Abstract
Ectopic expression of transcription factors can reprogram somatic cells to a pluripotent state. However, most of the studies used skin fibroblasts as the starting population for reprogramming, which usually take weeks for expansion from a single biopsy. We show here that induced pluripotent stem (iPS) cells can be generated from adult human adipose stem cells (hASCs) freshly isolated from patients. Furthermore, iPS cells can be readily derived from adult hASCs in a feeder-free condition, thereby eliminating potential variability caused by using feeder cells. hASCs can be safely and readily isolated from adult humans in large quantities without extended time for expansion, are easy to maintain in culture, and therefore represent an ideal autologous source of cells for generating individual-specific iPS cells.
Keywords: differentiation, pluripotency, reprogramming
Induced pluripotent stem (iPS) cells have been successfully derived from somatic cells with ectopic expression of transcription factors Oct4, Sox2, and either Klf4 and c-MYC (1, 2) or Nanog and Lin28 (3). More importantly, the generation of patient-specific and disease-specific (4) iPS cells has the potential to greatly impact the future of regenerative medicine, drug development, as well as our basic understanding of specific disease mechanisms. With the fast progress in this field, different reprogramming strategies have been developed, including using nonintegrating adenoviruses (5), two factors with small molecules (6), reprogramming with a polycistronic cassette containing all four factors (7, 8), excisable transposons (9, 10), and more recently, virus-free plasmid (11, 12). However, the majority of these studies use skin fibroblasts as the parental cells, which usually requires at least 4 weeks to expand from a single skin biopsy to get enough starting cells for reprogramming. The reprogramming efficiency of adult human fibroblasts using “Yamanaka” four factors (Oct4, Sox2, Klf4, and c-MYC) is also still very low at under 0.01% (1, 13–16). Although Zhao et al. reported improved reprogramming efficiency of adult human fibroblasts using p53 and UTF1 siRNAs (14), inhibition of p53, a potent tumor suppressor, may cause the reprogrammed iPS cells to be less safe for future application. Moreover, reprogramming adult human fibroblasts is a lengthy process that requires a minimum of 4 weeks to obtain expandable iPS colonies after transduction. Although a recent study reported ≈1% reprogramming efficiency using neonatal/juvenile human keratinocytes, the reprogramming efficiency of adult human keratinocytes is still unclear (16).
Another critical consideration within the iPS cell field is the more practical concern of accessibility of parental cells for reprogramming. In humans, many cell types such as hepatocytes and neural progenitors are not easily accessible without performing highly invasive procedures. Moreover, some sources of cells are rare and not available in large quantities, making them poor candidates for reprogramming in a clinical setting. For example, neural stem cells that can be reprogrammed with only a single factor, Oct4 (17), are a rare population and technically difficult to obtain.
Human adipose stem cells (hASCs) are a heterogeneous group of multipotent progenitor cells that can be readily derived from adipose tissue of adult humans in very large quantities by lipoaspiration (18, 19). These cells are multipotent stem cells and can differentiate into adipogenic, osteogenic, chondrogenic, and myogenic cell lineages (18, 20). hASCs may therefore possess a different genetic and epigenetic landscape that is more ideal for reprogramming than the terminally differentiated fibroblast cells. Here we report that hASCs obtained from four 40- to 65-year-old individuals can be reprogrammed into iPS cells. The appearance of embryonic stem (ES) cell-like colonies from reprogramming hASCs was ≈2-fold faster and ≈20-fold more efficient than from reprogramming human IMR90 fibroblasts using the Yamanaka four factors. Furthermore, iPS cells can be readily derived from hASCs on feeder-free surfaces using Matrigel-coated tissue culture dishes, thereby reducing the variability of reprogramming processes that may caused by using mouse feeder cells. Our results indicate that hASCs are an easily obtainable cell source that can be more efficiently reprogrammed into adult, individual-specific iPS cells.
Results
In our reprogramming experiments, we first isolated hASCs via lipoaspiration from four individuals between the ages of 40 and 65. The human fibroblast cell line IMR90 was used in parallel to compare the efficiency and length of time for the reprogramming process. Cells were first transduced with individual lentiviruses containing human Oct4, Sox2, Klf4, and c-MYC (Fig. S1) at a 1:1:1:1 ratio on day 0. Transduction was repeated on day 2 using the same batch of all four lentiviruses. The efficiency for each lentiviral transduction was greater than 50%. On day 3 after the first transduction, 50,000 cells were transferred onto mouse embryonic fibroblast (MEF) feeder layer, with the culture medium switched from the hASC growth medium to human embryonic stem (hES) cell growth medium mTeSR-1. We observed many small colonies of non-ES cell-like cells beginning on day 4 that had morphologies similar to the “background colonies” or “early colonies” described in previous studies (1, 21). These non-ES cell-like colonies expanded rapidly but lacked the typical characteristics of hES cells, such as defined boundaries and high nuclear-to-cytoplasm ratio within individual cells (Fig. S2).
From day 12–13, clearly recognizable, tightly packed colonies with morphologies similar to hES cells appeared. Previous studies have reported the isolation of human iPS cells based on both cell morphology and immunostaining of living cells with the embryonic surface marker TRA-1–81 (21). In our study, we compared the immunostaining of the hES cells and the reprogrammed ES cell-like colonies by TRA-1–81 and TRA-1–60, and found that TRA-1–60 showed comparable staining with that of TRA-1–81 for both hES cells and the ES cell-like colonies from reprogramming of hASCs (Fig. S3). TRA-1–60 has also been used in combination with endogenous Nanog expression to determine the success of reprogramming (13). We thus moved along using TRA-1–60 as the surface marker together with typical ES cell-like morphology to track the progression of the putative iPS colonies. We consistently observed TRA-1–60 positive colonies that appeared as early as on day 10, although these colonies were still too small to be recognized with brightfield microscopy. The number and size of the TRA-1–60 positive colonies increased over time after day 10. From day 15 to day 16, large ES cell-like colonies containing ≈400–500 cells could be isolated mechanically and transferred onto Matrigel for further expansion.
For each hASC line reprogrammed, we consistently observed ≈100 TRA-1–60 positive, ES cell-like colonies out of 50,000 cells on day 16 (Table 1). The number of non-ES cell-like colonies varied in each reprogramming and was higher than the number of ES cell-like colonies. Some non-ES cell-like colonies overgrew over time and detached from the feeder cells. Previous studies have reported calculating reprogramming efficiency based solely on the ES cell-like morphologies of the observed colonies with a success rate of ≈90–100% (6, 16). However, we did observe a few colonies that had ES cell-like morphologies but were negative for TRA-1–60/TRA-1–81 and some non-ES cell-like cell clumps with nonspecific TRA-1–60/TRA-1–81 staining (Fig. S4). Therefore in this study, we combined the ES cell-like morphology and TRA-1–60 immunostaining of living cells to improve the accuracy of determinations of our reprogramming efficiency. We believe that the combination of TRA-1–60 expression and ES cell-like morphology represents a more rigorous criteria, or at least comparable with previous criteria, for calculating reprogramming efficiency. Indeed, >90% TRA-1–60 positive ES cell-like colonies that we tested on day 18 posttransduction expressed the late pluripotency marker Nanog (Fig. S5), suggesting a high success rate of identifying reprogrammed colonies. Based on this method, the calculated efficiency of reprogramming was ≈0.2%. In contrast, reprogramming IMR90 cells carried out under the same conditions resulted in only approximately two to four recognizable TRA-1–60 positive colonies out of 50,000 cells 28 days after transduction. This low reprogramming efficiency using IMR90 fibroblasts is consistent with and comparable to the results reported by previous studies using Yamanaka four factors to reprogram human fibroblasts (1, 2, 14–16). To our knowledge, the highest efficiency of reprogramming adult human fibroblast cells with the Yamanaka four factors described in current literature is only ≈0.01%. Based on the appearance and number of TRA-1–60 positive ES cell-like colonies, our results indicate that reprogramming adult hASCs is more efficient and faster than reprogramming adult human fibroblasts.
Table 1.
TRA-1–60-positive ES-like colonies
| Cell line | Day 16 on MEFs (out of 5 × 104 cells) | Day 18 on Matrigel (out of 2 × 105 cells) |
|---|---|---|
| hASC-I1 | 73 ± 12 (n = 6) | 22 ± 5 (n = 5) |
| hASC-I2 | 96 ± 15 (n = 5) | 57 ± 10 (n = 4) |
| hASC-I3 | 110 ± 12 (n = 4) | 40 ± 7 (n = 4) |
| hASC-I4 | 87 ± 7 (n = 4) | 23 ± 4 (n = 3) |
| IMR90 | 2∼4 (n = 4) | 0 (n = 6) |
We also attempted to generate iPS cells from hASCs under feeder-free conditions. We seeded 2 × 105 hASCs from each of the four individuals directly on the Matrigel-coated surface within one well of a six-well tissue culture dish and transduced the cells with all four factors. TRA-1–60 positive colonies appeared as early as on day 12. Recognizable TRA-1–60 positive ES cell-like colonies surrounded by cobblestone-like cells originating from hASCs were observed as early as on day ≈13–14 under brightfield microscopy. Large TRA-1–60 positive, ES cell-like colonies could be visualized and were transferred to new Matrigel-coated dishes for further expansion from day ≈18–20. The developmental progression of a typical ES cell-like colony on Matrigel is shown in Fig. 1. We consistently obtained ≈20–70 TRA-1–60 positive ES cell-like colonies (Table 1) out of 2 × 105 seeded hASCs on Matrigel without feeder cells, which accounts for an efficiency of ≈0.01–0.03%. However, the initial density of the seeded cells seemed to be critical for successful reprogramming, because ES cell-like colonies were not observed when less than 1 × 105 hASCs were initially seeded. Because the hASC-derived ES cell-like colonies are morphologically indistinguishable from hES cells, we thus defined them as “hASC-iPS cells.”
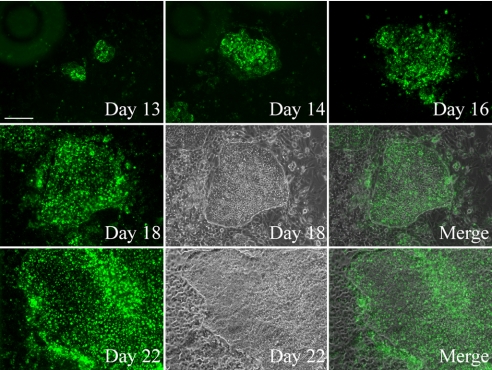
Fig. 1.
Tracking the appearance and growth of an ES cell-like colony by immunostaining the living cells with TRA-1–60 in feeder-free reprogramming. Adult hASCs were seeded on Matrigel-coated surface without MEF feeder cells. The living cells were stained repetitively with TRA-1–60 monoclonal antibodies and AlexaFluor 488 secondary antibodies over the indicated period. Day 18 and 22 images are presented both in fluorescent and phase contrast microscopy. Note that TRA-1–60 expression was specific for the ES cell-like colony. (Scale bar, 100 μm.)
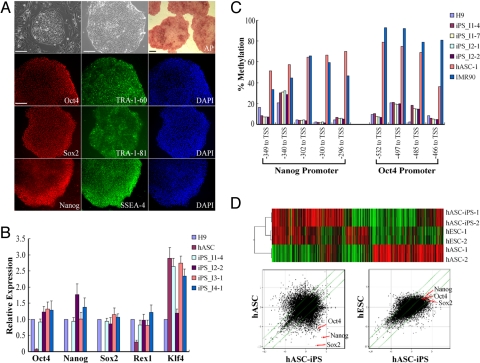
We next characterized the hASC-iPS cells that were isolated from the large ES cell-like TRA-1–60 positive colonies (≈>500–600 cells/colony) generated in feeder-free conditions. We chose to use feeder-free hASC-iPS cell lines for further analysis to eliminate the potential contaminating factors from feeder cells and also to ensure that our analysis was consistent among the different cell lines. We initially picked eight single colonies from feeder-free reprogramming hASCs of individual 1, and six out of the eight picked single colonies were successfully expanded in feeder-free culture condition. For individual 2, 3, and 4, we picked six single colonies from each individual. All total 18 colonies, but one was successfully expanded and cultured in feeder-free condition for extended time (Table S1). Immunostaining of different feeder-free hASC-iPS cell lines with alkaline phosphatase (AP), Oct4, Sox2, Nanog, TRA-1–60, TRA-1–81, and SSEA-4 indicated they are positive for typical hES cell markers (Fig. 2A). The expression level of several pluripotency genes in hASC-derived iPS cells was also analyzed by quantitative PCR (Fig. 2B). Of the four lines of hASC-derived iPS cells analyzed, Oct4, Sox2, Nanog, and Rex1 were expressed at comparable levels with those of H9 hES cells. In contrast, nonreprogrammed hASCs showed very low or no expression of these genes. Overall, our results indicate that the pluripotency gene expression level in hASC-derived iPS cells is similar with those in hES cells. Interestingly, the expression level of Klf4, one of the reprogramming factors, was found to be ≈3-fold higher in hASCs than in H9 hES cells (Fig. 2B). The hASC-iPS cells also had a normal karyotype after extended culture for 3 months with 46 chromosomes and no translocations (Fig. S6), indicating maintenance of chromosome stability overtime.
Fig. 2.
Characterization of hASC-iPS cells. (A) Immunostaining of hASC-iPS cell colonies with common hES cell markers. The two phase contrast microscopies show a typical hASC-iPS cell colony growing on MEF feeder cells and feeder-free Matrigel surface, respectively. (Scale bars, 100 μm.) (B) Quantitative-PCR analyzing pluripotency gene expression level within hASCs and hASC-iPS cells relative to those in H9 hES cells. iPS_I1–4 denotes iPS cell line #4 derived from individual 1. (C) Bisulphite pyrosequencing measuring methylation status within the promoter region of Oct4 and Nanog genes in H9 hES cells, hASC-iPS cells, hASCs, and IMR90 cells. TSS, transcription start site. (D) Microarray data comparing global gene expression profiles of hASCs, hASC-iPS cells, and hES cells. Upper panel, heat map and hierarchical clustering analysis by Pearson correlation showing hASC-iPS cells are similar to hES cells and distinct from hASCs. Lower panel, scatter plots comparing global gene expression patterns between hASCs, hASC-iPS cells, and hES cells. Highlighted are the pluripotency genes Oct4, Sox2, and Nanog (red arrows). The green diagonal lines indicated linear equivalent and 5-fold changes in gene expression levels between paired samples.
The promoter regions of pluripotency genes in reprogrammed somatic cells are often demethylated, causing increased expression of downstream genes. We therefore analyzed the methylation status of the Oct4 and Nanog promoter regions of hASCs and hASC-iPS cells by quantitative bisulphite pyrosequencing. All of the tested hASC-iPS cell lines shared a hypomethylation pattern similar to that of hES cells, whereas hASCs showed prominent methylation at these loci similar to that of IMR90 fibroblasts (Fig. 2C). These results demonstrate epigenetic remodeling of the Oct4 and Nanog promoters within the hASC-derived iPS cells and are indicative of successful reprogramming.
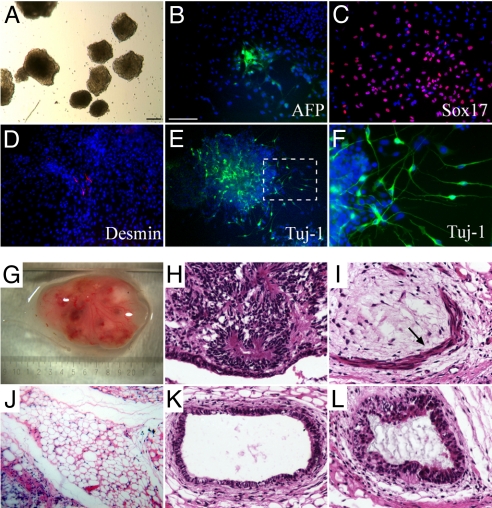
To further compare hASCs, hASC-iPS cells, and hES cells, whole genome expression profiling by microarray analysis was performed. hASC-iPS cells showed a high degree of similarity in their gene expression patterns and close Pearson correlation values with those of human ES cells, and were distinct from hASCs (Fig. 2D and Fig. S7). To demonstrate pluripotency of our hASC-iPS cells, we performed both in vitro (EB formation) and in vivo (teratoma formation) differentiation assays. Two individual lines of hASC-iPS cells from two patients were tested, and each readily differentiated into derivatives of the three embryonic germ layers in vitro (Fig. 3 A–F). We also injected eight different lines of hASC-iPS cells from the four human patients (two lines from each individual) into the dorsal flanks of immunodeficient athymic mice. From all of the eight lines injected, teratoma-like masses containing tissues of all three embryonic germ layers were observed 7–8 weeks after injection (Fig. 3 G–L and Fig. S8).
Fig. 3.
hASC-iPS cells are pluripotent. (A) hASC-iPS cells form EBs and can differentiate to cells of (B and C) endoderm [α-fetoprotein (AFP) and sox17], (D) mesoderm (desmin), and (E and F) ectoderm (Tuj-1 positive motor neurons) lineages. (F) Represents the enlarged view of the boxed area in (E). (Scale bars, 100 μm.) (G–L) Upon injection into nude mice, hASC-iPS cells form (G) teratoma in vivo, which contains tissues of all three embryonic germ layers, such as (H) neural epithelium (ectoderm), (I) smooth muscle (arrow) and (J) adipose tissue (mesoderm), and (K) gut epithelium and (L) respiratory epithelium (endoderm).
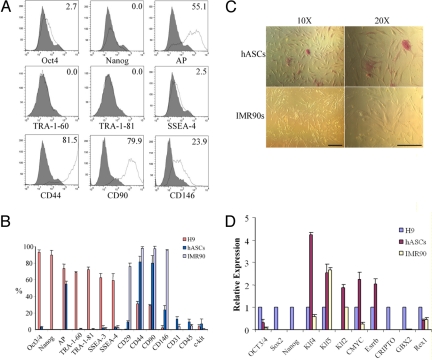
To understand the factors that may contribute to the faster and more efficient reprogramming of hASCs relative to IMR90 human fibroblasts, we analyzed the expression of a list of pluripotency and surface markers in hASCs and compared with those of hES cells and IMR90s. Fluorescence-activated cell sorting (FACS) analysis indicated that ≈55% of hASCs (P0 cells) were positive for the early pluripotency marker AP (Fig. 4 A and B), which agreed with a previous report that a subpopulation of multipotent adipose derived stromal cells express AP (22). Staining the AP activity of in vitro cultured hASCs confirmed that some of the hASCs (heterogeneous in nature) expressed AP at various level (Fig. 4C). In contrast, IMR90 cells did not express any AP activity as indicated by FACS analysis and AP staining (Fig. 4 B and C). This result suggested that a subpopulation of multipotent hASCs already have unique stem cell properties that significantly different from the unipotent fibroblast cells.
Fig. 4.
Comparison of pluripotency and stem cell marker expression in hASCs with those in hES cells and IMR90 cells. (A) Representative histograms of FACS analysis showing hASCs expressed AP and mesenchymal stem cell markers CD44, CD90, and CD146 but not any of the pluripotency markers, such as Oct4, Nanog, TRA-1–60, TRA-1–81, and SSEA-4. Numbers indicate percent of positive cells that expressing each respective marker. (B) Quantitative analysis of cell markers expression in hASCs, H9 hES cells, and IMR90 cells by FACS. (C) AP staining of hASCs and IMR90s cultured in dish. Some hASCs express high AP activity (upper panels) while IMR90s (lower panels) did not. (Scale bars, 100 μm.) (D) Quantitative-PCR analysis of expressions level of pluripotency genes and reprogramming factors in hASCs and IMR90 cells relative to those in H9 hES cells. Note that Klf2 data were normalized to that of IMR90 cells.
FACS analysis also indicated that individual hASCs expressed mesenchymal stem cell markers CD29, CD44, CD90, and CD146 (Fig. 4 A and B). hASCs did not express any of the pluripotency markers Oct4, Nanog, TRA-1–60, TRA-1–81, SSEA-3, and SSEA-4 (Fig. 4 A and B). Immunocytochemistry confirmed that individual hASCs did not express these pluripotency markers (Fig. S9). Interestingly, SSEA-3 was expressed when “colony-forming units-fibroblasts” (CFU-F) were formed (Fig. S9), but not in individual hASCs. We observed no pluripotency marker expression in IMR90 fibroblasts (Fig. 4B). Quantitative-PCR analysis of the expression level of certain pluripotency genes and reprogramming factors indicated that Klf4 (≈8-fold), Klf2 (≈2-fold), Esrrb (not detected in IMR90s), and c-MYC (≈9-fold) were expressed at a higher level in hASCs than those in IMR90s (Fig. 4D). Klf5 was expressed at a similar level in both hASCs and IMR90s, and was ≈2- to 3-fold higher than in hES cells (Fig. 4D). Klf4, Klf5, and Klf2 are the core Klf protein circuitry that regulates self-renewal of ES cells and Nanog expression (23). c-MYC itself is one of the reprogramming factors. Thus, these results overall indicated a clear difference at the gene expression level between hASCs and human fibroblasts.
Discussion
We have generated human iPS cells from hASCs isolated from adult human patients (between the ages of 40–65) with a faster speed and higher efficiency than comparable studies targeting adult human fibroblasts using Yamanaka four factors. Furthermore, we show that human iPS cells can be readily generated under feeder-free conditions using adult hASCs, which reduces the variability of reprogramming associated with using mouse feeder cells. hASC-derived iPS cells express ES cell markers and genes associated with pluripotency at similar levels to hES cells and are morphologically indistinguishable from their hES cell counterparts. Hypomethylation patterns within the Oct4 and Nanog promoter regions and global mRNA expression patterns are also similar to hES cell profiles. hASC-derived iPS cells can differentiate into cell types belonging to all three germ layers both in vitro and in vivo, indicating they are true pluripotent cells.
As stated previously, hASCs are progenitor cells capable of differentiating into multiple lineages, including osteogenic, myogenic, and adipogenic fates. Because hASCs retain this plasticity with regard to differentiation, it is likely that these cells have an epigenomic reulgatory pattern that is closer to pluripotent cells compared to terminally differentiated fibroblast cells. Thus, the unique epigenetic landscape of hASCs may present fewer barriers for reprogramming, resulting in higher efficiency and faster generation of iPS cells. Indeed, FACS analysis (Fig. 4 A and B) and AP staining (Fig. 4C) indicated that hASCs express AP activities, which is proposed as a most reliable early pluripotency marker of ES cells (24). Our results are consistent with a previous study showing that a subpopulation of CD146+, CD34−, CD45−, CD56− cells isolated from human adipose tissue express strong AP activity and are multipotent (22). Thus, this unique property of hASCs clearly differs from the unipotent fibroblast cells without AP activity and may lead to higher efficiency of and faster reprogramming. Furthermore, quantitative-PCR (Figs. 2B and 4D) and microarray (Fig. 2D) results show consistently high Klf4 expression within hASCs relative to not only fibroblasts, but also hES cells. Compared to IMR90 fibroblasts, hASCs also express higher level of pluripotency genes Klf2 and Esrrb, as well as the reprogramming factor c-MYC. hASCs also express Klf5 at a similar level with IMR90s. It is not a surprise to see relatively high Klf5 expression in IMR90s, because KLF5 has been shown to promote cell proliferation (25, 26) and present abundantly in epithelial cells (27). Klf4, Klf5, and Klf2 are the core KLF protein circuitry with redundant function in regulating self-renewal of ES cells and Nanog expression (23). Thus, the high endogenous expression level of Klf4, Klf2, Klf5, Esrrb, and c-MYC likely contributes toward the efficiency and speed by which hASCs can be reprogrammed.
Using hASCs as the parental cells for reprogramming has several advantages over other cell types such as neural stem cells, liver cells, and skin fibroblasts. First, the lipoaspiration procedure for isolating hASCs is relatively simple, fast, and safe. Second, it is easy to obtain a large quantity of hASCs as the starting population for reprogramming after a single lipoaspiration operation. Millions of hASCs can be derived on the same day of lipoaspiration, and the reprogramming can be performed immediately after the collected cells are seeded on culture dishes. In contrast, skin fibroblasts are typically derived from a small skin biopsy and require at least 4 weeks of culture and expansion to reach sufficient numbers for reprogramming. Third, unlike some cell types targeted for reprogramming, such as juvenile keratinocytes and neonatal fibroblasts, hASCs can be isolated from patients of all ages. This will have significant ramifications for clinical applications of iPS cells as it is more likely that older patients will require such therapies. In the future, these positive qualities of hASCs, combined with their faster reprogramming time as demonstrated in this study, could significantly reduce the time required for patients awaiting regenerative treatments. Feeder-free derivation of iPS cells from hASCs thus represents a more clinically applicable method for derivation of iPS cells compared to other cell types and should enable more efficient and rapid generation of patient-specific and disease-specific iPS cells.
Materials and Methods
Cell Culture and Maintenance of hASC-iPS Cells.
hASCs were maintained with Dulbecco's modified Eagle medium (DMEM) containing 10% FBS, Glutamax-I, 4.5 g/L glucose, 110 mg/L sodium pyruvate, 50 U/mL penicillin, and 50 μg/mL streptomycin at 37 °C, 95% air, and 5% CO2 in a humidified incubator. IMR90 human fibroblast cells were obtained from American Type Cell Culture (ATCC) and maintained with DMEM containing 10% FBS, L-glutamine, 4.5 g/L glucose, 100 U/mL penicillin, and 100 μg/mL streptomycin. All cells used for reprogramming were within passage two. Derived iPS cells were maintained either on MEF feeder layer or on Matrigel-coated tissue culture dishes (ES qualified; BD Biosciences) with mTESR-1 hES Growth Medium (Stemcell Technology).
Lentivirus Production and Transduction.
293FT cells (Invitrogen) were plated at ≈80% confluence per 100-mm dish and transfected with 12 μg each lentiviral vectors (Oct4, Sox2, Klf4, c-MYC) plus 8 μg packaging plasmids and 4 μg VSVG plasmids using Lipofectamine 2000 (Invitrogen) following the manufacturer's instructions. The resulting supernatant was collected 48 h after transfection, filtered through a 0.45-μm pore-size cellulose acetate filter (Whatman), and mixed with PEG-it Virus Concentration Solution (System Biosciences) overnight at 4 °C. Viruses were precipitated at 1,500 × g the next day and resuspended with Opi-MEM medium (Invitrogen).
Immunofluorescence and Alkaline Phosphatase Staining.
Cells were fixed with 2% formaldehyde in PBS for 2 min, permeabilized with 0.5% Triton X-100 in PBS for 10 min, and blocked with 5% BSA in PBS for 1 h. Cells were then stained with appropriate primary antibodies and AlexaFluor-conjugated secondary antibodies (Invitrogen). The primary antibodies for Oct3/4 (Santa Cruz Biotechnology), Sox2 (Biolegend), Klf4 (Abcam), c-MYC (Abcam), SSEA-3 (Chemicon), SSEA-4 (Chemicon), Tra-1–60 (Chemicon), Tra-1–81 (Chemicon), Nanog (Santa Cruz Biotechnology), Desmin (Sigma), Sox17 (R&D System), and Tuj-1 (Covance) were used in the staining. Alkaline phosphatase (AP) staining was performed using the Quantitative Alkaline Phosphatase ES Characterization kit (Chemicon) following the manufacturer's instruction.
Quantitative-PCR.
Total RNA and cDNA of each sample were prepared using the RNeasy Mini Plus kit (Qiagen) and the QuantiTect Reverse Transcription kit (Qiagen), respectively, following the manufacturer's instructions. Quantitative-PCR to measure mRNA expression levels was done with Taqman Gene Expression Assays (Applied Biosystems) using a SteponePlus Realtime-PCR System (Applied Biosystems) in the Protein and Nucleic Acid Facility at Stanford University School of Medicine.
In Vitro Differentiation.
hASC-iPS cells cultured on Matrigel were treated with collagenase type IV (Invitrogen) and transferred to ultra-low attachment plates (Corning Life Sciences) in suspension culture for 8 days with DMEM/F12 (1:1) containing 20% knockout serum (Invitrogen), 4.5 g/L L-glutamine, 1% nonessential amino acids, 0.1 mM 2-mercaptoethanol, 50 U/mL penicillin, and 50 μg/mL streptomycin. EBs were then seeded in 0.25% gelatin-coated tissue culture dish for another 8 days. Spontaneous differentiation of hASC-iPS cells into cells of mesoderm and endoderm lineages was then detected with appropriate markers by immunofluorescence. Differentiation into dopaminergic neurons was carried out by co-culture of hASC-iPS cells with PA6 cells as previously described for hES cells (28).
Teratoma Formation.
To form teratomas, ≈2 million hASC-iPS cells were harvested from Matrigel-coated culture dishes and injected s.c. to the dorsal flank of nude mice. After 6–8 weeks, tumors were dissected, and fixed with 10% formaldehyde in PBS. Parrafin embedded tissue sections were then generated and stained with hemotoxylin and eosin.
Bisulfite Pyrosequencing.
Briefly, 1,000 ng sample DNA was bisulfate-treated using the Zymo DNA Methylation kit (Zymo Research). The PCR was then performed with one of the PCR primers biotinylated to convert the PCR product to single-stranded DNA templates. The PCR products were sequenced by Pyrosequencing PSQ96 HS System (Biotage) following the manufacturer's instructions (Biotage). The methylation status of each locus was analyzed individually as a T/C SNP using QCpG software (Biotage).
Microarray Hybridization and Data Acquisition.
Total RNA samples were prepared using the RNeasy Mini Plus kit (Qiagen) from biological duplicate samples. Using Agilent Low RNA Input Fluorescent Linear Amplification kits, cDNA was reverse-transcribed, and cRNA then transcribed and fluorescently labeled with Cy5/Cy3. Cy3- and Cy5-labeled and amplified cRNA (825 ng) was hybridized to Agilent 4 × 44 K whole human genome microarrays (G4112F) and processed according to the manufacturer's instructions. The array was scanned using an Agilent G2505B DNA microarray scanner. The data were analyzed using GeneSpring GX 10.0 (Agilent Technologies) with multiple testing correction to identify genes that had statistically significant changes in expression between each group. For hierarchical clustering, we used Pearson correlation for similarity measure and for average linkage clustering.
Flowcytometry Analysis.
FACS analysis of the hASCs (P0), H9 hES cells, and IMR90 cells were carried out using a BD LSR analyzer (BD Biosciences) at the Stanford Shared FACS Facility and data were analyzed by FlowJo (Tree Star). Antibodies used in this study were phycoerythrin (PE)-conjugated anti-CD29, CD31, SSEA-4, Oct3/4, Nanog, CD44, CD45, and c-Kit, FITC-conjugated CD90, and SSEA-3 (all from BD PharMingen). PE-anti-alkaline phosphatase, AlexaFluor488-conjugated TRA-1–60, and TRA-1–81 were obtained from Millipore. FITC-conjugated anti-CD146 was obtained from ABD Serotec. Dead cells stained by propidium-iodide were excluded from the analysis. Isotype-identical antibodies (BD PharMingen) were used as controls.
SI.
Further methods are available in SI Methods.
Supplementary Material
Acknowledgments.
We thank Andrew J. Connolly for assistance with histological analysis and the Stanford Functional Genomics Facility for assistance with microarrays. This work was supported by the Mallinckrodt Foundation, an AHA Innovation grant, National Institutes of Health Director's New Innovator Award DP2OD004437 and Stanford Cardiovascular Institute (to J.C.W.); NIH R90 National Institutes of Health Grant 07010301, R21 DE018727, and R21 DE019274, California Institute for Regenerative Medicine Grant T1-00001 and RL1–00662-1, the Oak Foundation, and the Hagey Laboratory for Pediatric Regenerative Medicine (to M.T.L.).
Footnotes
The authors declare no conflicts of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0908450106/DCSupplemental.
References
- 1.Takahashi K, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 2.Park IH, et al. Reprogramming of human somatic cells to pluripotency with defined factors. Nature. 2008;451:141–146. doi: 10.1038/nature06534. [DOI] [PubMed] [Google Scholar]
- 3.Yu J, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 4.Park IH, et al. Disease-specific induced pluripotent stem cells. Cell. 2008;134:877–886. doi: 10.1016/j.cell.2008.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stadtfeld M, Nagaya M, Utikal J, Weir G, Hochedlinger K. Induced pluripotent stem cells generated without viral integration. Science. 2008;322:945–949. doi: 10.1126/science.1162494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huangfu D, et al. Induction of pluripotent stem cells from primary human fibroblasts with only Oct4 and Sox2. Nat Biotechnol. 2008;26:1269–1275. doi: 10.1038/nbt.1502. [DOI] [PubMed] [Google Scholar]
- 7.Shao L, et al. Generation of iPS cells using defined factors linked via the self-cleaving 2A sequences in a single open reading frame. Cell Res. 2009;19:296–306. doi: 10.1038/cr.2009.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carey BW, et al. Reprogramming of murine and human somatic cells using a single polycistronic vector. Proc Natl Acad Sci USA. 2009;106:157–162. doi: 10.1073/pnas.0811426106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Woltjen K, et al. piggyBac transposition reprograms fibroblasts to induced pluripotent stem cells. Nature. 2009;458:766–770. doi: 10.1038/nature07863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kaji K, et al. Virus-free induction of pluripotency and subsequent excision of reprogramming factors. Nature. 2009;458:771–775. doi: 10.1038/nature07864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Okita K, Nakagawa M, Hyenjong H, Ichisaka T, Yamanaka S. Generation of mouse induced pluripotent stem cells without viral vectors. Science. 2008;322:949–953. doi: 10.1126/science.1164270. [DOI] [PubMed] [Google Scholar]
- 12.Yu J, et al. Human induced pluripotent stem cells free of vector and transgene sequences. Science. 2009;324:797–801. doi: 10.1126/science.1172482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Soldner F, et al. Parkinson's disease patient-derived induced pluripotent stem cells free of viral reprogramming factors. Cell. 2009;136:964–977. doi: 10.1016/j.cell.2009.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhao Y, et al. Two supporting factors greatly improve the efficiency of human iPSC generation. Cell Stem Cell. 2008;3:475–479. doi: 10.1016/j.stem.2008.10.002. [DOI] [PubMed] [Google Scholar]
- 15.Nakagawa M, et al. Generation of induced pluripotent stem cells without Myc from mouse and human fibroblasts. Nat Biotechnol. 2008;26:101–106. doi: 10.1038/nbt1374. [DOI] [PubMed] [Google Scholar]
- 16.Aasen T, et al. Efficient and rapid generation of induced pluripotent stem cells from human keratinocytes. Nat Biotechnol. 2008;26:1276–1284. doi: 10.1038/nbt.1503. [DOI] [PubMed] [Google Scholar]
- 17.Kim JB, et al. Oct4-induced pluripotency in adult neural stem cells. Cell. 2009;136:411–419. doi: 10.1016/j.cell.2009.01.023. [DOI] [PubMed] [Google Scholar]
- 18.Guilak F, et al. Clonal analysis of the differentiation potential of human adipose-derived adult stem cells. J Cell Physiol. 2006;206:229–237. doi: 10.1002/jcp.20463. [DOI] [PubMed] [Google Scholar]
- 19.Zuk PA, et al. Human adipose tissue is a source of multipotent stem cells. Mol Biol Cell. 2002;13:4279–4295. doi: 10.1091/mbc.E02-02-0105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bunnell BA, Flaat M, Gagliardi C, Patel B, Ripoll C. Adipose-derived stem cells: isolation, expansion and differentiation. Methods. 2008;45:115–120. doi: 10.1016/j.ymeth.2008.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lowry WE, et al. Generation of human induced pluripotent stem cells from dermal fibroblasts. Proc Natl Acad Sci USA. 2008;105:2883–2888. doi: 10.1073/pnas.0711983105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Crisan M, et al. A perivascular origin for mesenchymal stem cells in multiple human organs. Cell Stem Cell. 2008;3:301–313. doi: 10.1016/j.stem.2008.07.003. [DOI] [PubMed] [Google Scholar]
- 23.Jiang J, et al. A core Klf circuitry regulates self-renewal of embryonic stem cells. Nat Cell Biol. 2008;10:353–360. doi: 10.1038/ncb1698. [DOI] [PubMed] [Google Scholar]
- 24.O'Connor MD, et al. Alkaline phosphatase-positive colony formation is a sensitive, specific, and quantitative indicator of undifferentiated human embryonic stem cells. Stem Cells. 2008;26:1109–1116. doi: 10.1634/stemcells.2007-0801. [DOI] [PubMed] [Google Scholar]
- 25.Zhang H, et al. Lysophosphatidic acid facilitates proliferation of colon cancer cells via induction of Kruppel-like factor 5. J Biol Chem. 2007;282:15541–15549. doi: 10.1074/jbc.M700702200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sun R, Chen X, Yang VW. Intestinal-enriched Kruppel-like factor (Kruppel-like factor 5) is a positive regulator of cellular proliferation. J Biol Chem. 2001;276:6897–6900. doi: 10.1074/jbc.C000870200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen C, et al. Human Kruppel-like factor 5 is a target of the E3 ubiquitin ligase WWP1 for proteolysis in epithelial cells. J Biol Chem. 2005;280:41553–41561. doi: 10.1074/jbc.M506183200. [DOI] [PubMed] [Google Scholar]
- 28.Vazin T, Chen J, Lee CT, Amable R, Freed WJ. Assesment of stromal-derived inducing activity in the generation of dopaminergic neurons from human embryonic stem cells. Stem Cells. 2008;26:1517–1525. doi: 10.1634/stemcells.2008-0039. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.