Abstract
Patients with alveolar rhabdomyosarcoma (ARMS) have poorer response to conventional chemotherapy and lower survival rates than those with embryonal RMS (ERMS). By high-throughput screening, we identified camptothecin as an ARMS-selective inhibitor. Camptothecin more efficiently inhibited proliferation and induced apoptosis in Rh30 (ARMS) than RD (ERMS) cells. Ectopic expression of the PAX3-FKHR (PF) fusion protein in RD cells significantly increased sensitivity, whereas siRNA knockdown of PF decreased sensitivity of Rh30 cells to camptothecin. The sensitization required a transcriptionally active PF, and camptothecin downregulated levels of PF protein. These findings suggest that it is feasible to develop agents that preferentially block the growth of ARMS.
Keywords: Alveolar rhabdomyosarcoma, camptothecin, differential cytotoxicity, PAX3-FKHR, apoptosis
1. Introduction
Rhabdomyosarcoma (RMS) is the most common soft tissue sarcoma (STS) in children [1]. RMS is a heterogeneous tumor that expresses skeletal muscle-specific markers. Two subtypes of RMS have been identified on the basis of histopathologic features – embryonal (ERMS) and alveolar (ARMS) – each with distinct clinical and genetic characteristics [2]. Most of the more aggressive ARMSs are associated with 2;13 or 1;13 chromosomal translocations, which generate PAX3-FKHR (PF) and PAX7-FKHR fusion products, respectively [3]. These translocations result in unique expression, function, and subcellular localization of the fusion proteins, and ultimately contribute to their oncogenic behavior by modifying cell growth, differentiation, and apoptosis pathways [4]. Overexpression of fusion genes is associated with poor prognosis, and the survival rate of patients with ARMS is much lower than those with ERMS [5, 6]. ARMS is very resistant to conventional chemotherapy and radiotherapy. Recent studies show that several target genes of PF such as TFAP2B, c-MET, MYCN, CXCR4, and PDGFR-A play important roles in ARMS tumorigenesis [7–11] and are potential therapeutic targets for treating ARMS in children [8, 10, 11]. Also, directly regulating the transcriptional activity of PF has been proposed as an alternative strategy to treat ARMS [12].
Camptothecin and its derivatives topotecan and irinotecan have been used in animal models and clinically to treat certain human cancers [13], and different human cancers vary in their sensitivities to camptothecin-based chemotherapy [14, 15]. In a clinical study, ARMS patients were shown to have a higher rate of initial response to topotecan than those with ERMS [16]. In vitro, sensitivity to camptothecin has been shown to vary significantly in a panel of breast and colon cancer cell lines [17, 18]. Although topoisomerase I is the target for camptothecin, cellular sensitivity to camptothecin can not be predicted by expression or activity levels of topoisomerase I, cellular accumulation of camptothecin, or the cellular level of the covalent complex between topoisomerase I, camptothecin and DNA [18]. Furthermore, none of the other factors studied so far, such as the doubling time of a cell or expression of MDR-1, Bcl-2, and BAX, or p53 status, can predict cellular sensitivity to camptothecin [19]. Recent studies have shown that camptothecin exerts its antitumor activity by interfering with other signaling pathways such as the phosphatidylinositol 3′-kinase (PI3K)/Akt signaling pathway [20] and MAPK signaling pathway [21] in addition to inhibiting topoisomerase I. At present, very little is known about the cellular parameters controlling the sensitivity or resistance of tumor cells to camptothecin.
In this study, we used high-throughput screening to identify compounds that specifically block the growth of ARMS. We screened a collection of approximately 5600 bioactive compounds against an Rh30 cell line (ARMS) and an RD cell line (ERMS) and identified camptothecin that was significantly more effective at inhibiting cell growth and inducing apoptosis in Rh30 cells than in RD cells. Ectopic expression of the fusion protein PF in RD cells significantly increased their sensitivity to camptothecin, whereas siRNA knockdown of PF decreased the sensitivity of Rh30 cells to camptothecin. The PF-mediated sensitization to camptothecin was dependent on the transcriptional activity of PF, and camptothecin inhibited PF activity by downregulating the protein levels of PF. Our findings suggest that it is feasible to develop agents that preferentially block the growth of ARMS.
2. Materials and Methods
2.1. Cell culture
Human RD cell line was obtained from the American Type Culture Collection (ATCC; Manassas, VA). The Rh30, Rh41 and JR-1 cell lines were kindly provided by Dr. Peter Houghton. Cells were grown in complete culture medium–Dulbecco’s modified Eagle’s medium (DMEM) (Invitrogen, Carlsbad, CA) supplemented with 10% fetal bovine serum (FBS) (HyClone, Logan, UT), 100 U/ml penicillin and 100 μg/ml streptomycin, 2 mM L-glutamine, and 1 mM sodium pyruvate (Invitrogen). RD/PF cells (RD cells stably expressing pcDNA3-PF plasmid) and RD/Vector cells (RD cells stably transfected with pcDNA3 vector plasmid) (generous gifts from Dr. Frederic Barr, University of Pennsylvania School of Medicine, Philadelphia) [10] were maintained in a complete culture medium containing 500 μg/ml of G-418. NIH3T3 and PF-ER/NIH3T3 (NIH3T3 cells stably expressing a PF-ER fusion protein, in which the ligand-binding domain of the estrogen receptor was fused to the C-terminus of PF; kindly provided by Dr. Frederic Barr) [22] were maintained in the complete culture medium containing 3 μg/ml of puromycin. To induce transcriptional activity of PF, PF-ER/NIH3T3 and NIH3T3 cells (as control) were pretreated with 100 ng/ml 4-hydroxytomaxifen (4-OHT) for 24 h before treatment with drugs. All cells were cultured in an incubator with a humidified atmosphere maintained at 5% CO2 and 95% air at 37°C. Cells were split every 3 days at 90–95% confluence. For all luminescence assays, phenol red-free DMEM was used.
2.2. Cell proliferation assay and high-throughput screening
Cells were plated into 384-well white Cultureplates (PerkinElmer) at a density of 1000 cells/well in a final volume of 25 μl. After 24-h incubation, compounds were added and incubated for another 48 h. Final DMSO concentration was kept constant at 0.1%. The CellTiter-Glo® Luminescent Cell Viability Assay (Promega, Madison, WI) was used to determine the number of viable cells by quantifying the ATP present, which indicates the presence of metabolically active cells. Luciferase activity was measured with an EnVision multilabel plate reader (PerkinElmer). Data (viable cells) are expressed as percentage of control (%), calculated using the following formula: % of control = 100% × (compound signal – medium alone signal)/(DMSO control signal – medium alone signal), wherein DMSO control represents 100% cell survival and medium alone (no cells) represents 0% cell survival (background). The compound library used for the screen contains 5600 bioactive compounds and has been previously described [23]. All screens were performed in a fully automatic robotic system designed for cell-based assays.
2.3. Apoptosis assay
Cells were plated and treated with compounds as described above. Apoptosis was determined by using Caspase-Glo® 3/7 Luminescent Assay (Promega), following manufacturer’s instructions. Data are expressed as percentage of activation of caspases 3 and 7 (% Activation), where DMSO control is set as 0% activation and the presence of 10 μM of camptothecin as 100% activation.
2.4. Western blot analysis
Cells treated either with DMSO (control) or drugs were washed once with cold PBS and then harvested by scraping. Cells were lysed with RIPA buffer containing protease inhibitor cocktail and phosphatase inhibitor cocktail (Thermo Scientific, Rockford, IL). Equal amounts of lysates (20 μg/lane) were loaded into each lane on an SDS-PAGE, and proteins were transferred onto a nitrocellulose membrane and analyzed by specific antibodies as described previously [23]. Anti-FKHR antibody (H-128; sc-11350) was obtained from Santa Cruz (Santa Cruz Biotechnology, Santa Cruz, CA).
2.5. Silencing of PF with siRNA
A PF-specific siRNA (PF siRNA) (CCUCUCACCUCAGAAUUCA) and a control non-targeting siRNA (NT siRNA) (CUACUAUACCGAUACUCCC) were synthesized as described previously [6]. Cells were transfected with 10 nM of siRNAs in a 6-well dish with Lipofectamine™ 2000 reagent according to manufacturer’s instructions. Six hours after the transfection, cells were plated and treated with compounds for cell proliferation assay as described above. To verify PF knockdown efficiency, cells treated with siRNAs under the same conditions were analyzed by Western blot.
2.6. Reverse transcription and polymerase chain reaction (RT-PCR)
Total RNA was extracted from control and drug-treated cells using the RNeasy Mini kit (Qiagen, Valencia, CA) followed by DNase treatment (Promega). Reverse transcription was performed using the QuantiTect reverse transcription kit (Qiagen) according to manufacturer’s instructions. PCR was performed using following primer pairs – PF sense primer: GCACTGTACACCAAAGCACG and PF anti-sense primer: AACTGTGATCCAGGGCTGTC; GAPDH sense primer: GTCAGTGGTGGACCTGACCT and GAPDH anti-sense primer: AGCGGTCTACATGGCAACTG. Conditions for PCR were 94°C for 5 min, 30 cycles at 94°C for 30 s, 55°C for 30 s, and 72°C for 1 min. The final extension step was at 72°C for 7 min. PCR products were then analyzed on a 1.5% agarose gel.
2.7. Luciferase reporter assay
RD/PF cells were co-transfected with a PF-responsive firefly luciferase reporter (pGL4.14– × PRS9-tk) and a constitutively-expressed Renilla luciferase reporter (PRL-TK, control for transfection efficiency and drug toxicity normalization) using FuGENE 6 according to manufacturer’s instructions. pGL4.14–6 × PRS9-tk was constructed by inserting 6 tandem copies of PRS9 [22] upstream of the herpes virus tk promoter (−105/+51) [24] inserted into pGL4.14 (Promega). Twenty-four hours after transfection, 5000 live cells were plated in each well of a 384-well culture plate and grown for an additional 24 h, and then drug at indicated concentrations was added followed by incubation for 16 h. The luciferase activity was measured using the Dual-Glo® Luciferase Assay System (Promega) according to manufacturer’s instructions. To minimize effect of difference in transfection efficiency and possible drug cytotoxicity, the firefly luciferase activity was normalized by Renilla luciferase activity. Data are presented as percentage inhibition (% Inhibition) of firefly luciferase activity in the presence of drug compared with a DMSO negative control (as 0% inhibition) and a positive control (10 μM camptothecin as 100% inhibition).
2.8. Label-free cell proliferation assay
The rate of cellular proliferation was determined with a real-time cell electronic sensing (RT-CES) system (ACEA Bioscience, San Diego, CA) [25], which measures cell viability by monitoring cell proliferation and morphology, using a dimensionless unit called the cell index that is based on the impedance changes caused by interaction of cells with microelectrodes. Cells were seeded into 96x e-Plate at a density of 5000 cells/well (Rh30) and 10,000 cells/well (RD) in a final volume of 100 μl cell culture medium (DMEM supplemented with 10% FBS, 2 mM L-glutamine). The sensor devices were placed into the 5% CO2 incubator. After about 20 h incubation (cell index reached about 1), 100 μl of compounds dissolved in cell culture medium (final 0.1% DMSO) were added and the cell index value was determined every 30 min automatically by the RT-CES system for up to 132 h.
2. 9. Statistical analysis
Results are expressed as the mean ± SD of 3 independent experiments. The Student’s t-test for the paired samples was used to determine statistical significance of difference between parameters. Differences were considered significant for p < 0.05 (*), 0.01 (**) or 0.001 (***) and non-significant for p > 0.05.
3. Results
3.1. Rh30 cells are more sensitive than RD cells to camptothecin and its derivative
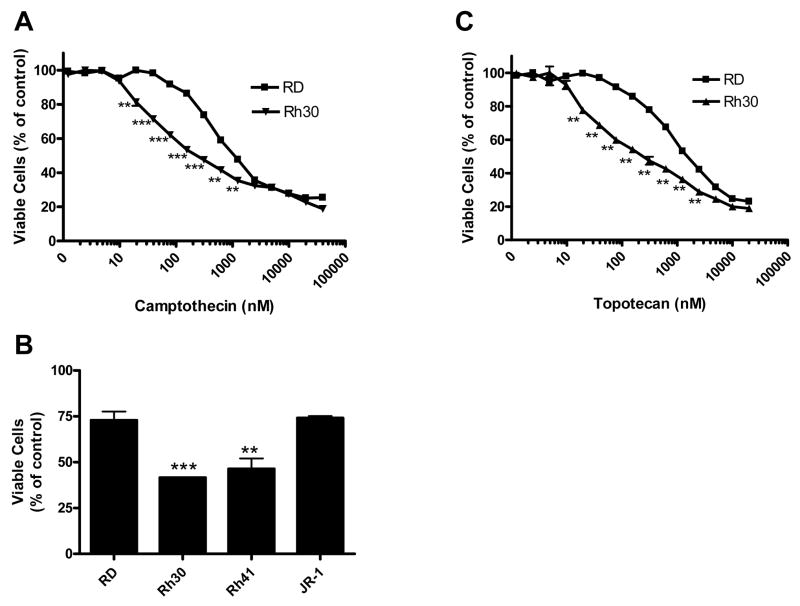
To identify compounds that preferentially block the growth of ARMS, we screened both RD cells (ERMS) and Rh30 cells (ARMS) in parallel under the same conditions against a collection of 5600 selected bioactive compounds by the CellTiter-Glo® assay. The bioactive compounds collection contains many cytotoxic compounds [23]. As expected, many compounds showed cytotoxicity to both Rh30 and RD cells (data not shown). Interestingly, camptothecin was identified as a compound that was significantly more effective in inhibiting cell growth in Rh30 cells than in RD cells at 15–1250 nM (p < 0.01) (Fig. 1A). Maximal difference in sensitivities of Rh30 and RD cells was observed between 40 and 250 nM. The enhanced cytotoxicity of camptothecin was also observed in ARMS cell line Rh41, but not in a different EMRS cell line JR-1 (Fig. 1B). To confirm these observations, we tested the camptothecin derivative topotecan. As seen for camptothecin, 15–1250 nM topotecan inhibited cell growth more effectively in Rh30 than in RD cells (p < 0.01) (Fig. 1C). These data demonstrate that camptothecin and its derivative inhibit cell proliferation more effectively in ARMS than in ERMS cells.
Fig. 1.
Differential inhibition of proliferation of ARMS and ERMS cells by camptothecin (A, B) and topotecan (C). Rh30, Rh41, JR-1 or RD cells were exposed to indicated concentrations of drugs (A, C) or 150 nM of camptothecin (B) for 48 h before determining viable cell number by the CellTiter-Glo® assay. Data represent 3 independent experiments, each performed in triplicate. Error bars indicate the standard error of the mean.
3.2. Rh30 cells are more sensitive than RD cells to camptothecin-induced apoptosis
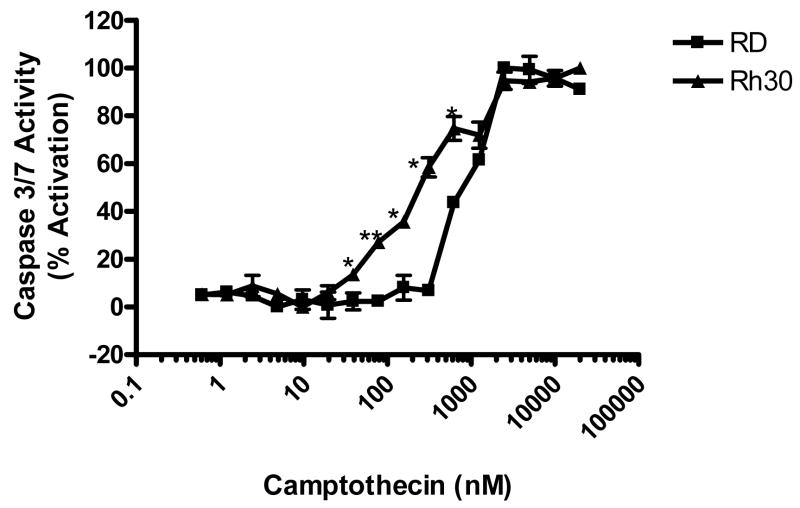
The basal activities of caspases 3 and 7 – a measure of apoptosis – were very low in both Rh30 and RD cells. There was no increase in activities of caspases 3 and 7 in RD cells at camptothecin concentrations below 300 nM (Fig. 2), however, there was a significant increase (p < 0.05) in activities of caspases 3 and 7 in Rh30 cells at camptothecin concentrations as low as 25 nM. The activities of caspases 3 and 7 were significantly (p < 0.05) higher in Rh30 than in RD for camptothecin concentrations of 25–750 nM (Fig. 2). These results agree well with data from the cell proliferation assay (Fig. 1A).
Fig. 2.
Differential activation of caspases 3 and 7 in Rh30 and RD cells by camptothecin. Rh30 and RD cells were exposed to indicated concentrations of camptothecin for 48 h before determining activation of caspases 3 and 7 by the Caspase-Glo® 3/7 assay. Data represent 3 independent experiments, each performed in triplicate. Error bars indicate the standard error of the mean.
3.3. Rh30 cells are more sensitive than RD cells to camptothecin in a dynamic and label-free cell-based assay
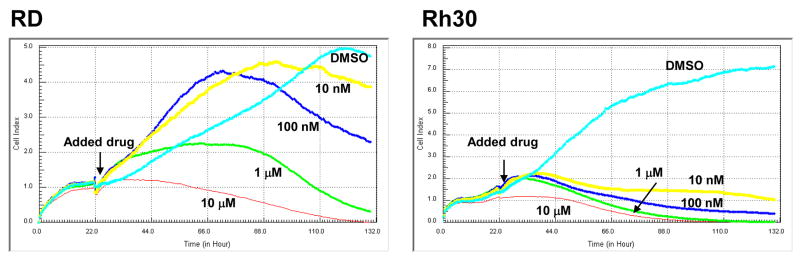
In order to monitor the cytotoxic effect of camptothecin in a real-time manner, we used the RT-CES system. In this system, cell index was used to measure cell proliferation (see Materials and Methods). As shown in Fig. 3, without camptothecin treatment (DMSO control), both RD and Rh30 cells continued to grow up to 110 h, at which time the cell index reached a saturation point. However, the cell index curves for RD and Rh30 cells were dramatically different in response to camptothecin treatment. Similar to the results from the cell proliferation assay (Fig. 1A), Rh30 cells are remarkably more sensitive than RD cells to camptothecin treatments at lower concentrations (10, 100 nM, and 1 μM), but not at higher concentration (10 μM) during the real-time monitoring. These results from real-time monitoring further demonstrate that Rh30 cells are more sensitive than RD cells to camptothecin.
Fig. 3.
Differential effect of camptothecin on cell growth as measured by RT-CES. The cells were seeded and incubated for 22 h before indicated concentrations of camptothecin or DMSO were added. The cell index value was determined every 30 min automatically by the RT-CES system for up to 132 h. Data shown is from a representative experiment.
3.4. Differential sensitivity of Rh30 and RD cells to camptothecin is dependent on PF
Topoisomerase I is the known target for camptothecin and its derivatives [13]. To examine whether the differential sensitivities of RD and Rh30 cells to camptothecin were due to differential expression levels of topoisomerase I, the basal expression level of topoisomerase I in RD and Rh30 was determined by Western blot with specific antibody. Rh30 and RD cells expressed topoisomerase I at similar levels and showed similar camptothecin-induced downregulation of topoisomerase I (data not shown). Thus, neither the basal expression level nor the camptothecin-mediated downregulation of topoisomerase I seemed to be responsible for the differential sensitivities of Rh30 and RD cells to camptothecin.
Camptothecin has been shown to exert its cytotoxicity through downregulating Akt in A549 cells [20]. However, we observed no significant difference in Akt phosphorylation between control and camptothecin-treated Rh30 and RD cells (Fig. S1A). In addition, Akt inhibitors did not cause differential cytotoxicity in Rh30 and RD cells (Fig. S1B). Thus, differential sensitivities of Rh30 and RD to camptothecin are unlikely mediated through downregulation of Akt activity.
It has been shown that disruption of p53 function in mouse embryonic fibroblasts sensitized cells to topotecan [26]. Rh30 harbors a p53 mutation. However, the mutated p53 in Rh30 is unlikely responsible for the enhanced sensitivity to topotecan, since overexpression of wild-type p53 further sensitizes Rh30 to topotecan [27]. Furthermore, camptothecin did not affect the nuclear localization of PF (data not shown).
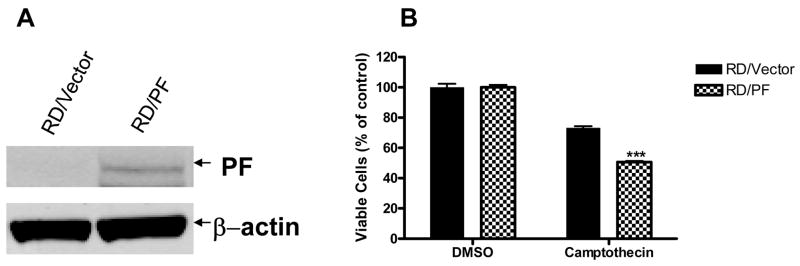
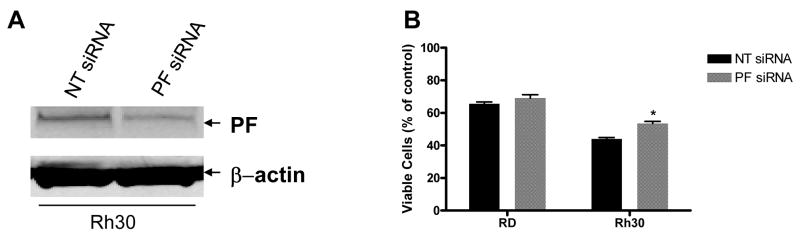
The unique PF fusion protein is expressed by the ARMS cell line Rh30 but not by the ERMS cell line RD [3]. We used this genetically defined difference to examine whether PF directly mediates the enhanced sensitivity of Rh30 to camptothecin. PF-negative RD cells stably expressing either vector alone (RD/Vector; as control) or ectopic PF (RD/PF) were treated with 150 nM camptothecin for 48 h and inhibition of cell growth was measured by the CellTiter-Glo® assay. The ectopic expression of PF in the RD/PF stable clone was confirmed as shown in Fig. 4A. As shown in Fig. 4B, ectopic expression of PF significantly sensitized RD cells to camptothecin-mediated inhibition of cell proliferation (49.4 ± 1.7%) compared with control RD/Vector cells (26.8 ± 2.8%) (p < 0.001). To verify that PF expression was directly responsible for this differential sensitivity to camptothecin, RD and Rh30 cells were transiently transfected with PF-specific siRNA (PF siRNA) or non-targeting control siRNA (NT siRNA) and then treated with 150 nM camptothecin for 48 h. As shown in Fig. 5A, the NT siRNA (negative control) did not affect expression of the PF protein in Rh30, but the PF siRNA reduced expression of the PF protein in Rh30 by approximately 50%. Reduction of PF expression in Rh30 cells by siRNA knockdown significantly decreased camptothecin-mediated inhibition of cell proliferation (46.6 ± 2.6%) in the CellTiter-Glo® assay compared with that of NT siRNA (55.9 ± 1.6%) (p < 0.05) (Fig. 5B). In a control experiment, PF siRNA had no significant effect on camptothecin-mediated inhibition of cell proliferation in RD cells (30.9 ± 4.1%) compared with NT siRNA (34.4 ± 2.4%) (Fig. 5B). Taken together, our results confirm that PF is at least partially responsible for the differential sensitivity of ARMS and ERMS cell lines to camptothecin-mediated inhibition of proliferation and apoptosis.
Fig. 4.
Ectopic expression of PF increases cell sensitivity to camptothecin. (A) Verification of PF expression by Western blot analysis. Whole cell lysate of RD/Vector or RD/PF cells were analyzed by Western blotting with FKHR-specific antibody and anti β-actin antibody (as loading control). (B) Cell proliferation assay. RD/Vector or RD/PF cells were treated with 0.1% DMSO or 150 nM camptothecin for 48 h. Cell viability was determined by the CellTiter-Glo® assay. Data represent 3 independent experiments, each performed in triplicate. Error bars indicate the standard error of the mean. Difference between the 2 cell lines was significant (p < 0.001) (***).
Fig. 5.
Knockdown of PF by siRNA decreases cell sensitivity to camptothecin. (A) Verification of PF expression by Western blot analysis. Whole cell lysate of Rh30 cells treated with NT siRNA or PF siRNA were analyzed by Western blotting with FKHR-specific antibody and anti β-actin antibody (as loading control). (B) Cell proliferation assay. After transfection with NT siRNA (control) or PF siRNA, cells were treated with 150 nM camptothecin for 48 h; cell viability was determined by the CellTiter-Glo® assay. Data represent 3 independent experiments, each performed in triplicate. Error bars indicate the standard error of the mean. Differences between PF siRNA and NT siRNA treatments in Rh30 cells was significant (p < 0.05) (*).
3.5. Transcriptional activity of PF is required for the enhanced sensitivity of cells to camptothecin
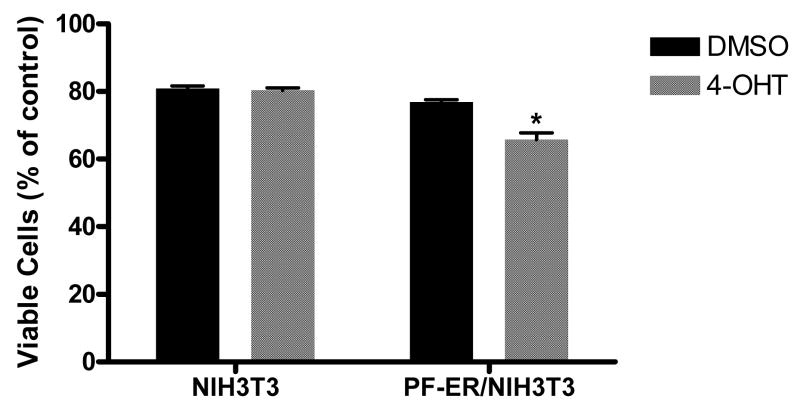
We used a cell system with inducible PF activity [22] to determine whether the transcriptional activity of PF is required for the enhanced sensitivity of cells to camptothecin. In this system, the PF-ER protein, constitutively expressed in the PF-ER/NIH3T3 cell line, is transcriptionally active only when cells are treated with 4-OHT, which binds ER and activates PF-ER [22]. 4-OHT treatment had no significant effect on camptothecin-mediated inhibition of cell proliferation of NIH3T3 cells (Fig. 6), but significantly enhanced the camptothecin-mediated inhibition of cell proliferation of PF-ER/NIH3T3 (30.9 ± 5.4 % and 19.5 ± 1.2% for 4-OHT and DMSO control, respectively) (p < 0.05). This result suggests that transcriptional activity of PF is required for it to increase the sensitivity of cells to camptothecin.
Fig. 6.
Active PF increases cell sensitivity to camptothecin. Cells were pretreated with DMSO or 100 ng/ml of 4-OHT for 24 h before exposure to 150 nM camptothecin for 48 h. Viable cells were measured by the CellTiter-Glo® assay. Data represent 3 independent experiments, each performed in triplicate. Error bars indicate the standard error of the mean. Difference was considered significant for p < 0.05 (*).
3.6. Camptothecin inhibits transcriptional activity of PF by downregulating PF protein level
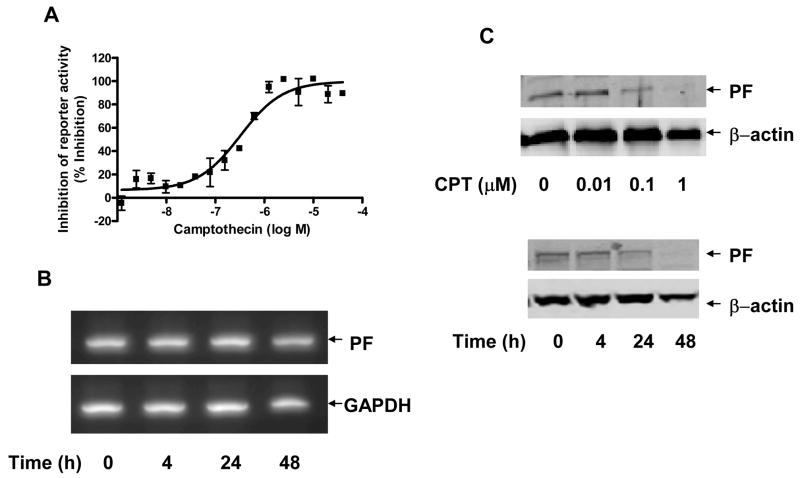
Since the transcriptional activity of PF at least partially contributed to the enhanced sensitivity of cells to camptothecin-mediated growth inhibition, we examined whether camptothecin could modulate the transcription activity of PF. The transcriptional activity of PF was measured in RD/PF cells transiently transfected with a PF-regulated luciferase reporter, pGL4.14–6 × PRS9-tk. As shown in Fig. 7A, camptothecin inhibited the luciferase reporter activity in a concentration-dependent manner with an IC50 of approximately 350 nM, indicating that camptothecin inhibited the transcriptional activity of PF. We then determined the effect of camptothecin treatment on the level of PF protein and mRNA. There was no significant change in mRNA levels of PF in Rh30 cells treated with 5 μM camptothecin up to 48 h (Fig. 7B). However, camptothecin downregulated the protein level of PF in both a dose-dependent (Fig. 7C, top panel) and time-dependent (Fig. 7C, bottom panel) manner. These results indicate that the transcriptional activity of PF may be inhibited because of camptothecin-mediated downregulation of the PF protein level.
Fig. 7.
Camptothecin attenuates transcriptional activity and protein level of PF. (A) Inhibition of transcriptional activity of PF by camptothecin. Dual luciferase assay was performed using PF-regulated luciferase reporter (pGL4.14-6 × PRS9-tk) in the presence of indicated concentrations of camptothecin in RD/PF cells. (B) No effect of camptothecin on mRNA levels of PF. Rh30 cells were treated with 5 μM camptothecin for indicated time periods. mRNA expression was analyzed by RT-PCR using PF-specific primers (see Materials and Methods). GAPDH was used as loading control. (C) Downregulation of PF protein level by camptothecin. Rh30 cells were treated with indicated concentrations of camptothecin for 48 h (top panel) or with 5 μM of camptothecin for indicated time period (bottom panel). Whole-cell lysate was analyzed by Western blotting with FKHR-specific antibody and anti β-actin antibody (as loading control).
4. Discussion
Camptothecin is a potent inhibitor of topoisomerase I, a key enzyme in DNA replication. It interferes with DNA replication by stabilizing covalent topoisomerase I-DNA complexes, thereby preventing religation of enzyme-linked single-strand DNA breaks [13–15]. Although camptothecin is known to mainly inhibit topoisomerase I, neither mRNA or protein levels of the enzyme nor cleavable DNA complex formation can predict tumor cell response to camptothecin in vitro [17–19]. In our study, although RD and Rh30 cells expressed similar levels of topoisomerase I (data not shown), they had significantly different sensitivities to camptothecin-mediated inhibition of cell proliferation and apoptosis. Hence, the differential sensitivities of the 2 cell lines to camptothecin may not be the consequence of differential induction of DNA damage. In addition to inhibiting topoisomerase I, camptothecin and its derivative topotecan have been shown to be cytotoxic by downregulating the PI3K-Akt signaling pathway [20]. In the lung cancer cell line A549, topotecan promotes Akt dephosphorylation and inactivation [20]. On the other hand, a camptothecin-resistant A549 clone exhibits significant increase in Akt kinase activity [20]. However, we observed no significant difference in the levels of phosphorylated Akt from control and camptothecin-treated Rh30 or RD cells (Fig. S1A). Therefore, neither topoisomerase I expression nor Akt activity could explain the significantly enhanced sensitivity of Rh30 to camptothecin and its derivative. In mouse embryonic fibroblasts, disruption of p53 function sensitized cells to topotecan [26]. However, the mutated p53 in Rh30 is unlikely responsible for the enhanced sensitivity to topotecan, because overexpression of wild-type p53 further sensitized Rh30 to topotecan [27]. Additionally, although RD cells (ERMS) and Rh41 (ARMS) cells express wild-type p53, we have found that Rh41 cells are significantly more sensitive than RD cells to camptothecin (Fig. 1B). These data indicate that camptothecin sensitivity may not be correlated to p53.
In the cell proliferation assay, Rh30 cells were significantly more sensitive than RD cells to camptothecin and topotecan only at 15–1250 nM (Fig. 1) but not at higher concentrations (>5 μM). Similar results were obtained by label-free RT-CES assay and in the apoptosis assay, suggesting that the increased sensitivity of Rh30 cells to camptothecin is likely owing to increased apoptosis. Ectopic expression of PF significantly enhanced the camptothecin sensitivity of RD cells. Knockdown of PF by specific siRNA verified the attenuation of sensitivity of both Rh30 cells and RD/PF cells (data not shown) to camptothecin. These results clearly demonstrate that PF is at least partly responsible for sensitizing cells to camptothecin inhibition. However, the precise mechanism responsible for the PF-dependent enhanced sensitivity to camptothecin remains undefined. It is unlikely that either topoisomerase I, p53, or Akt is involved. Interestingly, enhanced cellular sensitivity to chemical agents by fusion proteins created by chromosomal translocations is not unique to PAX3-FKHR. Recently, fusion oncogenic proteins MLL-AF4 and MLL-AF5 have been shown to enhance the sensitivity of MLL leukemia to inhibitors of GSK3 [28]. CDK inhibitor (CDKI) p27Kip1 is upregulated in MLL leukemia, and the inhibitory effect of GSK3 inhibitor on MLL leukemia is dependent on the increased level of p27Kip1. It is possible that the PF-mediated enhanced cellular sensitivity to camptothecin is dependent on altered expression of PF downstream target genes.
PF is a much stronger transcriptional activator than PAX3, and this increased activity may contribute to the increased proliferation rate and invasiveness of ARMS tumors [29]. We found that a transcriptionally active PF is required for the increased sensitivity to camptothecin, and camptothecin can inhibit this transcriptional activity. PF may potentiate camptothecin cytotoxicity through one or more of its target genes. The MYCN oncogene has been reported to be upregulated by ectopic expression of PF in ERMS cells [9]. MYCN and MYC also enhance camptothecin-induced apoptosis in human neuroblastoma cells and in rat fibroblast cells, respectively [30]. More recently, the kinase inhibitor PKC412 has been shown to be much more effective in inhibiting growth of ARMS than ERMS [12]. PKC412 inhibits the transcriptional activity of PF by regulating PF phosphorylation [12]. As seen for PKC412, in our study camptothecin did not affect nuclear localization of PF (data not shown) but interestingly downregulated the PF protein but not mRNA levels in a time- and dose-dependent manner. Therefore, camptothecin seems to inhibit transcriptional activity by directly downregulating the protein level of PF. PF has previously been shown to be ubiqutinated [31]. Because proteasome inhibitors are potent at inducing cell death in both ARMS and ERMS cells [32], we could not directly examine the effect of proteasome inhibitors on camptothecin-induced inhibition of ARMS cell proliferation. Future studies need to focus on whether camptothecin reduces the protein level of PF by modulating its ubiquitination status.
Interestingly, a clinical study indicated that ARMS patients had a higher rate of initial response to topotecan than those with ERMS [16]. Although the initial response and the treatment outcome have not been associated, these findings are worthy of further exploration in additional clinical trials. ARMS tumors are associated with a more aggressive disease pattern and a higher mortality rate than ERMS tumors and often fail to respond to current treatments such as chemotherapy, radiotherapy, or surgery. Alternative treatment agents therefore need to be developed. Several recent studies have focused on the development of small-molecule inhibitors against the downstream targets of PF, such as the Met receptor and PDGFR-A [8, 11]. However, directly targeting PF by either regulating its transcriptional activity or dowregulating its protein level may provide an effective alternative strategy to treat ARMS. Although the precise molecular mechanisms remain unknown, our observations, together with those reported for MLL leukemia [28], suggest that it is feasible to develop therapeutic agents to specifically block the growth of genetically defined tumors such as ARMS or MLL.
Supplementary Material
Fig. S1. Akt activity is not responsible for PF-mediated enhanced cellular sensitivity to camptothecin. (A) Effect of camptothecin on Akt phosphorylation. Rh30 and RD cells were treated with DMSO or 1 μM camptothecin for 24 h. Whole cell lysates were analyzed by Western blotting with specific antibody against phosphorylated Akt [p-Akt (S473)], Akt, or βactin (as loading control). (B) Inhibition of proliferation of Rh30 and RD cells by Akt inhibitors. Rh30 and RD cells were exposed to 10 μM of drugs as indicated for 48 h before determining viable cell number by the CellTiter-Glo® assay. Data represent 3 independent experiments, each performed in triplicate. Error bars indicate the standard error of the mean.
Acknowledgments
We thank Drs. Frederic Barr (University of Pennsylvania School of Medicine, Philadelphia) and Peter Houghton for cells, Dr. Wenwei Lin and Jing Wu for technical assistance, Drs. Satya Pondugula and David Bouck for reagents, other members of the Chen group for sharing reagents and for their valuable discussions, Dr. Anang Shelat for assistance with screening data analyses, and Cindy Nelson for assistance with compound management. We also thank Drs. Kip Guy and Peter Hougthon for critical review of the manuscript and Dr. Vani Shanker for editing the manuscript.
Role of the Funding Source
We acknowledge support from the American Lebanese Syrian Associated Charities (ALSAC), St. Jude Children’s Research Hospital, and National Cancer Institute grant P30CA027165.
Footnotes
Conflict of Interest Statement
None
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Raney RB, Anderson JR, Barr FG, Donaldson SS, Pappo AS, Qualman SJ, Wiener ES, Maurer HM, Crist WM. l. Rhabdomyosarcoma and undifferentiated sarcoma in the first two decades of life: a selective review of Intergroup Rhabdomyosarcoma Study Group experience and rationale for Intergroup Rhabdomyosarcoma Study V. J Pediatr Hematol Oncol. 2001;23:215–220. doi: 10.1097/00043426-200105000-00008. [DOI] [PubMed] [Google Scholar]
- 2.Linardic CM. PAX3-FOXO1 fusion gene in rhabdomyosarcoma. Cancer Lett. 2008;270:10–18. doi: 10.1016/j.canlet.2008.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barr FG. Fusions involving paired box and fork head family transcription factors in the pediatric cancer alveolar rhabdomyosarcoma. Curr Top Microbiol Immunol. 1997;220:113–129. doi: 10.1007/978-3-642-60479-9_7. [DOI] [PubMed] [Google Scholar]
- 4.Barr FG. The role of chimeric paired box transcription factors in the pathogenesis of pediatric rhabdomysarcoma. Cancer Res. 1999;59:1711–1715. [PubMed] [Google Scholar]
- 5.Sorensen PH, Lynch JC, Qualman SJ, Tirabosco R, Lim JF, Maurer HM, Bridge JA, Crist WM, Triche TJ, Barr FG. PAX3–FKHR and PAX7–FKHR gene fusions are prognostic indicators in alveolar rhabdomyosarcoma: a report from the children’s oncology group. J Clin Oncol. 2002;20:2672–2679. doi: 10.1200/JCO.2002.03.137. [DOI] [PubMed] [Google Scholar]
- 6.Kikuchi K, Tsuchiya K, Otabe O, Gotoh T, Tamura S, Katsumi Y, Yagyu S, Tsubai-Shimizu S, Miyachi M, Iehara T, Hosoi H. Effects of PAX3–FKHR on malignant phenotypes in alveolar rhabdomyosarcoma. Biochem Biophys Res Commun. 2008;365:568–574. doi: 10.1016/j.bbrc.2007.11.017. [DOI] [PubMed] [Google Scholar]
- 7.Ebauer M, Wachtel M, Niggli FK, Schafer BW. Comparative expression profiling identifies an in vivo target gene signature with TFAP2B as a mediator of the survival function of PAX3/FKHR. Oncogene. 2007;26:7267–7281. doi: 10.1038/sj.onc.1210525. [DOI] [PubMed] [Google Scholar]
- 8.Taulli R, Scuoppo C, Bersani F, Accornero P, Forni PE, Miretti S, Grinza A, Allegra P, Schmitt-Ney M, Crepaldi T, Ponzetto C. Validation of Met as a therapeutic target in alveolar and embryonal rhabdomyosarcoma. Cancer Res. 2006;66:4742–4749. doi: 10.1158/0008-5472.CAN-05-4292. [DOI] [PubMed] [Google Scholar]
- 9.Mercado GE, Xia SJ, Zhang C, Ahn EH, Gustafson DM, Laé M, Ladanyi M, Barr FG. Identification of PAX3-FKHR-regulated genes differentially expressed between alveolar and embryonal rhabdomyosarcoma: focus on MYCN as a biologically relevant target. Genes Chrom Cancer. 2008;47:510–520. doi: 10.1002/gcc.20554. [DOI] [PubMed] [Google Scholar]
- 10.Tomescu O, Xia SJ, Strezlecki D, Bennicelli JL, Ginsberg J, Pawel B, Barr FG. Inducible short-term and stable long-term cell culture systems reveal that the PAX3-FKHR fusion oncoprotein regulates CXCR4, PAX3, and PAX7 expression. Lab Invest. 2004;84:1060–1070. doi: 10.1038/labinvest.3700125. [DOI] [PubMed] [Google Scholar]
- 11.Taniguchi E, Nishijo K, McCleish AT, Michalek JE, Grayson MH, Infante AJ, Abboud HE, Legallo RD, Qualman SJ, Rubin BP, Keller C. PDGFR-A is a therapeutic target in alveolar rhabdomyosarcoma. Oncogene. 2008;27:6550–6560. doi: 10.1038/onc.2008.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Amstutz R, Wachtel M, Troxler H, Kleinert P, Ebauer M, Haneke T, Oehler-janne C, Fabbro D, Niggli FK, Schafer BW. Phosphorylation regulates transcriptional activity of PAX3/FKHR and reveals novel therapeutic possibilities. Cancer Res. 2008;68:3767–3775. doi: 10.1158/0008-5472.CAN-07-2447. [DOI] [PubMed] [Google Scholar]
- 13.Pommier Y. Topoisomerase I inhibitors: camptothecins and beyond. Nat Rev Cancer. 2006;6:789–806. doi: 10.1038/nrc1977. [DOI] [PubMed] [Google Scholar]
- 14.Beretta GL, Perego P, Zunino F. Mechanisms of cellular resistance to camptothecins. Curr Med Chem. 2006;13:3291–3305. doi: 10.2174/092986706778773121. [DOI] [PubMed] [Google Scholar]
- 15.Rasheed AZ, Rubin EH. Mechanisms of resistance to topoisomerase I-targeting drugs. Oncogene. 2003;22:7296–7304. doi: 10.1038/sj.onc.1206935. [DOI] [PubMed] [Google Scholar]
- 16.Pappo AS, Lyden E, Breneman J, Wiener E, Teot L, Meza J, Crist W, Vietti T. Up-front window trial of topotecan in previously untreated children and adolescents with metastatic rhabdomyosarcoma: An intergroup rhabdomyosarcoma study. J Clin Oncol. 2001;19:213–219. doi: 10.1200/JCO.2001.19.1.213. [DOI] [PubMed] [Google Scholar]
- 17.Goldwasser F, Bae I, Valenti M, Torres K, Pommier Y. Topoisomerase I-related parameters and camptothecin activity in the colon carcinoma cell lines from the National Cancer Institute anticancer screen. Cancer Res. 1995;55:2116–2121. [PubMed] [Google Scholar]
- 18.Desai SD, Li TK, Rodriguez-Bauman A, Rubin EH, Liu LF. Ubiquitin/26S proteasome-mediated degradation of topoisomerase I as a resistance mechanism to camptothecin in tumor cells. Cancer Res. 2001;61:5926–5932. [PubMed] [Google Scholar]
- 19.Davis PL, Shaiu WL, Scott GL, Iglehart JD, Hsieh TS, Marks JR. Complex response of breast epithelial cell lines to topoisomerase inhibitors. Anticancer Res. 1998;18:2919–2932. [PubMed] [Google Scholar]
- 20.Nakashio A, Fujita N, Rokudai S, Sato S, Tsuruo T. Prevention of phosphatidylinositol 3′-kinase-Akt survival signaling pathway during topotecan-induced apoptosis. Cancer Res. 2000;60:5303–5309. [PubMed] [Google Scholar]
- 21.Lee S, Lee HS, Baek M, Lee DY, Bang Y, Cho HN, Lee YS, Ha JH, Kim HY, Jeong D. MAPK signaling is involved in camptothecin-induced cell death. Mol Cells. 2002;14:348–354. [PubMed] [Google Scholar]
- 22.Xia SJ, Rajput P, Strzelecki DM, Barr FG. Analysis of genetic events that modulate the oncogenic and growth suppressive activities of the PAX3-FKHR fusion oncoprotein. Lab Invest. 2007;87:318–325. doi: 10.1038/labinvest.3700521. [DOI] [PubMed] [Google Scholar]
- 23.Lin WW, Wu J, Dong HQ, Bouck D, Zeng FY, Chen TS. Cyclin-dependent kinase 2 negatively regulates human pregnane X receptor-mediated CYP3A4 gene expression in HepG2 liver carcinoma cells. J Biol Chem. 2008;283:30650–30657. doi: 10.1074/jbc.M806132200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen T, Wang LH, Farrar WL. Interleukin-6 activates androgen receptor-mediated gene expression through a signal transducer and activator of transcription 3-dependent pathway in LNCaP prostate cancer cells. Cancer Res. 2000;60:2132–35. [PubMed] [Google Scholar]
- 25.Boyd JM, Huang L, Xie L, Moe B, Gabos S, Li XF. A cell-microelectronic sensing technique for profiling cytotoxicity of chemicals. Anal Chim Acta. 2008;615:80–87. doi: 10.1016/j.aca.2008.03.047. [DOI] [PubMed] [Google Scholar]
- 26.Tomicic MT, Christmann M, Kaina B. Topotecan-triggered degradation of topoisomerase I is p53-dependent and impacts cell survival. Cancer Res. 2008;65:8920–8926. doi: 10.1158/0008-5472.CAN-05-0266. [DOI] [PubMed] [Google Scholar]
- 27.Gibson AA, Harwood FG, Tillman DM, Houghton JA. Selective sensitization to DNA-damaging agents in a human rhabdomyosarcoma cell line with inducible wild-type p53 overexpression. Clin Cancer Res. 1998;4:145–152. [PubMed] [Google Scholar]
- 28.Wang Z, Smith KS, Murphy M, Piloto O, Somervaille TC, Cleary ML. Glycogen synthase kinase 3 in MLL leukaemia maintenance and targeted therapy. Nature. 2008;455:1205–1209. doi: 10.1038/nature07284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Davicioni E, Finckenstein FG, Shahbazian V, Buckley JD, Triche TJ, Anderson MJ. Identification of a PAX–FKHR gene expression signature that defines molecular classes and determines the prognosis of alveolar rhabdomyosarcomas. Cancer Res. 2006;66:6936–6946. doi: 10.1158/0008-5472.CAN-05-4578. [DOI] [PubMed] [Google Scholar]
- 30.Albihn A, Mo H, Yang Y, Henriksson M. Camptothecin-induced apoptosis is enhanced by MYC and involves PKCdelta signaling. Int J Cancer. 2007;121:1821–1829. doi: 10.1002/ijc.22866. [DOI] [PubMed] [Google Scholar]
- 31.Roeb W, Boyer A, Covenee WK, Arden KC. Guilt by association: PAX3-FOXO1 regulates gene expression through selective destabilization of the EGR1 transcription factor. Cell Cycle. 2008;7:837–841. doi: 10.4161/cc.7.7.5652. [DOI] [PubMed] [Google Scholar]
- 32.Bersani F, Taulli R, Accornero P, Morotti A, Miretti S, Crepaldi T, Ponzetto C. Bortezomib-mediated proteasome inhibition as a potential strategy for the treatment of rhabdomyosarcoma. Eur J Cancer. 2008;44:876–884. doi: 10.1016/j.ejca.2008.02.022. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Akt activity is not responsible for PF-mediated enhanced cellular sensitivity to camptothecin. (A) Effect of camptothecin on Akt phosphorylation. Rh30 and RD cells were treated with DMSO or 1 μM camptothecin for 24 h. Whole cell lysates were analyzed by Western blotting with specific antibody against phosphorylated Akt [p-Akt (S473)], Akt, or βactin (as loading control). (B) Inhibition of proliferation of Rh30 and RD cells by Akt inhibitors. Rh30 and RD cells were exposed to 10 μM of drugs as indicated for 48 h before determining viable cell number by the CellTiter-Glo® assay. Data represent 3 independent experiments, each performed in triplicate. Error bars indicate the standard error of the mean.