Abstract
We have developed a self-assembled nanoparticle (NP) that efficiently delivers small interfering RNA (siRNA) to the tumor by intravenous (IV) administration. The NP was obtained by mixing carrier DNA, siRNA, protamine, and lipids, followed by post-modification with polyethylene glycol and a ligand, anisamide. Four hours after IV injection of the formulation into a xenograft model, 70–80% of injected siRNA/g accumulated in the tumor, ~10% was detected in the liver and ~20% recovered in the lung. Confocal microscopy showed that fluorescent-labeled siRNA was efficiently delivered into the cytoplasm of the sigma receptor expressing NCI-H460 xenograft tumor by the targeted NPs, whereas free siRNA and non-targeted NPs showed little uptake. Three daily injections (1.2 mg/kg) of siRNA formulated in the targeted NPs silenced the epidermal growth factor receptor (EGFR) in the tumor and induced ~15% tumor cell apoptosis. Forty percent tumor growth inhibition was achieved by treatment with targeted NPs, while complete inhibition lasted for 1 week when combined with cisplatin. The serum level of liver enzymes and body weight monitoring during the treatment indicated a low level of toxicity of the formulation. The carrier itself also showed little immunotoxicity (IMT).
INTRODUCTION
Selective oncogene silencing, mediated by small interfering RNA (siRNA), shows promise for cancer treatment. However, the obstacles in successfully delivering siRNA hinder the therapeutic viability of this treatment.1–3 siRNA are susceptible to nuclease destruction and cannot penetrate the cell membrane when used in vivo due to the highly charged nucleic acid backbone. Although a variety of delivery systems have been developed for siRNA,2,4–16 the majority of the injected dose (ID) was taken up by the reticular endothelial system in the liver and spleen.17 This typically left only 2–5% of the ID/g tissue for the tumor;14,17 therefore, a more efficient delivery system still needs to be found.
Previously, we have shown that our nanoparticles (NPs) could efficiently deliver siRNA to the sigma receptor–expressing lung tumor cells (NCI-H1299), stimulate strong RNA interference effects and induce >80% apoptosis in vitro.18 The targeting ligand, anisamide, used in the formulation had a moderate affinity (estimated at Kd ~30 nmol/l) for sigma receptors and was shown to increase the intracellular delivery into prostate and lung cancer cells.18–20 In this study, we used a more aggressive lung cancer cell line, NCI-H460, to grow highly vasculatured tumors in vivo. This is a pre-requisite for the enhanced permeability and retention effect of any NP-based delivery systems.21 We hypothesized that the encapsulated siRNA is protected by the NPs and thus would accumulate in the tumor with an improved efficiency. The addition of anisamide ligand might also further increase the cellular uptake and enhance the RNA interference effect.
The therapeutic target in this study was the epidermal growth factor receptor (EGFR). EGFR is over-expressed in a variety of tumors and has been shown to be associated with many adverse features of tumor, including increased proliferation, decreased apoptosis, enhanced metastasis, and resistance to chemo- and radiation therapy.22,23 AntiEGFR therapy via tyrosine kinase inhibitors and monoclonal antibody has demonstrated great benefit for cancer patients.24–27 Silencing EGFR by RNA interference is an alternative to antiEGFR therapy and has already shown some promising results.28–35 EGFR silencing induces cell cycle arrest, apoptosis, tumor cell growth inhibition, and chemosensitization in vitro and in vivo.28–35 In the in vitro preliminary studies (methods described in Supplementary Materials and Methods), we showed that the targeted NP delivered a significantly higher amount of siRNA into NCI-H460 cells and showed a stronger gene silencing effect compared to non-targeted NP (Supplementary Figure S1). Targeted NP silenced the EGFR at the concentration of 120 nmol/l (Supplementary Figure S1b). The cytotoxicity of the NP was siRNA sequence and formulation dependent (Supplementary Figure S2). The cell death mechanism was confirmed to be apoptosis by means of immunostaining of the apoptosis inducing factor (AIF) (Supplementary Figure S2b).
RESULTS
Pharmacokinetic studies, tissue distribution, and intracellular uptake of siRNA
Free FAM-siRNA (fluorescein-labeled siRNA) was eliminated rapidly from the blood and the concentration was under the detection limit after a 40-minute time point (Figure 1). In 40 minutes, only 1% ID of free siRNA was detected in the blood. No significant difference in the pharmacokinetic (PK) profiles was observed between the tumor free and the tumor-bearing mice treated with free siRNA. NP significantly prolonged the circulation of siRNA and there was no difference in PK between the targeted and non-targeted NPs. NPs showed a rapid distribution phase, in which serum concentrations dropped to 1/10 within 40 minutes. After that, concentrations remained steady for at least 24 hours. Tumor-bearing mice cleared the NPs from the blood significantly quicker than the tumor free mice. In 2 minutes, almost a 100% ID of the NPs remained in the blood of the tumor free mice, while only a 25% ID remained in that of the tumor-bearing mice. At the terminal phase (80 minutes–24 hours), a 5–10% ID was detected in the blood of the tumor free mice, while only 1–2% ID was recovered in that of the tumor-bearing mice. The dose recoveries in the major organs of mice treated with free siRNA and siRNA in NP were ~30 and 60%, respectively.
Figure 1. Serum concentration profiles of FAM-siRNA in different formulations.
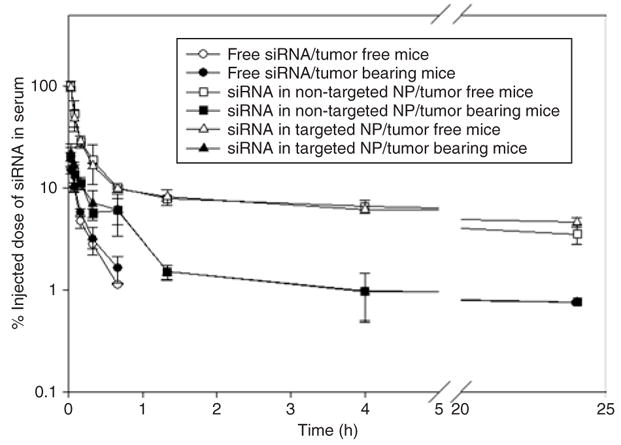
Data = mean ± SD, n = 4–8. NP, nanoparticle; siRNA, small interfering RNA.
The PK profiles were fitted with a non-compartment model using the WinNonlin program and the key PK parameters were obtained (Table 1). For free siRNA, the parameters obtained from the tumor free and the tumor-bearing mice were similar to each other. NPs significantly increased the terminal phase half-life (t1/2) 80-fold to 100-fold, and increased the area under the curve 15-fold to 100-fold and decreased the clearance 15-fold to 100-fold for siRNA. Again, there was no difference between the parameters of targeted and non-targeted NPs. However, the presence of tumor significantly changed some of the parameters of the NP. The area under the curve was reduced fourfold to sixfold, the clearance was increased fourfold to sixfold, and the steady state volume of distribution (Vss) was increased fivefold to sixfold in the tumor-bearing mice compared to the tumor free mice. The t1/2 and mean residence time remained unchanged in the presence of tumor.
Table 1.
Comparison of pharmacokinetic parameters of FAM-siRNA in different formulations in either NCI-H460 xenograft tumor-bearing mice or tumor free micea
|
t1/2 (λz) (h) |
AUC (h·μg/ml) |
CL (ml/h/kg) |
MRT (h) |
Vss (ml/kg) |
|
|---|---|---|---|---|---|
| Free siRNA/tumor-bearing mice | 0.30 | 1.15 | 1,040.01 | 0.35 | 360.65 |
| Free siRNA/tumor free mice | 0.25 | 1.10 | 1,087.15 | 0.24 | 265.55 |
| siRNA in non-targeted NP/tumor-bearing mice | 32.37 | 17.91 | 66.99 | 42.29 | 2,833.13 |
| siRNA in non-targeted NP/tumor free mice | 20.51 | 69.94 | 17.16 | 26.88 | 461.16 |
| siRNA in targeted NP/tumor-bearing mice | 32.37 | 18.08 | 66.36 | 41.90 | 2,780.09 |
| siRNA in targeted NP/tumor free mice | 36.56 | 110.31 | 10.88 | 50.91 | 553.86 |
Abbreviations: AUC, area under the curve; CL, clearance; h, hour; MRT, mean residence time; t1/2, half-life; Vss, steady state volume of distribution.
The serum concentration profiles (Figure 1) were analyzed with the WinNonlin program and the key parameters were obtained.
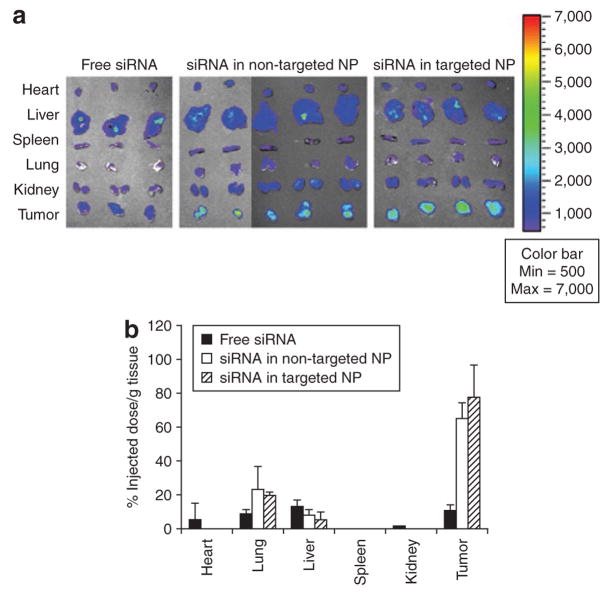
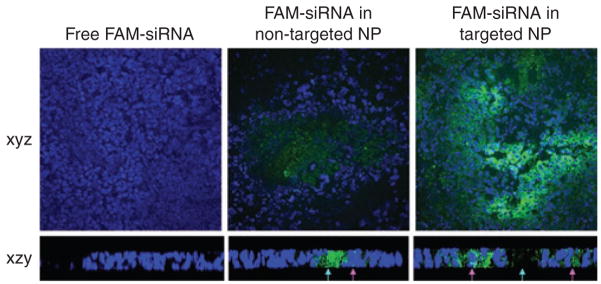
We employed Xenogen IVIS imaging system to visualize the FAM-siRNA distribution in major tissues in the mice 4 hours after intravenous (IV) injections. As shown in Figure 2a, fluorescence signals were hardly detected in the tissues collected from the mice treated with free siRNA. While the liver showed the strongest signal, it was still minimal. Among the tissues excised from the mice treated with NP, the NCI-H460 xenograft tumors showed the strongest signal and all other tissues showed only background to moderate signals. In the quantitative results (Figure 2b), the liver, lung, and tumor showed comparable uptake of free siRNA ranging from 10 to 20% ID/g. Other tissues showed minimal uptake of free siRNA (<5% ID/g). Non-targeted and targeted NP had similar tissue distribution patterns, in which 70–80% ID/g accumulated in the tumor, whereas both the lung and liver took up <20% ID/g. The uptake of siRNA formulated in NP was under the detection limit in the heart, spleen, and kidney. The bioavailability of siRNA in the tumor tissues was examined by confocal microscopy. As shown in Figure 3 (xyz images) and Supplementary Figure S3, the targeted NP showed strong cytosolic delivery of FAM-siRNA, while the non-targeted NP was less efficient in intracellular delivery and free siRNA showed no delivery. The percentages of fluorescence positive cells for free siRNA, non-targeted NP, and targeted NP were <1%, 5%, and 30%, respectively. The distribution of fluorescence in the tumor was heterogeneous. We used z-axis imaging to further address the intracellular delivery efficiency. Blue arrows indicate the extracellular space and pink arrows show the intracellular uptake of FAM-siRNA (Figure 3 xzy images and Supplementary Figure S4). A majority of the siRNA formulated in non-targeted NPs remained in extracellular space while the siRNA in targeted NPs was mostly internalized into the cells.
Figure 2. Tissue distribution study.
(a) Fluorescence signal of FAM-siRNA in different tissues detected by the Xenogen IVIS imaging system. (b) Tissue distribution of FAM-siRNA in different formulations. Data = mean ± SD, n = 3–4. NP, nanoparticle; siRNA, small interfering RNA.
Figure 3. Tumoral uptake of FAM-siRNA in different formulations.
Blue arrows indicate the extracellular space and pink arrows indicate the intracellular uptake of FAM-siRNA. Magnification = ×400 (xyz images), ×630 (xzy images). NP, nanoparticle; siRNA, small interfering RNA.
EGFR gene silencing, apoptosis induction, and tumor growth inhibition
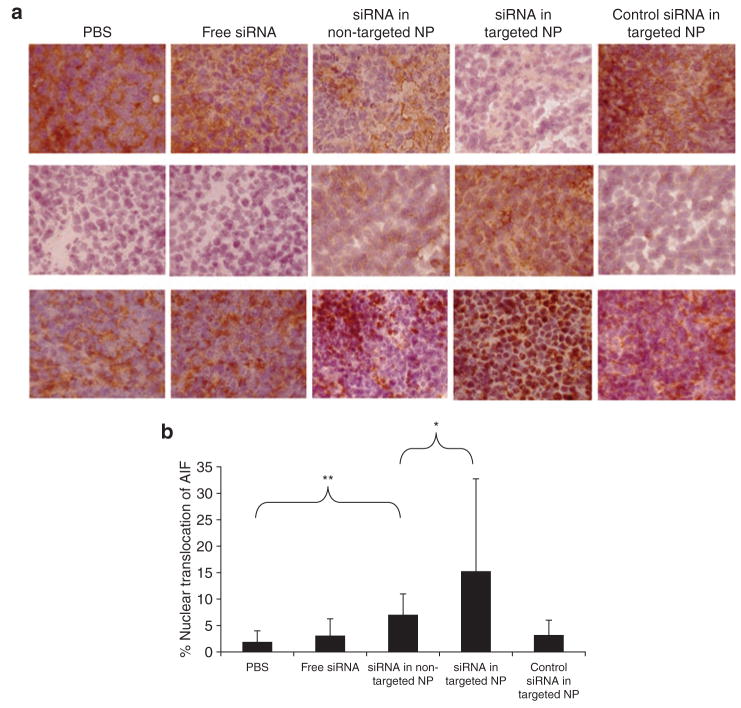
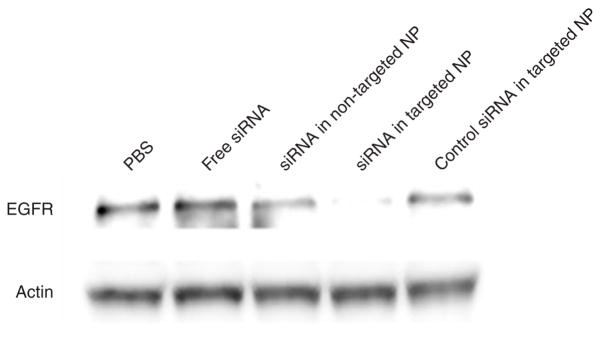
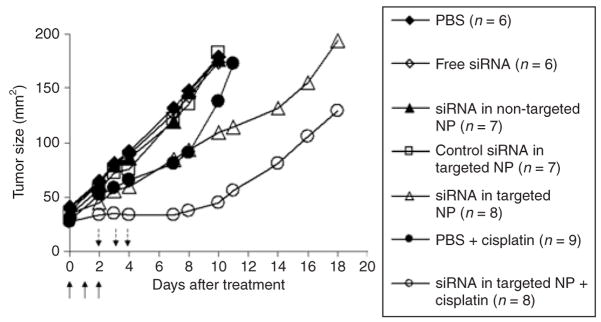
To examine the biological activities of siRNA, EGFR levels in the tumor were detected by immunohistochemistry (Figure 4a upper and Supplementary Figure S5) and Western blot analysis (Figure 5). The EGFR in NCI-H460 tumor was silenced by siRNA in targeted NPs. The non-targeted NPs showed only a partial effect, whereas free siRNA and the control siRNA in targeted NPs showed no effect. We also examined the apoptotic markers in the tumor (Figure 4a middle and bottom and Supplementary Figures S6 and S7). Prostate apoptotic response 4 levels were elevated after the treatment of siRNA in targeted NPs, while siRNA in non-targeted NPs showed only a moderate effect. Other control treatments had little effect. Immunostaining of the AIF showed similar results, in which siRNA in targeted NPs induced significant nuclear translocation of AIF (Figure 4a bottom and Supplementary Figure S7, P < 0.01). We calculated the percentage of AIF nuclear translocation to quantify the percentage of apoptosis. Tumor apoptosis in mice treated with phosphate- buffered saline, free siRNA, siRNA in non-targeted NPs, siRNA in targeted NPs, and control siRNA in targeted NPs were 2, 3, 7, 15, and 3%, respectively (Figure 4b). Three injections of siRNA in targeted NPs showed a partial inhibition of tumor growth (P < 0.01 at day 10) similar to that of intraperitoneal cisplatin (P < 0.05 at day 7) (Figure 6). However, 2 weeks later, the tumor growth rate became comparable to the untreated control. Other control treatments had no effects. Combination of siRNA (formulated in targeted NP) and cisplatin completely inhibited tumor growth for an entire week (P < 0.001 at day 7). Tumor growth, however, resumed after 1 week.
Figure 4. Immunohistochemical analysis of the tumor samples.
(a) Immunohistochemical staining on tumor sections: epidermal growth factor receptor (upper), prostate apoptotic response 4 (middle), and apoptosis inducing factor (AIF) (bottom). Magnification = ×200. (b) Quantitative analysis of nuclear translocation of AIF in the tumors treated with different formulations. **indicates P < 0.01, *indicates P < 0.05. NP, nanoparticle; PBS, phosphate-buffered saline; siRNA, small interfering RNA.
Figure 5. Western blot analysis of epidermal growth factor receptor (EGFR) in the NCI-H460 xenograft tumor after treatment with different formulations.
NP, nanoparticle; PBS, phosphate-buffered saline; siRNA, small interfering RNA.
Figure 6. NCI-H460 xenograft tumor growth inhibition by small interfering RNA (siRNA) in different formulations with or without the combination of cisplatin.
Solid arrows indicate the intravenous administrations of siRNA (1.2 mg/kg) and dash-line arrows indicate the intra-peritoneal injections of cisplatin (3 mg/kg). Data = mean, n = 6–9. SD of the data points is not shown for clarity.
Toxicity and immune response studies in normal mice
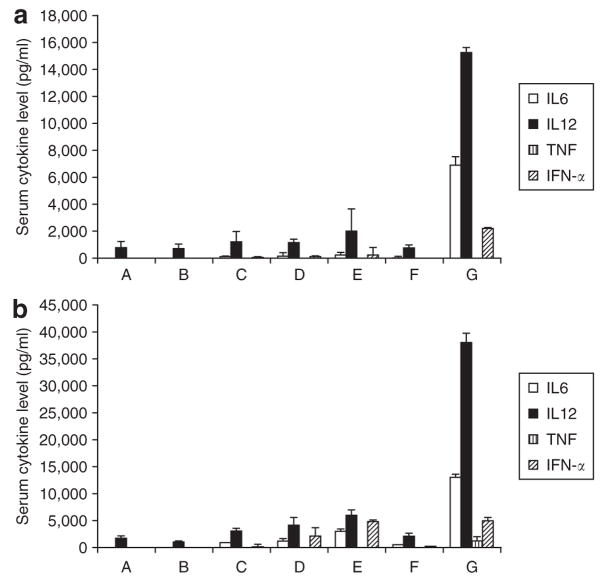
As shown in Figure 7a, siRNA alone (group B) or when formulated with calf thymus DNA into NPs induced a very mild production of inflammatory cytokines in the athymic nude mice (groups C–E). However, when plasmid DNA was co-formulated with siRNA (group G), the formulation became very immunogenic. In the C57BL/6 mice model (Figure 7b), free siRNA (group B) did not induce cytokine production, while siRNA formulated in NPs (groups C and D) induced a significant production of all analyzed cytokines, except for the tumor necrosis factor. Targeted NPs (group D) induced higher levels of cytokines compared to the non-targeted formulation (group C). Unexpectedly, targeted NPs containing murine double minute 2 (MDM2) siRNA (with no identifiable immunostimulatory motifs, group E) induced significantly higher levels of cytokines than the one containing EGFR siRNA (group D). Empty NP (group F) showed very mild (IMT) immunotoxicity. When using plasmid as the carrier DNA in the targeted NP formulation (group G), all inflammatory cytokines were induced to high levels. Additionally, serum levels of aspartate aminotransferase and alanine aminotransferase in mice treated with siRNA in different formulations were all in the normal range (Supplementary Figure S8) the method is described in the Supplementary Materials and Methods. Mouse body weight did not change significantly during the siRNA treatment (Supplementary Figure S9).
Figure 7. Serum cytokine analysis.
Serum cytokine levels in the (a) athymic nude mice and (b) C57BL/6 mice 2 hours after the injection of different formulations at the dose of 1.2 mg siRNA/kg (2.4 mg DNA/kg for group F). A, phosphate-buffered saline (PBS); B, epidermal growth factor receptor (EGFR) small interfering RNA (siRNA) in PBS; C, EGFR siRNA and calf thymus DNA in non-targeted nanoparticle (NP); D, EGFR siRNA and calf thymus DNA in targeted NP; E, MDM2 siRNA and calf thymus DNA in targeted NP; F, calf thymus DNA in targeted NP (empty NP); G, EGFR siRNA and plasmid DNA in targeted NP. Data = mean ± SD, n = 3–8.
DISCUSSION
The self-assembled NP without surface modification was developed earlier in our lab for plasmid DNA delivery.36 In this study, we reformulated the NP by replacing the plasmid DNA with a mixture of siRNA and calf thymus DNA. The resulting complex was further coated with polyethylene glycol-lipids for improved stability. The calf thymus DNA served as a carrier in the formulation, which reduced the particle size by 10–30% and increased delivery efficiency by 20–80% (data not shown). This may be due to improved core compaction provided by high molecular weight DNA. The calf thymus DNA also contains limited amounts of immunostimulating CpG motifs37 and thus provides a lower potential for IMT than plasmid, which was supported by Figure 7.
The PK results suggest that the majority of the free siRNA was eliminated from the blood before it accumulated in the tissues. On the other hand, the tumor appeared to be the major uptake tissue for NPs and contributed a high uptake (70–80% ID/g) (Figure 2b). Additionally, the liver and lung only took up 10–20% ID/g, which shows the high tumor selectivity of the NPs. The slightly positive charge of the NP (zeta potential 15–20 mV) may contribute to the relatively higher uptake in the lung, the first pass organ. The reason the NP delivered a majority of the dose to the tumor, instead of the liver, remains unknown at the present time. We hypothesize that the particle size of the NP (120–150 nm) is too big to penetrate the liver fenestrae (pore size ~100 nm)38 but still small enough to extravasate through the leaky capillaries in the tumor tissue (pore size ~400 nm).39 Similar results were reported by Pirollo et al.10,40 With a size of ~200 nm, their nanoimmunoliposome complex delivered siRNA specifically to primary and meta-static tumors with minimal liver uptake. The increase of Vss for NPs in the tumor-bearing mice as compared to that of the tumor free mice suggests that the tumor served as a distribution sink for NPs. The decrease in the area under the curve and increase in clearance are also consistent with this observation. The presence of the targeting ligand facilitated the internalization of siRNA by the tumor cells (Figure 2 and Supplementary Figures S3 and S4) but did not alter the PK (Figures 1 and 2). The results indicate that extravasation of the vector, which was mainly dependent on the enhanced permeability and retention effect, was the rate limiting factor. Enhanced intracellular uptake of the vector did not cause further clearance. The internalization of siRNA formulation in the tumor was uneven (Figure 2 and Supplementary Figure S3), suggesting that cells closer to the angiogenic capillaries are likely to take up more siRNA. However, the EGFR silencing in the entire tumor was nearly complete (Figures 4a upper and 5, and Supplementary Figure S5). This suggests only small amounts of siRNA, as long as properly delivered, are necessary for effective gene silencing. The in vivo RNA interference effect was highly dependent on the formulation and siRNA sequence. Only siRNA formulated in targeted NPs showed significant gene silencing activity (Figures 4a upper and 5, and Supplementary Figure S5). This targeted formulation also showed good efficiency in inducing apoptosis in the tumor (15%, P < 0.05) (Figure 4b), inhibiting tumor growth (~40%, P < 0.01 on day 10) and sensitizing the tumor to chemotherapy (Figure 6).
The cytokine induction effect was dependent on the model, formulation, and siRNA sequence. The results in Figure 7a showed that the immune response of the nude mice were not sensitive to siRNA but to CpG motifs. siRNA-mediated immune response is via toll-like receptor 7/8 (TLR7/8),41,42 suggesting that the toll-like receptor 7/8 may be not be expressed or only at a low level in the nude mice. On the other hand, the toll-like receptor 9 was shown to be a good immunostimulatory target by CpG oligonucleotides in nude mice,43 and, therefore, a formulation containing plasmid showed strong IMT (Figure 7a). In the C57BL/6 model, when siRNA was formulated into the NP, the immunostimulatory effect was strengthened (Figure 7b). Additionally, the presence of the targeting ligand in the formulation increased the IMT of siRNA. This is probably due to the increased cellular uptake through endosomal pathway. Surprisingly, MDM2 siRNA containing no identifiable inflammatory sequences induced higher levels of cytokines than EGFR siRNA containing the immunostimulatory sequence (5′-UGUGU-3′) (Figure 7b). MDM2 siRNA may contain some unknown, yet potent, sequences that induced the production of cytokines. Another possibility is that different toll-like receptor isoforms might be involved in the immunostimulatory effect of MDM2 siRNA, which is suggested by Wang and colleagues.44 More importantly, empty NP (group F) showed very mild IMT (Figure 7a and b), suggesting the carrier itself was safe.
Although several siRNA delivery systems have exhibited high activity against tumors in vivo,2,4–16 our NP formulation provides the advantages of an almost complete encapsulation efficiency (>95%), the highest tumor delivery efficiency so far reported (70–80% ID/g at 4 hours) (Figure 2b) and relatively easy preparation. Our carrier also showed low toxicity (Figure 7 and Supplementary Figures S8 and S9).
MATERIALS AND METHODS
Materials
NCI-H460 (human lung cancer cells) were obtained from American Type Culture Collection. Cells were maintained in Roswell Park Memorial Institute medium 1640 supplemented with 10% fetal bovine serum (Invitrogen, Carlsbad, CA), 100 U/ml penicillin, and 100 μg/ml streptomycin (Invitrogen, Carlsbad, CA). NCI-H460 cells were shown to be sigma receptor positive by immunostaining and Western blot (data not shown). Primary antibodies against EGFR (rabbit polyclonal), prostate apoptotic response 4 (mouse monoclonal), AIF (rabbit polyclonal) and actin/β-actin (mouse monoclonal) were purchased from Santa Cruz Biotechnologies (Santa Cruz, CA).
Experimental animals
Female athymic nude mice of age 6–8 weeks and female C57BL/6 mice of age 6–7 weeks (16–18 g) were purchased from Charles River Laboratories (Wilmington, MA). All work performed on animals was in accordance with and permitted by the University of North Carolina Institutional Animal Care and Use committee.
NP preparations
NPs were prepared by the method described previously,18,20 except that the DSPE-PEG amount was reduced to half. DSPE-PEG-anisamide for the modification of the NPs was synthesized according to the published method.19 Control siRNA that targets the sequence 5′-AATTCTCCGAACGTGTCACGT-3′ and EGFR siRNA that targets the sequence 5′-AACACAGTGGAGCGAATTCCT-3′ as described previously35 were synthesized in Dharmacon (Lafayette, CO). The control sequence does not match any human genome sequence. FAM-siRNA was also obtained from Dharmacon (Lafayette, CO).
PK study
NCI-H460 cells (5 × 106) were subcutaneously injected into the right flank of nude mice. When tumors reached the size of ~1 cm2, mice were IV injected with FAM-siRNA in different formulations at the dose of 1.2 mg/kg. At different time points, 50–100 μl of blood was collected from the tail artery and serum was isolated. Ten microliters of serum was mixed with 90 μl of lysis buffer (0.1% sodium dodecyl sulfate in phosphate- buffered saline) and incubated at 65 °C for 10 minutes, followed by the addition of 200 μl methanol and incubation at 65 °C for 10 minutes. The sample was centrifuged at 14,000 rpm for 5 minutes and 200 μl supernatant was transferred to a black 96-well plate (Corning, Corning, NY). The fluorescence intensity of the sample was measured by a plate reader (Bioscan, Washington, DC) at λex: 485 nm and λem: 535 nm. FAM-siRNA concentration in each sample was calculated from a standard curve. This extraction method provided 98% recovery of free FAM-siRNA and 48% recovery for FAM-siRNA containing NP. The FAM-siRNA concentration was corrected for the degree of sample recovery.
Tissue distribution and tumor uptake study
Mice with tumor size of ~1 cm2 were IV injected with FAM-siRNA in different formulations (1.2 mg/kg). Four hours later, mice were killed and tissues were collected and imaged by the IVIS Imaging System (Xenogen Imaging Technologies, Alameda, CA). To quantify the accumulated doses, the excised tissues were homogenized in 300 or 1,000 μl lysis buffer (1,000 μl for livers and 300 μl for others) and incubated at 65 °C for 10 minutes. One hundred micro-liter of supernatant was collected after centrifugation at 14,000 rpm for 10 minutes. FAM-siRNA in the supernatant was extracted and quantified by the method described in the PK study. The accumulated dose in each tissue was calculated from a standard curve obtained by spiking known amounts of FAM-siRNA or FAM-siRNA containing NP in tissues obtained from uninjected animals. Alternatively, tumors were sectioned (7 μm thick) by a cryostat (H/I Hacker Instruments & Industries, Winnsboro, SC). Sections were counterstained with 4′,6-diamidino-2-phenylindole and imaged using a Leica SP2 confocal microscope.
Immunohistochemistry and Western blot
NCI-H460 xenograft tumor-bearing mice (tumor size ~1 cm2) were IV injected with siRNA in different formulations (1.2 mg siRNA/kg, one injection per day for 3 days). One day after the third injection, mice were killed and tumor samples were collected. Tumors were frozen-sectioned and 7 μm thick section samples were immunostained with primary antibodies and visualized with kits from Dakocytomation [DakoCytomation Envision + Dual Link System-HRP (DAB+), DakoCytomation, Carpinteria, CA]. Samples were imaged by using a Nikon Microphot SA microscope. The percentage of AIF nuclear localization was calculated based on 30 random images obtained from three individual tumors. Total protein (40 μg) isolated from the tumors was loaded on a polyacrylamide gel and electrophoresed. Protein bands in the gel were then transferred to a polyvinylidene difluoride and the EGFR was probed by antibodies. The level of the target proteins in each lysate was detected by enhanced chemiluminescence using ChemiGlow (Alpha Innotech, San Leandro, CA) followed by detection by the AlphaImager (Alpha Innotech, San Leandro, CA).
Tumor growth inhibition study
NCI-H460 xenograft tumor-bearing mice (size 25–40 mm2) were IV injected with siRNA containing formulations at the dose of 1.2 mg/kg (one injection per day for 3 days). For the chemo-combination treatment, cisplatin (3 mg/kg) were intraperitoneally administered from the 3rd to 5th day after the first siRNA injection. Tumor growth in the treated mice was monitored thereafter. Mouse body weight was also monitored.
Cytokine induction studies
Female athymic nude mice or C57BL/6 mice were injected with siRNA in different formulations at the dose of 1.2 mg siRNA/kg. Two hours after the injection, blood samples were collected and serum was isolated. Tumor necrosis factor, interleukin-6, interleukin-12 (BD Biosciences, San Diego, CA) and interferon-α levels (PBL Biomedical Laboratories, Piscataway, NJ) were measured by enzyme-linked immunosorbent assay according to the manufacturer’s protocol. MDM2 siRNA (target sequence: 5′-GCUUCGGAACAAGAGACUC-3′) without any known immunostimulatory sequences (5′-GUCCUUCAA-3′ and 5′-UGUGU-3′)45,46 was used as a negative control. Calf thymus DNA alone was formulated into targeted NPs and injected into the mice at the dose of 2.4 mg DNA/kg. This formulation is also regarded as the empty NP (without siRNA). Targeted NPs containing EGFR siRNA and luciferase plasmid DNA were also prepared and injected.
Statistical analysis
All statistical analyses were performed by a two- tailed student t-test. Data were considered statistically significant when P value was less than 0.05.
Supplementary Material
Acknowledgments
We thank Joyeeta Sen [University of North Carolina (UNC)] for synthesizing the DSPE-PEG-anisamide and the Michael Hooker Microscopy Facility of the UNC for the microscopy images. This research was supported by the Industrial Technology Research Institute in Taiwan and by the UNC at Chapel Hill. We appreciate Weihsu Claire Chen’s (UNC) assistance in preparing the manuscript. The authors declare there is no conflict of interest.
References
- 1.Jana S, Chakraborty C, Nandi S, Deb JK. RNA interference: potential therapeutic targets. Appl Microbiol Biotechnol. 2004;65:649–657. doi: 10.1007/s00253-004-1732-1. [DOI] [PubMed] [Google Scholar]
- 2.Li SD, Huang L. Gene therapy progress and prospects: non-viral gene therapy by systemic delivery. Gene Ther. 2006;13:1313–1319. doi: 10.1038/sj.gt.3302838. [DOI] [PubMed] [Google Scholar]
- 3.Li W, Szoka FC., Jr Lipid-based nanoparticles for nucleic acid delivery. Pharm Res. 2007;24:438–449. doi: 10.1007/s11095-006-9180-5. [DOI] [PubMed] [Google Scholar]
- 4.Grzelinski M, Urban-Klein B, Martens T, Lamszus K, Bakowsky U, Hobel S, et al. RNA interference-mediated gene silencing of pleiotrophin through polyethylenimine-complexed small interfering RNAs in vivo exerts antitumoral effects in glioblastoma xenografts. Hum Gene Ther. 2006;17:751–766. doi: 10.1089/hum.2006.17.751. [DOI] [PubMed] [Google Scholar]
- 5.Halder J, Kamat AA, Landen CN, Jr, Han LY, Lutgendorf SK, Lin YG, et al. Focal adhesion kinase targeting using in vivo short interfering RNA delivery in neutral liposomes for ovarian carcinoma therapy. Clin Cancer Res. 2006;12:4916–4924. doi: 10.1158/1078-0432.CCR-06-0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hu-Lieskovan S, Heidel JD, Bartlett DW, Davis ME, Triche TJ. Sequence-specific knockdown of EWS-FLI1 by targeted, nonviral delivery of small interfering RNA inhibits tumor growth in a murine model of metastatic Ewing’s sarcoma. Cancer Res. 2005;65:8984–8992. doi: 10.1158/0008-5472.CAN-05-0565. [DOI] [PubMed] [Google Scholar]
- 7.Kim WJ, Chang CW, Lee M, Kim SW. Efficient siRNA delivery using water soluble lipopolymer for anti-angiogenic gene therapy. J Control Release. 2007;118:357–363. doi: 10.1016/j.jconrel.2006.12.026. [DOI] [PubMed] [Google Scholar]
- 8.McNamara JO, 2nd, Andrechek ER, Wang Y, Viles KD, Rempel RE, Gilboa E, et al. Cell type-specific delivery of siRNAs with aptamer-siRNA chimeras. Nat Biotechnol. 2006;24:1005–1015. doi: 10.1038/nbt1223. [DOI] [PubMed] [Google Scholar]
- 9.Medarova Z, Pham W, Farrar C, Petkova V, Moore A. In vivo imaging of siRNA delivery and silencing in tumors. Nat Med. 2007;13:372–377. doi: 10.1038/nm1486. [DOI] [PubMed] [Google Scholar]
- 10.Pirollo KF, Rait A, Zhou Q, Hwang SH, Dagata JA, Zon G, et al. Materializing the potential of small interfering RNA via a tumor-targeting nanodelivery system. Cancer Res. 2007;67:2938–2943. doi: 10.1158/0008-5472.CAN-06-4535. [DOI] [PubMed] [Google Scholar]
- 11.Santel A, Aleku M, Keil O, Endruschat J, Esche V, Fisch G, et al. A novel siRNA-lipoplex technology for RNA interference in the mouse vascular endothelium. Gene Ther. 2006;13:1222–1234. doi: 10.1038/sj.gt.3302777. [DOI] [PubMed] [Google Scholar]
- 12.Schiffelers RM, Ansari A, Xu J, Zhou Q, Tang Q, Storm G, et al. Cancer siRNA therapy by tumor selective delivery with ligand-targeted sterically stabilized nanoparticle. Nucleic Acids Res. 2004;32:e149. doi: 10.1093/nar/gnh140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Song E, Zhu P, Lee SK, Chowdhury D, Kussman S, Dykxhoorn DM, et al. Antibody mediated in vivo delivery of small interfering RNAs via cell-surface receptors. Nat Biotechnol. 2005;23:709–717. doi: 10.1038/nbt1101. [DOI] [PubMed] [Google Scholar]
- 14.Takeshita F, Minakuchi Y, Nagahara S, Honma K, Sasaki H, Hirai K, et al. Efficient delivery of small interfering RNA to bone-metastatic tumors by using atelocollagen in vivo. Proc Natl Acad Sci USA. 2005;102:12177–12182. doi: 10.1073/pnas.0501753102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang Y, Gao S, Ye WH, Yoon HS, Yang YY. Co-delivery of drugs and DNA from cationic core-shell nanoparticles self-assembled from a biodegradable copolymer. Nat Mater. 2006;5:791–796. doi: 10.1038/nmat1737. [DOI] [PubMed] [Google Scholar]
- 16.Yang R, Yang X, Zhang Z, Zhang Y, Wang S, Cai Z, et al. Retraction. Single-walled carbon nanotubes-mediated in vivo and in vitro delivery of siRNA into antigen-presenting cells. Gene Ther. 2007;14:920. doi: 10.1038/sj.gt.3302962. [DOI] [PubMed] [Google Scholar]
- 17.de Wolf HK, Snel CJ, Verbaan FJ, Schiffelers RM, Hennink WE, Storm G. Effect of cationic carriers on the pharmacokinetics and tumor localization of nucleic acids after intravenous administration. Int J Pharm. 2007;331:167–175. doi: 10.1016/j.ijpharm.2006.10.029. [DOI] [PubMed] [Google Scholar]
- 18.Li SD, Huang L. Targeted delivery of antisense oligodeoxynucleotide and small interference RNA into lung cancer cells. Mol Pharm. 2006;3:579–588. doi: 10.1021/mp060039w. [DOI] [PubMed] [Google Scholar]
- 19.Banerjee R, Tyagi P, Li S, Huang L. Anisamide-targeted stealth liposomes: a potent carrier for targeting doxorubicin to human prostate cancer cells. Int J Cancer. 2004;112:693–700. doi: 10.1002/ijc.20452. [DOI] [PubMed] [Google Scholar]
- 20.Li SD, Huang L. Surface-modified LPD nanoparticles for tumor targeting. Ann N Y Acad Sci. 2006;1082:1–8. doi: 10.1196/annals.1348.001. [DOI] [PubMed] [Google Scholar]
- 21.Brannon-Peppas L, Blanchette JO. Nanoparticle and targeted systems for cancer therapy. Adv Drug Deliv Rev. 2004;56:1649–1659. doi: 10.1016/j.addr.2004.02.014. [DOI] [PubMed] [Google Scholar]
- 22.Jones HE, Gee JM, Hutcheson IR, Knowlden JM, Barrow D, Nicholson RI. Growth factor receptor interplay and resistance in cancer. Endocr Relat Cancer. 2006;13 (suppl 1):S45–S51. doi: 10.1677/erc.1.01275. [DOI] [PubMed] [Google Scholar]
- 23.Nicholson RI, Gee JM, Harper ME. EGFR and cancer prognosis. Eur J Cancer. 2001;37 (suppl 4):S9–S15. doi: 10.1016/s0959-8049(01)00231-3. [DOI] [PubMed] [Google Scholar]
- 24.Byers LA, Heymach JV. Dual targeting of the vascular endothelial growth factor and epidermal growth factor receptor pathways: rationale and clinical applications for non-small-cell lung cancer. Clin Lung Cancer. 2007;8 (suppl 2):S79–S85. doi: 10.3816/clc.2007.s.006. [DOI] [PubMed] [Google Scholar]
- 25.Dassonville O, Bozec A, Fischel JL, Milano G. EGFR targeting therapies: monoclonal antibodies versus tyrosine kinase inhibitors. Similarities and differences. Crit Rev Oncol Hematol. 2007;62:53–61. doi: 10.1016/j.critrevonc.2006.12.008. [DOI] [PubMed] [Google Scholar]
- 26.Sathornsumetee S, Rich JN. New approaches to primary brain tumor treatment. Anticancer Drugs. 2006;17:1003–1016. doi: 10.1097/01.cad.0000231473.00030.1f. [DOI] [PubMed] [Google Scholar]
- 27.Wong SF. Cetuximab: an epidermal growth factor receptor monoclonal antibody for the treatment of colorectal cancer. Clin Ther. 2005;27:684–694. doi: 10.1016/j.clinthera.2005.06.003. [DOI] [PubMed] [Google Scholar]
- 28.Fan QW, Weiss WA. RNA interference against a glioma-derived allele of EGFR induces blockade at G2M. Oncogene. 2005;24:829–837. doi: 10.1038/sj.onc.1208227. [DOI] [PubMed] [Google Scholar]
- 29.Kang CS, Pu PY, Li YH, Zhang ZY, Qiu MZ, Huang Q, et al. An in vitro study on the suppressive effect of glioma cell growth induced by plasmid-based small interference RNA (siRNA) targeting human epidermal growth factor receptor. J Neurooncol. 2005;74:267–273. doi: 10.1007/s11060-004-8322-z. [DOI] [PubMed] [Google Scholar]
- 30.Kang CS, Zhang ZY, Jia ZF, Wang GX, Qiu MZ, Zhou HX, et al. Suppression of EGFR expression by antisense or small interference RNA inhibits U251 glioma cell growth in vitro and in vivo. Cancer Gene Ther. 2006;13:530–538. doi: 10.1038/sj.cgt.7700932. [DOI] [PubMed] [Google Scholar]
- 31.Nozawa H, Tadakuma T, Ono T, Sato M, Hiroi S, Masumoto K, et al. Small interfering RNA targeting epidermal growth factor receptor enhances chemosensitivity to cisplatin, 5-fluorouracil and docetaxel in head and neck squamous cell carcinoma. Cancer Sci. 2006;97:1115–1124. doi: 10.1111/j.1349-7006.2006.00287.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vollmann A, Vornlocher HP, Stempfl T, Brockhoff G, Apfel R, Bogdahn U. Effective silencing of EGFR with RNAi demonstrates non-EGFR dependent proliferation of glioma cells. Int J Oncol. 2006;28:1531–1542. [PubMed] [Google Scholar]
- 33.Wu X, Deng Y, Wang G, Tao K. Combining siRNAs at two different sites in the EGFR to suppress its expression, induce apoptosis, and enhance 5-fluorouracil sensitivity of colon cancer cells. J Surg Res. 2007;138:56–63. doi: 10.1016/j.jss.2006.06.027. [DOI] [PubMed] [Google Scholar]
- 34.Zhang M, Zhang X, Bai CX, Chen J, Wei MQ. Inhibition of epidermal growth factor receptor expression by RNA interference in A549 cells. Acta Pharmacol Sin. 2004;25:61–67. [PubMed] [Google Scholar]
- 35.Zhang X, Chen ZG, Choe MS, Lin Y, Sun SY, Wieand HS, et al. Tumor growth inhibition by simultaneously blocking epidermal growth factor receptor and cyclooxygenase-2 in a xenograft model. Clin Cancer Res. 2005;11:6261–6269. doi: 10.1158/1078-0432.CCR-04-2102. [DOI] [PubMed] [Google Scholar]
- 36.Li S, Huang L. In vivo gene transfer via intravenous administration of cationic lipid-protamine-DNA (LPD) complexes. Gene Ther. 1997;4:891–900. doi: 10.1038/sj.gt.3300482. [DOI] [PubMed] [Google Scholar]
- 37.Schwartz DA, Quinn TJ, Thorne PS, Sayeed S, Yi AK, Krieg AM. CpG motifs in bacterial DNA cause inflammation in the lower respiratory tract. J Clin Invest. 1997;100:68–73. doi: 10.1172/JCI119523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Braet F, Wisse E. Structural and functional aspects of liver sinusoidal endothelial cell fenestrae: a review. Comp Hepatol. 2002;1:1. doi: 10.1186/1476-5926-1-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Monsky WL, Fukumura D, Gohongi T, Ancukiewcz M, Weich HA, Torchilin VP, et al. Augmentation of transvascular transport of macromolecules and nanoparticles in tumors using vascular endothelial growth factor. Cancer Res. 1999;59:4129–4135. [PubMed] [Google Scholar]
- 40.Pirollo KF, Zon G, Rait A, Zhou Q, Yu W, Hogrefe R, et al. Tumor-targeting nanoimmunoliposome complex for short interfering RNA delivery. Hum Gene Ther. 2006;17:117–124. doi: 10.1089/hum.2006.17.117. [DOI] [PubMed] [Google Scholar]
- 41.Agrawal S, Kandimalla ER. Role of Toll-like receptors in antisense and siRNA [corrected] Nat Biotechnol. 2004;22:1533–1537. doi: 10.1038/nbt1042. [DOI] [PubMed] [Google Scholar]
- 42.Robbins MA, Rossi JJ. Sensing the danger in RNA. Nat Med. 2005;11:250–251. doi: 10.1038/nm0305-250. [DOI] [PubMed] [Google Scholar]
- 43.Gekeler V, Gimmnich P, Hofmann HP, Grebe C, Rommele M, Leja A, et al. G3139 and other CpG-containing immunostimulatory phosphorothioate oligodeoxynucleotides are potent suppressors of the growth of human tumor xenografts in nude mice. Oligonucleotides. 2006;16:83–93. doi: 10.1089/oli.2006.16.83. [DOI] [PubMed] [Google Scholar]
- 44.Wang L, Smith D, Bot S, Dellamary L, Bloom A, Bot A. Noncoding RNA danger motifs bridge innate and adaptive immunity and are potent adjuvants for vaccination. J Clin Invest. 2002;110:1175–1184. doi: 10.1172/JCI15536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hornung V, Guenthner-Biller M, Bourquin C, Ablasser A, Schlee M, Uematsu S, et al. Sequence-specific potent induction of IFN-alpha by short interfering RNA in plasmacytoid dendritic cells through TLR7. Nat Med. 2005;11:263–270. doi: 10.1038/nm1191. [DOI] [PubMed] [Google Scholar]
- 46.Judge AD, Sood V, Shaw JR, Fang D, McClintock K, MacLachlan I. Sequence-dependent stimulation of the mammalian innate immune response by synthetic siRNA. Nat Biotechnol. 2005;23:457–462. doi: 10.1038/nbt1081. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.