Abstract
MicroRNAs (miRNA/miR) are a class of small non-coding RNAs implicated in the pathogenesis of various malignancies. In the current study, using micro(RNA)arrays, we found a ubiquitous loss of miR-126 expression in colon cancer lines when compared to normal human colon epithelia. Reconstitution of miR-126 in colon cancer cells resulted in a significant growth reduction as evidenced in clonogenic assays. A search for miR-126 gene targets revealed p85β, a regulatory subunit involved in stabilizing and propagating the PI3K signal, as one of the potential substrates. Restoration of miR-126 in cancer cells induced a ≥3-fold reduction in p85β protein levels, with no concomitant change in p85α, a gene that is functionally related to p85β but not a supposed target of miR-126. Additionally, using reporter constructs, we show that the p85β-3′ UTR is directly targeted by miR-126. Furthermore, this miR-126 mediated reduction of p85β was accompanied by a substantial reduction in phosphorylated AKT levels in the cancer cells, suggesting an impairment in PI3K signaling. Finally, in a panel of matched normal colon and primary colon tumors, each of the tumors demonstrated miR-126 down-regulation together with an increase in the p85β protein level. Taken together, we propose that miR-126 regulates PI3K signaling partly by targeting p85β, and that the loss of miR-126 may provide a selective growth advantage during colon carcinogenesis.
Keywords: miR-126, colon cancer, AKT, p85β, microarray
Introduction
MicroRNAs (miRNA/miR) are a group of small non-coding 18–22 nucleotide RNAs, which have been suggested to play an important role in development and disease by regulating gene expression (Gartel and Kandel, 2008). miRNAs bind to complementary sites in the 3′-untranslated region (UTR) of target mRNAs and subsequently either degrade the transcript or block its translation, depending on whether the miRNA is completely or partially matched to the target gene sequence, respectively (Gartel and Kandel, 2008). Recently, miRNAs have been shown to be aberrantly expressed in various cancers including breast, colon, lung, brain, thyroid, pancreas and hematologic malignancies (Gartel and Kandel, 2008). In addition, there is a growing body of evidence suggesting that some miRNAs may have oncogenic or tumor suppressor functions (Gartel and Kandel, 2008). Nevertheless, the mechanisms involved in transcriptional regulation of miRNAs, in addition to their target genes, remain mostly unknown.
In the current study, we identify miR-126 as being frequently lost in colon cancers and as normally functioning as a growth suppressor in colon cells. Moreover, we provide experimental evidence suggesting that miR-126 regulates phosphatidylinositol 3-kinase (PI3K) signaling by targeting the phosphatidylinositol 3-kinase regulatory subunit beta (p85β).
Materials and Methods
Cell culture and Microarray
The Vaco series colon cancer cell lines were cultured in low serum (2%) as described (Willson et al., 1987). LS174T and DLD1 cell lines with an inducible inhibition of the β-catenin signaling (van de Wetering et al., 2002) were obtained from Hans Clevers (Hubrecht Institute, Uppsalalaan, The Netherlands). The colon cancer cell line, FET that has retained TGF-β responsiveness and autocrine activity (Wu et al., 1992) was obtained from Michael Brattain (Roswell Park Cancer Institute, Buffalo, NY). Total RNA, which includes small nucleic acid fractions, was extracted from the cell lines, tissues and human colon crypt epithelia (Chen et al., 2005) using mirVana miRNA Isolation Kit (Ambion, Austin, TX). miRNAs were enriched from 50μg of total RNA from cell lines and miRNA-certified Firstchoice normal colon RNA (Ambion), using the FlashPAGE fractionator (Ambion). The microarray protocol was then carried out as per the manufacturer’s instructions using mirVana miRNA Labeling kit and Bioarray chips (Ambion). Briefly, the isolated miRNAs were spiked with controls, appended with 3′ Amine-modified tails and were labeled with Cy5 fluorescent dye (Amersham Biosciences, Piscataway, NJ). Following post-labeling cleanup, the samples were hybridized on to array slides, sealed in hybridization chambers (Corning, Acton, MA) and incubated in a water bath at 42°C, overnight. The slides were then washed, and dried using a quick centrifugation method. Images were acquired using the Genepix 4000B array scanner (Molecular Devices, Sunnyvale, CA) at 600 PMT, 100% gain and 5μm resolution. The raw data was then normalized using background-subtracted Mean Intensity values and the ratios of cancer vs normal were calculated using log2 transformed normalized intensities (Bioarray data analysis, Ambion).
Real Time PCR
miRNA expression levels were quantified using mirVana qRT-PCR miRNA detection protocol (Ambion). Briefly, 500ng of total RNA from cell lines and human colon crypt epithelia (Chen et al., 2005) was used for miRNA-specific reverse-transcription followed by PCR amplification. 5S-rRNA was used as a normalization control. Each PCR reaction was carried out in triplicate in a 25μl volume using SYBR Green Assay Master Mix (Applied Biosystems, Foster City, CA) for 3min at 95°C, followed by 40 cycles of 95°C for 15s and 60°C for 30s in a Bio-Rad I Cycler (Bio-Rad Laboratories, Hercules, CA). miRNA levels were quantified based on the ratio of miRNA/5S-rRNA using the formula, 2(CTmicroRNA–CT5S). 2μg of total RNA was reverse transcribed to assay for p85β, p85α and β2-microglobulin gene expression using primers p85β (F:5′-CAT TTC AAG GGA GGA GGT GA-3′, R:5′-AGC TTA TTG TTC CCG CCT TT-3′), p85α (F:5′-GCT GAA TGG TAC TGG GGA GA-3′, R:5′-TAC CAA AAA GGT CCC GTC TG-3′) and β2-microglobulin (F:5′-GTG CTC GCG CTA CTC TCT CTT TC-3′, R:5′-CTT CAA TGT CGG ATG GAT GAA AC-3′). Each PCR reaction was carried out in triplicate in a 25μl volume using the SYBR Green Assay as described above for 40 cycles of 95°C for 15s and 60°C for 20s and 72°C for 20s and mRNA levels quantified using the formula as mentioned above. The gene expressions of p85β and p85α were normalized to the levels of β2-microglobulin.
MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) Assay
Cells were seeded at 1.0×105 cells/well of a six-well plate and transfected the next day with 12.5nM, 25nM and 50nM concentrations of a double stranded RNA miR-126 precursor molecule (AM17100, Ambion) or a precursor negative control miRNA with a scrambled sequence (AM17110, Ambion,) using 5 μL of LipofectAMINE 2000 (Invitrogen, Carlsbad, CA) as per the manufacturer’s instructions. 72 hours after transfection, cells were washed with minimum essential medium (MEM) without phenol red (Invitrogen). 2 mL MEM with 80 μL of MTT (5 mg/mL, Sigma-Aldrich, St Louis, MO) were added to each well, and plates were incubated at 37°C for 4 hours. After medium was removed, MTT crystals were solubilized in acidic isopropanol and subjected to centrifugation to pellet the cellular debris. Spectrophotometric absorbance of each sample was measured at 590 nm using a spectrophotometer (Beckman, Corona, CA).
Clonogenic Assay
Cells were seeded at 1.0×105 cells/well of a six-well plate and transfected the next day with 12.5nM, 25nM and 50nM concentrations of a double stranded RNA miR-126 precursor molecule or control miRNA. 18 hours after transfection, the cells were washed in PBS and trypsinized. Cells were then seeded in six-well plates at 400 cells/well in 2 mL medium. Colonies were grown for 10 days, stained as previously described (Guda et al., 2007) and quantified using Alpha imager (Alpha Innotech, San Leandro, CA).
Western Blots
V429 and V703 cells were seeded at 1.0×105 cells/well of a six-well plate and transfected the next day with 50nM concentration of a double stranded RNA miR-126 precursor molecule or control miRNA. Total cell lysates were prepared 48 hours after miRNA transfection, by incubating at 4°C for 30 min in RIPA buffer (20 mM Tris-Cl (pH 7.5), 150 mM NaCl, 1% Igepal CA-630, 0.5% sodium deoxycholate, 1 mM EDTA) supplemented with Complete Mini protease inhibitor cocktail (Roche, Indianapolis, IN) and phosphate inhibitor cocktail (Sigma, St. Louis, MO). After 10 min of centrifugation at 14000 r.p.m. at 4°C, the clarified supernatants were resolved on 4–12% gradient SDS-PAGE gels, followed by electrotransfer onto Immobilon™-P PVDF membranes (Millipore, Bedford, MA). Protein lysates from homogenized frozen matched normal colon mucosa and primary colon tumors were prepared in a similar way as described above. For analysis of protein levels, blots were blocked with either 5% BSA (for p85β, p85α, AKT and phospho-AKT blots) or 5% milk (for β-actin) in TTBS (0.05% Tween-20 in TBS), then probed with antibodies against p85β (1:50, Abcam, Cambridge, MA), p85α (1:1000, Calbiochem, CA), phospho-AKT (Ser473) (1:1000, Cell Signaling, MA), AKT (pan) (1:1000, Cell Signaling, MA) or β-actin (1:2000, Abcam), and detected with horseradish peroxidase–conjugated secondary antibody (1:1500; Jackson Immuno Research Laboratories, West Grove, PA) and Enhanced Chemiluminescence Plus (Amersham Biosciences, Piscataway, NJ). The signals were visualized using radiographic films and image densitometry was performed with Alpha Imager.
P85 β 3′-UTR Luciferase Reporter Assay
A 0.6kb region that includes the 3′-UTR segment of p85β predicted to interact with miR-126 (Figure 3A) was PCR amplified from human genomic DNA, using primers: Forward-WT 5′-CTT CTT ACG CGT CTC CAG CCT GGG TAA CAG AG-3′ and Reverse-WT 5′-ACC ACC AAG CTT CCT GTA TGA CCT TGG GCA CT-3′. The PCR conditions included, 3min at 95°C, 25 cycles of 95°C for 15s, 60°C for 30s and 72°C for 45s. The mutant segment was generated by replacing the 8-nucleotide target sequence of p85β 3′-UTR (Figure 3A) with a complimentary sequence, wild-type p85β: 5′-aggcaggttttgtACGGTACGt-3′, mutant p85β: 5′-aggcaggttttgtTGCCATGCt-3′, by a 2-step process. Briefly, two PCR fragments were amplified using the Forward-WT and 5′-GCA TGG CAA CAA AAC CTG CCT CCC AGC T-3′ primer sets, and 5′-TGC CAT GCT TGT TAT TGA TAT GAT ATA AAA CAT C-3′ and Reverse-WT primer sets, using the 0.6kb wild-type sequence as a template. The two fragments were then annealed, extended and PCR amplified using Forward-WT and Reverse-WT primer sets. The wild-type and mutant 3′-UTR segments were then inserted into the MluI and HindIII sites of pMIR-REPORT™ Luciferase vector immediately downstream from the stop codon of the luciferase gene (Ambion). For the luciferase reporter assay, SW480 cells were co-transfected using LipofectAMINE (Invitrogen) in 6-well plates with 0.76 μg of the luciferase report vector and 0.04 μg of the control vector containing Renilla luciferase, pRL-CMV (Promega, Madison WI), as well as with 12.5nM, 25nM and 50nM concentrations of a double stranded RNA miR-126 precursor molecule or a precursor negative control miRNA with a scrambled sequence (Ambion) using LipofectAMINE 2000 (Invitrogen) transfection reagent. Firefly and Renilla luciferase activities were measured consecutively according to the manufacturer’s instruction (Promega) using a Microtiter plate Luminometer (Dynex technologies, Chantilly, VA).
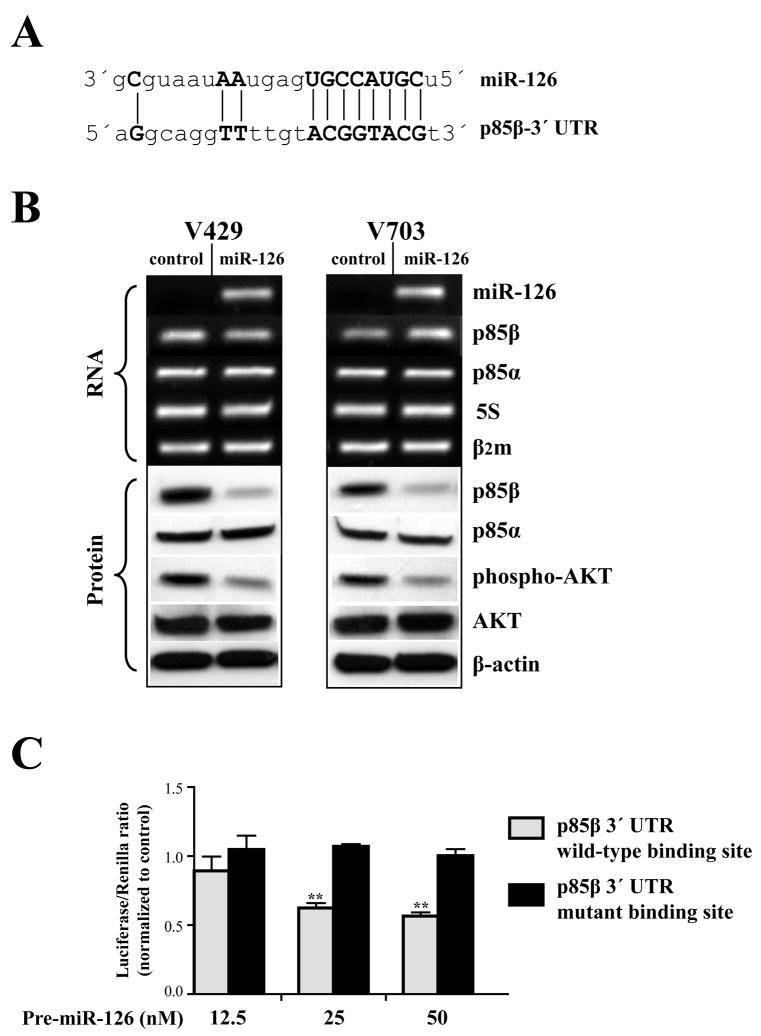
Figure 3. miR-126 represses PI3K signaling by targeting p85β.
A) Sequence alignment of miR-126 with 3′ UTR of p85β (Griffiths-Jones et al., 2006). Note the complimentarity of miR-126 seed sequence to p85β 3′ UTR displayed as vertical lines. B) Rows 1–5 show the RNA levels analyzed by qRT-PCR and rows 6–10 display the protein levels determined by Western blots after miR-126 induction. Row 1 demonstrates the successful restoration of miR-126 in cell lines transfected with its precursor molecule (Ambion). Note the reduction in p85β (row 6) and phospho-AKT (row 8) proteins in miR-126 treated cells. β–actin, β2microglobulin (β2m) and 5S-rRNA were used as loading controls for protein, mRNA and miRNA respectively. All analyses were performed 48 hrs after miR-126 transfection. C) Dual luciferase assays of luciferase reporter constructs carrying a wild-type p85β 3′ UTR verses a 3′ UTR mutation in the potential miR-126 binding site. Note the dose-dependent reduction in the luciferase activity of wild-type p85β when compared to the mutant p85β following miR-126 induction. (**) indicates significant differences between the means of wild-type and mutant p85β 3′ UTR luciferase activities within each group (t-test, p<0.005). All analyses were performed in triplicates after 48 hr incubation and values expressed as the ratio of luciferase/renilla activities and normalized to the control miRNA precursor within the respective groups.
Results and Discussion
miR-126 is Frequently Lost in Colon Cancers
We determined the differential expression of 262 miRNAs in representative colon cancer cell lines using mirVana miRNA Bioarrays (Ambion). As shown in Table 1, a total of 45 miRNAs were aberrantly expressed (≥2-fold change) in cancer lines relative to the normal colon (Ambion). More than half of the miRNAs identified as altered in colon cancers in the microarrays had also previously been so detected using different techniques (Bandres et al., 2006; Cummins et al., 2006; Volinia et al., 2006; Gartel and Kandel, 2008). We, however, particularly noted the down-regulation of miR-126 in colon cancers, as this had not been previously described. miR-126 was down-regulated in 4 of 4 colon cancer cell lines tested using microarrays. As miR-126 has recently been shown to be also lost in a majority of primary breast (Tavazoie et al., 2008) and lung (Yanaihara et al., 2006) tumors, this suggested a possible general role for the loss of miR-126 in cancer. To first confirm whether miR-126 expression in normal colon was derived from colonic epithelial cells, we examined for miR-126 levels in RNA derived from purified colonic epithelial crypts pooled from five individuals (Chen et al., 2005). Additionally, we extended our analysis of colon cancer cell lines to a total of twelve lines. As illustrated in Figure 1, miR-126 was robustly expressed in purified colon crypt epithelial cells; whereas, all colon cancer lines showed a marked loss of miR-126 expression. Moreover, analysis of homogenates from primary colon cancers showed that in all cases miR-126 was down-regulated by at least 2-fold when compared to matched normal colon mucosa (Figure 4). Residual miR-126 expression in these tumor tissues may be likely derived from non-malignant normal/stromal elements in the tumors. Our results indicate that miR-126 down-regulation is a recurrent event during colon carcinogenesis.
Table 1.
miRNA Signatures in Colon Cancer Cell Lines
| Down-regulated miRNAs in colon cancer cells | Up-regulated miRNAs in colon cancer cells | |
|---|---|---|
| miR-96 | miR-93 | *miR-19a |
| miR-485_5p | *miR-92 | *miR-18a |
| miR-422b | miR-520h | *miR-183 |
| *miR-342 | miR-508 | *miR-182 |
| *miR-214 | miR-505 | *miR-181b |
| *miR-199a | miR-449 | miR-181a |
| *miR-195 | *miR-429 | miR-181c |
| miR-150 | miR-384 | *miR-17_5p |
| *miR-145 | miR-373 | *miR-148a |
| *miR-143 | miR-34c | *miR-141 |
| *miR-133a | miR-326 | miR-130b |
| miR-126 | *miR-25 | miR-128a |
| *miR-125b | miR-224 | *miR-106b |
| miR-100 | miR-210 | *miR-106a |
| *miR-200a | let-7d | |
| *miR-19b | ||
Four representative colon cancer cell lines (V9m, V855, V410, V478) were utilized as the discovery set for determining the miRNA profiles. The table lists all the miRNAs that are differentially expressed (≥2-fold, log2 values) in at least 3 of 4 colon cancer lines, relative to normal colon. More than 50% of these miRNAs (*) have been previously reported to be altered in a similar way in primary colon tumors (Bandres et al., 2006; Cummins et al., 2006; Volinia et al., 2006; Grady et al., 2008), thereby providing an independent validation of our experimental approach.
Figure 1. Loss of miR-126 in colon cancer cells.
miR-126 expression levels in colon cancer cell lines were shown as fold changes relative to the normal colon epithelial crypts (NCC) pooled from 5 individuals. Note the marked reduction of miR-126 expression in 12 of 12 cancer lines. Amplification products of the qRT-PCR reaction from NCCs were cloned, sequenced and confirmed as miR-126 (data not shown). 5S-rRNA was used as a normalization control.
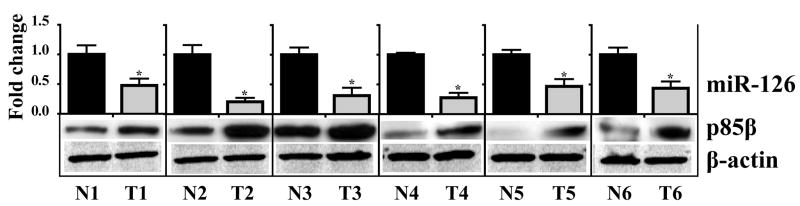
Figure 4. Inverse correlation between miR-126 and p85β in primary colon tumors.
The upper panel shows the qRT-PCR quantification of miR-126 and lower panel shows the western blot analysis of p85β protein levels in matched colon normal (N) and primary colon tumor (T) pairs. miR-126 in tumors is expressed as fold change relative to their matched normals. Note the significant downregulation of miR-126 (≥2-fold, t-test p<0.05) and overexpression of p85β protein in each of the colon tumors. 5S-rRNA and β–actin protein were used as normalization controls for miR-126 and p85β protein, respectively.
miR-126 Impedes the Growth of Cancer Cells
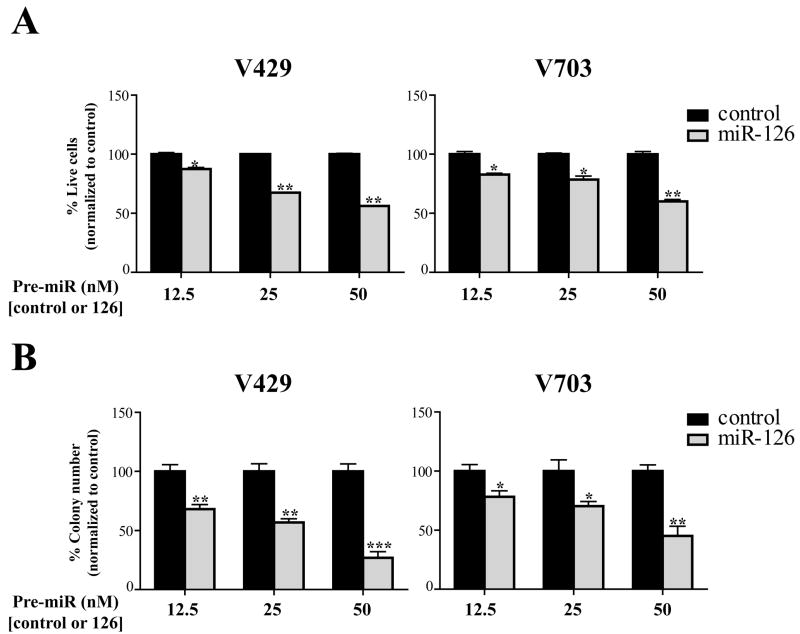
To investigate the functional significance of the loss of miR-126 in colon cancers, we performed MTT and clonogenic assays in cancer cells after restoring miR-126 expression. As demonstrated in Figure 2, both V429 and V703 cell lines transfected with miR-126 showed a dose-dependent reduction in cell (Figure 2A) and colony numbers (Figure 2B) when compared to the negative control. These findings in colon cancer cells are in agreement with a recent report showing the growth suppressive effects of miR-126 in breast cancers (Tavazoie et al., 2008), and thereby further support a general role for miR-126 down-regulation in promoting tumor development.
Figure 2. miR-126 suppresses the growth of colon cancer cells.
A) Quantification of cell growth in V429 and V703 cell lines transfected with a control or miR-126 precursor molecule at 3 different concentrations (Ambion). The bar graphs represent the percentage of live cells, normalized to the control group, at 72 hrs after control or miR-126 transfection. Data was obtained from 3 independent wells and each experiment repeated 3 times. Note the dose-dependent suppression of cell growth by miR-126 in both V429 and V703 cells.
B) Quantification of colony numbers in cell lines. The bar graphs represent colony numbers expressed as a percentage of the colony counts observed in the control group for each concentration. The colony counts for control and miR-126 groups at different doses were obtained from 3 independent wells and each experiment was repeated 3 times. Note the dose-dependent reduction in the colony number following miR-126 reconstitution in both cell lines. Statistical analysis was performed using an unpaired t test and the error bars indicate standard error of the means. (*) represents significant differences (*p<0.05, **p<0.005, ***p<0.0005) between means of the control verses miR-126 treated groups for each concentration.
The PI3K Signaling Pathway is a Target of miR-126-mediated Growth Suppression
To understand possible mechanism(s) that may underlie miR-126-mediated growth suppression, we performed miRanda scan and identified several potential gene targets of miR-126 (Griffiths-Jones et al., 2006). We narrowed the list by restricting to genes that had the best p-value based on the miRanda statistical model (Griffiths-Jones et al., 2006), and also to those that were integral to pathways often deregulated in colon cancer. Based on the above criteria, we identified p85β, a fundamental component of the PI3K signaling network, as a candidate target gene of interest. Figure 3A illustrates the in-silico pairing of the miR-126 seed sequence from 5′ nucleotides 2–9 that is suggested to be an important region for target recognition by vertebrate miRNAs (Griffiths-Jones et al., 2006), to sequences of the p85β 3′ UTR (nucleotides 3897–3904, NM_005027).
p85β is one of the regulatory subunits of class IA PI3Ks. Mammalian PI3K is composed of a heterodimer of a p110 catalytic subunit (p110α, p110β, p110δ)with a p85 regulatory subunit (p85α, p85β, p55γ, p55α, p50α). p85s stabilize the thermally labile p110s, but also conformationally inhibit their catalytic activity. Upon cellular stimulation, the Src-homology 2 (SH2) domains of p85s mediate the recruitment of p110s to membrane-proximal protein substrates, thereby activating p110 and initiating signaling through a series of pathway elements including PIP3, PDK1, and AKT, with activated AKT phosphorylating a wide array of proteins that induce cell proliferation, survival and increased motility (Vanhaesebroeck and Waterfield, 1999; Osaki et al., 2004). Based on the fact that p85 is indispensable for p110 recruitment and subsequent activation (Vanhaesebroeck and Waterfield, 1999), we hypothesized that a targeted degradation of p85β by miR-126 may impair the downstream signaling cascade.
At first, we tested the validity of miRanda target prediction (Griffiths-Jones et al., 2006) by analyzing p85β status in the cancer cells after miR-126 induction. As shown in Figure 3B, miR-126 substantially reduced p85β protein (≥3-fold) with no apparent degradation of the transcript, thereby indicating a translational repression of the p85β mRNA by miR-126. Additionally, no alterations were evident in p85α (Figure 3B), a gene that is functionally related to p85β, but not a supposed target of miR-126 (Griffiths-Jones et al., 2006). To further demonstrate that p85β is a direct target of miR-126, we constructed luciferase reporters, as described in Materials and Methods, encoding either a wild-type p85β 3′ UTR sequence or a p85β 3′ UTR sequence mutant at the predicted miR-126 recognition site (Figure 3A). As shown in Figure 3C, miR-126 significantly reduced the luciferase activity of the wild-type p85β when compared to the mutant p85β and this reduction in luciferase activity was dependent on the concentration of miR-126. These findings indicate that p85β is a direct miR-126 target, and that the decrease in p85β protein observed in the cell lines is a miR-126-mediated effect. Additionally, analysis of a panel of matched normal colon mucosa and primary colon cancers show that miR-126 is down-regulated in each of these tumors, and that miR-126 down-regulation is in each instance accompanied by an increase in p85β protein level (Figure 4). This observation provides further support for p85β being an in vivo target of miR-126.
Next, we evaluated the activity of the PI3K pathway by analyzing the status of phosphorylated AKT in the cancer cells after miR-126 restoration. As revealed in Figure 3B, miR-126 significantly reduced the levels of phospho-AKT, without altering total AKT protein. It is therefore likely that miR-126 targeting of p85β normally provides a negative regulation of the PI3K-AKT signaling pathway. This observation, reinforcing the important role of p85 subunits in mediating PI3K activity (Yu et al., 1998), is consistent with findings from p85α and p85β knockout mice (Luo et al., 2006), with studies examining the association of p85β expression and PI3K activity in human brain tissue (Dwivedi et al., 2007), and with studies employing genetic manipulation of p85s in cell lines (Yu et al., 1998; Haneline et al., 2006). Of note, frequent somatic mutations in the p110α catalytic subunit, as described in various cancers including colon, has been suggested to contribute to tumor development via upregulation of the PI3K signaling pathway (Samuels et al., 2004). A recent study on the functional aspects of these hot-spot mutations, that occur specifically in the kinase domain of p110α, show that their gain of function activity is highly dependent on the interaction with the p85 regulatory subunit (Zhao and Vogt, 2008). These findings underscore the requirement of p85s in mediating the activity of mutant p110α. Incidentally, we identified a Y1021C missense mutation within the kinase domain of p110α in the V429 cell line (data not shown). Despite the presence of a mutant p110α, our functional analyses in the V429 cell line show that a targeted down-regulation of p85β by miR-126 could in fact impede the PI3K downstream signaling cascade (Figure 3B). These observations are consistent with findings from the previous report (Zhao and Vogt, 2008) and further emphasize the essential role of p85 subunits in the tumorigenic process.
Lastly, we investigated possible mechanisms that might account for the down-regulation of miR-126 in colon cancers. miR-126 is located in a region (9q34.3) that is not commonly lost in colon tumors (Platzer et al., 2002). Accordingly, we analyzed whether miR-126 loss could be downstream of either Wnt pathway activation or loss of TGF-β signaling, two of the most common alterations in signaling pathways in colon cancers. Results however showed no apparent regulation of miR-126 by Wnt, as assessed in cells carrying an inducible dominant negative TCF4 (van de Wetering et al., 2002), or by TGF-β as assessed in a rare colon cancer cell line that has retained responsiveness to TGF-β (Wu et al., 1992) (data not shown).
The PI3K-AKT pathway plays a central oncogenic role in inducing cell proliferation and tumor development, as evidenced by the occurrence of frequent mutations in the p110α catalytic subunit in cancers, and overexpression of the phosphorylated forms of AKT in various malignancies, including colon (Osaki et al., 2004; Samuels et al., 2004). In this report, we uncover yet another important aspect of this pathway by providing evidence for a non-coding RNA-mediated regulation of PI3K signaling. We propose that miR-126 modulates the activity of PI3K at the level of signal initiation by limiting p85β levels in normal colon epithelium. A loss of miR-126 during tumorigenesis would abolish this check point, contributing to amplification of PI3K signal. We suggest these findings may similarly apply to other cancer types that have also shown frequent loss of miR-126 (Tavazoie et al., 2008; Yanaihara et al., 2006).
Acknowledgments
Supported by: Public Health Service Grants CA116867 (to S.D.M.) and a gift from the National Colon Cancer Research Alliance (to S.D.M.). S.D.M. is an investigator of the Howard Hughes Medical Institute
We thank Jim Lutterbaugh and Min Yan for technical assistance with human tissues.
References
- Bandres E, Cubedo E, Agirre X, Malumbres R, Zarate R, Ramirez N, Abajo A, Navarro A, Moreno I, Monzo M, Garcia-Foncillas J. Identification by Real-time PCR of 13 mature microRNAs differentially expressed in colorectal cancer and non-tumoral tissues. Molecular cancer. 2006;5:29. doi: 10.1186/1476-4598-5-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen WD, Han ZJ, Skoletsky J, Olson J, Sah J, Myeroff L, Platzer P, Lu S, Dawson D, Willis J, Pretlow TP, Lutterbaugh J, Kasturi L, Willson JK, Rao JS, Shuber A, Markowitz SD. Detection in fecal DNA of colon cancer-specific methylation of the nonexpressed vimentin gene. Journal of the National Cancer Institute. 2005;97:1124–1132. doi: 10.1093/jnci/dji204. [DOI] [PubMed] [Google Scholar]
- Cummins JM, He Y, Leary RJ, Pagliarini R, Diaz LA, Jr, Sjoblom T, Barad O, Bentwich Z, Szafranska AE, Labourier E, Raymond CK, Roberts BS, Juhl H, Kinzler KW, Vogelstein B, Velculescu VE. The colorectal microRNAome. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:3687–3692. doi: 10.1073/pnas.0511155103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dwivedi Y, Rizavi HS, Teppen T, Zhang H, Mondal A, Roberts RC, Conley RR, Pandey GN. Lower phosphoinositide 3-kinase (PI 3-kinase) activity and differential expression levels of selective catalytic and regulatory PI 3-kinase subunit isoforms in prefrontal cortex and hippocampus of suicide subjects. Neuropsychopharmacology. 2007 doi: 10.1038/sj.npp.1301641. in press. [DOI] [PubMed] [Google Scholar]
- Gartel AL, Kandel ES. miRNAs: Little known mediators of oncogenesis. Seminars in cancer biology. 2008;18:103–110. doi: 10.1016/j.semcancer.2008.01.008. [DOI] [PubMed] [Google Scholar]
- Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic acids research. 2006;34:D140–144. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guda K, Natale L, Markowitz S. An improved method for staining cell colonies in clonogenic assays. Cytotechnology. 2007;54:85–88. doi: 10.1007/s10616-007-9083-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haneline LS, White H, Yang FC, Chen S, Orschell C, Kapur R, Ingram DA. Genetic reduction of class IA PI-3 kinase activity alters fetal hematopoiesis and competitive repopulating ability of hematopoietic stem cells in vivo. Blood. 2006;107:1375–1382. doi: 10.1182/blood-2005-05-1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo J, Sobkiw CL, Hirshman MF, Logsdon MN, Li TQ, Goodyear LJ, Cantley LC. Loss of class IA PI3K signaling in muscle leads to impaired muscle growth, insulin response, and hyperlipidemia. Cell metabolism. 2006;3:355–366. doi: 10.1016/j.cmet.2006.04.003. [DOI] [PubMed] [Google Scholar]
- Osaki M, Oshimura M, Ito H. PI3K-Akt pathway: its functions and alterations in human cancer. Apoptosis. 2004;9:667–676. doi: 10.1023/B:APPT.0000045801.15585.dd. [DOI] [PubMed] [Google Scholar]
- Platzer P, Upender MB, Wilson K, Willis J, Lutterbaugh J, Nosrati A, Willson JK, Mack D, Ried T, Markowitz S. Silence of chromosomal amplifications in colon cancer. Cancer research. 2002;62:1134–1138. [PubMed] [Google Scholar]
- Samuels Y, Wang Z, Bardelli A, Silliman N, Ptak J, Szabo S, Yan H, Gazdar A, Powell SM, Riggins GJ, Willson JK, Markowitz S, Kinzler KW, Vogelstein B, Velculescu VE. High frequency of mutations of the PIK3CA gene in human cancers. Science (New York, NY) 2004;304:554. doi: 10.1126/science.1096502. [DOI] [PubMed] [Google Scholar]
- Tavazoie SF, Alarcon C, Oskarsson T, Padua D, Wang Q, Bos PD, Gerald WL, Massague J. Endogenous human microRNAs that suppress breast cancer metastasis. Nature. 2008;451:147–152. doi: 10.1038/nature06487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Wetering M, Sancho E, Verweij C, de Lau W, Oving I, Hurlstone A, van der Horn K, Batlle E, Coudreuse D, Haramis AP, Tjon-Pon-Fong M, Moerer P, van den Born M, Soete G, Pals S, Eilers M, Medema R, Clevers H. The beta-catenin/TCF-4 complex imposes a crypt progenitor phenotype on colorectal cancer cells. Cell. 2002;111:241–250. doi: 10.1016/s0092-8674(02)01014-0. [DOI] [PubMed] [Google Scholar]
- Vanhaesebroeck B, Waterfield MD. Signaling by distinct classes of phosphoinositide 3-kinases. Experimental cell research. 1999;253:239–254. doi: 10.1006/excr.1999.4701. [DOI] [PubMed] [Google Scholar]
- Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, Prueitt RL, Yanaihara N, Lanza G, Scarpa A, Vecchione A, Negrini M, Harris CC, Croce CM. A microRNA expression signature of human solid tumors defines cancer gene targets. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willson JK, Bittner GN, Oberley TD, Meisner LF, Weese JL. Cell culture of human colon adenomas and carcinomas. Cancer research. 1987;47:2704–2713. [PubMed] [Google Scholar]
- Wu SP, Theodorescu D, Kerbel RS, Willson JK, Mulder KM, Humphrey LE, Brattain MG. TGF-beta 1 is an autocrine-negative growth regulator of human colon carcinoma FET cells in vivo as revealed by transfection of an antisense expression vector. The Journal of cell biology. 1992;116:187–196. doi: 10.1083/jcb.116.1.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanaihara N, Caplen N, Bowman E, Seike M, Kumamoto K, Yi M, Stephens RM, Okamoto A, Yokota J, Tanaka T, Calin GA, Liu CG, Croce CM, Harris CC. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer cell. 2006;9:189–198. doi: 10.1016/j.ccr.2006.01.025. [DOI] [PubMed] [Google Scholar]
- Yu J, Zhang Y, McIlroy J, Rordorf-Nikolic T, Orr GA, Backer JM. Regulation of the p85/p110 phosphatidylinositol 3′-kinase: stabilization and inhibition of the p110alpha catalytic subunit by the p85 regulatory subunit. Molecular and cellular biology. 1998;18:1379–1387. doi: 10.1128/mcb.18.3.1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L, Vogt PK. Helical domain and kinase domain mutations in p110alpha of phosphatidylinositol 3-kinase induce gain of function by different mechanisms. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:2652–2657. doi: 10.1073/pnas.0712169105. [DOI] [PMC free article] [PubMed] [Google Scholar]