Abstract
Oxidative damage to cell macromolecules by reactive oxygen species is associated with numerous diseases and aging. In Drosophila, RNAi-mediated silencing of the mitochondrial antioxidant manganese superoxide dismutase (SOD2) throughout the body dramatically reduces life span, accelerates senescence of locomotor function, and enhances sensitivity to applied oxidative stress. Here, we show that Sod2 knock-down in the musculature alone is sufficient to cause the shortened life span and accelerated locomotor declines observed with knock-down of Sod2 throughout the body, indicating that Sod2 deficiency in muscle is central to these phenotypes. Knock-down of Sod2 in the muscle also increased caspase activity (a marker for apoptosis) and caused a mitochondrial pathology characterized by swollen mitochondria, decreased mitochondrial content and reduced ATP levels. These findings indicate that Sod2 plays a crucial role in the musculature in Drosophila and that the consequences of Sod2 loss in this tissue extend to the viability of the organism as a whole.
Keywords: Sod2, muscle, mitochondria
Introduction
The electron transport chain (ETC) in mitochondria is the primary source of reactive oxygen species (ROS) in eukaryotes, with 0.1% or more of oxygen entering the chain being univalently reduced to superoxide [1, 2]. Superoxide escaping from the ETC and other ROS derived from superoxide represent a significant oxidative threat to mitochondria and cells as a whole [3]. Superoxide dismutase (SOD, EC 1.15.1.1) enzymes are antioxidants that protect cells from oxidative stress by catalyzing the dismutation of superoxide to oxygen and hydrogen peroxide. Hydrogen peroxide is subsequently reduced to water by catalase (EC 1.11.1.6) or a number of peroxidases [4].
The fruit fly, Drosophila melanogaster, possesses the three major eukaryotic SOD isoforms: SOD1 found predominantly in the cytosol, SOD2 in the mitochondrial matrix, and SOD3 in the extracellular space [5]. The localization of SOD2 to the mitochondrial matrix places it in close proximity to the ETC, suggesting that this enzyme has an important role in protecting cells from superoxide produced by mitochondrial respiration. Accordingly, complete loss of SOD2 activity in mice results in a number of pathological conditions including dilated cardiomyopathy, accumulation of lipid in liver and skeletal muscle, motor disturbances with neurodegeneration (in a mixed C57BL/6 and 129Sv background), and a very short life span [6–8]. At the cellular level, Sod2 mutant mice exhibit increased oxidative damage to lipid in addition to genomic and mitochondrial DNA [9, 10], decreased mitochondrial respiration rates, deficits in ETC activity reminiscent of mitochondrial disease [11] and elevated hepatic apoptosis [9]. In Drosophila, knock-out or knock-down of Sod2 throughout the body also severely shortens life span [12–15]. Furthermore, Sod2 deficient flies exhibit accelerated declines in olfactory and locomotor behavior with age in addition to neurodegeneration associated with elevated apoptosis [14, 16]. All of these studies underscore the importance of SOD2 in protecting cells and tissues from oxidative damage.
In this study we investigated whether Sod2 functions within specific tissues to support normal life span and locomotor function. We find that expression of Sod2 RNAi transgenes in the musculature, but not the nervous system, dramatically shortens life span and accelerates loss of locomotor behavior across age. Flies with knock-down of Sod2 in muscle exhibit mitochondrial pathology, reduced ATP content and elevated caspase activity, supporting the hypothesis that mitochondrial dysfunction in the musculature is central to the shortened life span and accelerated decline in locomotor function in these animals.
Materials and methods
Fly stocks and husbandry
Flies were reared to adulthood and housed for all aging studies at 25°C and 55% relative humidity under a 12 hour light–dark cycle on a sugar: yeast: cornmeal: agar medium (10%: 2%: 3.3%: 1% w/v) supplemented with 0.2% Tegosept (Sigma Chemical Co., St. Louis, MO) and active yeast. Sod2 was knocked-down by using the Gal4/UAS system [17] to express Sod2 inverted repeat (Sod2-IR) transgenes previously described [13]. The UAS-Sod2-IR24, UAS-Sod2-IR15 and daughterless-Gal4 (da-Gal4) lines were provided by John Phillips (University of Guelph). Other fly lines were from the following sources: Mef2-Gal4, Sunita Gupta Kramer (Rutgers); Appl-Gal4; Lawrence Goldstein (University of California, San Diego); D42-Gal4, Gabrielle Bouilanne (University of Toronto); GMH5-Gal4, R. J. Wessells (University of Michigan); 24B-Gal4, repo-Gal4 and elav-Gal4, Drosophila Stock Center (Bloomington, IN); 91Y-Gal4 and 188Y-Gal4, J. Douglas Armstrong (University of Edinburgh); UAS-Shibirets1, Ron Davis (Baylor College of Medicine), Flies with a second chromosome harboring the UAS-lacZ and UAS-Sod2-IR24 transgenes were generated by recombination and confirmed by PCR using the primers 5′-GAACACGTCGCTAAGCGAAAGC-3′ and 5′-GCGAAATTTTACGGGCCACGAAC-3′ to detect UAS-Sod2-IR and the primers 5′-CTGACGGGCTCCAGGAGTC-3′ and 5′-CTACCGGATTGATGGTAGTGGTC-3′ to detect UAS-lacZ.
SOD activity
Groups of 25 adult males (0–3 days old) per genotype were collected under brief CO2 anesthesia and homogenized in extraction buffer (50 mM potassium phosphate/0.1 mM EDTA/2% Triton-X-100, pH 7.8) on ice. To rupture mitochondria, samples were sonicated for 20 sec and then incubated at 4°C for 45 min. Ruptured mitochondrial samples were centrifuged at 16,000 × g for 15 min at 4°C and the resulting supernatant was stored at 4°C. Protein concentration was determined using the Lowry method (DC Protein Assay, Bio-Rad). Samples containing equal amounts of protein were electrophoresed by Discontinuous Native PAGE (4% stacking gel (pH 6.8), 20% resolving gel (pH 8.8)) in sample buffer (0.5 M Tris-HCl/50% glycerol/0.01% bromophenol blue) at 80–100 V. SOD activity was measured colorimetrically using a modified version of an “in-gel” assay previously described [13]. Briefly, gels containing electrophoresed protein samples were incubated in 2.5 mM nitroblue tetrazolium and 50 mM potassium phosphate buffer for 20 min in the dark under gentle agitation. Gels were washed briefly in 50 mM phosphate buffer and then incubated in 28 mM N,N,N′,N′-tetramethylethylenediamine/28 μM riboflavin/50 mM potassium phosphate for 15 min in the dark with gentle agitation. Following a brief wash in 50 mM potassium phosphate buffer, gels were exposed to white light for ~15 min for full color development. In-gel SOD activity was quantified by densitometry using Alpha Imager software (Alpha Innotech Corp., San Leandro, CA).
Life span analysis
Adult males (1–3-days old, 100–200 flies per genotype) were collected under brief CO2 anesthesia and transferred to fresh food vials at a density of 25 flies per vial. Flies were transferred to fresh food vials every 2–4 days. Dead and surviving flies were counted during each transfer to fresh food.
Negative geotaxis
Groups of 25 male flies (1–3 days old) were collected under brief CO2 anesthesia and allowed to recover at least 18 hours at 25°C and 55% relative humidity prior to assay. Negative geotaxis (bang-induced climbing) was measured in Rapid Iterative Negative Geotaxis (RING) assays as previously described [18]. Flies were transferred to a RING apparatus and rested for 1 min. Negative geotaxis was initiated by sharply rapping the apparatus on a table three times in rapid succession. The flies’ positions in the tubes were captured in digital images taken 4 sec after initiating the behavior. Data (distance climbed for each fly) were extracted from the images using Scion Image as described [18]. The performance of flies in a single vial in 5 consecutive trials was averaged to generate a single datum. Five vials of flies per genotype were tested to generate N=5 for each genotype in individual experiments. After each RING test, flies were transferred to fresh food vials and housed until the next assessment of negative geotaxis.
Determination of decline-time 50 (DT50, time required for negative geotaxis to decline to 50% of the youngest flies) was performed as described previously [19]. DT50 values, interpolated from second-order curve fits using Prism 4.02 (GraphPad Software, San Diego, CA), were determined for each vial of flies individually and then compiled for each genotype.
Temperature-sensitive Shibire (Shits1) was expressed using the Gal4/UAS system [17]. Shits1 encodes a dominant-negative dynamin that blocks neurotransmission at chemical synapses at 31°C [20, 21]. Adult flies expressing Shits1 (containing a Gal4 driver and two UAS-Shits1 transgenes) and control flies (containing the UAS-Shits1 transgenes but no Gal4 driver) were collected and allowed to recover as above. Negative geotaxis was first assessed in RING assays performed at 25°C. Flies were allowed to recover for 1 hour, transferred to 31°C for 30 minutes, and then tested again in RING assays at the elevated (i.e. restrictive) temperature. The effect of Shits1 expression on RING was quantitated as percent negative geotaxis remaining at the restrictive temperature [(negative geotaxis at 31°C) ÷ (negative geotaxis at 25°C) × 100%] for each genotype.
Expression patterns of Gal4 drivers and β-galactosidase histology
Gal4 expression was assessed by using a UAS-lacZ reporter gene in a Sod2 knock-down background. Flies with a double-transgenic UAS-lacZ, UAS-Sod2-IR24 second chromosome (generated by recombination) were mated to Gal4 driver stocks to produce progeny with concurrent expression of lacZ and knock-down of Sod2.
The spatial expression patterns of da-Gal4, Mef2-Gal4 and 24B-Gal4 across age were assessed by β-galactosidase histology. Briefly, adult flies were collected under brief CO2 anesthesia, embedded in Tissue-Tek O.C.T. (Sakura Finetak USA, Inc.), transferred in embedding medium to a specimen holder and frozen at −40°C. Whole-body tissue sections (15μM) were cut using a Hacker Bright Cryostat (model OTC5000, GMI, Ramsey, MN) at −20°C. Sections were transferred to slides, allowed to air dry for 1 hour at room temperature and then fixed in 2% glutaraldehyde for 20 min. Slides were washed twice for 5 min in phosphate-buffered saline (PBS) and placed at 37°C until dry. β-galactosidase (EC 3.2.1.23) activity was visualized by adding pre-warmed X-gal substrate to the slides for 30 min. Slides were washed twice for 5 min in PBS and coverslips mounted in 70% glycerol in PBS. Digital images were obtained using a Zeiss Axioplan-2 microscope, Axiocam CCD camera, and Axiovision software (Carl Zeiss, Germany).
Quantitative assessment of whole-body lacZ expression across age was performed by spectrophotometric assessment of β-galactosidase activity as previously described [22]. Three adult males were homogenized in extraction buffer (50 mM potassium phosphate, 1 mM MgCl2, 0.5 μg/ml leupeptin, 0.5 μg/ml aprotenin, 0.7 μg/ml pepstatin A, pH 7.2) at the indicated ages. Homogenates were centrifuged at 16,000 × g for 5 min. β-galactosidase activity measurements (i.e. change in absorbance at 562 nM over 5 min in a Ultrospec 2000 spectrophotometer (Pharmacia Biotech, Piscataway, NJ)) in the resulting supernatants were initiated by addition of 100 μM chlorophenol red-β-D-galactopyranoside. β-galactosidase activity was expressed as the absorbance change per minute per milligram of homogenate protein. β-galactosidase activity in control flies containing either the UAS or Gal4 transgenes alone was negligible.
Transmission electron microscopy
Thoraces were dissected from adult male flies under anesthesia and immediately fixed in 1% glutaraldehyde/2% paraformaldehyde/0.2 M sodium cacodylate (pH 7.4), osmicated (1% osmium tetroxide), stained (1% uranyl acetate) and dehydrated in an ethanol series with microwave assistance. Fixed and dehydrated samples were briefly incubated in propylene oxide and then embedded in propylene oxide/Embed 812 resin. Thin sections (80–100 nm) were transferred to copper grids and visualized using a JEOL 1010 transmission electron microscope (JEOL USA Inc., Peabody, MA). Mitochondrial content in representative images was quantitated as the fraction of total image area occupied by mitochondria. Four to eight representative low magnification images were analyzed for each genotype at each age using Scion Image software resulting in a total image field of 0.65–1.3 mm2 analyzed.
ATP content
ATP content was assessed using the ATP Bioluminescent Assay Kit (Sigma Chemical Co., St. Louis, MO). For each sample, five thoraces were dissected from adult males under CO2 anesthesia, homogenized in 6 M guanine hydrochloride and heated at 95°C for 5 min to eliminate endogenous ATPase activity. Homogenates were centrifuged at 16,000 × g for 15 min at room temperature and the resulting supernatant was diluted 10-fold in sample buffer (0.2 M glycine, 50 mM MgCl2, 4 mM EDTA, pH 7.4). Luminescence was measured following addition of ATP Assay Kit reagents according to the manufacturer’s instructions in a Wallac 1420 Victor V plate reader (Perkin-Elmer, Waltham, MA). ATP concentrations of thorax homogenates were interpolated from standard curves using ATP standards.
Caspase activity
Caspase activity was assessed as the hydrolysis of the peptide substrate Ac-asp-glu-val-asp-7-amido-4-methylcoumarin (Ac-DEVD-AMC) essentially as described [23]. Briefly, two thoraces were homogenized in lysis buffer (50 mM Hepes, pH 7.5/100 mM NaCl/1 mM EDTA/0.1% CHAPS/10% sucrose/5 mM DTT/0.5% Triton X-100/4% glycerol) and centrifuged for 5min at 13,000 × g at 4°C to generate a supernatant. The supernatant was incubated with 25 mM Ac-DEVD-AMC for 1 h at 27°C and fluorescence of AMC product was determined using a Spectra Max Fluorescent Microplate Reader (Molecular Devices, Sunnyvale, CA) with excitation and emission set at 360 nm and 460 nm, respectively. Protease-specific caspase product was determined by subtracting fluorescence of a duplicate sample pre-treated with caspase inhibitor Ac-DEVD-CHO (2.5 mM) for 15 min from each sample.
Statistical analyses
Statistical treatment of data (one- and two-way ANOVA, Bonferroni’s multiple comparison tests, Tukey’s HSD multiple comparison tests, log-rank tests) were performed with JMP 5.01 (SAS Institute, Cary, NC, USA) and Prism 4.02 (GraphPad Software, San Diego, CA, USA).
Results
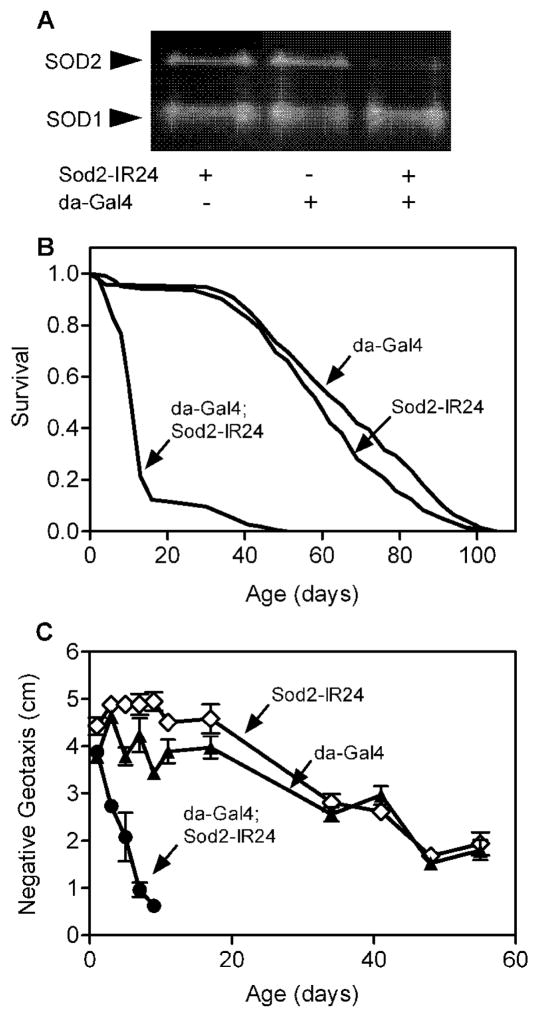
Knock-down of Sod2 in Drosophila via RNA interference (RNAi) using GAL4 to ubiquitously express a UAS-Sod2-inverted repeat (UAS-Sod2-IR) transgene substantially reduces SOD2 enzymatic activity, shortens life span and elicits hallmarks of mitochondrial dysfunction that are consistent with oxidative damage [13]. We confirmed that ubiquitous GAL4-driven expression of Sod2-IR knocked-down SOD2 (but not SOD1) activity (Fig. 1A) and decreased the life span of males (Fig. 1B) and females [15]. Additionally, ubiquitous expression of Sod2-IR caused a rapid age-dependent loss of negative geotaxis (i.e. bang-induced climbing, a locomotor behavior [24]) across age in males (Fig. 1C) and females [15]. Interestingly, negative geotaxis in 1-day-old Sod2 knock-down and control flies was indistinguishable, indicating that newly emerged flies with ubiquitous expression of Sod2-IR are not globally sick. This finding is consistent with a previous report showing that ubiquitous Sod2 knock-down does not affect eclosion of adult flies possibly because oxygen metabolism or ROS signaling are fundamentally different during development and adulthood [13].
Figure 1.
Ubiquitous expression of Sod2 RNAi reduces SOD2 activity, shortens life span and accelerates locomotor senescence. (A) Protein extracts (45 μg total protein/lane) from adult males (~2 days old) were electrophoresed on native polyacrylamide gels. SOD activity was visualized using an “in-gel” assay as described in materials and methods. (B) Mean and median life span in flies expressing Sod2-IR via da-Gal4 (da-Gal4; Sod2-IR24) were reduced compared to control flies harboring da-Gal4 or Sod2-IR24 transgenes alone (log-rank test, p < 0.0001, n = 150 flies per genotype). (C) Age-related loss of negative geotaxis was accelerated in flies expressing Sod2-IR via da-Gal4 (da-Gal4; Sod2-IR24) compared to da-Gal4 and Sod2-IR24 control flies (two-way ANOVA, p < 0.0001, n = 5 vials of 25 adult males per genotype). Negative geotaxis data are mean ± SEM.
Expression of Sod2-IR in the nervous system has modest effects on locomotor function and life span
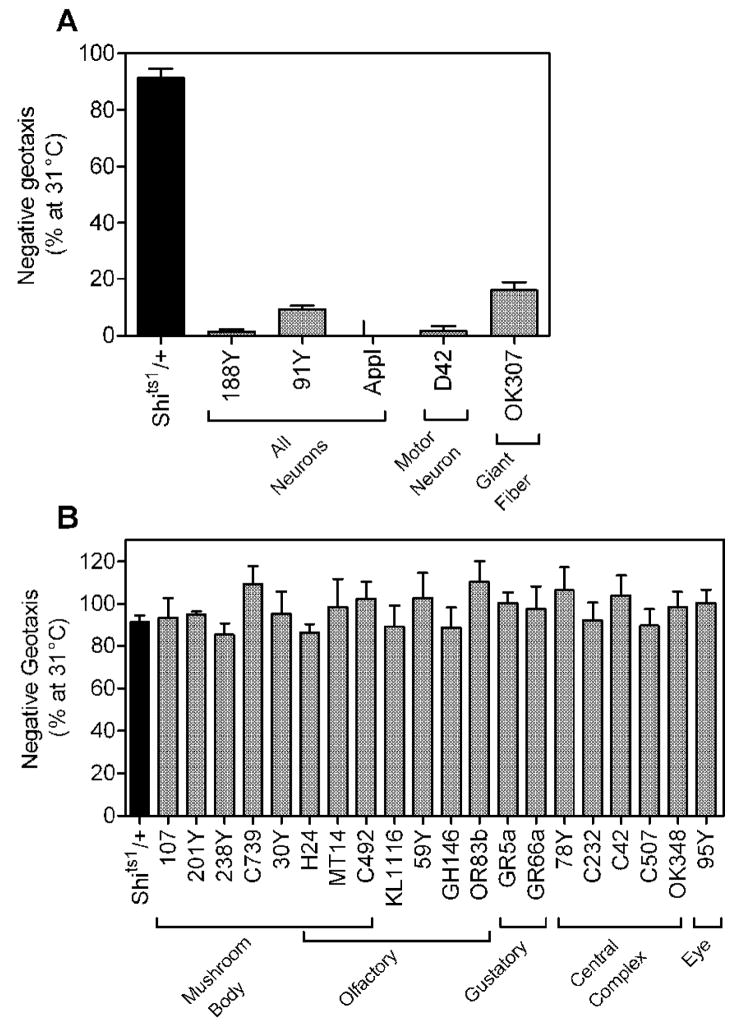
We reasoned that expression of Sod2-IR in the nervous system might accelerate age-related impairment in negative geotaxis and shorten life span. Tyrosine hydroxylase-Gal4 (TH-Gal4) expresses in dopaminergic neurons required for negative geotaxis [25], making it a useful tool for exploring the consequences of Sod2-IR expression in regions of the nervous system relevant to the behavior. To identify additional Gal4 lines for neurons involved in negative geotaxis, we assessed negative geotaxis in flies with expression of Shits1 (a temperature-sensitive dominant-negative dynamin that blocks neurotransmission [20, 26, 27] via a number of different Gal4 drivers. Flies with pan-neural expression of Shits1 via 188Y-Gal4, 91Y-Gal4 [28] and Appl-Gal4 [29] were largely paralyzed and had little or no negative geotaxis behavior remaining at the restrictive temperature (31°C), consistent with inhibition of neurotransmission throughout the nervous system (Fig. 2A). Expression of Shits1 by the spatially restricted Gal4 drivers D42 (motorneurons, [30] and OK307 (giant fiber neurons, [31]) also caused substantial defects in negative geotaxis at the restrictive temperature (Fig. 2A). In contrast, control flies with the UAS-Shits1 transgene and no Gal4 driver (Shits1/+) or flies with expression of Shits1 in mushroom body, olfactory, gustatory, central complex or eye neurons via a number of other Gal4 drivers had normal negative geotaxis at the restrictive temperature (Fig. 2B). Together, these studies indicate that 91Y, 188Y, Appl, D42, OK307 and TH express Gal4 in neurons required for negative geotaxis.
Figure 2.
Gal4 drivers for neurons involved in negative geotaxis. There was a significant overall effect of genotype (i.e. Gal4 driver used) on behavior (one-way ANOVA, p < 0.0001, n = 4–5 (representing 100–125 flies) for Gal4;Shits1 flies, n = 35 (representing 875 flies) for Shits1/+ control). (A) Expression of Shits1 via the Gal4 drivers 91Y, 188Y, Appl, D42 and OK307 caused a temperature-sensitive disruption of negative geotaxis (Tukey HSD multiple comparison, p < 0.05). (B) Negative geotaxis was not significantly affected by temperature in flies expressing Shits1 via all other Gal4 drivers (Tukey HSD multiple comparison). Data (mean ± S.E.M., compiled from 7 experiments) are the percent negative geotaxis observed at 25°C remaining at 31°C.
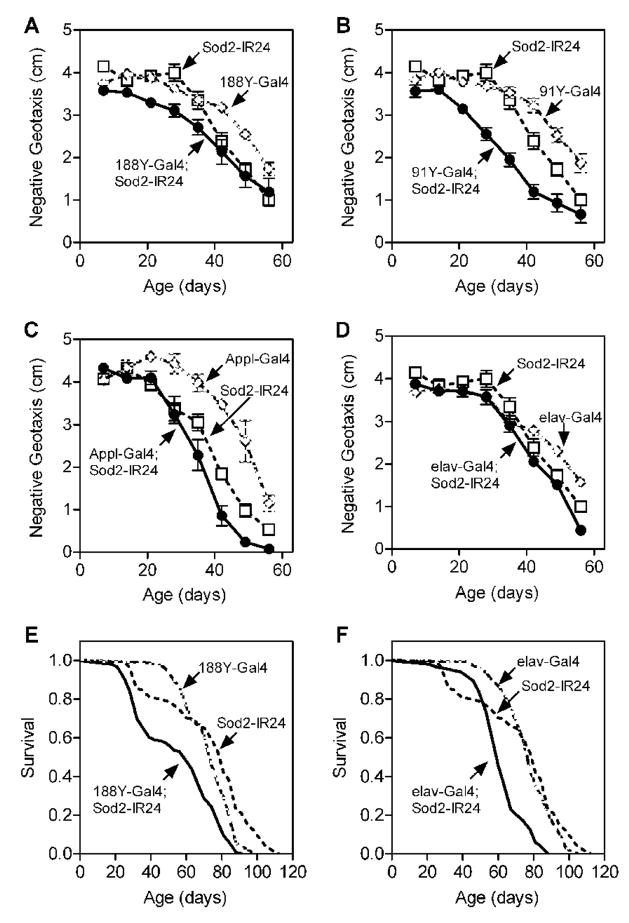
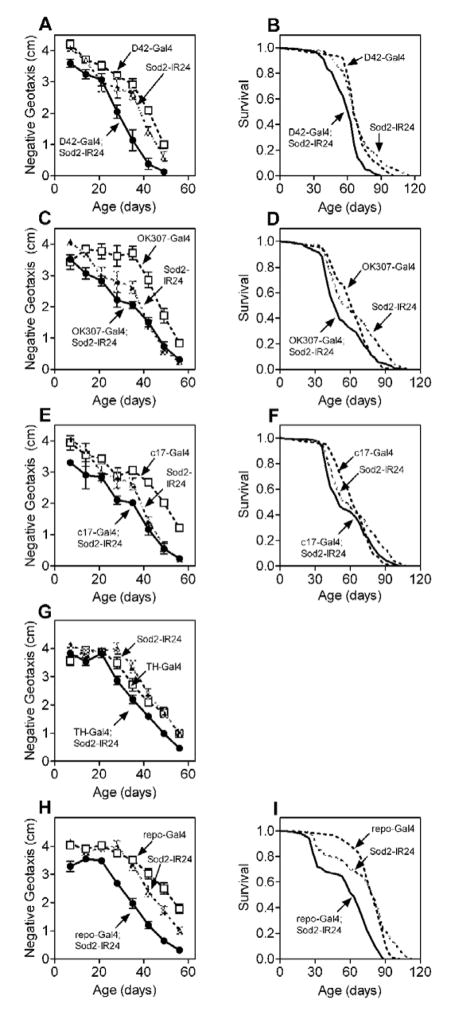
Expression of Sod2-IR via the pan-neuronal Gal4 drivers 188Y, 91Y and Appl caused modest but statistically discernable impairments in negative geotaxis across age compared to control flies harboring either the Gal4 or Sod2-IR transgene alone (Fig 3A–C). Expression of Sod2-IR using another pan-neuronal Gal4 driver, elav-Gal4 [32], likewise caused a subtle but statistically significant defect in negative geotaxis with age (Fig. 3D). Expression of Sod2-IR via 188Y-Gal4 and elav-Gal4 also decreased life span modestly (Fig. 3E and F). Additionally, expression of Sod2-IRin motor (D42-Gal4), dopaminergic (TH-Gal4) or giant fiber (OK307-Gal4 and C17-Gal4, [31]) neurons required for negative geotaxis or throughout the glia (repo-Gal4, [33]) caused subtle impairments in negative geotaxis across age and/or marginally reduced life span (Fig. 4). The detrimental effects of nervous system Sod2-IR expression on negative geotaxis and life span, however, were substantially smaller than those observed following ubiquitous Sod2-IR expression (Table).
Figure 3.
Nervous system Sod2-IR expression has modest effects on locomotor senescence and life span. Sod2-IR expression via the pan-neuronal drivers 188Y-Gal4 (A), 91Y-Gal4 (B), Appl-Gal4 (C) or elav-Gal4 (D) significantly impaired negative geotaxis behavior across age (individual two-way ANOVAs for effect of age and genotype, p < 0.0001 for both factors, n = 5–10 vials of 25 males per genotype). Tukey’s honestly significant difference (HSD) post-test revealed that all Sod2 knock-down lines performed significantly worse across age than either of their control groups (p < 0.05). Negative geotaxis data (mean ± SEM) are compiled from two independent experiments except in (D) which is from one experiment. Life span was also reduced in flies (150 per genotype) following Sod2-IR expression via 188Y-Gal4 (E) or elav-Gal4 (F). See Table for percent decrease in median life span.
Figure 4.
Neuronal Sod2 knock-down impairs locomotor function across age and shortens survival. Sod2-IR24 was expressed specifically in motor neurons via D42-Gal4 (A and B), giant fiber neurons via OK307-Gal4 (C and D) or c17-Gal4 (E and F), dopaminergic neurons via TH-Gal4 (G) and throughout glia via repo-Gal4 (H and I). Negative geotaxis performance across age (A, C, E, G, I) was significantly worse in flies expressing Sod2-IR24 via all Gal4 drivers except OK307-Gal4 in (C) (individual two-way ANOVAs for effect of age and genotype, p<0.0001, n = 5–10 vials of 25 male flies for both factors in all data sets, followed by Tukey’s HSD post-test (p < 0.05, or n.s. for OK307-Gal4;Sod2-IR24). Life span (B,D,F and I) was significantly reduced in all Sod2 knock-down genotypes (log-rank test p ≤ 0.0263, 150 flies per genotype). Survival was not assessed for Sod2-IR24 expression via TH-Gal4.
Table.
Accelerated loss of negative geotaxis and shortened life span in flies with Gal4-driven expression of Sod2-IR. The negative geotaxis decline time50 (DT50) is the time required for negative geotaxis performance to decline by 50% of original values. Percent decrease in DT50 or median life span for each Sod2 knock-down line was calculated compared to the poorest performing control containing Gal4 or Sod2-IR24 alone. n.d. = not determined.
| Gal4 driver | Tissue specificity | % ↓ in negative geotaxis DT50 | % ↓ in median life span |
|---|---|---|---|
| da-Gal4 | ubiquitous | 89 | 79 |
| Actin-Gal4 | ubiquitous | n.d. | 88 |
| elav-Gal4 | pan-neuronal | 5 | 23 |
| 188Y-Gal4 | pan-neuronal | 2 | 19 |
| 91Y-Gal4 | pan-neuronal | 19 | n.d. |
| Appl-Gal4 | pan-neuronal | 14 | n.d. |
| D42-Gal4 | motor neurons | 17 | 5 |
| TH-Gal4 | dopaminergic neurons | 15 | n.d. |
| c17-Gal4 | giant fiber system | 0 | 12 |
| OK307-Gal4 | giant fiber system | 0 | 19 |
| repo-Gal4 | pan-glial | 14 | 22 |
| Mef2-Gal4 | pan-muscle | 94 | 93 |
| 24B-Gal4 | pan-muscle | 89 | 95 |
| GMH5-Gal4 | cardiac muscle | 26 | 11 |
Expression of Sod2-IR in the musculature severely curtails life span and impairs locomotor function across age
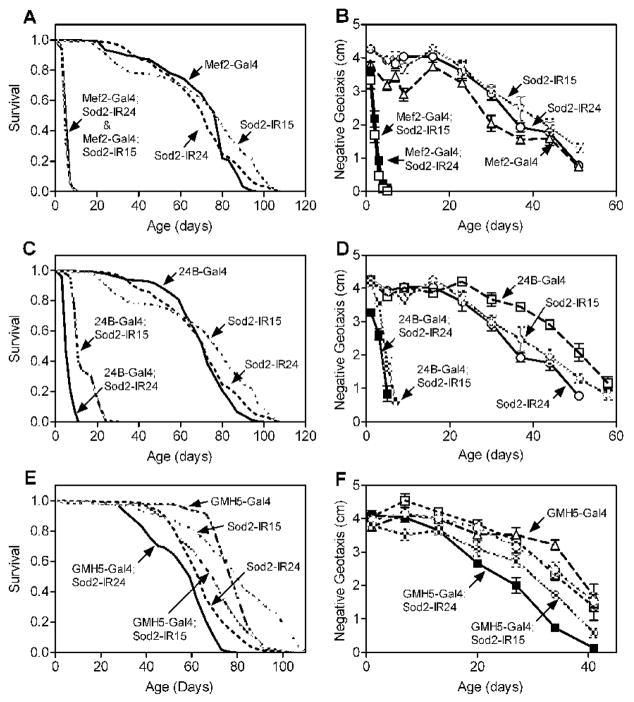
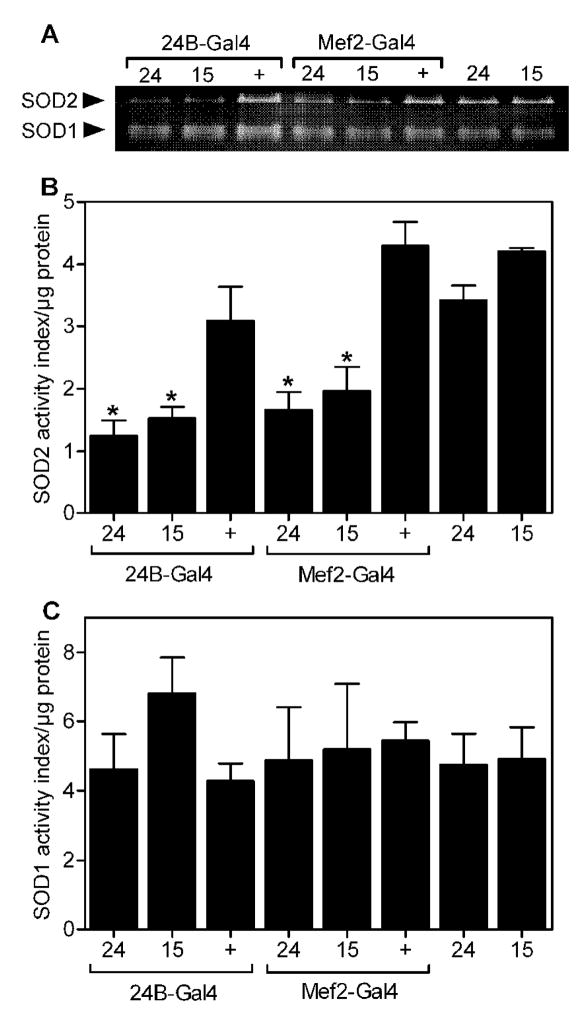
In stark contrast to the modest effects of Sod2-IR expression in the nervous system, expression of two independent Sod2-IR transgenes (Sod2-IR15 and Sod2-IR24) in the musculature using Mef2-Gal4 [34] dramatically reduced life span (Fig. 5A) and greatly accelerated the age-related impairment in negative geotaxis (Fig. 5B) compared to control flies with either the Gal4 driver alone or the Sod2-IR transgene alone. Sod2-IR expression in the musculature via 24B-Gal4 [35] also shortened life span substantially (Fig. 5C) and hastened the age-related loss of locomotor function (Fig. 5D). Expression of Sod2-IR via Mef2-Gal4 or 24B-Gal4 reduced SOD2 activity in whole body extracts without significantly impacting the activity of SOD1 (Fig. 6), confirming that expression of Sod2-IR knocks-down the function of Sod2. Interestingly, the life span and locomotor effects of Sod2-IR expression in the musculature via Mef2-Gal4 and 24B-Gal4 were comparable to or possibly more severe than with the ubiquitous drivers Actin-Gal4 and da-Gal4 (Table).
Figure 5.
Expression of Sod2-IR in muscle reduces life span and accelerates age-related locomotor impairment. Two independent Sod2-IR transgenes, Sod2-IR15 and Sod2-IR24, were expressed via Mef2-Gal4 (A and B), 24B-Gal4 (C and D), or GMH5-Gal4 (E and F). (A, C and E) Life span was significantly shortened by expression of Sod2-IR via all three Gal4 drivers (log-rank test, p < 0.0001, n = 150 flies per genotype). (B, D and F) Age-related locomotor impairment was accelerated in flies with Sod2-IR expression driven by all three Gal4 lines (two-way ANOVA; effect of age and genotype, p < 0.0001; Tukey’s HSD multiple comparison, p < 0.05; n = 5 vials of 25 flies per genotype). Data are representative of two independent experiments.
Figure 6.
Sod2-IR expression in the musculature knocks-down SOD2 activity. (A) Whole-body SOD activity. SOD activity was measured in whole-body extracts using an in gel assay described in materials and methods. Each lane contained 45 μg total protein from adult males (~2 days old). Flies had the 24B-Gal4, Mef2-Gal4 or no Gal4 driver along with the Sod2-IR24 (24), Sod2-IR15 (15), or no Sod2-IR transgene (+). (B) Quantitation of whole-body SOD activity by densitometry. Whole-body SOD2 activity was significantly reduced in flies with Sod2-IR24 and Sod2-IR15 expression driven by 24B-Gal4 and Mef2-Gal4 compared to controls with the Gal4 or the Sod2-IR transgenesalone (individual one-way ANOVAs, p = 0.0126, n = 3 groups of 25 flies per genotype, followed by Tukey’s HSD post-test, * p < 0.05). (C) Quantitation of whole-body SOD1 activity. Whole-body SOD1 activity was not affected by genotype (individual one-way ANOVAs, not significant, n = 3 groups of 25 flies per genotype). Data in B and C are mean ± SEM.
We expressed Sod2-IR in cardiac muscle to determine whether Sod2 function in this tissue might be important for life span and locomotor behavior across age. Median life span was reduced by ~11% (Fig. 5E) and age-related loss of locomotor behavior was significantly accelerated (Fig. 5F) by expression of Sod2-IR driven by the cardiac Gal4 line GMH5 [36]. The life span and locomotor effects of GMH5-driven Sod2-IR expression, however, were considerably less severe than observed with Mef2-Gal4 and 24B-Gal4 (Table).
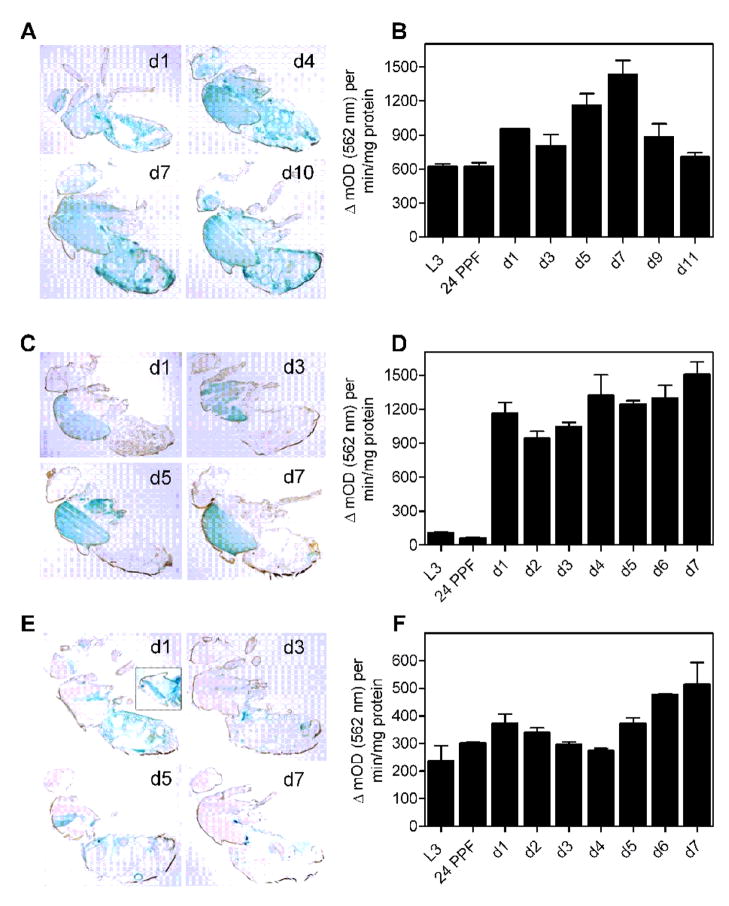
We examined lacZ reporter expression in whole-body adult cryosections to characterize the expression patterns for da-Gal4, Mef2-Gal4 and 24B-Gal4 (Fig. 7A, C and E). Gal4drivers often exhibit age-dependent patterns of expression [22, 37] and could, in principle, be sensitive to the enhanced oxidative stress in flies with knock-down of Sod2. We therefore assessed da-Gal4, Mef2-Gal4 and 24B-GAL4 expression patterns across the shortened adult life span of flies that concurrently expressed UAS-lacZ and UAS-Sod2-IR24 transgenes. As expected [13, 29], da-Gal4 drove expression of lacZ throughout the body (Fig. 7A). Mef2-Gal4 drove expression mainly in the thoracic muscle (Fig. 7C) while 24B-Gal4 drove lacZ expression in thoracic muscle, abdominal muscle and fat body (Fig. 7E and inset for d1). This is largely consistent with the muscle-specific expression patterns previously reported for these two Gal4 drivers [34, 38–40]. At the level of resolution possible with this approach, we did not observe a substantive change in lacZ expression across the short life span of Sod2 knock-down flies driven by the three Gal4 drivers (Fig. 7A, C and E). The spatial patterns of expression in da-Gal4, Mef2-Gal4 and 24B-Gal4 drivers overlap in the musculature, but do not appreciably change with age in short-lived Sod2 knock-down animals.
Figure 7.
Spatial and temporal expression patterns of Gal4 in Sod2 knock-down flies. Gal4 expression was assessed in flies harboring the indicated Gal4 driver, a Sod2-IR24 transgene and a UAS-lacZ reporter. Whole-body sagittal cryosections stained for β-galactosidase activity (blue) for da-Gal4 (A), Mef2-Gal4(C) and 24B-Gal4 (E) at the indicated ages in days. (A) da-Gal4 expressed ubiquitously at all ages examined. (C) Mef2-Gal4 expressed strongly in indirect flight muscle and to a lesser extent in other muscles across age. (E) 24B-Gal4 expressed in the indirect flight muscle and fat body across age. Expression of 24B-Gal4 throughout thoracic muscle was observed upon incubating tissue sections for longer periods in X-gal substrate (E, D1 panel inset). lacZ expression driven by da-Gal4 (B), Mef2-Gal4(D) and 24B-Gal4 (F) were quantified in whole-body extracts of 3rd instar larvae (L3), pupae 24 hrs post-puparium formation (24 PPF) and adults at the indicated ages in days. There was a significant effect of age on Gal4 expression levels for all three Gal4 drivers (individual ANOVAs, p < 0.0001, n = 3 groups of 3 flies per genotype). Data in B, D and F are mean ± SEM.
We quantified lacZ reporter activity in whole body extracts of Sod2 knock-down animals throughout development and adulthood to characterize the temporal expression patterns of the da-Gal4, Mef2-Gal4 and 24B-Gal4 drivers. Whole body lacZ reporter activity was dynamic across age for all three drivers. Expression of lacZ driven by da-Gal4 was easily detectable during late larval and pupal stages, increased to a peak of expression at day 7 of adulthood, and then declined by day 11 of adulthood (close to the maximum life span of flies expressing Sod2-IR ubiquitously) to a level similar to that found in larvae and pupae (Fig. 7B). Mef2-Gal4 drove minimal lacZ expression during larval and pupal stages, but thereafter drove very strong expression in adults that increased out to the near-maximal life span of 7 days in flies expressing Sod2-IR via Mef2-Gal4 (Fig. 7D). 24B-Gal4 expressed lacZ at moderate levels during development with peaks of expression at days 1 and 7 of adulthood (Fig. 7F). Although the three Gal4 drivers have distinct temporal patterns of expression, they share the common feature of robust expression during adulthood.
To address whether normal expression of SOD2 in muscles is limiting for life span and age-related locomotor impairment, we assessed survival and negative geotaxis across age in flies overexpressing Sod2 using the Gal4 drivers Mef2 and 24B. Overexpression of Sod2 in the musculature did not extend life span or ameliorate age-related locomotor impairment (data not shown). In fact, Sod2 overexpression driven by Mef2-Gal4 resulted in a slight decrease in life span and acceleration of age-related locomotor impairment.
Cellular consequences of Sod2 knock-down
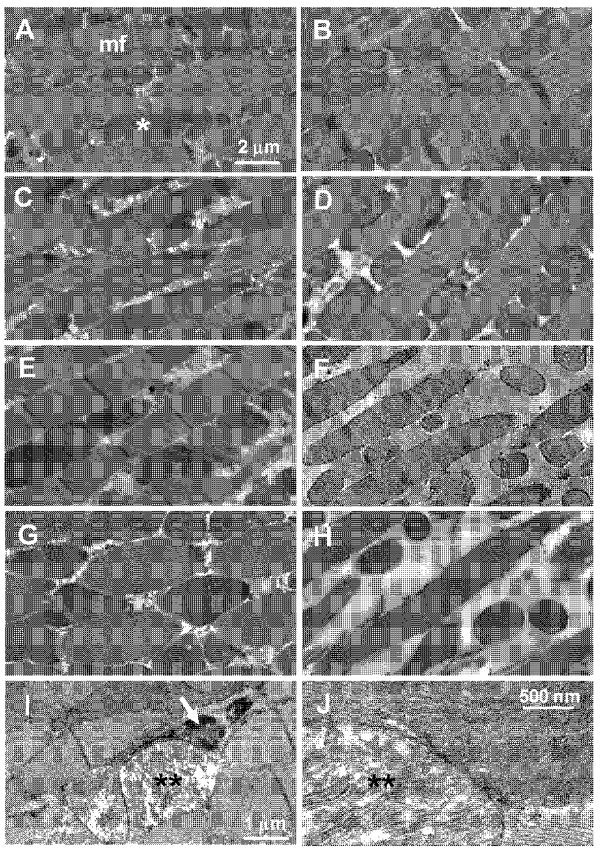
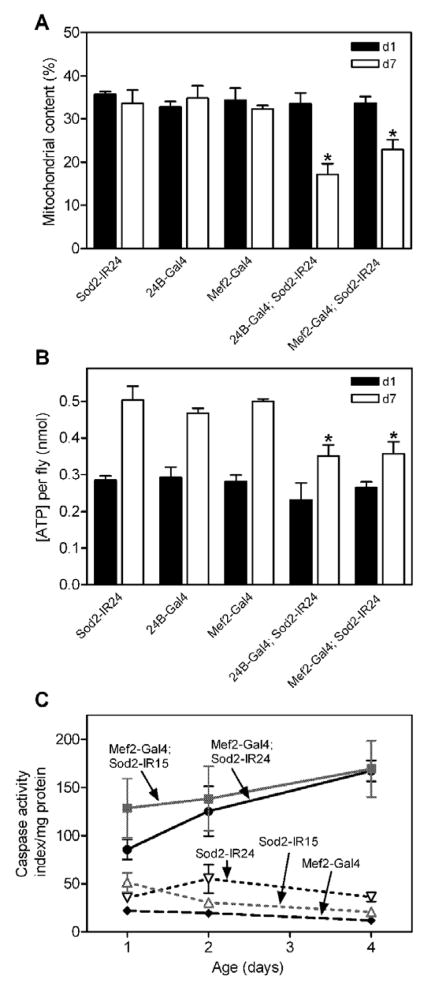
To gain insight into the cellular consequences of knocking-down Sod2, we examined the ultrastructure of indirect flight muscle (IFM) by transmission electron microscopy (TEM). IFM ultrastructure was indistinguishable at 1-day (Fig. 8A and C) and 7-days (Fig. 8B and D) of age in control flies. The IFM in control animals at both ages had a regular arrangement of myofibrils interspersed by rows of densely-packed mitochondria. The IFM from flies expressing Sod2-IR in the musculature via 24B-Gal4 (Fig. 8E) or Mef2-Gal4 (Fig. 8G) also had largely normal ultrastructure at 1-day of age, although there were occasional clusters of swollen mitochondria (Fig. 8I, J). The swollen mitochondria in 1-day old Sod2 knock-down flies exhibited a rarified arrangement of cristae and in some cases were associated with lysosomes (Fig. 8I, arrow), indicating that the damaged mitochondria were being removed by autophagy. By 7 days of age, the IFM in flies with Sod2 knock-down in muscle had an obvious reduction in mitochondrial content yet they retained normal myofibril ultrastructure (Fig. 8F and H). Quantitative assessment of TEM images revealed that while mitochondrial content was maintained in control flies out to 7 days of age, it was reduced by 32–49% in Sod2 knock-down animals during this same time (Fig. 9A).
Figure 8.
Knock-down of Sod2 causes mitochondrial pathology in indirect flight muscles (IFMs). Images are TEM analysis of IFM in 1-day-old (A, C, E, G, I and J) and 7-day-old (B, D, F and H) flies. (A and B), control Sod2-IR24 alone. (C and D), control 24B-Gal4 alone. (E, F, I and J), 24B-Gal4; Sod2-IR24. (G and H), Mef2-Gal4; Sod2-IR24. Representative myofibrils are indicated by mf and representative mitochondria are indicated by an asterisk (*) in panel A. IFMs of 1-day-old Sod2 knock-down flies contained swollen mitochondria with a rarified arrangement of cristae (I and J, **). Swollen mitochondria were often engulfed in autophagolysosomes (I, arrow).
Figure 9.
Sod2 knock-down in muscle causes loss of mitochondrial content, decreased ATP and increased caspase activity. (A) Quantitation of mitochondrial content in IFMs at 1 day (black bars) and 7 days (open bars) of age in controls (Sod2-IR24, 24B-Gal4 or Mef2-Gal4 alone) or Sod2 knock-down flies (Sod2-IR24; 24B-Gal4 and Sod2-IR24; Mef2-Gal4). Two-way ANOVA indicated significant overall effects of age (p < 0.001) and genotype (p < 0.01) with an interaction (p < 0.01) between these two factors (n = 4–8 images per genotype). Bonferroni’s multiple comparison post-tests revealed a significant reduction in mitochondrial content between controls and Sod2 knock-down flies at 7 days of age (* p < 0.05). (B) Thoracic ATP content in control and Sod2 knock-down flies at 1 (black bars) and 7 (open bars) days of age. Age (p < 0.0001) and genotype (p < 0.01) had significant effects on thoracic ATP content (two-way ANOVA, n = 3 groups of 5 flies per genotype). Sod2 knock-down genotypes had significantly less ATP than controls at 7 days of age (Bonferroni’s post-tests, * p < 0.05). (C) Caspase activity (nM AMC/sec/mg protein) in control (Sod2-IR24, Sod2-IR15 or Mef2-Gal4 alone) and Sod2 knock-down flies (Mef2-Gal4; Sod2-IR15 and Mef2-Gal4; Sod2-IR24) flies. Overall, caspase activity was significantly elevated in both Sod2 knock-down genotypes relative to control flies (two-way ANOVA, p < 0.0001, n = 4 groups of 2 flies per genotype). Bonferroni’s post-tests revealed that caspase activity was increased in both Sod2 knock-down genotypes at all ages (p< 0.05). Data (mean ± SEM) are representative of 2 independent experiments.
Ubiquitous disruption of Sod2 function through mutations in mice [11] and flies [14] or RNAi-mediated knock-down of Sod2 in flies [13] causes oxidative damage to and decreased function of mitochondrial enzymes important in energy metabolism. We measured ATP content in extracts from Sod2 knock-down flies to determine whether decreased SOD2 activity in the musculature impaired mitochondrial function. We focused on ATP content in the thorax because this tissue is predominantly composed of muscle in Drosophila. ATP content increased significantly from day 1 to day 7 of adulthood in control flies harboring a Sod2-IR transgene alone, 24B-Gal4 alone, or Mef2-Gal4 alone (Fig. 9B) as expected from previous reports [41]. ATP content at 1 day of age was indistinguishable in control flies and in flies with knock-down of Sod2 in the muscle (Fig. 9B). By 7-days of age, however, Sod2 knock-down flies had a 25–30% reduction in thoracic ATP content compared to age-matched controls (Fig. 9B).
Decreased SOD2 activity causes an elevation in apoptosis in mouse liver, retina and heart [9, 42–44] and also in the fly brain [14]. We therefore determined whether knock-down of Sod2 in the musculature altered caspase activity (a marker of apoptosis) and, if so, whether caspase activity changed with age in Sod2 knock-down flies. As expected from previous studies [23], control flies had low levels of caspase activity in thoraces out to 4 days of age (Fig. 9C). In Sod2 knock-down flies, caspase activity was significantly elevated at all ages examined.
Discussion
SOD2 is a critical antioxidant localized to mitochondria [5]. Whole-body loss of Sod2 function through RNAi-mediated silencing or knock-out of the endogenous Sod2 gene, dramatically shortens life span and causes behavioral defects in flies [12–15], and mice [6–8]. Here, we used tissue-specific expression of Sod2-IR transgenes in Drosophila to address whether these effects on life span and behavior are due to loss of Sod2 function throughout the body or within specific tissues. We focused on the nervous system and the musculature due to their high metabolic activity with attendant generation of ROS [4].
Expression of Sod2-IR transgenes throughout the nervous system, in specific subsets of neurons required for negative geotaxis behavior, or in glia modestly shortened life span and accelerated age-related loss of negative geotaxis. The effects of nervous system expression of Sod2-IR, however, were considerably less severe than observed in flies with whole-body Sod2-IR expression (Table). While other interpretations are possible, these data suggest that the nervous system may not be a key site of Sod2 function within the context of life span and locomotor function across age in flies.
Consistent with the possibility that tissues outside of the nervous system are important for Sod2 function, expression of Sod2-IR in the musculature via two different Gal4 lines (Mef2 and 24B) dramatically shortened life span and accelerated the age-dependent loss of locomotor behavior (Table). The effect of Sod2-IR expression via these two Gal4 drivers was equal to or even greater than the effect of ubiquitous Sod2 knock-down (Table). Importantly, we confirmed that Mef2-Gal4 and 24B-Gal4 drive expression in the thoracic flight muscle and found that both were expressed throughout the short life span of Sod2 knock-down flies. Together, these data indicate that Sod2 functions within the musculature to protect flies from premature death and loss of locomotor behavior.
Previous studies point toward Sod2 being important for normal muscle function and integrity. For example, Sod2 knock-out mice exhibit reduced activity of ETC components and lipid accumulation in skeletal muscle [7, 11]. Additionally, Sod2 knock-out mice display a dilated cardiomyopathy [6, 7]. These studies, however, do not explicitly address the importance of Sod2 function in the muscle for whole organism vitality and vigor. Our studies extend these previous reports by demonstrating that normal life span and locomotor function depend on expression of Sod2 within the musculature in Drosophila.
The effects of Sod2 knock-down in the musculature led us to propose that overexpression of this enzyme in this tissue might extend life span or preserve locomotor behavior across age. Overexpression of Sod2 in muscle, however, had no effect or possibly even negative effects on these two measures. One possible interpretation of these data is that Sod2 in Drosophila muscle is not limiting for life span or locomotor senescence. An extrapolation of our findings is that the life span-extending effect of ubiquitous Sod2 overexpression [45] might be due to increased SOD2 activity in a tissue other than or in addition to the musculature. Our data, however, are also consistent with other interpretations. For example, increased longevity was obtained via ubiquitous Sod2 overexpression conditionally during adulthood only [45] whereas in the GAL4 system used here, Sod2 overexpression occurs during development and adulthood. Hence, it is possible that overexpression of Sod2 during development has negative physiological consequences that counteract any positive effects of Sod2 overexpression on aging during adulthood. Additionally, it is possible that overexpression of Sod2 beyond a level providing maximal benefit might have negative consequences on aging. Since ROS can act as important signaling molecules [46] in addition to driving oxidative damage, any negative outcomes associated with Sod2 overexpression could be due to inhibition of ROS signaling. Additional experiments are required to address these possibilities.
Sod2 knock-out mice exhibit dilated cardiomyopathy, a severe degenerative state thought to be an important contributor to the perinatal mortality in these animals [6, 7]. Interestingly, patients with hereditary hemochromatosis that carry a Sod2 allele associated with reduced enzymatic activity were found to be at a higher risk for developing cardiomyopathy [47]. Although expression of Sod2-IR in Drosophila cardiac muscle shortened life span and accelerated the loss of locomotor function across age, these effects were rather modest compared to knocking-down Sod2 ubiquitously or in the flight muscle (Table). Hence in flies, Sod2 function within cardiac muscle is important for normal life span and locomotor function across age, but other muscles such as thoracic flight muscle are likely to be more important sites of SOD2 activity.
Sod2 knock-out mice display swollen mitochondria in nervous and muscle tissue [6, 43, 48], although this phenotype is not universally observed [7]. Knock-down of Sod2 in the musculature of flies caused mitochondrial swelling in 1-day-old animals that was followed by a decrease in both mitochondrial and ATP content by 7 days of age. Loss of Sod2 in muscle, therefore, causes a progressive decrease in mitochondrial functional capacity that manifests during adulthood in Drosophila.
There is a strong link between oxidative stress and apoptosis [49]. For example, exposure of cells or intact animals to pro-oxidant chemicals (i.e. exogenous oxidative stress) induces apoptosis [23, 50, 51]. Oxidative stress derived from endogenous sources can also activate apoptosis. Sod2 knock-out mice have elevated apoptosis in liver, retina and heart [9, 42, 44, 52] and Sod2 null flies have increased apoptosis in the central brain [14]. Our studies indicate that knock-down of Sod2 in the musculature is sufficient to induce a large increase in apoptosis in Drosophila, which could be an important mechanism related to the short life span and possibly other phenotypes in Sod2 knock-down flies. Additionally, our studies support the notion that induction of apoptosis by knock-down of Sod2 can occur in a tissue autonomous fashion and therefore does not require loss of Sod2 throughout the body.
Acknowledgments
This work was supported by grant AG024259 from the National Institutes for Aging (M.G.) and grant MOP79519 from the Institute of Aging of the Canadian Institutes of Health Research (L.S.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Fridovich I. Mitochondria: are they the seat of senescence? Aging cell. 3:13–16. doi: 10.1046/j.1474-9728.2003.00075.x. [DOI] [PubMed] [Google Scholar]
- 2.Chance B, Sies H, Boveris A. Hydroperoxide metabolism in mammalian organs. Physiological reviews. 59:527–605. doi: 10.1152/physrev.1979.59.3.527. [DOI] [PubMed] [Google Scholar]
- 3.Balaban RS, Nemoto S, Finkel T. Mitochondria, oxidants, and aging. Cell. 120:483–495. doi: 10.1016/j.cell.2005.02.001. [DOI] [PubMed] [Google Scholar]
- 4.Halliwell B, Gutteridge JM. Free Radicals in Biology and Medicine. Oxford: Oxford University Press; 1999. [Google Scholar]
- 5.Landis GN, Tower J. Superoxide dismutase evolution and life span regulation. Mechanisms of ageing and development. 126:365–379. doi: 10.1016/j.mad.2004.08.012. [DOI] [PubMed] [Google Scholar]
- 6.Lebovitz RM, Zhang H, Vogel H, Cartwright J, Jr, Dionne L, Lu N, Huang S, Matzuk MM. Neurodegeneration, myocardial injury, and perinatal death in mitochondrial superoxide dismutase-deficient mice. Proceedings of the National Academy of Sciences of the United States of America. 93:9782–9787. doi: 10.1073/pnas.93.18.9782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li Y, Huang TT, Carlson EJ, Melov S, Ursell PC, Olson JL, Noble LJ, Yoshimura MP, Berger C, Chan PH, Wallace DC, Epstein CJ. Dilated cardiomyopathy and neonatal lethality in mutant mice lacking manganese superoxide dismutase. Nature genetics. 11:376–381. doi: 10.1038/ng1295-376. [DOI] [PubMed] [Google Scholar]
- 8.Huang TT, Yasunami M, Carlson EJ, Gillespie AM, Reaume AG, Hoffman EK, Chan PH, Scott RW, Epstein CJ. Superoxide-mediated cytotoxicity in superoxide dismutase-deficient fetal fibroblasts. Archives of biochemistry and biophysics. 344:424–432. doi: 10.1006/abbi.1997.0237. [DOI] [PubMed] [Google Scholar]
- 9.Kokoszka JE, Coskun P, Esposito LA, Wallace DC. Increased mitochondrial oxidative stress in the Sod2 (+/−) mouse results in the age-related decline of mitochondrial function culminating in increased apoptosis. Proceedings of the National Academy of Sciences of the United States of America. 98:2278–2283. doi: 10.1073/pnas.051627098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Van Remmen H, Ikeno Y, Hamilton M, Pahlavani M, Wolf N, Thorpe SR, Alderson NL, Baynes JW, Epstein CJ, Huang TT, Nelson J, Strong R, Richardson A. Life-long reduction in MnSOD activity results in increased DNA damage and higher incidence of cancer but does not accelerate aging. Physiological genomics. 16:29–37. doi: 10.1152/physiolgenomics.00122.2003. [DOI] [PubMed] [Google Scholar]
- 11.Esposito LA, Melov S, Panov A, Cottrell BA, Wallace DC. Mitochondrial disease in mouse results in increased oxidative stress. Proceedings of the National Academy of Sciences of the United States of America. 96:4820–4825. doi: 10.1073/pnas.96.9.4820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Duttaroy A, Paul A, Kundu M, Belton A. A Sod2 null mutation confers severely reduced adult life span in Drosophila. Genetics. 165:2295–2299. doi: 10.1093/genetics/165.4.2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kirby K, Hu J, Hilliker AJ, Phillips JP. RNA interference-mediated silencing of Sod2 in Drosophila leads to early adult-onset mortality and elevated endogenous oxidative stress. Proceedings of the National Academy of Sciences of the United States of America. 99:16162–16167. doi: 10.1073/pnas.252342899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Paul A, Belton A, Nag S, Martin I, Grotewiel MS, Duttaroy A. Reduced mitochondrial SOD displays mortality characteristics reminiscent of natural aging. Mechanisms of ageing and development. 128:706–716. doi: 10.1016/j.mad.2007.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bhandari P, Jones MA, Martin I, Grotewiel MS. Dietary restriction alters demographic but not behavioral aging in Drosophila. Aging cell. 6:631–637. doi: 10.1111/j.1474-9726.2007.00320.x. [DOI] [PubMed] [Google Scholar]
- 16.Piazza N, Hayes M, Martin I, Duttaroy A, Grotewiel M, Wessells R. Multiple measures of functionality exhibit progressive decline in a parallel, stochastic fashion in Drosophila Sod2 null mutants. Biogerontology. 2009 doi: 10.1007/s10522-008-9210-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development (Cambridge, England) 118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- 18.Gargano JW, Martin I, Bhandari P, Grotewiel MS. Rapid iterative negative geotaxis (RING): a new method for assessing age-related locomotor decline in Drosophila. Experimental gerontology. 40:386–395. doi: 10.1016/j.exger.2005.02.005. [DOI] [PubMed] [Google Scholar]
- 19.Martin I, Gargano JW, Grotewiel MS. A proposed set of descriptors for functional senescence data. Aging cell. 4:161–164. doi: 10.1111/j.1474-9726.2005.00155.x. [DOI] [PubMed] [Google Scholar]
- 20.Kitamoto T. Conditional modification of behavior in Drosophila by targeted expression of a temperature-sensitive shibire allele in defined neurons. J Neurobiol. 47:81–92. doi: 10.1002/neu.1018. [DOI] [PubMed] [Google Scholar]
- 21.Kitamoto T. Conditional disruption of synaptic transmission induces male-male courtship behavior in Drosophila. Proceedings of the National Academy of Sciences of the United States of America. 99:13232–13237. doi: 10.1073/pnas.202489099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Seroude L, Brummel T, Kapahi P, Benzer S. Spatio-temporal analysis of gene expression during aging in Drosophila melanogaster. Aging cell. 1:47–56. doi: 10.1046/j.1474-9728.2002.00007.x. [DOI] [PubMed] [Google Scholar]
- 23.Zheng J, Edelman SW, Tharmarajah G, Walker DW, Pletcher SD, Seroude L. Differential patterns of apoptosis in response to aging in Drosophila. Proceedings of the National Academy of Sciences of the United States of America. 102:12083–12088. doi: 10.1073/pnas.0503374102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rhodenizer D, Martin I, Bhandari P, Pletcher SD, Grotewiel M. Genetic and environmental factors impact age-related impairment of negative geotaxis in Drosophila by altering age-dependent climbing speed. Experimental gerontology. 43:739–748. doi: 10.1016/j.exger.2008.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Friggi-Grelin F, Coulom H, Meller M, Gomez D, Hirsh J, Birman S. Targeted gene expression in Drosophila dopaminergic cells using regulatory sequences from tyrosine hydroxylase. J Neurobiol. 54:618–627. doi: 10.1002/neu.10185. [DOI] [PubMed] [Google Scholar]
- 26.Chen MS, Obar RA, Schroeder CC, Austin TW, Poodry CA, Wadsworth SC, Vallee RB. Multiple forms of dynamin are encoded by shibire, a Drosophila gene involved in endocytosis. Nature. 351:583–586. doi: 10.1038/351583a0. [DOI] [PubMed] [Google Scholar]
- 27.van der Bliek AM, Meyerowitz EM. Dynamin-like protein encoded by the Drosophila shibire gene associated with vesicular traffic. Nature. 351:411–414. doi: 10.1038/351411a0. [DOI] [PubMed] [Google Scholar]
- 28.Armstrong JD. Fly Trap Lines. www.fly-trap.org; www.fly-trap.org.
- 29.Kapahi P, Zid BM, Harper T, Koslover D, Sapin V, Benzer S. Regulation of lifespan in Drosophila by modulation of genes in the TOR signaling pathway. Curr Biol. 14:885–890. doi: 10.1016/j.cub.2004.03.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Parkes TL, Elia AJ, Dickinson D, Hilliker AJ, Phillips JP, Boulianne GL. Extension of Drosophila lifespan by overexpression of human SOD1 in motorneurons. Nature genetics. 19:171–174. doi: 10.1038/534. [DOI] [PubMed] [Google Scholar]
- 31.Fayyazuddin A, Zaheer MA, Hiesinger PR, Bellen HJ. The nicotinic acetylcholine receptor Dalpha7 is required for an escape behavior in Drosophila. PLoS Biol. 2006;4:e63. doi: 10.1371/journal.pbio.0040063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Olofsson B, Page DT. Condensation of the central nervous system in embryonic Drosophila is inhibited by blocking hemocyte migration or neural activity. Developmental biology. 279:233–243. doi: 10.1016/j.ydbio.2004.12.020. [DOI] [PubMed] [Google Scholar]
- 33.Lievens JC, Iche M, Laval M, Faivre-Sarrailh C, Birman S. AKT-sensitive or insensitive pathways of toxicity in glial cells and neurons in Drosophila models of Huntington’s disease. Human molecular genetics. 17:882–894. doi: 10.1093/hmg/ddm360. [DOI] [PubMed] [Google Scholar]
- 34.Ranganayakulu G, Elliott DA, Harvey RP, Olson EN. Divergent roles for NK-2 class homeobox genes in cardiogenesis in flies and mice. Development (Cambridge, England) 125:3037–3048. doi: 10.1242/dev.125.16.3037. [DOI] [PubMed] [Google Scholar]
- 35.van der Plas MC, Pilgram GS, Plomp JJ, de Jong A, Fradkin LG, Noordermeer JN. Dystrophin is required for appropriate retrograde control of neurotransmitter release at the Drosophila neuromuscular junction. J Neurosci. 26:333–344. doi: 10.1523/JNEUROSCI.4069-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wessells RJ, Fitzgerald E, Cypser JR, Tatar M, Bodmer R. Insulin regulation of heart function in aging fruit flies. Nature genetics. 36:1275–1281. doi: 10.1038/ng1476. [DOI] [PubMed] [Google Scholar]
- 37.Seroude L. GAL4 drivers expression in the whole adult fly. Genesis. 34:34–38. doi: 10.1002/gene.10128. [DOI] [PubMed] [Google Scholar]
- 38.Deng H, Dodson MW, Huang H, Guo M. The Parkinson’s disease genes pink1 and parkin promote mitochondrial fission and/or inhibit fusion in Drosophila. Proceedings of the National Academy of Sciences of the United States of America. 105:14503–14508. doi: 10.1073/pnas.0803998105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Roper K, Mao Y, Brown NH. Contribution of sequence variation in Drosophila actins to their incorporation into actin-based structures in vivo. Journal of cell science. 118:3937–3948. doi: 10.1242/jcs.02517. [DOI] [PubMed] [Google Scholar]
- 40.Thompson EC, Travers AA. A Drosophila Smyd4 homologue is a muscle-specific transcriptional modulator involved in development. PLoS ONE. 2008;3:e3008. doi: 10.1371/journal.pone.0003008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vernace VA, Arnaud L, Schmidt-Glenewinkel T, Figueiredo-Pereira ME. Aging perturbs 26S proteasome assembly in Drosophila melanogaster. Faseb J. 21:2672–2682. doi: 10.1096/fj.06-6751com. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Van Remmen H, Williams MD, Guo Z, Estlack L, Yang H, Carlson EJ, Epstein CJ, Huang TT, Richardson A. Knockout mice heterozygous for Sod2 show alterations in cardiac mitochondrial function and apoptosis. American journal of physiology. 2001;281:H1422–1432. doi: 10.1152/ajpheart.2001.281.3.H1422. [DOI] [PubMed] [Google Scholar]
- 43.Sandbach JM, Coscun PE, Grossniklaus HE, Kokoszka JE, Newman NJ, Wallace DC. Ocular pathology in mitochondrial superoxide dismutase (Sod2)-deficient mice. Investigative ophthalmology & visual science. 42:2173–2178. [PubMed] [Google Scholar]
- 44.Strassburger M, Bloch W, Sulyok S, Schuller J, Keist AF, Schmidt A, Wenk J, Peters T, Wlaschek M, Lenart J, Krieg T, Hafner M, Kumin A, Werner S, Muller W, Scharffetter-Kochanek K. Heterozygous deficiency of manganese superoxide dismutase results in severe lipid peroxidation and spontaneous apoptosis in murine myocardium in vivo. Free radical biology & medicine. 38:1458–1470. doi: 10.1016/j.freeradbiomed.2005.02.009. [DOI] [PubMed] [Google Scholar]
- 45.Sun J, Folk D, Bradley TJ, Tower J. Induced overexpression of mitochondrial Mn-superoxide dismutase extends the life span of adult Drosophila melanogaster. Genetics. 2002;161:661–672. doi: 10.1093/genetics/161.2.661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Buetler TM, Krauskopf A, Ruegg UT. Role of superoxide as a signaling molecule. News Physiol Sci. 19:120–123. doi: 10.1152/nips.01514.2003. [DOI] [PubMed] [Google Scholar]
- 47.Valenti L, Conte D, Piperno A, Dongiovanni P, Fracanzani AL, Fraquelli M, Vergani A, Gianni C, Carmagnola L, Fargion S. The mitochondrial superoxide dismutase A16V polymorphism in the cardiomyopathy associated with hereditary haemochromatosis. Journal of medical genetics. 41:946–950. doi: 10.1136/jmg.2004.019588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Melov S, Schneider JA, Day BJ, Hinerfeld D, Coskun P, Mirra SS, Crapo JD, Wallace DC. A novel neurological phenotype in mice lacking mitochondrial manganese superoxide dismutase. Nature genetics. 18:159–163. doi: 10.1038/ng0298-159. [DOI] [PubMed] [Google Scholar]
- 49.Buttke TM, Sandstrom PA. Oxidative stress as a mediator of apoptosis. Immunol Today. 15:7–10. doi: 10.1016/0167-5699(94)90018-3. [DOI] [PubMed] [Google Scholar]
- 50.Ran Q, Liang H, Gu M, Qi W, Walter CA, Roberts LJ, 2nd, Herman B, Richardson A, Van Remmen H. Transgenic mice overexpressing glutathione peroxidase 4 are protected against oxidative stress-induced apoptosis. The Journal of biological chemistry. 279:55137–55146. doi: 10.1074/jbc.M410387200. [DOI] [PubMed] [Google Scholar]
- 51.Choi WS, Kruse SE, Palmiter RD, Xia Z. Mitochondrial complex I inhibition is not required for dopaminergic neuron death induced by rotenone, MPP+, or paraquat. Proceedings of the National Academy of Sciences of the United States of America. 105:15136–15141. doi: 10.1073/pnas.0807581105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Justilien V, Pang JJ, Renganathan K, Zhan X, Crabb JW, Kim SR, Sparrow JR, Hauswirth WW, Lewin AS. SOD2 knockdown mouse model of early AMD. Investigative ophthalmology & visual science. 48:4407–4420. doi: 10.1167/iovs.07-0432. [DOI] [PMC free article] [PubMed] [Google Scholar]