Abstract
Exposure to altered microgravity during space travel induces changes in the brain and these are reflected in many of the physical behavior seen in the astronauts. The vulnerability of the brain to microgravity stress has been reviewed and reported. Identifying microgravity-induced changes in the brain proteome may aid in understanding the impact of the microgravity environment on brain function. In our previous study we have reported changes in specific proteins under simulated microgravity in the hippocampus using proteomics approach. In the present study the profiling of the hypothalamus region in the brain was studied as a step towards exploring the effect of microgravity in this region of the brain. Hypothalamus is the critical region in the brain that strictly controls the pituitary gland that in turn is responsible for the secretion of important hormones. Here we report a 2-dimensional gel electrophoretic analysis of the mouse hypothalamus in response to simulated microgravity. Lowered glutathione and differences in abundance expression of seven proteins were detected in the hypothalamus of mice exposed to microgravity. These changes included decreased superoxide dismutase-2 (SOD-2) and increased malate dehydrogenase and peroxiredoxin-6, reflecting reduction of the antioxidant system in the hypothalamus. Taken together the results reported here indicate that oxidative imbalance occurred in the hypothalamus in response to simulated microgravity.
Keywords: Brain, Hypothalamus, Microgravity
Introduction
Space flight has substantial effect in the brain; such changes are reflected in orthostatic intolerance and vestibular-related changes. The changes were seen in balance, eye movements and reflex control of cardiovascular, respiratory and gastrointestinal functions [1, 2]. These activities are directly or indirectly related to the changes that occur due to the activity of the brain under zero gravity in space. Previous studies showed that a population of neurons in the rat brain that discharge as a function of the rat’s head direction in a gravitationally horizontal plane and is dependent on an intact vestibular system [3]. The sensitivity of the mammalian central nervous system to gravitational influences involves both direct and indirect factors. It has been shown earlier that changes in the gravitational environment might represent a useful tool to investigate the neurobiological and behavioral responses to stressors and may provide insights into the mechanisms underlying development and plasticity of the brain, the heart, and the lung [1, 4–6]. The neurophysiological basis for adaptive phenomena in recurrent vestibular stimulation has been found to reside partly in the vestibular nuclei of the medulla, and not to require integrity of connections with higher vestibular centers, such as vestibular ganglion. A study on gene expression analysis under altered gravitational fields showed molecular changes in vestibular nuclei that controls somatic functions in rats during the NASA Neurolab Mission (STS-90) [7].
Effort towards understanding the gene and protein expression changes under microgravity has revealed the involvement of oxidative stress induced distorted signaling events [2]. Moreover microarray analysis showed changes in the pattern of global gene expression in skeletal muscle of rats that had been in space [8]. These studies have reported distorted expression of cytoskeleton genes and alterations in several oxidative stress-inducible genes [8]. Additional changes detected in skeletal muscle of rats exposed to space flight included down regulation of genes regulating cell proliferation, growth factor cascades, cell cycle, and signal transduction proteins.
Recent studies reported changes in the proteome of the hippocampus in mice subjected to simulated microgravity [9]. These changes included cytoskeleton proteins and proteins involved in metabolism. Induction of oxidative stress by microgravity has been detected both in simulated microgravity as well as in space [10, 11]. In addition oxidative stress has been argued as a potential cause of gene expression changes under microgravity. Oxidative stress induced NF κB in mouse brain has been reported in terrestrial model of microgravity [10]. The oxidative stress altered several signal transduction pathways and therefore was considered as one of the major factors that can modulate cellular signaling [12]. We previously demonstrated generation of oxidative stress in different regions of mouse brain and the responses were discrete among different regions [10]. It therefore argues that increase in oxidative stress could elicit different changes specific to the different regions of the brain and thus induce confined proteome alterations.
The pituitary gland may have regulatory functions, but it is under control of the hypothalamus. Some of the neurons within the hypothalamus neurosecretory neurons—secrete hormones that tightly control secretion of hormones from the anterior pituitary. The hypothalamus is therefore a critical region in the brain that controls the master gland, the pituitary; changes in the hypothalamus would therefore have physiological consequences. This study complements the proteomic analysis of effects of microgravity on the hippocampus.
Experimental Procedures
Simulated Microgravity Exposure to Mice
A total of 12 mice were divided equally into two groups one representing control and the other microgravity. The control mice were maintained in normal cages with free access to drinking water and diet. To simulate microgravity, mice were suspended by their tail in the center of the cage but allowed to touch the floor with their front paws, and left for 7 days with free access to drinking water and food [9, 10, 13]. At the end of the experiment, the mice were euthanized by carbon dioxide asphyxiation. Each brain was dissected out as described by our standardized procedures and, the hypothalamus was removed followed by snap frozen in liquid nitrogen [9, 10]. The tissue was at −80°C for further experiments. We have previously used this tail suspension model to simulate microgravity and have shown changes in the proteome of the mouse brain hippocampus [9]. Analysis of the same model also showed changes in cytokine levels and reactive oxygen species in the mouse brain and signaling events linked with ROS [10, 13]. Some of the changes detected using this model were similar to those detected in mice flown in space. The tail suspension method mimics microgravity by causing fluid to shift into the brain similar to what is observed in real space flight conditions [9, 10].
Glutathione (GSH) Assay
GSH was assayed in hypothalamus tissue extract using a kit from Sigma (CS0260). In brief the tissue was first deproteinized by homogenization in 5% 5-sulfosalicylic acid solution and centrifuged for 10 min at 10,000× g; GSH was estimated in the collected supernatant, by allowing GSH to reduce 5,5′-dithiobis (2-nitrobenzoic acid) (DTNB) to TNB; the yellow TNB formed was measured at 412 nm in a spectrophotometer at room temperature.
Two-dimensional Gel Electrophoresis
Two-dimensional gel electrophoresis was performed as detailed by Sarkar et al. [9]. In brief the hypothalamus was suspended in isoelectrofocusing buffer containing 20 mM Tris, 7 M urea, 2 M thiourea, 4% CHAPS, 10 mM 1,4-dithioerythritol, 0.5% ampholyte [3–10] and a mixture of protease inhibitors (1 tablet CompleteTM Roche Diagnostics) per 10 ml of suspension buffer). The suspension was sonicated for approximately 30 s and centrifuged at 100,000× g for 30 min. The protein content in the supernatant was determined using the Bradford reagent (BioRad).
The dissolved proteins were resolved by isoelectric focusing in 11 cm immobilized pH gradient (IPG) strips (BioRad), containing a 3–10 pH gradient as described elsewhere [9]. For the second dimension, separation was performed by electrophoresis at 150 V in 12.5% SDS polyacrylamide gels. The gels were fixed in 40% methanol, containing 10% acetic acid for 2 h followed by staining with Sypro Ruby (BioRad) overnight for 12 h. Excess dye was removed from the gels by destaining with 10% methanol and 7% acetic acid. Gels were scanned on Pro-Express equipped with CCD scanner (Perkin Elmer) and the images were captured as 16-bit tiff files for further analysis. We quantified the 2-DE protein patterns and mutually matched them by visual analysis because the protein patterns did not show any complex profile changes when control and microgravity exposed hippocampus was observed. The visually differentiated spots were manually selected and picked up using genomic solutions ProPic spot cutter.
MALDI-TOF-MS
MALDI TOF MS was performed as described by Sarkar et al. [9, 14] and Deshane et al. [9, 14]. In brief, gel spots of interest were excised and destained in 30% acetonitrile in 50 mM ammonium bicarbonate and dried in a Speedvac evaporator. The samples were subsequently resuspended in trypsin and allowed to digest overnight and MALDI TOF analysis was performed according to established procedures [9]. Peptide matching and protein searches were performed using MASCOT search engine at http://matrixscience.com [14, 15]. The non-redundant NCBI database was used in conjunction with MASCOT to determine the putative protein identifications. Search parameters allowed for oxidation of methionine, carbamidomethylation of cysteine, one missed trypsin cleavage, and 50 ppm mass tolerance before searching database. The MS/MS data were recorded using automated MS to MS/MS data dependent scanning, switching at a threshold of six counts. The peptide masses were compared with the theoretical peptide masses of all available proteins from all species. Monoisotopic masses were used and a mass tolerance of 0.0025% was allowed. Unmatched peptides or missed cleavage sites were not considered for protein identification.
Immunoblotting
Western blot analysis was performed as described [9]. In brief, hypothalamus tissue was homogenized in 2DE buffer and was adjusted with SDS-PAGE sample buffer so that the final sample for loading contained 50–75 μg, and boiled for 3 min at 95°C in a water bath. The proteins were resolved by electrophoresis in 10% SDS-PAGE and transferred to nitrocellulose membrane. The membranes were blocked in 5% skim milk followed by incubation with primary antibodies specific for superoxide dismutase-2 (Santa Cruz, SC-18503). The blots were then incubated with anti-rabbit horse radish-peroxidase conjugated antibody and bands were visualized by incubating the blots with ECL reagent (Amersham, NJ) followed by exposing to X-ray film.
Results
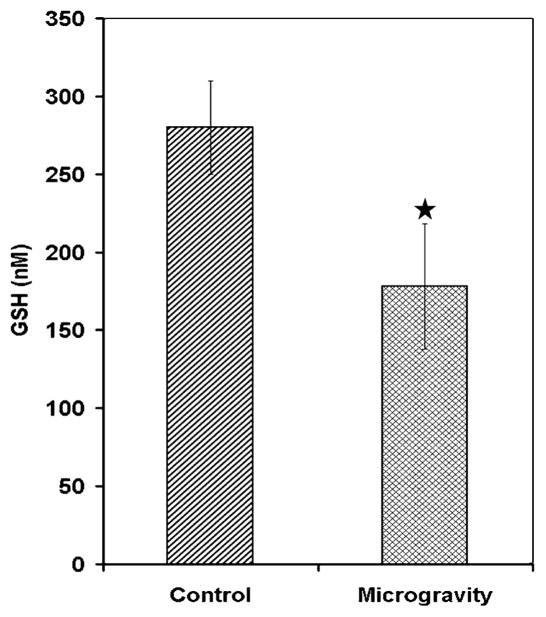
Microgravity is a hostile environment for living species the microgravity environment induces stress as evidenced by data obtained from animal studies performed in real space shuttle missions and also in simulated microgravity conditions [2, 8, 9]. The ground-based model for studying microgravity used in this study causes shifting of fluids in the brain, similar to that experienced by astronauts in space. This treatment is stressful, but has been extensively used to mimic microgravity on earth and therefore is an accepted model for microgravity research. In this model increases in oxidative stress and lipid per oxidation have been demonstrated in different brain regions [10]. Therefore to study localized oxidant status we estimated glutathione levels in the hypothalamus. The results indicated that glutathione was significantly decreased in the hypothalamus of mice exposed to simulated microgravity (Fig. 1). This is consistent with previous studies that reported similar decreases in GSH under simulated microgravity in the brain stem and frontal cortex and thus indicated induction of oxidative stress in other brain regions [10]. Decrease in GSH in the present study therefore also indicated indirectly generation of oxidative stress in the hypothalamus under simulated microgravity. The decreases in GSH can possibly lead to lipid per oxidation as a loss of antioxidants within the cells in hypothalamus. We therefore determined the level of lipid per oxidation in the hypothalamus but no significant difference was detected between the control and microgravity (Data not shown).
Fig. 1.
Microgravity decreases glutathione in hypothalamus in simulated microgravity exposed mice. Hypothalamus homogenate from control and simulated microgravity exposed mice for 7 days was deproteinized and centrifuged to remove the protein. The supernatant was treated with DTNB and the TNB formed was measured at 412 nm in a spectrophotometer. Values are mean ± SD of six mice and is a representative of three independent experiment and, star indicates significance level at P < 0.01
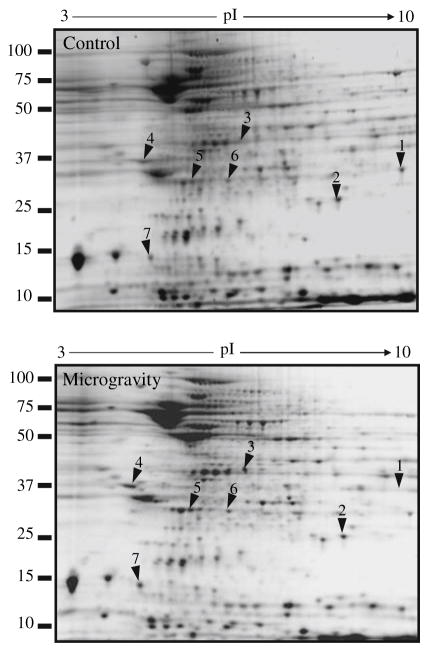
Loss of antioxidant defense in simulated microgravity is a reflection of oxidative stress [16]. To investigate whether simulated microgravity affects the protein expression profile in hypothalamus, we resolved the proteins in hypothalamus homogenate by 2DE. The gels which we used resolved proteins with molecular masses between 100 kD and 10 kD and pIs between 3 and 10. We acquired the 2DE protein patterns then assessed them visually because the protein patterns between control and microgravity exposed hippocampus were strikingly similar. Therefore spots that were visually different between the microgravity and the control were selected for further analysis. For comparing protein expression differences between the microgravity and the paired control samples we selected seven spots that were common across four sets of independent experiments where each sample was analyzed on triplicate gels (Table 1 and Fig. 2). All the spots indicated in Fig. 2 were significantly different between the control and microgravity exposed hypothalamus. These spots were subjected to peptide mass fingerprinting analysis by MALDI-TOF-MS as described in the materials and method section. The MOWSE scores above 60 are usually considered to be reliable peptide sequence. Therefore we have presented all the matched peptide with a MOWSE score of above 60. Among the seven proteins five were increased and two were decreased in simulated microgravity compared to control brain. Two proteins were significantly decreased in microgravity-glutathione S-transferase (Spot 1) and superoxide dismutase-2 (SOD-2) (Spot 2) (Fig. 2). The detected increase in two of the proteins is discussed in the context of microgravity environment in the hypothalamus. Malate dehydrogenase usually responds to oxidative stress as shown earlier in Alzheimer’s disease [17]. In the present study the increase in malate dehydrogenase (Spot 3) may be a counter active response to the increase in oxidative stress. The other stress-related protein that was more abundant in the hypothalamus of mice subjected to microgravity was ubiquitin carboxy-terminal hydrolase L1 (UCLH-1) (Spot 5). UCH-L1 is thought to regulate ubiquitin hydrolysis of larger ubiquitin conjugates after stress stimuli; ubiquitin in turn is thought to be a stress protein that plays an important role in protecting cells under stress conditions [18]. Peroxiredoxin (PRX) (Spot 6) is an antioxidant protein that plays a critical role in protecting against endogenously produced peroxides in both prokaryotes and eukaryotes. PRX was more abundant in hypothalamus from microgravity-treated brain as compared to control [19]. Therefore the changes in the protein indicated by this 2D gel analysis reflected increase in stress within the hypothalamus due to simulated microgravity. Stress increases in simulated microgravity and is evidenced by alterations in stress-related proteins [20]. Our results extend earlier findings by reflecting more specific proteins involved in this stress signaling.
Table 1.
Differentially expressed proteins in hypothalamus from control and microgravity simulated mice
| Spot # | Protein name | Accession # | Peptide sequence | MOWSe score | Predicted molecular weight (Dalton) |
|---|---|---|---|---|---|
| Control | |||||
| 1 | Glutathione S-transferase | giI6754084 | RPWFAGDK | 107 | 25953 |
| 2 | Superoxide dismutase-2, mitochondrial | giI31980762 | RDFGSFEK | 117 | 24588 |
| Microgravity | |||||
| 3 | Malate dehydrogenase 1, NAD (soluble) | giI37589957 | GEFITTVQQR | 72 | 36488 |
| 4 | Tyrosine 3-monooxygenase/tryptophan 5-monooxygenase alpha activation protein | giI 31981925 | NLLSVAYK | 92 | 29170 |
| 5 | Ubiquitin carboxy-terminal hydrolase L1 | giI 6755929 | EFTEREQGEVR | 69 | 24822 |
| 6 | Peroxiredoxin 6 | giI30267702 | LAPEFAKR | 86 | 24855 |
| 7 | Unnamed protein product | giI12852348 | EGVLYVGSK | 108 | 12425 |
Sequences obtained from MALDI-TOF-MS spectra are shown with their corresponding MOWSE score. Proteins identified in control and simulated microgravity of mice hippocampus are shown separately. The accession numbers indicated are from National Center for Biotechnology Information (NCBI)
Fig. 2.
Representative 2DE protein patterns from control and microgravity exposed mouse hypothalamus. Control mice were maintained under normal condition in cages while other group of mice was kept for microgravity exposure by suspending them by their tail for 7 days as described in materials and methods. Hypothalamus was dissected and homogenized in 2DE extraction buffer. Approximately 150 μg of protein was used for 2DE and stained with Sypro Ruby. Images were captured and differentially expressed proteins were identified by visual analysis. Differentially expressed proteins spots are marked on the images with arrows
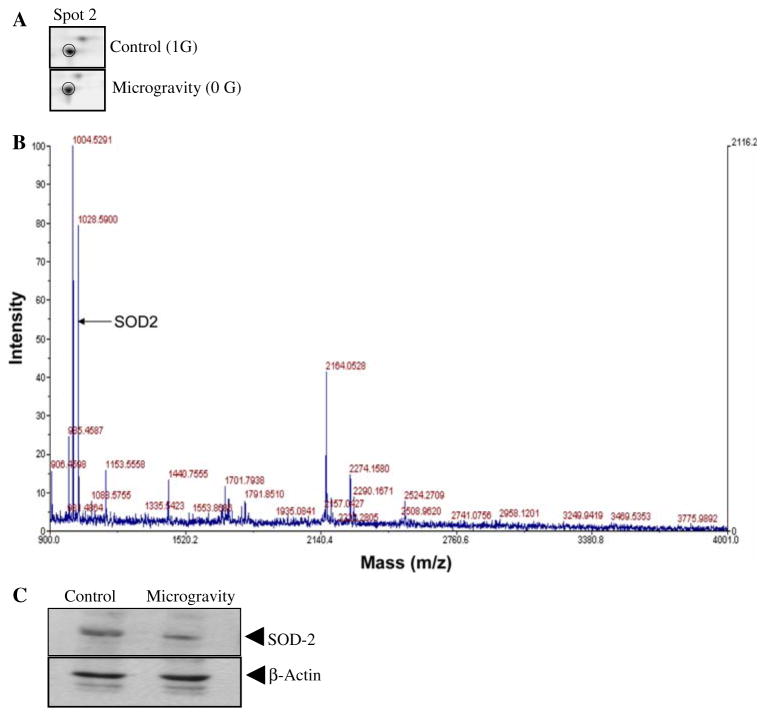
We validated the changes in amount of SOD-2 in the same hypothalamus tissue that was subjected to proteomics analysis (Fig. 3a). The enlarged illustration shows loss of SOD-2 in hypothalamus of mice subjected to simulated microgravity (Fig. 3a). The MALDI-TOF MS spectrum identifying SOD-2 is shown in Fig. 3b, also shown is the matching peptide determined by MASCOT. To validate the protein identity we performed immunoblot analysis with SOD-2-specific antibody (SC-18503; Santa Cruz Biotech-nology Inc. CA). This analysis showed loss of SOD-2 protein in hypothalamus exposed to simulated microgravity (Fig. 3c). The results confirmed the proteomics data that SOD-2 may be down regulated in modeled microgravity. Thus the changes in the proteome under simulated microgravity reflect loss of oxidative stress-related proteins; this is not surprising because analysis of this same model showed increase in lipid peroxidation has been demonstrated and this can be attributed to failure of antioxidant defense systems in the brain.
Fig. 3.
2DE profile of superoxide dismutase-2 in control and mice kept in simulated microgravity (a–c). (a) The panel shows enlarged regions of the 2DE protein profile representing changes in protein amount of Superoxide dismutase-2, (b) Superoxide dismutase-2 identification by peptide fingerprinting, (c) Western blot analysis of Superoxide dismutase-2 in control and in mice kept in simulated microgravity for 7 days. Beta-actin protein immunoblot was used for equal loading of samples
Discussion
Space travel produces profound changes of neuronal activity due to microgravity reflecting stress in the brain. Such stress results from cephalic fluid shift and increased capillary pressure in the head. These changes become manifested as oxidative stress in different regions of the brain. It was previously demonstrated that hippocampus is significantly affected by simulated microgravity and that this is correlated with differential expression of several proteins. Since brain regions are specific to different functions it is therefore expected that the proteomic response will vary across regions. The present study illustrates the power of global screening methodologies to evaluate the involvement of certain proteins in response to simulated microgravity in hypothalamus of mice brain. Our results clearly showed a significant decrease in GSH amount in hypothalamus and this indicates generation of oxidative stress in simulated microgravity. The reduced GSH and its redox forms, glutathione disulfide (GSSG) and glutathionylated proteins are biomarkers of oxidative stress [21]. Therefore the oxidative stress generated in the hypothalamus in microgravity supports earlier findings showing generation of oxidative stress under similar conditions. The increase in oxidative stress at its downstream effect mainly culminates in lipid per oxidation further deteriorating to tissue degradation. However, in the present study simulated microgravity induced no significant change in lipid per oxidation level in the hypothalamus. It has been argued that increase in lipid per oxidation requires increased synthesis of proteins and lipids. However, it is reported that protein synthesis decreases in simulated microgravity and this could possibly argue for no change in lipid per oxidative products in the hypothalamus [22].
To get additional insight into the changes incurred due to microgravity we resolved the proteome of the hypothalamus by 2D gels. We used a pI range of 3–10 to resolve the proteins in first dimension by isoelecrtric focusing followed by separation of the focussed protein in 12.5% homogenous gels. There were seven proteins that were significantly different in the hypothalamus between the control and microgravity treated animals. Five of these proteins were higher in the brains of the animals subjected to microgravity; this might be an adaptive response to the microgravity stress. Preliminary microarray analysis of 4,673 human genes revealed a 2-fold increase in the expression of 95 genes in HepG2 cells exposed to microgravity, whereas 10 genes were down regulated 2-fold in these same cells [23]. Consistent with these observations the present proteomics analysis of the hypothalamus showed more increase in protein expression compared to the loss of total protein. This observation is in contrast to the proteomic analysis of the hippocampus under similar conditions, which showed more loss of proteins as compared to control [9]. The observed difference can be attributed to several factors that include the response of the specific region of the brain, the antioxidant defense capacity and the amount of stress induced in hypothalamus by microgravity. Similar finding is reported for oxidative stress and lipid per oxidation that varied across different region of the brain in simulated microgravity [10].
The decrease in GST and SOD-2 implicates partial loss of antioxidant defense, since both GST and SOD-2 are involved in oxidative defense mechanisms. SOD-2 is an inducible protein that is thought to be important in protecting the brain from oxidative damage; its expression has been demonstrated to be critical in modulating the response of neurons to various kinds of stress [24]. The decrease in SOD-2 and GSH in microgravity was consistent with the idea that oxidative stress might be increased in hypothalamus in microgravity. In addition the significant decrease in GST also supports occurrence of oxidative assault in the hypothalamus under simulated microgravity. Taken together these observations are in agreement with previous observation that has shown increase in oxidative stress in microgravity both in terrestrial model and in actual space flown rats. MDHs catalyzes the interconversion of oxaloacetate and malate linked to the oxidation/reduction of dinucleotide coenzymes. Oxaloacetate plays a crucial role in many metabolic pathways including operation of the tricarboxylic acid cycle, glyoxylate bypass, amino acid synthesis, gluconeogenesis, maintenance of oxidation/reduction balance, and facilitation of the exchange of metabolites between cytoplasm and sub cellular organelles [25]. Since it is known that in microgravity the redox state is altered and to counteract such changes the MDH may have increased in the hypothalamus [10, 11]. The other protein that was significantly up-regulated in hypothalamus exposed to microgravity was peroxiredoxins. Peroxiredoxins (PRXs) are a recently characterized family of antioxidant enzymes that are reported to control cytokine-induced peroxide levels mediating signal transduction in mammalian cells [26]. The increase in the PRXs therefore once again argues for a counter active defense against oxidative stress. Brain is a highly protected organ and any potentially damaging alteration in the cellular signaling will be counteracted. However, taken together the present observations it indicates that microgravity may alter the proteome in the hypothalamus. The changes indicate the vulnerability of the hypothalamus to the stress generated by the microgravity. The changes due to microgravity in hypothalamus observed in the terrestrial model do provide a useful model for studying the effect of microgravity environment. Now that some of these molecular effects have been identified, they define a model that can be used to design counter-strategies against the effects of microgravity.
Acknowledgments
This work was supported by NASA funding NCC 9-165; NIH-NCMHHD 1P20MD001822; NASA NSTI NNA06CB14H.
References
- 1.Ishihara A, Ohira Y, Roy RR, Nagaoka S, Sekiguchi C, Hinds WE, Edgerton VR. Effects of 14 days of spaceflight and nine days of recovery on cell body size and succinate dehydrogenase activity of rat dorsal root ganglion. Neuroscience. 1997;81:275–279. doi: 10.1016/s0306-4522(97)00097-3. [DOI] [PubMed] [Google Scholar]
- 2.Nichols HL, Zhang N, Wen X. Proteomics and genomics of microgravity. Physiol Genom. 2006;26:163–171. doi: 10.1152/physiolgenomics.00323.2005. [DOI] [PubMed] [Google Scholar]
- 3.Taube JS, Stackman RW, Calton JL, Oman CM. Rat head direction cell responses in zero-gravity parabolic flight. J Neurophysiol. 2004;92:2887–2997. doi: 10.1152/jn.00887.2003. [DOI] [PubMed] [Google Scholar]
- 4.Harm DL, Jennings RT, Meck JV, Powell MR, Putcha L, Sams CP, Schneider SM, Shackelford LC, Smith SM, Whitson PA. Invited review: gender issues related to spaceflight: a NASA perspective. J Appl Physiol. 2001;91:2374–2383. doi: 10.1152/jappl.2001.91.5.2374. [DOI] [PubMed] [Google Scholar]
- 5.Feuilloley M, Yon L, Kawamura K, Kikuyama S, Gutkowska J, Vaudry H. Immunocytochemical localization of atrial natriuretic factor (ANF)-like peptides in the brain and heart of the treefrog Hyla japonica: effect of weightlessness on the distribution of immunoreactive neurons and cardiocytes. J Comp Neurol. 1993;330:32–47. doi: 10.1002/cne.903300104. [DOI] [PubMed] [Google Scholar]
- 6.Nikawa T, Ishidoh K, Hirasaka K, Ishihara I, Ikemoto ME, Kominami KE, Nonaka I, Ogawa T, Adams GR, Baldwin KM, Yasui N, Kishi K, Takeda S. Skeletal muscle gene expression in space-flown rats. FASEB J. 2004;18:522–524. doi: 10.1096/fj.03-0419fje. [DOI] [PubMed] [Google Scholar]
- 7.Pompeiano O, d’Ascanio P, Centini C, Pompeiano M, Cirelli CG, Tononi G. Immediate early gene expression in the vestibular nuclei and related vegetative areas in rats during space flight. Acta Otolaryngol Suppl. 2001;545:120–126. doi: 10.1080/000164801750388289. [DOI] [PubMed] [Google Scholar]
- 8.Taylor WE, Bhasin S, Lalani R, Datta A, Gonzalez-Cadavid NF. Alteration of gene expression profiles in skeletal muscle of rats exposed to microgravity during a spaceflight. J Gravit Physiol. 2002;9:61–70. [PubMed] [Google Scholar]
- 9.Sarkar P, Sarkar S, Ramesh V, Hayes BE, Thomas RL, Wilson BL, Kim H, Barnes S, Kulkarni A, Pellis N, Ramesh GT. Proteomic analysis of mice hippocampus in simulated microgravity environment. J Proteome Res. 2006;5:548–553. doi: 10.1021/pr050274r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wise KC, Manna SK, Yamauchi K, Ramesh V, Wilson BL, Thomas RL, Sarkar S, Kulkarni A, Pellis NR, Ramesh GT. Activation of nuclear transcription factor-kappaB in mouse brain induced by a simulated microgravity environment. In Vitro Cell Dev Biol Anim. 2005;41:118–123. doi: 10.1290/0501006.1. [DOI] [PubMed] [Google Scholar]
- 11.Stein TP. Space flight and oxidative stress. Nutrition. 2002;18:867–871. doi: 10.1016/s0899-9007(02)00938-3. [DOI] [PubMed] [Google Scholar]
- 12.Won SJ, Kim DY, Gwag BJ. Cellular and molecular pathways of ischemic neuronal death. J Biochem Mol Biol. 2002;35:67–86. doi: 10.5483/bmbrep.2002.35.1.067. [DOI] [PubMed] [Google Scholar]
- 13.Felix K, Wise KC, Manna SK, Yamauchi K, Wilson BL, Thomas RL, Kulkarni AD, Pellis NR, Ramesh GT. Altered cytokine expression in tissues of mice subjected to simulated microgravity. Mol Cell Biochem. 2004;266:79–85. doi: 10.1023/b:mcbi.0000049136.55611.dd. [DOI] [PubMed] [Google Scholar]
- 14.Deshane J, Chaves L, Sarikonda KV, Isbell S, Wilson L, Kirk M, Grubbs C, Barnes S, Meleth S, Kim H. Proteomics analysis of rat brain protein modulations by grape seed extract. J Agric Food Chem. 2004;29:7872–7883. doi: 10.1021/jf040407d. [DOI] [PubMed] [Google Scholar]
- 15.Berndt P, Hobohm U, Langen H. Reliable automatic protein identification from matrix-assisted laser desorption/ionization mass spectrometric peptide fingerprints. Electrophoresis. 1999;20:3521–3526. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3521::AID-ELPS3521>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 16.Butterfield DA, Poon HF, St Clair D, Keller JN, Pierce WM, Klein JB, Markesbery WR. Redox proteomics identification of oxidatively modified hippocampal proteins in mild cognitive impairment: insights into the development of Alzheimer’s disease. Neurobiol Dis. 2006;22:223–232. doi: 10.1016/j.nbd.2005.11.002. [DOI] [PubMed] [Google Scholar]
- 17.Korolainen MA, Goldsteins G, Nyman TA, Alafuzoff I, Koistinaho J, Pirttila T. Oxidative modification of proteins in the frontal cortex of Alzheimer’s disease brain. Neurobiol Aging. 2006;27:42–53. doi: 10.1016/j.neurobiolaging.2004.11.010. [DOI] [PubMed] [Google Scholar]
- 18.Harada T, Harada C, Wang YL, Osaka H, Amanai K, Tanaka K, Takizawa S, Setsuie R, Sakurai M, Sato Y, Noda M, Wada K. Role of ubiquitin carboxy terminal hydrolase-L1 in neural cell apoptosis induced by ischemic retinal injury in vivo. Am J Pathol. 2004;164:59–64. doi: 10.1016/S0002-9440(10)63096-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Han JY, Song KD, Shin JH, Han BK, Park TS, Park HJ, Kim JK, Lillehoj HS, Lim JM, Kim H. Identification and characterization of the peroxiredoxin gene family in chickens. Poult Sci. 2005;84:1432–1438. doi: 10.1093/ps/84.9.1432. [DOI] [PubMed] [Google Scholar]
- 20.Sarkar S, Wise KC, Manna SK, Ramesh V, Yamauchi K, Thomas RL, Wilson BL, Kulkarni AD, Pellis NR, Ramesh GT. Activation of activator protein-1 in mouse brain regions exposed to simulated microgravity. In Vitro Cell Dev Biol Anim. 2006;42:96–99. doi: 10.1290/0512083.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rossi R, Dalle-Donne I, Milzani A, Giustarini D. Oxidized forms of glutathione in peripheral blood as biomarkers of oxidative stress. Clin Chem. 2006;52:1406–1414. doi: 10.1373/clinchem.2006.067793. [DOI] [PubMed] [Google Scholar]
- 22.Maddaiah VT. Glutathione correlates with lipid peroxidation in liver mitochondria of triiodothyronine-injected hypophysectomized rats. FASEB J. 1990;4:1513–1518. doi: 10.1096/fasebj.4.5.2307329. [DOI] [PubMed] [Google Scholar]
- 23.Khaoustov VI, Risin D, Pellis NR, Yoffe B. Microarray analysis of genes differentially expressed in HepG2 cells cultured in simulated microgravity: preliminary report. In Vitro Cell Dev Biol Anim. 2001;37:84–88. doi: 10.1290/1071-2690(2001)037<0084:MAOGDE>2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 24.Cassarino DS, Bennett JP., Jr An evaluation of the role of mitochondria in neurodegenerative diseases: mitochondrial mutations and oxidative pathology, protective nuclear responses, and cell death in neurodegeneration. Brain Res Brain Res Rev. 1999;29:1–25. doi: 10.1016/s0165-0173(98)00046-0. [DOI] [PubMed] [Google Scholar]
- 25.Goward CR, Nicholls DJ. Malate dehydrogenase: a model for structure, evolution, and catalysis. Protein Sci. 1994;3:1883–1888. doi: 10.1002/pro.5560031027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wood ZA, Schroder E, Robin Harris J, Poole LB. Structure, mechanism and regulation of peroxiredoxins. Trends Biochem Sci. 2003;28:32–40. doi: 10.1016/s0968-0004(02)00003-8. [DOI] [PubMed] [Google Scholar]