Abstract
The Rho family GTPases are stringently regulated through the action of a large family of GTPase activating proteins (GAPs) that stimulate their relatively weak intrinsic GTP hydrolyzing activity. The p190RhoGAPs, which include the p190A and p190B proteins are potent and widely expressed GAPs acting on both Rho and Rac GTPases. We have observed that several acidic phospholipids inhibit the RhoGAP activity and promote the RacGAP activity of p190 proteins. In liposome binding assays we have demonstrated that binding of p190A to phospholipids is controlled by electrostatic interactions. Using mapping techniques, we determined that a small polybasic peptide stretch within p190A is a common site for both the phospholipid binding and PKC phosphorylation. Moreover, PKC-mediated phosphorylation of two amino acids (serine-1221 and threonine-1226) within this region of p190A prevents the binding and susbstrate specificity regulation by phospholipids. Transfection of COS-7 cells with mutant forms of p190A either unable to bind to phospholipids or unable to become phosphorylated induced distinct morphological changes. Together, these findings reveal a novel GAP regulatory mechanism in which phosphorylation indirectly alters GTPase substrate preference by affecting the interaction with acidic phospholipids. Our observations provide a potential mechanism of Rac/Rho antagonism described in several cellular functions.
Cellular signaling often involves the assembly of molecular complexes stabilized by both protein-protein and protein-lipid interactions. Binding of proteins to membrane phospholipids (PLs) can occur by different mechanisms based either on highly specific recognition or on electrostatic forces. Most studies have focused on the interaction of identified protein domains (such as the PH-, PX- or FYVE domains) with specific phosphoinositides (1). Various species of phosphatidylinositolmono- bis- or trisphosphates (PIPs) are present in variable but generally very low amounts (typically < 0.05%) in different intracellular membranes, and reversible association and dissociation of the recognized proteins is directed by the rapid turnover of the relevant PIP (2). A significant increase (up to 20-fold) in the level of certain PIP species has been reported upon cell stimulation, resulting in rapid redistribution of distinct proteins between cytosol and plasma membrane. Specific binding of proteins to other acidic phospholipids such as phosphatidylserine (PS) or phosphatidic acid (PA) has also been reported (3).
In addition to specific PL-binding, electrostatic forces have also been shown to direct protein-lipid interaction on the membrane interface (4). This type of interaction generally involves the more abundant species of acidic phospholipids such as PS, phosphatidylinositol (PI) and phosphatidylinositol 4,5-bisphosphate (PIP2) and a polybasic motif of the protein. Binding of the protein is controlled by the charge density on the membrane surface that can be modified, e.g., by hydrolysis of PIP2 or redistribution of PS between the inner and outer layer of the plasma membrane (5). However, protein association or dissociation can also occur without any change in the membrane PL composition by altering the charge density on the protein via phosphorylation or protein-protein interaction, as has been demonstrated in detail for the PKC substrate myristoylated alanine-rich C-kinase substrate (MARCKS) (6). Recently, several studies addressed the role of electrostatic interactions in the intracellular localization of signaling proteins such as Src kinases or the small GTPases K-Ras, Rac1, RhoB and other members of the Rab and Arf family (5,7,8,9).
Small GTPases demonstrate a relatively weak intrinsic ability to hydrolyze GTP that can be significantly enhanced by GTPase activating proteins (GAPs). There are many potential GAPs for each subfamily of the small GTPases but those acting on Rho family proteins are especially numerous (10,11). In addition to the conserved GAP catalytic domain, these proteins contain a variety of other domains that mediate additional protein interactions or confer additional levels of regulation upon individual GAPs (12).
Several GAPs specific for the Arf subfamily of small GTPases were shown to be regulated by PLs. These include highly specific interactions with and enhancement of the ArfGAP activity by PIP3 or defined isoforms of PIP2 (13-15). Recently we have reported a different type of regulation of a GAP by PLs. Thus, the substrate preference of p190A, a GAP specific for the small GTPases Rho and Rac, is changed by exposure to acidic PLs such as PS, PI or PIP2 (16). In the present report we identify the region of p190A responsible for the PL effect and show that the lipid association and substrate specificity of p190A are regulated by electrostatic forces, and can be reversibly altered by phosphorylation by protein kinase Cα (PKC).
EXPERIMENTAL PROCEDURES
Materials
L-α-Phosphatidylserine (PS), L-α-Phosphatidylinositol (PI) and L-α-Phosphatidylcholine (PC) were purchased from Sigma. Protein Kinase C Alpha Isoenzyme (PKCα) was from Sigma and Upstate, respectively. PfuTurbo DNA Polymerase was from Stratagene, and DpnI was from Fermentas. [γ-32P]GTP and [γ-32P]ATP (both 185 TBq/mmol) were obtained from the Institute of Isotopes, Budapest, Hungary.
Preparation of Proteins
Prenylated baculovirus-produced Rac1 and RhoA were isolated from the membrane fraction of Sf9 cells and affinity purified on a nickel-Sepharose column as previously described (16). The p190A RhoGAP protein constructs p190(1135), p190(1191), p190(1252) were constructed by first performing PCR on RcHAp190 RhoGAP (17), and then cloned in-frame into the EcoRI site of pGEX-4T-1 vector and produced as glutathione S-transferase fusion proteins in Escherichia coli. The p190(1191) mutations were generated by site-directed mutagenesis using the QuikChangeTM site-directed mutagenesis kit (Stratagene) and following the manufacturer's instructions. The ΔPBR mutant of full-length p190A was created by overlap extension PCR followed by a three-part ligation. This plasmid was used as template for production of p190(1191)ΔPBR by PCR followed by sub-cloning into the EcoRI site of pGEX-4T-1 vector (18). All of the mutant clones were verified by restriction enzyme mapping and DNA sequencing. The primers used for the different protein constructs are listed in Table 1 of the Supporting Information.
Plasmids for transfection
p190FL-WT-GFP and p190FL-ΔPBR-GFP were constructed by first performing PCR on RcHAp190 RhoGAP or RcHAp190 RhoGAPΔPBR (17) and then subcloning the BspEI and EcoRI digested PCR product into the EGFP-C1 vector. The mutations in the p190FL-DMG-FP plasmid were generated by site-directed mutagenesis using the QuikChangeTM site-directed mutagenesis kit (Stratagene) and following the manufacturer's instructions. The primers used for the PCR and for the mutagenesis are listed in Table 1 of the Supporting Information.
Measurement of GTP hydrolysis by small GTPases
In most experiments the nitrocellulose filter binding assay was used to measure GTPase and GAP activities as previously described (16). Some findings were reproduced by the charcoal-precipitation method (19) or using [α-32P]GTP.
Preparation of lipid vesicles
Lipids were dissolved in chloroform (PI and PC) or a chloroform/methanol (95:5) mixture (PS) and dried in a nitrogen atmosphere. To prepare liposomes, lipids were rehydrated at room temperature in 200 μl of buffer (25 mM HEPES, 50 mM NaCl, pH 7.5) followed by vortexing until the mixture was homogeneously opalescent.
Liposome binding assays
Multilamellar vesicles (MLVs) were prepared as described above. The protein was centrifuged at 90 000 × g for 20 min to remove all aggregates. One μl of supernatant was added to 50 μl liposomes and binding was performed at 30°C for 15 minutes. The vesicles were centrifuged at 90 000 × g for 20 min. Pellet and supernatant were separated and the pellet was resuspended in 25 mM HEPES, 50mM NaCl, pH 7.5. Both pellet and supernatant were subjected to SDS-PAGE. The protein was detected by Coomassie staining. NIH Image J software was used for densitometry analysis.
In vitro PKC phosphorylation assay
PKCα phosphorylation in vitro was measured in 24 μl of a solution containing 20 mM MOPS buffer, pH 7.2, 25 mM β-glycerophosphate, 1 mM DTT, 1 mM CaCl2, 10μM ATP, 10μM GTP. For each reaction, 100 ng p190A RhoGAP was incubated in the presence of 50 ng PKC and 37 kBq [γ-32P]ATP. PKCα was activated with low PS concentration (0.02 μg/μl). Bisindolylmaleimide (BIM) was used as a specific PKC inhibitor at 1 μM concentration. Following incubation at room temperature for 20 minutes, the reactions were terminated by the addition of 6 μl 5x concentrated Laemmli sample buffer. The samples were boiled for 5 min and subjected to SDS–PAGE in 11% gels. The gels were Coomassie Blue-stained, dried, and subjected to phosphoimager analysis on a Bio-Rad GS-525 molecular image system using Multi-Analyst software (version 1.1). PKCα phosphorylation in the nitrocellulose filter binding assay was performed as described above, but in the presence of 2 mM ATP and absence of radioactive ATP.
Cell culture and transfection
COS7 cells were plated on glass coverslips in 6-well plates and grown in DMEM with 10% fetal bovine serum, 50 units/ml penicillin, and 50 μg/ml streptomycin at 37°C in a 5% CO2 incubator. Transient overnight transfection was performed using TurboFect™ (Fermentas) according to the manufacturer's instructions. COS7 cells were transfected with wild type or mutant p190A-GFP.
RESULTS
Identification of the region of p190A responsible for lipid-regulated GAP activity
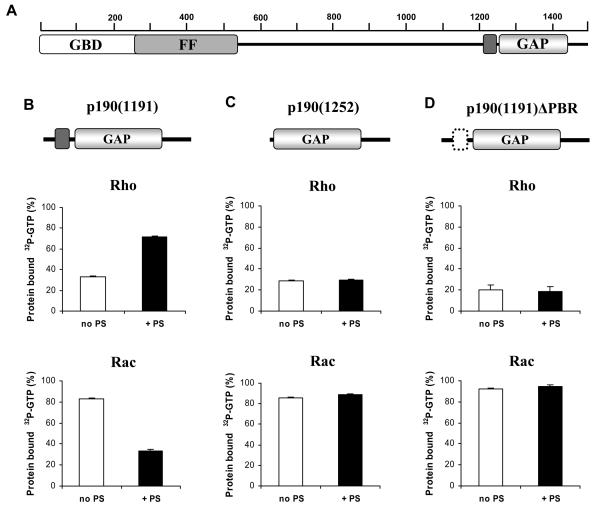
In a previous study, we observed that certain acidic phospholipids, such as PS and PI, inhibit the RhoGAP activity and promote the RacGAP activity of full-length p190A and p190B proteins in in vitro GAP assays. Neutral phospholipids were ineffective and none of the phospholipids had an effect on the endogenous GTPase activity of either Rac1 or RhoA. The same results were obtained when a C-terminal fragment of p190A (amino acids 1135-1499) was investigated (16), indicating that regulation of p190's GAP activity by phospholipids requires a region of the protein close to or within the GAP catalytic domain (Fig. 1A).
Fig. 1. Effect of PS on the RhoGAP and RacGAP activity of three truncated fragments of p190A.
A. Domain structure of p190A protein. B.-D. GAP activity of the three indicated p190A proteins, p190(1191) (B), p190(1252) (C), and p190(1191)ΔPBR (D) against the Rho and Rac GTPases is shown. An increase of radioactivity remaining bound to the protein indicates a decrease in the GAP activity. Where indicated, PS was present at a concentration of 0.1 μg/μl. Mean ± SEM of 72 independent experiments is shown.
Next, we prepared shorter fragments of p190A and investigated the effect of PS on their GAP activity towards RhoA and Rac1 (Fig. 1B-D). In the case of fragment p190(1191), we observed a significant inhibition of the RhoGAP activity by PS. In contrast, when tested with Rac1, PS vesicles promoted the RacGAP activity of this fragment (Fig. 1B). Significantly, these effects were not observed when using [α-32P]GTP instead of [γ-32P]GTP, demonstrating that the observed effects were on GAP activity as opposed to nucleotide binding. Thus, fragment p190(1191) behaved like p190(1135): in the presence of PS, the protein exhibited a decrease in RhoGAP activity and an increase in RacGAP activity. In contrast, fragment p190(1252) was completely insensitive to phospholipids: neither the RhoGAP nor RacGAP activity was modified by PS (Fig. 1C). Similar results were obtained with both fragments when PS was substituted by PI (data not shown). Thus, phospholipid regulation of p190A seems to require a protein domain between amino acids 1191 and 1252.
This region of the p190A protein sequence contains a peptide stretch (amino acids 1213-1236) in which 11 out of 24 amino acids are of basic character (7 arginines and 4 lysines) (Fig. 3A). To examine a potential role of this polybasic region (PBR), we prepared the fragment p190(1191) with a specific PBR deletion (p190(1191)ΔPBR; Fig. 1D). Similar to the p190(1252) protein, the p190(1191)ΔPBR protein was also insensitive to incubation with PS (Fig. 1D) or PI (not shown). Thus, the PBR situated between amino acids 1213 and 1236 is required for phospholipid regulation of the substrate preference of the p190A catalytic domain.
Fig. 3. Identification of the PKC phosphorylation sites of p190A.
A. PBR: the polybasic region of p190A, comprised of amino acids 1213-1236. The three putative phosphorylation sites are underlined. B. Investigation of the PKC-catalyzed phosphorylation of three truncated fragments of p190A. C. Investigation of the PKC-catalyzed phosphorylation of single, double and triple amino acid mutants of p190(1191). In panels B and C the upper row shows the phosphoimager data, and the lower row shows the Commassie Blue staining of the same gels. One representative out of 10 similar experiments is shown. Where indicated, the PKC inhibitor BIM was present at a concentration of 10 μM.
Binding of p190A to phospholipids
Next, we investigated whether modification of the GAP activity of p190A by PS can be ascribed to direct binding of the protein to phospholipids. Therefore, we performed a liposome binding assay in which soluble and lipid-bound proteins are separated by centrifugation and analyzed by SDS-PAGE. Addition of PS vesicles to p190(1191) resulted in almost complete redistribution of the protein from the supernatant into the pellet (Fig. 2A). In contrast to PS, only negligible redistribution occurred when liposomes were prepared from phosphatidylcholine (PC) (see below). As an additional control, we tested the potential binding of the applied small GTPases Rac1 and RhoA to PS vesicles, but no protein could be detected in the pellet independently of whether the small GTPase was in a GTP- or GDP-bound form (data not shown).
Fig. 2. Binding of p190A to phospholipids.
A. Binding of p190(1191) and p190(1191)ΔPBR to PS liposomes (S: supernatant; P: pellet). Result of one representative liposome binding assay out of 11 similar ones. The protein was stained with Comassie Blue. B. Binding of p190(1191), p190(1191)ΔPBR and p1191(1252) to PS liposomes. Bars represent the ratio of protein in the pellet to the total protein amount (pellet + supernatant), as determined by densitometry. Mean ± SEM of 5 independent experiments. C. and D. Binding of p190(1191) to PC liposomes containing varying proportions of PS or PI. Mean ± SEM of 6 (PS) or 5 (PI) independent experiments.
Next, we examined the region of the p190A protein responsible for binding to acidic phospholipids. As shown in Fig. 2A and B, association of the mutants p190(1252) and p190(1191)ΔPBR with PS vesicles exceeded the control value only marginally. Thus, the polybasic stretch of p190A is required for binding of the protein to acidic phospholipids.
We also examined the strength and specificity of the binding of p190A to phospholipid vesicles of varying composition. First we tested the binding of p190A to mixed liposomes containing PC and increasing amounts of PS (Fig. 2C). Addition of 25% PS resulted in a moderate increase of the binding of p190A to lipid vesicles, and the majority of the protein became lipid-bound only when the proportion of PS was 50%. Maximal binding was observed with 75% PS in the lipid mixture. Similar data were obtained when the mixed liposomes contained PI (Fig. 2D). However, no specific binding of p190A could be detected to PIP2-containing vesicles or to any PIP, PIP2 or PIP3 species on planar lipid blots (data not shown).
These lipid binding experiments indicate that p190A is able to associate with several acidic phospholipids, but that high-specificity recognition is not seen with any particular phospholipid species. Moreover, the polybasic PBR sequence of p190A is responsible for binding of the protein to acidic phospholipids.
Phosphorylation of p190A by PKCα
A variety of cellular proteins contain polybasic regions embedded with serines and threonines, and phosphorylation of these amino acids - typically by PKC - has been found to reduce the electrostatic attraction of these regions for phospholipid membranes (4). We have previously demonstrated that p190A is phosphorylated by PKC both in vitro and in vivo in epithelial cells (20). Subsequently, we found that the PKC isoform α was the most efficient among the various PKC enzyme isoforms for phosphorylating both p190A and p190B proteins. The PBR of p190A contains two serines and one threonine which are candidate PKC phosphorylation sites based on Scansite software predictions (Fig. 3A). Therefore, a potential role of these amino acids in PKC phosphorylation was investigated. As shown in Fig. 3B, purified p190(1191) is intensely phosphorylated by PKCα in vitro and phosphorylation is inhibited by the PKC inhibitor BIM. In contrast, neither p190(1252) nor the mutant p190(1191)ΔPBR can be detectably phosphorylated by PKCα, despite similar expression levels. Thus, phosphorylation by PKCα, like lipid binding, also requires the PBR of p190A.
Next, we carried out site-directed mutagenesis of each potentially phosphorylatable amino acid within the PBR. As summarized in Fig. 3C, all possible single-mutant and double-mutant proteins were phosphorylated, although the intensity of phosphorylation varied slightly among the mutants. Significantly, the triple-mutant protein (S1221A, T1226A, S1236A) is efficiently expressed, but it is not detectably phosphorylated. Thus, PKCα is able to phosphorylate all three amino acids (S1221, T1226 and S1236) within the PBR of p190A.
Phosphorylation by PKC affects lipid binding and GAP activity of p190A
Phosphorylation of three amino acids within the PBR of p190A is predicted to alter the charge density of this region and consequently it might influence electrostatic interactions with the protein. Therefore we examined the role of PKC-mediated phosphorylation of p190A on lipid binding. Typically, in the liposome binding assay, ~90% of p190(1191) was associated with PS vesicles, whereas binding of the phosphorylated protein to PS vesicles was reduced to ~30% (Fig. 4A). Similar results were obtained when liposomes were prepared with PI instead of PS. Thus, phosphorylation of p190A by PKC is able to prevent the association of the protein with acidic phospholipids.
Fig. 4. Effect of PKC phosphorylation on lipid binding (A) and on the GAP activity (B-D) of p190A.
A. Binding of non-phosphorylated (black bar) or phosphorylated (hatched bar) p190(1191) to PS liposomes. Mean ± SEM of 6 independent experiments. B-D. RacGAP and RhoGAP activity of non-phosphorylated and PKC-phosphorylated p190(1191) was tested in the absence of phospholipids (B) or in the presence of 0.1 μg/μl PS (C, D). Mean ± SEM of 24 independent experiments is shown.
Next we investigated the effect of PKC phosphorylation on the GAP activity of p190A. In the absence of added lipids, the state of phosphorylation did not affect the GAP activity of p190A (Fig. 4B). In contrast, the effect of PS on the substrate preference of p190(1191) is strongly counteracted by prior PKC-catalyzed phosphorylation (Fig. 4C and D). In the presence of PS, non-phosphorylated p190(1191) has poor RhoGAP, but very good RacGAP activity whereas phosphorylation by PKCα enhances the RhoGAP activity and impairs the RacGAP activity of p190A. Reversal of RhoGAP and RacGAP activity by PKCα can be completely prevented both by the PKC-inhibitor BIM or by omission of ATP from the phosphorylation reaction (Figures S1 and S2 in the Supporting Information). These findings indicate that PKCα-mediated phosphorylation of p190A does not affect GAP activity per se, but by decreasing the association of the protein with acidic phospholipids, it is able to modulate the GTPase substrate preference indirectly.
Role of the different PKC phosphorylation sites in lipid binding and GAP activity of p190A
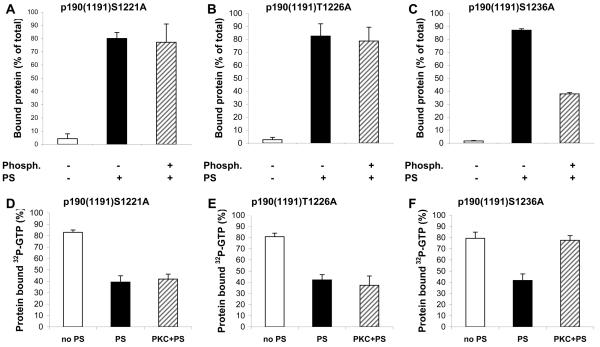
Next, we wanted to determine whether phosphorylation of all three PKC sites is required for regulation of lipid binding and substrate preference of p190A or whether there is any difference in the contribution of the individual sites. Therefore, we examined the lipid binding properties and GAP activity of the different p190A mutants (Fig. 5). Mutation of S1221 or T1226 to alanine resulted in proteins which were able to bind to PS vesicles even in the phosphorylated state (Fig. 5A and B). In agreement with the lipid binding data, we did not observe any change in the GAP activity of S1221A and T1226A upon phosphorylation by PKCα (Fig. 5D and E). All of the double-mutants as well as the triple-mutant protein behaved similarly to S1221A and T1226A. In contrast, the S1236A mutant protein behaved like the wild-type protein: phosphorylation by PKCα prevented its lipid binding (Fig. 5C) and reversed the effect of PS on its GAP activity (Fig. 5F).
Fig. 5. Effect of PKC phosphorylation on lipid binding (A-C) and on the GAP activity (D-F) of single amino acid mutants of p190(1191).
A.-C. Binding of non-phosphorylated (black bar) or PKC-phosphorylated (hatched bar) mutants of p190(1191) to PS liposomes. Mean ± SEM of 6 independent experiments. D.-F. RacGAP activity of non-phosphorylated and PKC-phosphorylated mutants of p190(1191) was tested in the absence of phospholipids (white bars) or in the presence of 0.1 μg/μl PS (black and hatched bars). Mean ± SEM of 11 independent experiments is shown.
We also generated and tested phospho-mimicking substitution mutants of S1221 and T1226 by replacing the phosphorylation sites with aspartate residues. Similar to the wild-type protein, PS also enhanced the RacGAP activity of these mutants. However, phosphorylation of p190(1191)S1221D and p190(1191)T1226D had only very weak effect on the GAP activity of the proteins containing the phospho-mimicking mutation (Fig. S3 in the Supporting Information).
Overall, the significance of the three PKC-phosphorylation sites appears to be distinct: lipid binding and GAP activity are only affected if phosphate groups are added both to S1221 and T1226, whereas the presence or absence of the additional negative charge on S1236 has little influence. However, the negative charge contributed by aspartate seems to have a weaker effect than addition of a phosphate group in the same position, in agreement with the fact that phosphate contributes two negative charges but aspartate provides only one negative charge.
Relation of lipid binding and PKC- phosphorylation of p190A to cell morphology
In order to gain information on the potential physiological significance of lipid binding and phosphorylation of p190A, we carried out transfection experiments with both the wild-type and mutant full-length (FL) proteins (Fig. 6). In agreement with earlier observations (18,21), transfection of wild-type (WT) p190A (p190FL-WT-GFP) induced a typical morphological change: the cells rounded up and developed long extensions. Typically, this dendritic phenotype was seen in about half of the transfected cells, whereas the other half of the cells remained spread. It is important to note that there was no visible difference in the expression level of p190A-GFP in the two population of cells (compare the left and right side of Fig. 6A).
Fig. 6. Effect of transfection with wild-type or mutant p190A on cell morphology.
COS-7 cells were transiently transfected with the GFP-fusion protein of A. Full-length wild type p190A (p190FL-WT-GFP), B. The double phosphorylation site mutant p190A (p190FL-DM-GFP) or C. the PBR-deleted mutant of p190A (p190FL-ΔPBR-GFP). One representative experiment out of 5 is shown. D. Statistical analysis of the data of 5 independent experiments.
When transfection was carried out with p190A-GFP containing alanine both at positions 1221 and 1226 (“double-mutant”, p190FL-DM-GFP; Fig. 6B), most of the cells showed the dendritic phenotype. Statistical analysis of 5 independent experiments revealed a prevalence of 88% for the dendritic phenotype (Fig. 6D). In contrast, when transfection was carried out with the ΔPBR mutant (p190FL-ΔPBR-GFP; Fig. 6C) 92% of the cells showed the spread morphology. In pull-down experiments carried out with the Rac-binding domain (RBD) of p-21 activated kinase (PAK) we did not see convincing differences in the amount of active, GTP-bound Rac in lysates of cell populations transfected with wild-type or the two mutants of p190A. This finding is consistent with our suggestion that alteration of the substrate preference of p190A GAP is a local event, regulating the Rac/Rho activity at distinct cellular locations but not disturbing the overall balance of the cell.
The ΔPBR mutation of p190A prevents its binding to acidic phospholipids whereas mutation of both S1221 and T1226 to alanine prevents the phosphorylation of the protein by PKC, hence allows its lipid binding. The clearly distinct phenotypes observed with these two mutants indicate the physiological significance of this region of p190A in determining its effects on cell morphology. The half-dendritic, half-spread morphology of the cells transfected with wild-type p190A may indicate the partially phosphorylated state of p190A in resting cells.
DISCUSSION
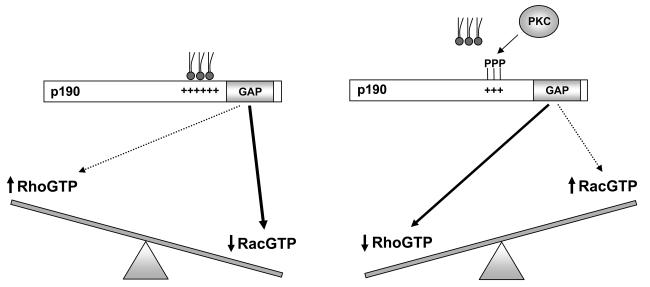
The results of these studies indicate that the GTPase substrate preference of p190A depends on the interaction of the protein with membrane lipids. As shown in the model of Fig. 7, p190A demonstrates greater RacGAP activity when associated with acidic PLs whereas the RhoGAP activity predominates when it is dissociated from the lipids. Binding to PLs occurs via electrostatic interactions between the acidic headgroups of PLs and the PBR situated amino-terminally with respect to the GAP catalytic domain of p190A. Significant association of the protein with PL vesicles requires a relatively high proportion (25 to 50%) of acidic PL in the lipid mixture (Fig. 2). This is in contrast to the behaviour of ArfGAPs, where addition of 3 to 5% of the critical PIP to a neutral lipid mixture significantly increased the binding of the protein (13-15). However, the sensitivity of p190A to acidic PLs corresponds to the reported proportion of these lipids at various intracellular locations (1,2,5). The apparently weaker binding of p190A to acidic PLs renders the association more easily reversible. Indeed, our results indicate that addition of two phosphate groups to the PBR of p190A is sufficient for dissociation of the protein from PL vesicles and reversal of the substrate preference (Fig. 5). Our results are in good agreement with earlier observations made on MARCKS: 90% of that protein was attached to PL vesicles when the proportion of PS reached 20%, (6) but phosphorylation of three amino acids was required for its dissociation from the lipid surface (4).
Fig. 7.
Model of the interaction of phospholipids and phosphorylation in the regulation of p190A substrate preference.
Phosphorylation of p190A by PKCα does not alter the GAP activity of the soluble protein (Fig. 4B). Instead, PKC-phosphorylation affects the binding of p190A to phospholipids and modulates its GAP activity indirectly, via effects on lipid association. We have recently reported related findings in which the effect of phosphorylation of p190A by glycogen synthase kinase-3-beta (GSK-3β) was investigated (18). However, phosphorylation of four amino acids within the C-terminal end of the GAP domain by GSK-3β resulted in a significant decrease of both the RhoGAP (18) and the RacGAP (Lévay et al. unpublished data) activity of p190A. Together with the present studies, these findings reveal multiple levels of phosphorylation-mediated regulation of p190A GAP catalytic activity at distinct sites.
The p190B protein has an overall sequence homology of 70% to p190A. p190B also has a polybasic region positioned N-terminally to the GAP domain, containing 13 lysines in a stretch of 24 amino acids. The corresponding region of p190A contains 4 lysines and 7 arginines. Consistent with the similar structure, we observed similar effect of PS on the Rac- and RhoGAP activity of p190B as shown for p190A (16). However, the polybasic region of p190B contains no serine or threonine, thus PKC-regulation of the lipid binding and GTPase preference seems to be a specific property of p190A.
Antagonism of Rac and Rho activity has been demonstrated to be required for several vital biological functions such as neurite outgrowth or directed cell migration. p190GAPs seem to play a significant role in this process (20-25). Recent findings demonstrated that activated Rac1 was able to recruit p190B to the plasma membrane and enhance its RhoGAP activity and this crosstalk between Rac and Rho GTPases was required for control of cell shape by integrin signaling (23). This mechanism seems not to function with p190A (23) although p190A was also shown to be translocated to the cell membrane upon physiological stimuli such as the fibronectin surface or by PKC-activating phorbol esters (20,22,24,25). The observations that tyrosine phosphorylation and complex formation with p120RasGAP or p120-catenin are required for translocation of p190A (25,26) suggest that p190A may become part of a larger protein complex at the plasma membrane. The size and varied domain structure of p190A definitely allow multiple interactions with proteins and lipids at the same time. PKCα has also been suggested to play a role in regulation of the distribution of p190A, however the mechanism has not been clarified (20,22,24). PKCα may have multiple effects: translocation of p190A could be affected indirectly, by promoting its interaction with other proteins, whereas direct phosphorylation of p190A regulates the lipid binding of the PBR, hence the substrate preference of the protein. Reversible alteration of the RacGAP and RhoGAP activity of p190A directed by PKC-catalyzed phosphorylation may thus represent another mechanism of Rac/Rho antagonism and may be a key element in the molecular organization of cell shape or directed cell movement. The distinct morphological changes observed in cells transfected with mutant forms of p190A substantiate the physiological relevance of our in vitro data.
On the basis of our biochemical experiments, we propose the following model (Fig. 7). In its native, unphosphorylated form, the PBR of p190A is able to establish electrostatic interaction with acidic phospholipid surfaces and in this state p190A functions predominantly as a RacGAP. Phosphorylation of the critical amino acids in position 1221 and 1226 by PKC largely prevents the association of the PBR of p190A with membrane lipids, and promotes the RhoGAP activity of the protein. Thus, phosphorylation or dephosphorylation of the critical amino acids of p190A switches rapidly and reversibly the RhoGAP and RacGAP activity, resulting in opposite changes in the local concentration of activated Rac and Rho proteins.
Taken together, our observations reveal a novel biochemical regulatory mechanism in which the coordinated actions of phosphorylation and lipid binding are able to influence the balance between Rho and Rac GTPase activities.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank to Dr. M.C. Dagher for providing prenylated Rac protein, to Drs. R. Sordella and S. Field for help with phosphorylation assays and lipid binding, respectively, to Profs. A. Faragó and S. Grinstein for stimulating discussions, to Dr. G. Szanda and P. Koncz for help with confocal microscopy and Dr. P. Várnai for advises and critical reading of the manuscript.
ABBREVIATIONS
- BIM
bisindolylmaleimide
- GAP
GTPase activating protein
- GSK-3β
glycogen synthase kinase-3-beta
- MARCKS
myristoylated alanine-rich C-kinase substrate
- PBR
polybasic region
- PBS
phosphate buffered saline
- PC
phosphatidylcholine
- PI
phosphatidylinositol
- PIP
phosphatidylinositol-phosphate
- PKC
protein kinase C
- PL
phospholipids
- PS
phosphatidylserine
Footnotes
Experimental work has been supported by grants from FIRCA [1 R03 TW006421-01A1], the Hungarian Research Fund [OTKA NK62221 and K75084] and NIH [RO1 CA62142 to J.S].
SUPPORTING INFORMATION AVAILABLE
The primers used for different DNA constructs (Table S1) and figures showing the ineffectiveness of PKC in the presence of the inhibitor BIM (Fig. S1) and in the absence of ATP (Fig. S2) and data obtained with the phosphomimicking mutants p190(1191)S1221D and p190(1191)T1226D (Fig. S3). This material is available free of charge via the internet at http://pubs.acs.org
REFERENCES
- 1.Lemmon MA. Membrane recognition by phospholipid-binding domains. Nature Rev. Mol. Cell. Biol. 2008;9:99–111. doi: 10.1038/nrm2328. [DOI] [PubMed] [Google Scholar]
- 2.Yeung T, Grinstein S. Lipid signaling and the modulation of surface charge during phagocytosis. Immunol. Rev. 2007;219:17–36. doi: 10.1111/j.1600-065X.2007.00546.x. [DOI] [PubMed] [Google Scholar]
- 3.Stace CL, Ktistakis NT. Phosphatidic acid- and phosphatidylserine-binding proteins. Biochim. Biophys. Acta. 2006;1761:913–926. doi: 10.1016/j.bbalip.2006.03.006. [DOI] [PubMed] [Google Scholar]
- 4.McLaughlin S, Murray D. Plasma membrane phosphoinositide organization by protein electrostatics. Nature. 2005;438:605–611. doi: 10.1038/nature04398. [DOI] [PubMed] [Google Scholar]
- 5.Yeung T, Terebiznik M, Yu L, Silvius J, Abidi WM, Philips M, Levine T, Kapus A, Grinstein S. Receptor activation alters inner surface potential during phagocytosis. Science. 2006;313:347–351. doi: 10.1126/science.1129551. [DOI] [PubMed] [Google Scholar]
- 6.Kim J, Shishido T, Jiang X, Aderem A, McLaughlin S. Phosphorylation, high ionic strength, and calmodulin reverse the binding of MARCKS to phospholipid vesicles. J. Biol. Chem. 1994;269:28214–28219. [PubMed] [Google Scholar]
- 7.Yeung T, Gilbert GE, Shi J, Silvius J, Kapus A, Grinstein S. Membrane phosphatidylserine regulates surface charge and protein localization. Science. 2008;319:210–213. doi: 10.1126/science.1152066. [DOI] [PubMed] [Google Scholar]
- 8.Heo WD, Inoue T, Park WS, Kim ML, Park BO, Wandless TJ, Meyer T. PI(3,4,5)P3 and PI(4,5)P2 lipids target proteins with polybasic clusters to the plasma membrane. Science. 2006;314:1458–1461. doi: 10.1126/science.1134389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Homchaudhuri L, Polverini E, Gao W, Harauz G, Boggs J. Influence of membrane surface charge and post-translational modifications to myelin basic protein on its ability to tether the Fyn-SH3 domain to a membrane in vitro. Biochemistry. 2009;48:2385–2393. doi: 10.1021/bi8022587. [DOI] [PubMed] [Google Scholar]
- 10.Bernards A. GAPs galore! A survey of putative Ras superfamily GTPase activating proteins in man and Drosophila. Biochim Biophys Acta. 2003;1603:47–82. doi: 10.1016/s0304-419x(02)00082-3. [DOI] [PubMed] [Google Scholar]
- 11.Tcherkezian J, Lamarche-Vane N. Current knowledge of the large RhoGAP family of proteins. Biol. Cell. 2007;99:67–86. doi: 10.1042/BC20060086. [DOI] [PubMed] [Google Scholar]
- 12.Bernards A, Settleman J. GAP control: regulating the regulators of small GTPases. Trends Cell Biol. 2004;14:377–385. doi: 10.1016/j.tcb.2004.05.003. [DOI] [PubMed] [Google Scholar]
- 13.Miura K, Jacques KM, Stauffer S, Kubosaki A, Zhu K, Hirsch DS, Resau J, Zheng Y, Randazzo PA. ARAP1: a point of convergence for Arf and Rho signaling. Mol. Cell. 2002;9:109–119. doi: 10.1016/s1097-2765(02)00428-8. [DOI] [PubMed] [Google Scholar]
- 14.Krugmann S, Anderson KE, Ridley SH, Risso N, McGregor A, Coadwell J, Davidson K, Eguinoa A, Ellson CD, Lipp P, Manifava M, Ktistakis N, Painter G, Thuring JW, Cooper MA, Lim ZY, Holmes AB, Dove SK, Michell RH, Grewal A, Nazarian A, Erdjument-Bromage H, Tempst P, Stephens LR, Hawkins PT. Identification of ARAP3, a novel PI3K effector regulating both Arf and Rho GTPases, by selective capture on phosphoinositide affinity matrices. Mol. Cell. 2002;9:95–108. doi: 10.1016/s1097-2765(02)00434-3. [DOI] [PubMed] [Google Scholar]
- 15.Brown MT, Andrade J, Radhakrishna H, Donaldson JG, Cooper JA, Randazzo PA. ASAP1, a phospholipid-dependent arf GTPase-activating protein that associates with and is phosphorylated by Src. Mol. Cell. Biol. 1998;18:7038–7051. doi: 10.1128/mcb.18.12.7038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ligeti E, Dagher MC, Hernandez SE, Koleske AJ, Settleman J. Phospholipids can switch the GTPase substrate preference of a GTPase-activating protein. J. Biol. Chem. 2004;279:5055–5058. doi: 10.1074/jbc.C300547200. [DOI] [PubMed] [Google Scholar]
- 17.Hu KQ, Settleman J. Tandem SH2 binding sites mediate the RasGAP-RhoGAP interaction: a conformational mechanism for SH3 domain regulation. EMBO J. 1997;3:473–483. doi: 10.1093/emboj/16.3.473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jiang W, Betson M, Mulloy R, Foster R, Lévay M, Ligeti E, Settleman J. p190A RhoGAP is a glycogen synthase kinase-3-beta substrate required for polarized cell migration. J. Biol. Chem. 2008;283:20978–20988. doi: 10.1074/jbc.M802588200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ligeti E, Pizon V, Wittinghofer A, Gierschik P, Jakobs KH. GTPase activity of small GTP-binding proteins in HL-60 membranes is stimulated by arachidonic acid. Eur. J. Biochem. 1993;216:813–820. doi: 10.1111/j.1432-1033.1993.tb18202.x. [DOI] [PubMed] [Google Scholar]
- 20.Brouns MR, Matheson SF, Hu KQ, Delalle I, Caviness VS, Silver J, Bronson RT, Settleman J. The adhesion signaling molecule p190 RhoGAP is required for morphogenetic processes in neural development. Development. 2000;127:4891–4903. doi: 10.1242/dev.127.22.4891. [DOI] [PubMed] [Google Scholar]
- 21.Tatsis N, Lannigan DA, Macara IG. The function of the p190 Rho GTPase-activating protein is controlled by its N-terminal GTP binding domain. J. Biol. Chem. 1998;273:34631–34638. doi: 10.1074/jbc.273.51.34631. [DOI] [PubMed] [Google Scholar]
- 22.Bass MD, Morgan MR, Roach KA, Settleman J, Goryachev AW, Humphries MJ. p190RhoGAP is the convergence point of adhesion signals from α5β1 integrin and syndecan-4. J. Cell Biol. 2008;181:1013–1026. doi: 10.1083/jcb.200711129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bustos RI, Forget MA, Settleman JE, Hansen SH. Coordination of Rho and Rac GTPase function via p190B RhoGAP. Curr Biol. 18:1606–1611. doi: 10.1016/j.cub.2008.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bass MD, Roach KA, Morgan MR, Mostafavi-Pour Z, Schoen T, Muramatsu T, Mayer U, Ballestrem C, Spatz JP, Humphries MJ. Syndecan-4-dependent Rac1 regulation determines directional migration in response to the extracellular matrix. J. Cell Biol. 2007;177:527–538. doi: 10.1083/jcb.200610076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bradley WD, Hernandez SE, Settleman J, Koleske AJ. Integrin signaling through Arg activates p190RhoGAP by promoting its binding to p120RasGAP and recruitment to the membrane. Mol. Biol. Cell. 2006;17:4827–4836. doi: 10.1091/mbc.E06-02-0132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wildenberg GA, Dohn MR, Carnahan RH, Davis MA, Lobdell NA, Settleman J, Reynolds AB. p120-catenin and p190RhoGAP regulate cell-cell adhesion by coordinating antagonism between Rac and Rho. Cell. 2006;127:1027–1039. doi: 10.1016/j.cell.2006.09.046. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.