this complex results in downstream trimethylation of the coding region and gene activation, we performed ChIP experiments using an anti-H3K4 trimethylation antibody. Accordingly, and despite the fact that Myf5 is already activated in myoblasts, H3K4 trimethylation in the immediate 53 and coding regions of Myf5 in myoblasts with ectopic Pax7 expression (Pax7–FLAG) was increased by 250–300% above that seen in EGFP cells, consistent with a propagation of the methylation signal by the HMT complex downstream from the Pax7 binding site. Significantly, in myoblasts where Pax7 expression (and Myf5, Fig. 4b) was reduced (siRNA–Pax7), the level of H3K4 trimethylation was markedly diminished (Fig. 4h).
Taken together, our data support a model whereby specific binding of Pax7 to dimethylated-H3K4 regulatory elements in target genes leads to the recruitment of HMT core complexes, strong H3K4 trimethylation of their regulatory elements, and ensuing activation of gene expression. This is consistent with structural data showing an interaction between the Wdr5–Ash2L core complex and histone H3 (refs 21–25), and answers the question of how this generic complex is recruited to specific genes. It also corroborates other genomic data27–29 indicating that recruitment of this complex and subsequent trimethylation of the 5′ promoter and coding regions result in target gene activation. Finally, it defines a specific role and molecular mechanism for Pax7 in myogenic stem cells1–3,11–16 in regulating Myf5 and other target genes. Together, these experiments indicate that Pax7 enforces satellite cell commitment by recruiting a HMT complex to Myf5, resulting in transcriptional activation. Notably, Pax-family genes are essential for the embryonic specification of diverse tissues; thus, Pax recruitment of HMT complexes could be a conserved mechanism for seeding lineage-specific gene-expression programmes during development.
METHODS
Generation of cell lines
C2C12 myoblasts and 10T1/2 fibroblasts were cultured in DMEM with 10% fetal calf serum and 1% penicillin/streptomycin. Primary myoblasts were isolated from adult C57BL/6 mice and cultured using two previously described methods1,20. Pax7– or Pax3–FLAG were cloned onto the pHAN backbone (with puromycin resistance driven from a distinct SV40 promoter). Control virus expressed puromycin-resistance alone. Additionally, full-length mouse Pax7d was fused in-frame to a C-terminal TAP tag and cloned into the pBRIT retroviral plasmid to make pBRIT–Pax7–CTAP (Fig. 2a). A plasmid comprising only the TAP tag was used as a control. Retrovirus was produced by transient co-transfection as described19; or using Lipofectamine 2000 (Invitrogen) into Phoenix-Eco cells (a gift from the Nolan lab, Stanford University Medical Center). Viral supernatant containing 8 μg ml−1 polybrene was used to infect C2C12 cells. Stable lines were created by antibiotic selection (1.0–1.5 μg ml−1 puromycin; Sigma).
Microarray analysis
Triplicate pools of C2C12 cells stably infected with retro-virus expressing Pax7d, or with empty-control retrovirus were produced. Total RNA was purified from cultured cells using the RNeasy Mini kit (Qiagen) according to the manufacturer’s instructions. Samples were hybridized to MOE430A GeneChips (Affymetrix,) at the Ottawa Genome Centre (Ontario, Canada) and analysed as previously described19. Criteria used to derive Tables 1 and S1 included: log-fold change of greater than one (that is, 2-fold cut-off); significant change (Increase or Marginal Increase call); detectable expression above background (non-Absent call) for the higher signal, and consistency between replicate samples. Raw microarray data is available from the StemBase, Ontario Genomics Innovation Centre (http://www.scgp.ca:8080/StemBase/) as experiment E102 (samples S137, S310) and from the Gene Expression Omnibus, National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/geo/) under series accession no. GSE3224 (GSM72628, -30, -32, -34…6).
Primers
PCR primers for all RT- and real-time PCR were designed using the online Primer3 software (http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www.cgi). All primer sequence specifications are provided in Supplementary Information, Table S2.
Real-time RT-PCR
Total RNA was isolated as described for microarray samples and used as a template for first-strand reverse transcription (RT) using the RNA PCR Core kit (Perkin Elmer) with random hexamer primers. SYBR Green real-time PCR reactions were performed in triplicate using an MX4000 PCR machine (Stratagene) with fold-change normalized against GAPDH β-Actin, or tubulin. Primer specificity was validated by denaturation curve analysis (55–94 °C) and direct sequencing of the PCR products. Amplification-curve plotting and calculation of Ct values were performed using the MX4000 software (v4.20; Stratagene), with further calculations performed using Microsoft Excel. Each experiment was performed independently on at least three occasions.
Tandem affinity purification
Whole cell extracts were prepared from Pax7–CTAP and HisFLAG–tag C2C12 cells, corresponding to approximately 200 × 100 mm plates. Cells were homogenized in 10 mM Tris-Cl pH 7.9, 0.1 M NaCl, 1.5 mM MgCl2, 0.18% NP-40, on ice. One volume of 50 mM Tris-Cl pH 7.9, 0.6 M NaCl, 1.5 mM MgCl2, 25% glycerol was then added, and the mixture was homogenized again before being treated with benzonase nuclease (Novagen) for 30 min at 4 °C. The lysate was centrifuged, dialysed against 10 mM Tris-Cl pH 7.9, 0.1 M NaCl, 0.1 mM EDTA, 10% glycerol and cleared by centrifugation. Pax7–CTAP or HisFLAG-tag complexes were immunoprecipitated with M2-agarose (Sigma) at 4 °C then eluted with TEV protease (Invitrogen) and FLAG peptide (Sigma) in 20 mM Tris-Cl pH 7.9, 0.1 M NaCl, 0.1 mM EDTA, 0.1% NP40. The eluate was incubated with ProBond resin (Invitrogen) and complexes eluted with 100 mM EDTA, dialysed against 10 mM Tris-Cl pH 7.9, 0.1 M NaCl, 0.1 mM EDTA, 10% glycerol and concentrated on a Centricon column (Amicon).
Mass spectrometry
Samples processed for MALDI-TOF were subjected to SDS–PAGE and silver-stained as described previously34. Bands were excised directly from the gel and analysed at the Ottawa Genome Centre. MALDI-MS/MS spectra were acquired using a QSTAR XL tandem mass spectrometer (ABI/Sciex) with a MALDI-2 source, α-Cyano-4-hydroxycinnamic acid (Agilent, Palo Alto, CA) matrix and Analyst QS version 1.1, build 9865. Spectra were searched against the NCBI database using Mascot daemon version 2.0.5 on an in-house Mascot server version 2.0.04.
Co-immunoprecipitation analysis
Mouse primary myoblasts were used for immunoprecipitation analysis. For differentiation protocols, cells were maintained for 7 days in DMEM + 2% horse serum. Nuclear extracts were produced as follows: cells were re-suspended in hypotonic lysis buffer (10 mM Hepes pH 7.6, 1.5 mM MgCl2, 10 mM KCl, 0.5 mM DTT and protease inhibitors), lysed using a 27-gauge needle and the nuclei collected by centrifugation. Nuclei were re-suspended in 20 mM Hepes pH 7.6, 1.5 mM MgCl2, 420 mM NaCl, 2 mM DTT, 0.2 mM EDTA, 25% glycerol, plus protease inhibitors, and lysed using a 27-gauge needle. The nuclear suspension was agitated for 60 min at 4 °C, cleared by centrifugation, and adjusted to 20 mM Hepes pH 7.6, 1.5 mM MgCl2, 150 mM NaCl, 2 mM DTT, 0.2 mM EDTA, 20% glycerol, plus protease inhibitors, for subsequent use. Primary antibodies (concentrated α-Pax7 hybridoma supernatant, DSHB and α-Wdr5, kindly donated by Winship Herr; α-Ash2L, kindly donated by Marjorie Brand; control normal IgG, (Upstate) were either incubated at a concentration of 1–2 μg 500 μg−1 nuclear extract overnight at 4 °C, before being collected with Protein G/A; or alternatively, were cross-linked to Protein G/A via DMP before incubation with nuclear extract. Immuno-complexes were washed and subjected to SDS–PAGE and western blotting with the antibodies listed above and α-Erk1/2 (Chemicon).
HMT activity assay
Mouse primary myoblast nuclear extracts were immuno-precipitated with α-Pax7 or control IgG as described above and incubated with 2 μg core histones (Upstate) in the presence of 3H-adenosyl-L-methionine (PerkinElmer), for 60 mins at 30 °C, spotted onto P81 phosphocellulose squares (Upstate), and read in a Beckman LS 6500 scintillation counter. Counts per minute for the α-Pax7 immunoprecipitations were normalized against the background counts observed with the IgG immunoprecipitations. Three independent experiments were performed and each sample was read in triplicate. In complementary experiments the HMT assay samples were subjected to SDS–PAGE and treated in one of two ways: (1) Gels were stained in 0.25% Coomassie Brilliant Blue R250, 50% methanol, 10% acetic acid; de-stained in 50% methanol, 10% acetic acid, amplified with Enlightning (NEN Life Sciences), dried and exposed for fluorography; (2) Gels were transferred to nitrocellulose membrane and subjected to western blotting with antibodies raised against dimethylated H3K4, trimethylated H3K4 and dimethylated H3K9 (Upstate).
ChIP analysis
Pax7–FLAG, Ash2L and H3K4 trimethyl protein-DNA complexes were crosslinked with 1% formaldehyde and sonicated. Processing of samples was performed according to the ChIP kit manufacturer’s protocols (Upstate). DNA was recovered using PCR purification columns (Invitrogen). The immunoprecipitated DNA was subjected to real-time PCR assays with each primer pair, and normalized against primers for GAPDH as a control locus representing a control fragment that was immunoprecipitated non-specifically to produce robust and reproducible results. This strategy is validated by noting that, for example, Pax7–FLAG samples showed little or no enrichment at flanking loci.
Each DNA sample was analysed by real-time PCR in triplicate, in at least two independent experiments. PCR specificity was validated by denaturation curve analysis and direct sequencing of the products.
Additional methods
Information on western blots, Pax-7 domain deletion construction, histone pull-down assays, FACS of satellite cells, siRNA transfection and immunocytochemistry is available in Supplementary Information S5.
Supplementary Material
Acknowledgments
The authors are indebted to Doug Borris and Murray Smith for mass spectrometry analysis; Jeff Baker and Joyce Li for technical assistance; and to Marjorie Brand, Jennifer McCann, Mark Gillespie and Dave Picketts for critical input. This work was supported by grants to M.A.R. from the National Institutes of Health, the HHMI, the Canadian Institutes of Health Research, the Muscular Dystrophy Association and the CRC Program.
References
- 1.Kuang S, Kuroda K, Le Grand F, Rudnicki MA. Asymmetric self-renewal and commitment of satellite stem cells in muscle. Cell. 2007;129:999–1010. doi: 10.1016/j.cell.2007.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Montarras D, et al. Direct isolation of satellite cells for skeletal muscle regeneration. Science. 2005;309:2064–2067. doi: 10.1126/science.1114758. [DOI] [PubMed] [Google Scholar]
- 3.Seale P, et al. Pax7 is required for the specification of myogenic satellite cells. Cell. 2000;102:777–786. doi: 10.1016/s0092-8674(00)00066-0. [DOI] [PubMed] [Google Scholar]
- 4.Steward MM, et al. Molecular regulation of H3K4 trimethylation by ASH2L, a shared subunit of MLL complexes. Nature Struct Mol Biol. 2006;13:852–854. doi: 10.1038/nsmb1131. [DOI] [PubMed] [Google Scholar]
- 5.Wysocka J, Myers MP, Laherty CD, Eisenman RN, Herr W. Human Sin3 deacetylase and trithorax-related Set1/Ash2 histone H3-K4 methyltransferase are tethered together selectively by the cell-proliferation factor HCF-1. Genes Dev. 2003;17:896–911. doi: 10.1101/gad.252103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Roguev A, et al. The Saccharomyces cerevisiae Set1 complex includes an Ash2 homologue and methylates histone 3 lysine 4. Embo J. 2001;20:7137–7148. doi: 10.1093/emboj/20.24.7137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yokoyama A, et al. Leukemia proto-oncoprotein MLL forms a SET1-like histone methyltransferase complex with menin to regulate Hox gene expression. Mol Cell Biol. 2004;24:5639–5649. doi: 10.1128/MCB.24.13.5639-5649.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Milne TA, et al. MLL associates specifically with a subset of transcriptionally active target genes. Proc Natl Acad Sci USA. 2005;102:14765–14770. doi: 10.1073/pnas.0503630102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Milne TA, et al. Menin and MLL cooperatively regulate expression of cyclin-dependent kinase inhibitors. Proc Natl Acad Sci USA. 2005;102:749–754. doi: 10.1073/pnas.0408836102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hughes CM, et al. Menin associates with a trithorax family histone methyltransferase complex and with the hoxc8 locus. Mol Cell. 2004;13:587–597. doi: 10.1016/s1097-2765(04)00081-4. [DOI] [PubMed] [Google Scholar]
- 11.Relaix F, Rocancourt D, Mansouri A, Buckingham M. A Pax3/Pax7-dependent population of skeletal muscle progenitor cells. Nature. 2005;435:948–953. doi: 10.1038/nature03594. [DOI] [PubMed] [Google Scholar]
- 12.Gros J, Manceau M, Thome V, Marcelle C. A common somitic origin for embryonic muscle progenitors and satellite cells. Nature. 2005;435:954–958. doi: 10.1038/nature03572. [DOI] [PubMed] [Google Scholar]
- 13.Kassar-Duchossoy L, et al. Pax3/Pax7 mark a novel population of primitive myogenic cells during development. Genes Dev. 2005;19:1426–1431. doi: 10.1101/gad.345505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Relaix F, et al. Pax3 and Pax7 have distinct and overlapping functions in adult muscle progenitor cells. J Cell Biol. 2006;172:91–102. doi: 10.1083/jcb.200508044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kuang S, Charge SB, Seale P, Huh M, Rudnicki MA. Distinct roles for Pax7 and Pax3 in adult regenerative myogenesis. J Cell Biol. 2006;172:103–113. doi: 10.1083/jcb.200508001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bennicelli JL, Advani S, Schafer BW, Barr FG. PAX3 and PAX7 exhibit conserved cis-acting transcription repression domains and utilize a common gain of function mechanism in alveolar rhabdomyosarcoma. Oncogene. 1999;18:4348–4356. doi: 10.1038/sj.onc.1202812. [DOI] [PubMed] [Google Scholar]
- 17.Zammit PS, et al. Muscle satellite cells adopt divergent fates: a mechanism for self-renewal? J Cell Biol. 2004;166:347–357. doi: 10.1083/jcb.200312007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Berkes CA, Tapscott SJ. MyoD and the transcriptional control of myogenesis. Semin Cell Dev Biol. 2005;16:585–595. doi: 10.1016/j.semcdb.2005.07.006. [DOI] [PubMed] [Google Scholar]
- 19.Ishibashi J, Perry RL, Asakura A, Rudnicki MA. MyoD induces myogenic differentiation through cooperation of its NH2- and COOH-terminal regions. J Cell Biol. 2005;171:471–482. doi: 10.1083/jcb.200502101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sabourin LA, Girgis-Gabardo A, Seale P, Asakura A, Rudnicki MA. Reduced differentiation potential of primary MyoD−/− myogenic cells derived from adult skeletal muscle. J Cell Biol. 1999;144:631–643. doi: 10.1083/jcb.144.4.631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dou Y, et al. Regulation of MLL1 H3K4 methyltransferase activity by its core components. Nature Struct Mol Biol. 2006;13:713–719. doi: 10.1038/nsmb1128. [DOI] [PubMed] [Google Scholar]
- 22.Ruthenburg AJ, et al. Histone H3 recognition and presentation by the WDR5 module of the MLL1 complex. Nature Struct Mol Biol. 2006;13:704–712. doi: 10.1038/nsmb1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Couture JF, Collazo E, Trievel RC. Molecular recognition of histone H3 by the WD40 protein WDR5. Nature Struct Mol Biol. 2006;13:698–703. doi: 10.1038/nsmb1116. [DOI] [PubMed] [Google Scholar]
- 24.Han Z, et al. Structural basis for the specific recognition of methylated histone H3 lysine 4 by the WD-40 protein WDR5. Mol Cell. 2006;22:137–144. doi: 10.1016/j.molcel.2006.03.018. [DOI] [PubMed] [Google Scholar]
- 25.Wysocka J, et al. WDR5 associates with histone H3 methylated at K4 and is essential for H3 K4 methylation and vertebrate development. Cell. 2005;121:859–872. doi: 10.1016/j.cell.2005.03.036. [DOI] [PubMed] [Google Scholar]
- 26.Martin C, Zhang Y. The diverse functions of histone lysine methylation. Nature Rev Mol Cell Biol. 2005;6:838–849. doi: 10.1038/nrm1761. [DOI] [PubMed] [Google Scholar]
- 27.Bernstein BE, et al. Methylation of histone H3 Lys 4 in coding regions of active genes. Proc Natl Acad Sci USA. 2002;99:8695–700. doi: 10.1073/pnas.082249499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liang G, et al. Distinct localization of histone H3 acetylation and H3-K4 methylation to the transcription start sites in the human genome. Proc Natl Acad Sci USA. 2004;101:7357–7362. doi: 10.1073/pnas.0401866101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Santos-Rosa H, et al. Active genes are tri-methylated at K4 of histone H3. Nature. 2002;419:407–411. doi: 10.1038/nature01080. [DOI] [PubMed] [Google Scholar]
- 30.Bernstein BE, et al. Genomic maps and comparative analysis of histone modifications in human and mouse. Cell. 2005;120:169–181. doi: 10.1016/j.cell.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 31.Rea S, et al. Regulation of chromatin structure by site-specific histone H3 methyl-transferases. Nature. 2000;406:593–599. doi: 10.1038/35020506. [DOI] [PubMed] [Google Scholar]
- 32.Bajard L, et al. A novel genetic hierarchy functions during hypaxial myogenesis: Pax3 directly activates Myf5 in muscle progenitor cells in the limb. Genes Dev. 2006;20:2450–2464. doi: 10.1101/gad.382806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Buchberger A, Freitag D, Arnold HH. A homeo-paired domain-binding motif directs Myf5 expression in progenitor cells of limb muscle. Development. 2007;134:1171–1180. doi: 10.1242/dev.02798. [DOI] [PubMed] [Google Scholar]
- 34.Shevchenko A, Wilm M, Vorm O, Mann M. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal Chem. 1996;68:850–858. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.