Abstract
The 5’→3’ exoribonucleases (XRNs) comprise a large family of conserved enzymes in eukaryotes with crucial functions in RNA metabolism and RNA interference1–5. XRN2, or Rat1 in yeast6, functions primarily in the nucleus and also plays an important role in transcription termination by RNA polymerase II (Pol II)7–14. Rat1 exoribonuclease activity is stimulated by the protein Rai115, 16. Here we report the crystal structure at 2.2 Å resolution of S. pombe Rat1 in complex with Rai1, as well as the structures of Rai1 and its murine homolog DOM3Z alone at 2.0 Å resolution. The structures reveal the molecular mechanism for the activation of Rat1 by Rai1 and for the exclusive exoribonuclease activity of Rat1. Biochemical studies confirm these observations, and show that Rai1 allows Rat1 to more effectively degrade RNAs with stable secondary structure. There are large differences in the active site landscape of Rat1 compared to related and PIN (PilT N-terminus) domain-containing nucleases17–20. Unexpectedly, we identified a large pocket in Rai1 and DOM3Z that contains highly conserved residues, including three acidic side chains that coordinate a divalent cation. Mutagenesis and biochemical studies demonstrate that Rai1 possesses pyrophosphohydrolase activity towards 5’ triphosphorylated RNA. Such an activity is important for mRNA degradation in bacteria21, but ours is the first demonstration of this activity in eukaryotes and suggests that Rai1/DOM3Z may have additional important functions in RNA metabolism.
Keywords: RNA metabolism, pol II termination, RNA pyrophosphohydrolase, nonsense mediated decay, RNAi, torpedo model, mRNA processing, protein complex
The XRNs share two regions of sequence conservation, corresponding to residues 1–390 and 590–751 in S. pombe Rat1 22 (Fig. 1a, Supplemental Fig. 1). The segment between them and the segment following the second region are poorly conserved (Fig. 1a). The XRNs display processive and exclusive exoribonuclease activity towards RNA substrates with a 5’ monophosphate, while being essentially inactive towards RNAs with a 5’ triphosphate 15, 23. They require divalent cations (Mg2+ or Mn2+) for activity, and contain seven conserved acidic residues in the first region that are essential for function 7, 24, 25. It has been suggested that these acidic residues may be located in the active site of XRNs, equivalent to those in other Mg2+-dependent nucleases such as T4 RNase H 25. However, XRNs and RNase H share no sequence homology besides these motifs. No structural information is currently available on any of the XRNs.
Figure 1. Structure of the Rat1-Rai1 complex.
(a). Domain organization of S. pombe Rat1, S. cerevisiae Rat1, human XRN2 and human XRN1. The first conserved region is colored in cyan, the second in magenta, and the linker segment between them in gray. A poorly conserved segment in the C-terminus that is also observed in our structure is shown in yellow. (b). Schematic drawing of the structure of S. pombe Rat1-Rai1 complex. The structure of Rat1 is colored as in panel a, and the structure of Rai1 is in green. The active site of Rat1 is indicated with the red star, and the red arrow points to the opening of the Rai1 active site pocket. A bound divalent cation in the active site of Rai1 is shown as a gray sphere. (c). Molecular surface of the active site region of Rat1, colored as in panel a. (d). Good surface complementarity at the interface between Rat1 and Rai1. Rat1 is shown as a molecular surface, and residues in the interface with Rai1 are colored in light blue and yellow for the first conserved region and the C-terminal segment, respectively. Rai1 is shown as stick models, with carbon atoms in black. All the structure figures were produced with Pymol 29 or Grasp 30.
Rai1 has strong sequence homologs in other fungal species, including S. pombe 22. A weak sequence homolog, known as DOM3Z, exists in mammals 16, although it probably does not associate with XRN2 13. The Rai1 proteins share no recognizable sequence homology with other proteins.
We have determined the crystal structure of the S. pombe Rat1-Rai1 complex at 2.2 Å resolution. The expression constructs contain residues 1–885 of Rat1 (101 kD) and full-length Rai1 (41 kD), respectively. Although the last 106 residues of Rat1 are not included in the construct, deletion of the C-terminal 125 residues does not affect cell viability 22. The refined structure has excellent agreement with the crystallographic data and the expected geometric parameters (Supplemental Table 1). The majority of the residues (90%) are in the most favored region of the Ramachandran plot.
The structure of Rat1 shows that its two conserved regions constitute a single, large domain (Fig. 1b, Supplemental text and Supplemental Fig. 2). The active site of Rat1 is formed primarily by residues in the first region, which has several weak structural homologs in the Protein Data Bank. All of these homologs are nucleases, including RNase H (Supplemental Fig. 3) 17, 18. Like RNase H, Rat1 contains a cluster of acidic residues in the active site (Supplemental Fig. 4) 25, although there are differences in the positions of some of the equivalent acidic residues (Supplemental Fig. 5). The structural analysis suggests that Rat1 shares the same catalytic mechanism as these related nucleases 26.
The second conserved region of Rat1 introduces large differences in the overall landscape of the active site as compared to other related nucleases. This region makes few direct contribution to the active site but generally surrounds it, such that the active site of Rat1 is mostly a pocket (Fig. 1c, Supplemental Fig. 2 and Supplemental Fig. 4), while the equivalent region in the other nucleases is much more open (Supplemental Fig. 3). Therefore, an endonuclease activity for Rat1 is incompatible with the structure of its active site, providing the molecular basis for the exclusive exonuclease activity of the XRNs.
Helix αD in the first conserved region is exceptionally long (Supplemental Fig. 1), and its C-terminal end is projected 30 Å away from the rest of the structure (Fig. 1b and Supplemental Fig. 2). We have named this feature of the Rat1 structure the ‘tower’ domain. The N-terminal region of this helix contributes several conserved residues to the active site (Supplemental Fig. 4). In comparison, the XRN1 homologs have a deletion in the C-terminal region of this helix (Supplemental Fig. 1), suggesting that the tower domain may be unique to XRN2 and serve XRN2-specific functions, for example Pol II termination.
Surprisingly, the poorly conserved segment following the second conserved region (Fig. 1a) has intimate interactions with the first conserved region (Fig. 1b, Supplemental Fig. 2). Most importantly, residues 840–863 form a long loop that traverses the bottom of the first region, and are located in the center of the interface with Rai1 (see below). A deletion mutant lacking the last 204 residues of S. pombe Rat1 (and therefore missing this C-terminal loop) is non-functional 22, and our biochemical data showed that deletion of this loop abolished interaction with Rai1 (Supplemental text). This C-terminal segment is located on the opposite face of Rat1 from the active site (Fig. 1b) and does not make any direct contribution to it (Supplemental Fig. 4), suggesting that it may instead be important for stabilizing the conformation of Rat1 for catalysis (see below).
Rai1 is bound about 30 Å away and on the opposite face from the active site of Rat1 (Fig. 1b). The primary interactions involve the poorly conserved C-terminal segment of Rat1 and the β8-αE segment and strand β4 of Rai1 (Fig. 1d, Supplemental Fig. 6). Approximately 800 Å2 of the surface area of Rat1 and Rai1 are buried in this interface, and the two surfaces at the interface are highly complementary to each other, with a shape correlation score of 0.76 27, consistent with the stability of the Rat1-Rai1 complex (see below) 15, 16.
Our structural data, together with earlier biochemical data, indicate that the activating effect of Rai1 is due at least in part to its stabilization of Rat1. Rat1 alone is relatively unstable in vitro 15 and loses nuclease activity upon pre-incubation, while the Rat1-Rai1 complex is more stable and can retain most of its activity 16. This should produce higher apparent activity for the Rat1-Rai1 complex than Rat1 alone in exoribonuclease assays. Although Rai1 does not make any direct contribution to the Rat1 active site, the C-terminal loop at the center of the interface is essential for Rat1 activity 22, suggesting that Rai1 could also stimulate Rat1 by indirectly affecting its catalysis.
Residues in the Rat1-Rai1 interface are generally conserved among the fungal proteins (Supplemental Fig. 1, Supplemental Fig. 7), consistent with the fact that this interaction has been observed in S. cerevisiae and S. pombe 15, 16, 22. However, these residues in Rai1 are not conserved in the mammalian homolog, as there is a 7-residue deletion between β8 and αE in DOM3Z (Supplemental Fig. 7). Our structure of DOM3Z alone confirms that the β8-αE region has a different conformation (Supplemental Fig. 8), and there is no evidence that DOM3Z associates with mammalian XRN2 by gel filtration (unpublished data).
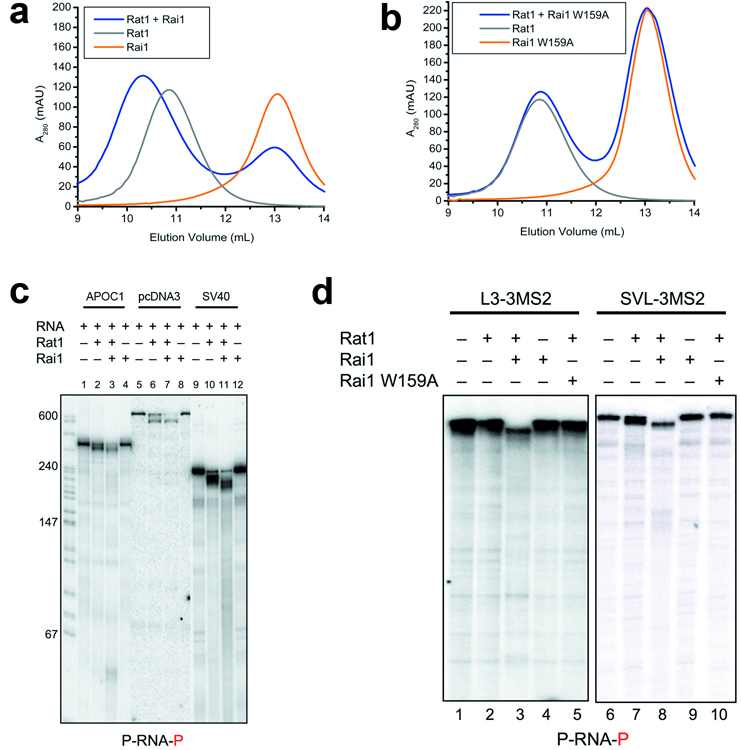
To assess the importance of individual residues for the stability of the Rat1-Rai1 complex, we introduced mutations in the interface and characterized their effects on the complex using gel filtration chromatography. The presence of wild-type Rai1 gave rise to a clear shift of the Rat1 peak (Fig. 2a), corresponding to the formation of the Rat1-Rai1 complex, while mutation of Rai1 residues in the interface, including W159A, R164A and W204A (Fig. 1d and Supplemental Fig. 6), greatly reduced the interactions with wild-type Rat1 (Fig. 2b). Similarly, mutation of Rat1 residues in the interface also abolished interaction with wild-type Rai1 (data not shown). The migration behavior of the mutants alone was similar to that of the wild-type protein (Fig. 2b), suggesting that the mutation did not disrupt the structure of Rai1. As a control, we created the K256A mutant of Rai1. This residue is outside the interface with Rat1, and the mutation did not affect the interaction (data not shown).
Figure 2. Biochemical and functional characterization of the Rat1-Rai1 interaction.
(a). Gel filtration profiles for wild-type Rat1 (full-length) alone, wild-type Rai1 (full-length) alone, and their mixture (with Rai1 present in roughly 2-fold molar excess). (b). Gel filtration profiles for wild-type Rat1 alone, W159A mutant of Rai1 alone, and their mixture. (c). Cleavage of three different 5’ phosphorylated, 3’ labeled RNA substrates by Rat1 and the Rat1-Rai1 complex. The RNA substrate is indicated at the bottom of the figure, with the radiolabeled phosphate group shown in red. (d). Cleavage of two other RNA substrates, each with three MS2 binding sites, by Rat1 and the Rat1-Rai1 complex.
To assess the functional significance of the Rat1-Rai1 interaction, we next carried out exoribonuclease assays with Rat1 alone and with Rat1 plus Rai1. We initially used three different RNA substrates (at 1 nM concentration), each labeled at the 3’-end and containing a 5’ monophosphate. Significantly, Rat1 alone (50 nM) degraded all three substrates, but only partially, and in each case generated major intermediates that were 30–100 nts smaller than the substrate (Fig. 2c, lanes 2, 6, 10). This may reflect stalling of Rat1 caused by secondary structure features in the substrates, as suggested by previous results 15, 28 (Supplemental text). Strikingly however, all three RNAs were degraded significantly more efficiently in the presence of equal molar amounts of Rai1, with roughly 60% of the substrate completely digested for each of the RNAs (Fig. 2c, lanes 3, 7, 11), while Rai1 alone did not show any ribonuclease activity (Fig. 2c, lanes 4, 8, 12).
To provide evidence that Rat1 stalling was indeed due to secondary structure, we used two additional substrates containing three MS2 binding sites, which form stable stem-loops, very near the 5’-end (1 nM concentration). With both substrates, Rat1 alone (50 nM) displayed at best weak activity, consistent with a block at the 5’ MS2 hairpins (Fig. 2d, lanes 2 and 7; note that the SV40 and SVL-3MS2 substrates are identical except for the MS2 sites). In contrast, significant fractions (roughly 60%) of both RNAs were completely digested in the presence of equal molar amounts of Rai1 (Fig. 2d, lanes 3 and 8), while the W159A mutation abolished the stimulatory effects of Rai1 (Fig. 2d, lanes 5 and 10). Overall, these experiments indicate that Rai1 allows Rat1 to more effectively degrade RNAs with stable secondary structures, and confirm the functional importance of the Rat1-Rai1 interface.
The structures of Rai1 and DOM3Z contain two highly-twisted, mixed β-sheets, and several helices cover up some of the exposed surfaces of the two β-sheets (Fig. 3a). Consistent with the unique sequences of these proteins, the structures do not appear to have a close homolog in the Protein Data Bank, and they may represent a new protein fold.
Figure 3. Structure of Rai1.
(a). Schematic drawing of the structure of S. pombe Rai1. Strands in the large β-sheet are shown in green, and those in the small β-sheet in cyan. Side chains of some of the conserved residues in the large pocket in the structure are shown in black, and the pocket is highlighted in light pink. A bound divalent cation is shown as a gray sphere. The arrow indicates the interface region with Rat1. (b). Molecular surface of Rai1, showing the large pocket in the structure. (c). Final 2Fo–Fc electron density (in light blue) at 2.2 Å resolution for the divalent cation and its ligands in the large pocket in Rai1, contoured at 1.5σ. Omit Fo–Fc electron density for the cation and the two water molecules is shown in green, contoured at 3σ. (d). Schematic drawing of the detailed interactions between GDP (in light gray) and DOM3Z (side chains in black). (e). Panel d after 90° rotation around the horizontal axis.
Unexpectedly, the structures suggest a catalytic function for these proteins. Strikingly, there is a large pocket in their surface (Fig. 3b), and many of the side chains in this pocket are highly conserved among Rai1 homologs (Supplemental Fig. 7). Most importantly, there is a cluster of four acidic residues (Glu150, Glu199, Asp201, and Glu239 in Rai1) at the bottom of the pocket (Fig. 3a), and three of them (Glu150, Asp201, and Glu239), together with the main chain carbonyl of Leu240 and two water molecules, form the octahedral coordination sphere of a cation (Fig. 3c, Supplemental Fig. 9). In a Rat1-Rai1 crystal soaked with 50 mM MnCl2, a strong difference electron density feature is observed at this position, confirming that it is a binding site for divalent cations.
The structural analysis therefore suggests that this pocket could be an active site of Rai1 and DOM3Z. This active site is distinct from the interface with Rat1, which involves a separate surface area of Rai1 (Fig. 1b and Fig. 3a). In addition, the conformation of the active site region of DOM3Z is essentially the same (Supplemental Fig. 9), even though it cannot interact with Rat1, providing further evidence that this active site may function independently of Rat1 binding. However, no enzymatic activity has been reported for Rai1 16.
During the course of examining the effects of Rai1 on Rat1 activity, we obtained the first evidence that Rai1 possesses catalytic activity. Specifically, when we used as a substrate a 3’ labeled RNA containing a 5’ triphosphate (1 nM), Rat1 alone (50 nM) displayed very weak activity (Fig. 4a, lane 2). Strikingly, addition of Rai1 (50 nM) resulted in efficient degradation of this substrate (Fig. 4a, lane 3). To determine whether this required the active site of Rai1, we mutated two of the conserved acidic residues in this region, E199A/D201A. Significantly, the double mutation abolished the degradation of the RNA substrate with a 5’ triphosphate (Fig. 4a, lane 7), but did not disrupt the stimulatory effect on hydrolysis of the RNA with a 5’ monophosphate (Supplemental Fig. 10, lane 7). As a control, the K256A mutation, located outside the Rat1-Rai1 interface and the active site of Rai1, did not affect hydrolysis of either substrate (Fig. 4a, lane 5; Supplemental Fig. 10, lane 5).
Figure 4. Biochemical evidence for pyrophosphohydrolase activity of Rai1.
(a). Cleavage of 5’ triphosphorylated, 3’ labeled RNA substrate by Rat1 and the Rat1-Rai1 complex. Rat1 does not exhibit ribonuclease activity towards this RNA in the absence of Rai1. The E199A/D201A mutant does not enable this ribonuclease activity. (b). Pre-incubation of 5’ triphosphorylated RNA with wild-type Rai1, but not E199A/D201A mutant, allows Rat1 to degrade the substrate. (c). Rai1 can release pyrophosphate from RNA with 5’ triphosphate. The assays were carried out in the absence (lanes 1–6) or presence (lanes 7–10) of Rat1. Human Dcp2 decapping protein was used as a negative control. The pyrophosphate marker is indicated on the left and was generated by RNA polymerase during in vitro transcription, and could be clearly distinguished from free phosphate. (d). Quantification of the percent pyrophosphate generated by Rai1 from four independent experiments. The error bars represent standard error of the mean.
These experimental data suggest that Rai1 could possess RNA 5’ pyrophosphohydrolase activity, which would convert the 5’ triphosphate group to a single phosphate, making the RNA a substrate for Rat1. To obtain additional evidence for this, we pre-incubated a 3’ labeled RNA substrate containing a 5’ triphosphate with wild-type Rai1 or the E199A/D201A double mutant (50 nM) and then repurified the RNA. Subsequent exoribonuclease assays with Rat1 demonstrated that the RNA pre-incubated with wild-type Rai1 was now efficiently degraded, while the RNA pre-incubated with the mutant was not (Fig. 4b). Longer pre-incubation with Rai1 resulted in more of the RNA substrate being degraded by Rat1 (Fig. 4b), also consistent with Rai1 having a catalytic activity on its own.
To obtain direct evidence for the release of pyrophosphate from 5’ triphosphorylated RNA, we used a substrate carrying a 32P label at the γ position (at 50 nM concentration). Strikingly, both the wild-type Rai1 and the W159A mutant (50 nM) were able to hydrolyze the triphosphate linkage and release pyrophosphate (Fig. 4c, lanes 4 and 5). In contrast, the E199A/D201A double mutant at the same concentration lacked detectable activity (Fig. 4c, lane 6) as did Rat1 (lane 3) and the Dcp2 decapping enzyme (lane 2), confirming that the pyrophosphohydrolase activity is due to Rai1. Notably, Rat1 (50 nM) displayed a stimulatory effect on Rai1-mediated pyrophosphohydrolase activity, and this stimulation was not observed with the W159A mutant (Figs. 4c lanes 8 and 9, 4d). Pre-incubation experiments confirmed that this stimulation was not due to simple stabilization of Rai1 by Rat1 (data not shown). Instead, the stimulatory effect of Rat1 could be due to its RNA binding property, bringing the substrate near the active site of Rai1. These data confirm that Rai1 possesses an intrinsic RNA 5’ pyrophosphohydrolase activity. In comparison, Rai1 displayed no pyrophosphohydrolase activity towards GTP (Supplemental Fig. 11), suggesting that this activity is limited to RNA substrates (Fig. 3b).
To define the substrate binding mode in the active site of Rai1 or DOM3Z, we soaked their crystals with various nucleotides and observed the binding of GDP in the active site of DOM3Z (Supplemental Fig. 12). Only the diphosphate group of GDP has strong electron density. The α phosphate is situated near the N-terminus of helix αB, having favorable interactions with the dipole of this helix (Fig. 3d). It also interacts with the side chain of conserved Arg132 (Arg94 in S. pombe Rai1, Supplemental Fig. 7). The β phosphate interacts with Arg132 and the conserved Gln280 in helix αF (Gln263 in Rai1). In comparison, although the ribose has van der Waals interactions with the conserved Trp131 (Trp93 in Rai1, Fig. 3e), its hydroxyls as well as the guanine base are not recognized by conserved residues. The conserved Glu234 residue (Glu199 in Rai1) is located near the phosphates of GDP and could serve a catalytic role in the pyrophosphohydrolase reaction.
Our structural and biochemical studies have shown that Rai1 possesses catalytic activity, acting as a pyrophosphohydrolase on RNA substrates with a 5’ triphosphate. While an RNA 5’ pyrophosphohydrolase activity is important for mRNA degradation in bacteria 21, ours is the first demonstration of such an activity in eukaryotes. Rai1 shares neither sequence nor structural homology with the bacterial 5’ pyrophosphohydrolase RppH. Rai1 and DOM3Z homologs therefore have two distinct functions: (1) stimulation of Rat1 exoribonuclease activity, by a mechanism independent of Rai1 catalytic activity, and (2) a pyrophosphohydrolase activity towards selected RNA substrates. The former is involved in Pol II termination and RNA metabolism, and appears to be restricted to the fungal Rai1 homologs. The latter likely applies to both the fungal Rai1 and the animal DOM3Z homologs, and this activity appears to be unrelated to Pol II termination. Our studies have identified a novel eukaryotic enzyme that is likely to have important catalytic functions in RNA metabolism. For example, Rai1/DOM3Z may be part of an RNA quality control system in the nucleus, promoting the degradation of 5’ triphosphorylated (i.e., uncapped) RNA.
Methods summary
Crystallography
S. pombe Rat1 and Rai1 were over-expressed separately in E. coli. The bacterial cells were mixed together and lysed, and the Rat1-Rai1 complex was purified. Crystals were obtained by the sitting-drop vapor diffusion method, and the structures were determined by the seleno-methionyl single-wavelength anomalous diffraction method.
Rat1 exoribonuclease assays
RNA substrates were produced by in vitro transcription, purified, and labeled at the 5’ or 3’-end. Exoribonuclease assays were performed at 37 °C for 15 min. The RNA products were isolated and fractionated by 6% PAGE in 7 M urea. The assays were repeated several times to ensure reproducibility.
Pyrophosphohydrolase assays
An RNA substrate with 5’ triphosphate labeled at the γ position was produced by in vitro transcription in the presence of γ-32P-GTP. The products from the pyrophosphohydrolase reaction were resolved by polyethyleneimine-cellulose thin layer chromatography plates developed in 1.5 M KH2PO4 (pH 7.5).
Full Methods and any associated references are available in the online version of the paper at www.nature.com/nature.
Methods
Protein expression and purification
DNA fragment corresponding to residues 1–885 of S. pombe Rat1 was amplified from the genome and inserted into the pET24d vector (Novagen). The recombinant Rat1 protein carries a hexa-histidine tag at the C-terminus. Full-length S. pombe Rai1 was cloned into the pET26b vector, and the recombinant protein does not carry any affinity tag. The plasmids were transformed separately into E. coli BL21 (DE3) Rosetta cells. After induction with 0.3 mM IPTG, the cells were allowed to grow at 20°C for 14–16 hours. Cells expressing Rat1 and Rai1 were mixed and lysed by sonication, and the Rat1-Rai1 complex was purified by Ni-NTA (Qiagen) and gel filtration (Sephacryl S-300, GE Healthcare) chromatography. Purified protein was concentrated to 10 mg/ml in a buffer containing 20 mM Tris (pH 7.5), 200 mM NaCl, 2 mM DTT and 5% (v/v) glycerol, flash frozen in liquid nitrogen and stored at −80°C. The C-terminal His-tag on Rat1 was not removed for crystallization.
The selenomethionyl protein sample was produced in minimal media supplemented with specific amino acids to inhibit endogenous methionine biosynthesis, and the bacteria were grown in the presence of selenomethionine 31. The purification procedure is the same as for the native protein except that the DTT concentration in the storage buffer was increased to 10 mM.
To study Rai1 alone, S. pombe Rai1 was cloned into the pET26b vector and over-expressed in E. coli. The recombinant protein, with a C-terminal His-tag, was purified following the same protocol as that for the Rat1-Rai1 complex.
Mouse DOM3Z was cloned into vector pET28a. The recombinant protein contains a n N-terminal His-tag. Native and SeMet proteins were expressed and purified following the same protocol as for the Rat1-Rai1 complex.
Protein crystallization
Plate-like crystals of the Rat1-Rai1 complex were obtained with the sitting-drop vapor diffusion method at 23 °C. The reservoir solution contained 0.3 M sodium malonate (pH 5.0), 10 mM DTT and 14 % (w/v) PEG 3350. Prior to crystallization setup the protein was supplemented with 60 mM of Gly-Gly-Gly (Hampton research). The quality of the crystals was improved by micro seeding. The crystals belong to space group P21212, and there is one Rat1-Rai1 hetero-dimer in the asymmetric unit.
Crystals of Rai1 alone were obtained with the sitting-drop vapor diffusion method at 23 °C. The reservoir solution contained 0.2 M sodium citrate tribasic (pH 5.0) and 20 % (w/v) PEG 3350. The crystals belong to space group C2, and there is one Rai1 molecule in the asymmetric unit.
Crystals of DOM3Z were obtained with the sitting-drop vapor diffusion method at 23 °C. Two different crystal forms were characterized. Rod-like crystal form 1 was obtained with a reservoir buffer containing 0.1 M Hepes (pH 7.5) and 20% (w/v) PEG8000. The crystals belong to space group P21, and there is one DOM3Z molecule in the asymmetric unit. Rod-like crystal form 2 was obtained with a reservoir buffer containing 25% (w/v) PEG3350 and 0.2 M KH2PO4. 10 mM DTT was added to the reservoir for the SeMet protein. The crystals belong to space group P212121, and there is one molecule in the asymmetric unit.
The crystals were cryo-protected by the reservoir solution supplemented with 25% (v/v) glycerol and flash-frozen in liquid nitrogen for data collection.
To obtain the GDP complex of DOM3Z, the crystals (in form 2) were soaked with reservoir buffer supplemented with 25 mM GDP and 10 mM MgCl2 over night.
Data collection and structure determination
The structures of the Rat1-Rai1 complex and DOM3Z were determined by the seleno-methionyl anomalous diffraction method 32. For the Rat1-Rai1 complex, a single-wavelength anomalous diffraction (SAD) data set to 2.5 Å resolution was collected on a SeMet-substituted crystal at the NSLS beamline X4A. A native reflection data set to 2.2 Å resolution was collected at the X4C beamline. The diffraction data were processed and scaled with the HKL package 33. The data processing statistics are summarized in Table 1. The diffraction pattern of this crystal is anisotropic beyond 2.5 Å resolution, contributing to the relatively lower completeness of the data at the high resolution shell.
The Se atoms were located with the program BnP 34, and the reflections were phased with the program Solve 35. The majority of the residues in the complex was built automatically by the program Resolve, and the model was completed by manual building with the program O 36. The structure refinement was carried out with the programs CNS 37 and Refmac 38, and water molecules were added with the program ARP 39.
Free-enzyme crystals of the Rat1-Rai1 complex were soaked overnight in a solution containing 50 mM MnCl2, and a diffraction data set to 2.5 Å resolution was collected at the X4C beamline. After structure refinement with the protein atoms only, a strong peak (12σ in the difference electron density map) was observed at the divalent cation position in the putative active site of Rai1, which was modeled as a Mn2+ ion.
A native diffraction data set to 2.0 Å resolution for Rai1 alone was collected at the X4C beamline. The structure was solved by the molecular replacement method with the program Molrep 40, and the structure refinement was carried out with Refmac 38.
For DOM3Z, a SAD data set to 2.6 Å resolution was collected on a SeMet crystal in crystal form 2 on NSLS beamline X29A. Native data sets for the free enzyme in crystal form 1 to 2.0 Å resolution and the GDP complex (in crystal form 2) to 2.6 Å resolution were collected at NSLS beamlines X4A and X4C, respectively. The diffraction data were processed by the HKL package (Table 1) 33. The Se atoms in were located by BnP 34, and the reflections were phased with the program Solve 35. The atomic model was completed with the program O 36. The structure of crystal form 1 was solved by the molecular replacement method with the program Molrep 40. The structure refinement was carried out with the programs CNS 37 and Refmac 38, and water molecules were added with the program ARP 39.
All diffraction data were collected near the Se edge, with X-ray wavelength of 0.98 Å.
Rat1-Rai1 interactions
Rat1 and Rai1 mutants were made with the QuikChange kit (Stratagene) and verified by sequencing. The mutant proteins were expressed and purified following the same protocol as that for the wild-type protein. All the proteins carry a hexa-histidine tag at the N-terminus.
Analytical gel filtration experiments were carried out on a Superose-12 10/30 column (GE Healthcare), with a buffer containing 20 mM Tris (pH 7.5) and 200 mM NaCl. 400 µg of Rat1 and Rai1 were mixed and diluted to a final volume of 120 µl with the gel filtration buffer. The mixture was incubated on ice for 30 minutes before being loaded onto the column. The proteins were also run separately on the column to determine their migration behavior alone.
Rat1 exoribonuclease assays
pGEM SVL was linearized with DraI and transcribed with Sp6 RNA polymerase (Promega) for 2h at 37 °C. The RNA transcripts were gel purified using standard procedures. The 5’ termini of unlabeled SVL pre-mRNA were dephosphorylated by calf intestinal phosphatase (New England Biolabs) for 1h at 37 °C. SVL pre-mRNAs were subsequently labeled at their 5’-ends with T4 polynucleotide kinase (NEB) and γ-32P ATP at 37°C for 1h, or at their 3’-ends with 32P Cp and T4 RNA ligase (Promega) for 1h at 37°C. Exoribonuclease assays were performed at 37 °C for 15 min. The reaction mixtures contained ~1 ng labeled pre-mRNA, 50 mM NaCl, 5 mM MgCl2, 20 mM Tris (pH 8.0), 0.5 mM DTT, 500 ng BSA and the indicated amounts of recombinant Rat1 and Rai1. The RNA products were isolated and fractionated by 6% PAGE in 7 M urea. The data were analyzed by Phosphor Imager. The assays were repeated several times to ensure reproducibility.
pGEM APOC1 was cut with PstI and in vitro transcribed by T7 RNA polymerase (Promega). pcDNA3 was cut with NdeI and in vitro transcribed by Sp6 RNA polymerase (Promega).
Rai1 pre-incubation experiments
SVL pre-mRNA carrying 5’ triphosphate and 3’ 32P phosphate was incubated for 15, 45 and 60 min at 30°C with wild-type Rai1 or the E199A/D201A double mutant. The treated RNA was purified by phenol/chloroform extraction and ethanol precipitation, and used in exoribonuclease assays with Rat1.
Pyrophosphohydrolase assays
The pcDNA3 polylinker (pcP) was amplified by PCR with a forward primer corresponding to the SP6 promoter primer and a 3′ primer consisting of the T7 promoter sequences containing 16 cytosines at the 5’ end. The PCR product was used as template to transcribe RNA with SP6 RNA polymerase in the presence of γ-32P-GTP as described 41. The resulting RNA is exclusively labeled at the 5′-end at the first phosphate within the 5´ triphosphate and contains a 3′ terminal 16 guanosines to stabilize the 3′-end. The indicated recombinant proteins were incubated with 32P 5’-end-labeled RNA in RNA decapping buffer (10 mM Tris-HCl (pH 7.5), 100 mM KOAc, 2 mM MgOAc, 0.5 mM MnCl2 and 2 mM DTT) at 37 °C for 30 minutes as previously descried 42. The reaction products were resolved by polyethyleneimine-cellulose thin layer chromatography plates developed in 1.5 M KH2PO4 (pH 7.5) and exposed to a PhosphorImager. Molecular Dynamics PhosphorImager (Storm860) and ImageQuant-5 software were used for product quantifications. The data were analyzed with GraphPad Instat software.
Supplementary Material
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Acknowledgements
We thank Randy Abramowitz and John Schwanof for setting up the X4A and X4C beamlines and Howard Robinson for setting up the X29A beamline at the NSLS; Songtao Jia for providing the S. pombe genome. This research is supported in part by grants from the NIH to LT (GM077175), JLM (GM28983) and MK (GM67005).
Footnotes
The authors declare no competing financial interests.
References
- 1.Parker R, Song H. The enzymes and control of eukaryotic mRNA turnover. Nat. Struct. Mol. Biol. 2004;11:121–127. doi: 10.1038/nsmb724. [DOI] [PubMed] [Google Scholar]
- 2.Newbury SF. Control of mRNA stability in eukaryotes. Biochem. Soc. Trans. 2006;34:30–34. doi: 10.1042/BST20060030. [DOI] [PubMed] [Google Scholar]
- 3.Bousquet-Antonelli C, Presutti C, Tollervey D. Identification of a regulated pathway for nuclear pre-mRNA turnover. Cell. 2000;102:765–775. doi: 10.1016/s0092-8674(00)00065-9. [DOI] [PubMed] [Google Scholar]
- 4.Gatfield D, Izaurralde E. Nonsense-mediated messenger RNA decay is initiated by endonucleolytic cleavage in Drosophila. Nature. 2004;429:575–578. doi: 10.1038/nature02559. [DOI] [PubMed] [Google Scholar]
- 5.Gazzani S, Lawrenson T, Woodward C, Headon D, Sablowski R. A link between mRNA turnover and RNA interference in Arabidopsis. Science. 2004;306:1046–1048. doi: 10.1126/science.1101092. [DOI] [PubMed] [Google Scholar]
- 6.Amberg DC, Goldstein AL, Cole CN. Isolation and characterization of RAT1: an essential gene of Saccharymyces cerevisiae required for the efficient nucleocytoplasmic trafficking of mRNA. Genes Develop. 1992;6:1173–1189. doi: 10.1101/gad.6.7.1173. [DOI] [PubMed] [Google Scholar]
- 7.Kim M, et al. The yeast Rat1 exonuclease promotes transcription termination by RNA polymerase II. Nature. 2004;432:517–522. doi: 10.1038/nature03041. [DOI] [PubMed] [Google Scholar]
- 8.West S, Gromak N, Proudfoot NJ. Human 5'->3' exonuclease Xrn2 promotes transcription termination at co-transcriptional cleavage sites. Nature. 2004;432:522–525. doi: 10.1038/nature03035. [DOI] [PubMed] [Google Scholar]
- 9.Connelly S, Manley JL. A functional mRNA polyadenylation signal is required for transcription termination by RNA polymerase II. Genes Develop. 1988;2:440–452. doi: 10.1101/gad.2.4.440. [DOI] [PubMed] [Google Scholar]
- 10.Luo W, Bentley D. A ribonucleolytic Rat torpedoes RNA polymerase II. Cell. 2004;119:911–914. doi: 10.1016/j.cell.2004.11.041. [DOI] [PubMed] [Google Scholar]
- 11.Buratowski S. Connections between mRNA 3' end processing and transcription termination. Curr. Opin. Cell Biol. 2005;17:257–261. doi: 10.1016/j.ceb.2005.04.003. [DOI] [PubMed] [Google Scholar]
- 12.Luo W, Johnson AW, Bentley DL. The role of Rat1 in coupling mRNA 3'-end processing to transcription termination: implications for a unified allosterictorpedo model. Genes Develop. 2006;20:954–965. doi: 10.1101/gad.1409106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kaneko S, Rozenblatt-Rosen O, Meyerson M, Manley JL. The multifunctional protein p54nrb/PSF recruits the exonuclease XRN2 to facilitate pre-mRNA 3' processing and transcription termination. Genes Develop. 2007;21:1779–1789. doi: 10.1101/gad.1565207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.West S, Proudfoot NJ. Human Pcf11 enhances degradation of RNA polymerase II-associated nascent RNA and transcriptional termination. Nucl. Acid Res. 2008;36:905–914. doi: 10.1093/nar/gkm1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stevens A, Poole TL . 5'-exonuclease-2 of Saccharomyces cerevisiae. Purification and features of ribonuclease activity with comparison to 5'-exonuclease-1. J. Biol Chem. 1995;270:16063–16069. doi: 10.1074/jbc.270.27.16063. [DOI] [PubMed] [Google Scholar]
- 16.Xue Y, et al. Saccharomyces cerevisiae RAI1 (YGL246c) is homologous to human DOM3Z and encodes a protein that binds the nuclear exoribonuclease Rat1p. Mol. Cell. Biol. 2000;20:4006–4015. doi: 10.1128/mcb.20.11.4006-4015.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mueser TC, Nossal NG, Hyde CC. Structure of bacteriophage T4 RNase H, a 5' to 3' RNA-DNA and DNA-DNA exonuclease with sequence similarity to the RAD2 family of eukaryotic proteins. Cell. 1996;85:1101–1112. doi: 10.1016/s0092-8674(00)81310-0. [DOI] [PubMed] [Google Scholar]
- 18.Devos JM, Tomanicek SJ, Jones CE, Nossal NG, Mueser TC. Crystal structure of bacteriophage T4 5' nuclease in complex with a branched DNA reveals how Flap endonuclease-1 family nucleases bind their substrates. J. Biol. Chem. 2007;282:31713–31724. doi: 10.1074/jbc.M703209200. [DOI] [PubMed] [Google Scholar]
- 19.Glavan F, Behm-Ansmant I, Izaurralde E, Conti E. Structures of the PIN domains of SMG6 and SMG5 reveal a nuclease within the mRNA surveillance complex. EMBO J. 2006;25:5117–5125. doi: 10.1038/sj.emboj.7601377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Clissold PM, Ponting CP. PIN domains in nonsense-mediated mRNA decay and RNAi. Curr. Biol. 2000;10:R888–R890. doi: 10.1016/s0960-9822(00)00858-7. [DOI] [PubMed] [Google Scholar]
- 21.Deana A, Celesnik H, Belasco JG. The bacterial enzyme RppH triggers messenger RNA degradation by 5' pyrophosphate removal. Nature. 2008;451:355–358. doi: 10.1038/nature06475. [DOI] [PubMed] [Google Scholar]
- 22.Shobuike T, Tatebayashi K, Tani T, Sugano S, Ikeda H. The dhp1+ gene, encoding a putative nuclear 5'->3' exoribonuclease, is required for proper chromosome segregation in fission yeast. Nucl. Acid Res. 2001;29:1326–1333. doi: 10.1093/nar/29.6.1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kenna M, Stevens A, McCammon M, Douglas MG. An essential yeast gene with homology to the exonuclease-encoding XRN1/KEM1 gene also encodes a protein with exoribonuclease activity. Mol. Cell. Biol. 1993;13:341–350. doi: 10.1128/mcb.13.1.341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Page AM, Davis K, Molineux C, Kolodner RD, Johnson AW. Mutational analysis of exoribonuclease I from Saccharomyces cerevisiae. Nucl. Acid Res. 1998;26:3707–3716. doi: 10.1093/nar/26.16.3707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Solinger JA, Pascolini D, Heyer W-D. Active-site mutations in the Xrn1p exoribonuclease of Saccharomyces cerevisiae reveal a specific role in meiosis. Mol. Cell. Biol. 1999;19:5930–5942. doi: 10.1128/mcb.19.9.5930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang W, Lee JY, Nowotny M. Making and breaking nucleic acids: Two-Mg2+-ion catalysis and substrate specificity. Mol. Cell. 2006;22:5–13. doi: 10.1016/j.molcel.2006.03.013. [DOI] [PubMed] [Google Scholar]
- 27.Lawrence MC, Colman PM. Shape complementarity at protein/protein interfaces. J. Mol. Biol. 1993;234:946–950. doi: 10.1006/jmbi.1993.1648. [DOI] [PubMed] [Google Scholar]
- 28.Poole TL, Stevens A. Structural modifications of RNA influence the 5' exoribonucleolytic hydrolysis by XRN1 and HKE1 of Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun. 1997;235:799–805. doi: 10.1006/bbrc.1997.6877. [DOI] [PubMed] [Google Scholar]
- 29.DeLano WL. San Carlos, CA: DeLano Scientific; 2002. [Google Scholar]
- 30.Nicholls A, Sharp KA, Honig B. Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins. 1991;11:281–296. doi: 10.1002/prot.340110407. [DOI] [PubMed] [Google Scholar]
- 31.Doublie S, et al. Crystallization and preliminary X-ray analysis of the 9 kDa protein of the mouse signal recognition particle and the selenomethionyl-SRP9. FEBS Lett. 1996;384:219–221. doi: 10.1016/0014-5793(96)00316-x. [DOI] [PubMed] [Google Scholar]
- 32.Hendrickson WA. Determination of macromolecular structures from anomalous diffraction of synchrotron radiation. Science. 1991;254:51–58. doi: 10.1126/science.1925561. [DOI] [PubMed] [Google Scholar]
- 33.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Method Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 34.Weeks CM, Miller R. The design and implementation of SnB v2.0. J. Appl. Cryst. 1999;32:120–124. [Google Scholar]
- 35.Terwilliger TC. SOLVE and RESOLVE: Automated structure solution and density modification. Meth. Enzymol. 2003;374:22–37. doi: 10.1016/S0076-6879(03)74002-6. [DOI] [PubMed] [Google Scholar]
- 36.Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Cryst. 1991;A47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 37.Brunger AT, et al. Crystallography & NMR System: A new software suite for macromolecular structure determination. Acta Cryst. 1998;D54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 38.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Cryst. 1997;D53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 39.Lamzin VS, Perrakis A, Wilson KS. Crystallography of biological macromolecules. In: Crystallography of biological macromolecules. International tables for crystallography. Rossmann MG, Arnold E, editors. Vol. F. Dordrecht: Kluwer Academic Publishers; 2001. pp. 720–721. [Google Scholar]
- 40.Vagin AA, Teplyakov A. An approach to multi-copy search in molecular replacement. Acta Cryst. 2000;D56:1622–1624. doi: 10.1107/s0907444900013780. [DOI] [PubMed] [Google Scholar]
- 41.Jiao X, Wang Z, Kiledjian M. Identification of an mRNA-decapping regulator implicated in X-linked mental retardation. Mol. Cell. 2006;24:713–722. doi: 10.1016/j.molcel.2006.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Piccirillo C, Khanna R, Kiledjian M. Functional characterization of the mammalian mRNA decapping enzyme hDcp2. RNA. 2003;9:1138–1147. doi: 10.1261/rna.5690503. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.