Abstract
The control of translation and mRNA degradation plays a key role in the regulation of eukaryotic gene expression. In the cytosol, mRNAs engaged in translation are distributed throughout the cytosol, while translationally inactive mRNAs can accumulate in P bodies, in complex with mRNA degradation and translation repression machinery, or in stress granules, which appear to be mRNAs stalled in translation initiation. Here we discuss how these different granules suggest a dynamic model for the metabolism of cytoplasmic mRNAs wherein they cycle between different mRNP states with different functional properties and subcellular locations.
Keywords: mRNA decay, Translational repression, P bodies and Stress granules
Introduction
The translation and decay of mRNAs play key roles in the control of eukaryotic gene expression. In eukaryotes, degradation of mRNAs is usually initiated by shortening of the 3′ poly A tail (deadenylation), which generally, at least in yeast, leads to the removal of 5′ cap structure by a Dcp1/Dcp2 decapping enzyme followed by 5′- 3′ exonucleolytic digestion by Xrn1 (Reviewed in[1–3]). Alternately, deadenylated mRNA can also be degraded from 3′ to 5′ by the exosome. Eukaryotic cells contain multiple mRNA deadenylases with the major one being the Ccr4/Pop2/Not complex.
The processes of translation and mRNA degradation are often in competition. This was first suggested by the observation that removal of the cap structure, which stimulates translation initiation, was a key step in mRNA decapping [4]. In addition, at least in yeast, inhibition of translation initiation by strong secondary structures in the 5′UTR, a poor AUG context or mutations in initiation factors increases the rates of deadenylation and decapping[5]. Moreover, the cap binding protein eIF4E, known to stimulate translation initiation, inhibits the decapping enzyme, Dcp1/Dcp2, both in vivo and in vitro [6,7]. Finally, many mRNA specific regulatory factors, (e.g. miRNAs or PUF proteins), both repress translation and accelerate deadenylation and decapping [3,8–10].These results suggest that prior to decapping translation initiation factors are exchanged for decapping factors, thereby assembling a distinct “decapping” mRNP no longer capable of translation initiation [11].This is also supported by the observation that some decapping activators also function as translational repressors[12,13]. Thus, mRNA decapping appears to occur in two steps, first inhibition of translation initiation and exchange of translation factors for the general repression/degradation machinery, and a second step whereby the mRNA is actually degraded.
The rate of decapping is enhanced by decapping activators including Dhh1, Pat1, the Lsm1-7 complex, and the Edc1-3 proteins, some of which can either repress translation initiation and/or directly enhance the rate of catalysis by Dcp1/Dcp2 [12–15]. Furthermore, the rate of decapping of specific mRNAs can be stimulated by mRNA specific binding proteins or miRNAs, that then recruit components of the general mRNA decapping/translation repression machinery (reviewed in [3,16]).
Non-translating mRNAs can assemble into P bodies
The competition between mRNPs engaged in translation and those assembled with the decapping machinery correlates with the location of the mRNPs. Specifically, while translating mRNAs are distributed throughout the cytosol, mRNAs complexed with the decapping machinery can concentrate in RNA-protein granules referred to as P bodies (reviewed in [3,16,17]). P bodies are dynamic complexes whose assembly is dependent on, and proportional to, the pool of untranslating mRNA [18–20]. However, not every translationally inactive mRNA accumulates in P bodies. For example, mRNAs whose translation is inhibited by a 5′ stem-loop, which blocks ribosome loading, or by puromycin, which triggers release of elongating ribosomes, do not accumulate in P bodies, unless they contain elements that recruit P body components[21*,22*]. Although their complete composition is not yet known, P bodies include the decapping enzymes Dcp1/Dcp2, the activators of decapping Dhh1/RCK/p54, Pat1, Scd6/RAP55, Edc3, the Lsm1–7 complex, and the exonuclease, Xrn1 (reviewed in [16,17,23]). In addition, P bodies can also contain mRNAs and proteins involved in Nonsense Mediated Decay, as well as components of the miRNA repression machinery [3,9].
Studies in yeast suggest that P bodies assemble by formation of a translationally repressed mRNP, with some hierarchal interactions, which are then aggregated into larger structures by specific protein-protein interaction domains. For example, accumulation of Lsm1–7 or Dcp1 in yeast P bodies is dependent on the presence of Pat1 or Dcp2, respectively [24]. In yeast, aggregation of mRNPs into P bodies has been shown to be primarily dependent on a self- interaction domain (Yjef-N) in the Edc3 protein and glutamine/asparagine (Q/N) rich prion-like domain in the Lsm4 C-terminus [25**– 27*]. Because the YjeF domain of Edc3 is conserved, it is likely that Edc3 will contribute to assembly of metazoan P-bodies. However, since depletion of Edc3 does not block P body assembly in S2 cells [21*], one anticipates that Q/N domains, and possibly other mechanisms, will also play a role in metazoan P body assembly. Interestingly, multiple proteins in metazoan P bodies contain Q/N rich domains including GW182, which functions in miRNA mediated repression, and Ge-1/Hedls, a component of the metazoan decapping enzyme [25**]. Moreover, depletion of either of these proteins lead to decreased P bodies in Human and Drosophila cells [21*, 28–30].
What are Stress Granules?
Non-translating mRNAs can also form a second cytoplasmic RNP granule referred to as stress granules. Stress granules are dynamic and appear to be composed of mRNAs that are stalled in the process of translation initiation. Stress granules contain non-translating mRNAs complexed with a subset of translation initiation factors (eIF4E, eIF4G, eIF4A, eIF3, and eIF2), the 40S ribosomal subunit and the poly (A) binding protein (Pab1) [31].These have been primarily studied in mammalian cells and are usually seen under stress where translation initiation is inhibited. An unresolved issue is whether the mRNAs in stress granules are primarily exiting translation, entering translation, or both. The complete composition of stress granules is unknown but they include RNA binding proteins such as TIA-R, TIA-1 and G3BP. TIA-1 and G3BP are known to contain self aggregation domains, which appear to play an important role in the aggregation of stress granules [32, 33].
A cytoplasmic granule of similar composition and assembly mechanism has been reported in yeast cells under glucose deprivation conditions and is referred to as either EGP bodies or yeast stress granules [34–37]. The composition of yeast stress granules include the orthologues of the mammalian stress granule components TIA-R, TIA-1 and Ataxin-2 (yeast proteins Pub1, Ngr1 and Pbp1 respectively.) They also contain Pab1 and translation initiation factors such as eIF4E and eIF4G. Similar to mammalian stress granules, their assembly is blocked by cycloheximide and promoted by eIF2α phosphorylation. Pub1 and Pbp1, whose mammalian orthologues affect stress granule assembly, are key in the assembly of these granules as well. Unlike the mammalian stress granules, these yeast granules do not contain Prt1 (an eIF3 subunit) or eIF2, and are not as readily induced by a variety of stresses [35**], which might be a consequence of what step in translation initiation is limiting under certain conditions (see below).
Dynamics of mRNPs in the cytoplasm
Several observations argue that cytoplasmic mRNAs cycle between polysomes, P bodies and stress granules. First, inhibition of translation initiation by drugs, stresses, or mutations leads to loss of mRNAs from polysomes and a corresponding increase of mRNAs in P bodies and stress granules [18,23,38]. Second, trapping mRNAs in polysome by blocking translation elongation decreases P bodies and stress granules even during continued stress in mammalian cells, which suggests that mRNAs in these compartments are in dynamic equilibrium with polysomes [18,39-41]. This is consistent with the dynamic nature of P bodies and stress granules based on fluorescence recovery after photobleaching (FRAP) studies [38,42]. Third, P bodies and stress granules physically interact; often docking together in mammalian cells during stress [38,43,44] or partially overlapping in yeast [34–36*]. Fourth, mRNAs within P bodies in both yeast and mammalian cells can be shown to return to translation [45,46].
An unresolved issue is the mechanisms and directionality of mRNA movement between P-bodies and stress granules. In mammalian cells, stress granules can assemble in a manner spatially distinct from P-bodies, and during heat shock, stress granules form prior to P-bodies [38]. These observations suggest that mRNAs exiting translation may first accumulate in stress granules and then later be transferred to P-bodies [31, 38]. In contrast, during glucose deprivation in yeast, stress granules form after P-bodies, primarily assemble on pre-exisitng P-bodies, and are dependent on existing P-bodies for their efficient assembly [35**]. This suggests that yeast mRNAs exiting translation first form a P body mRNP, and then mRNAs which are targeted for re-entry into translation, remodel their mRNP to load translation initiation factors, thereby forming the type of mRNP seen to accumulate in stress granules. One realistic possibility is that mRNAs may exchange in a bidirectional manner between stress granules and P-bodies and the specific mRNA, cell type, or condition, may affect the predominant flow of bulk mRNA.
The movement of mRNAs between polysomes, stress granules and P bodies implies transitions between different mRNP states through specific re-arrangements and exchanges of proteins on individual mRNAs. This may be facilitated by RNA helicases as well as being influenced by post-translational modifications of the key RNA binding proteins (reviewed in [47]). Although, P bodies and stress granules represent microscopically visible aggregates of different mRNPs, the simplest model is that these transitions can occur independent of the larger aggregates too. An important area of future work will be to determine how mRNPs in either stress granules or P-bodies are remodeled to affect their fate and how that impinges on the control of gene expression.
A Working Model: The mRNA Cycle
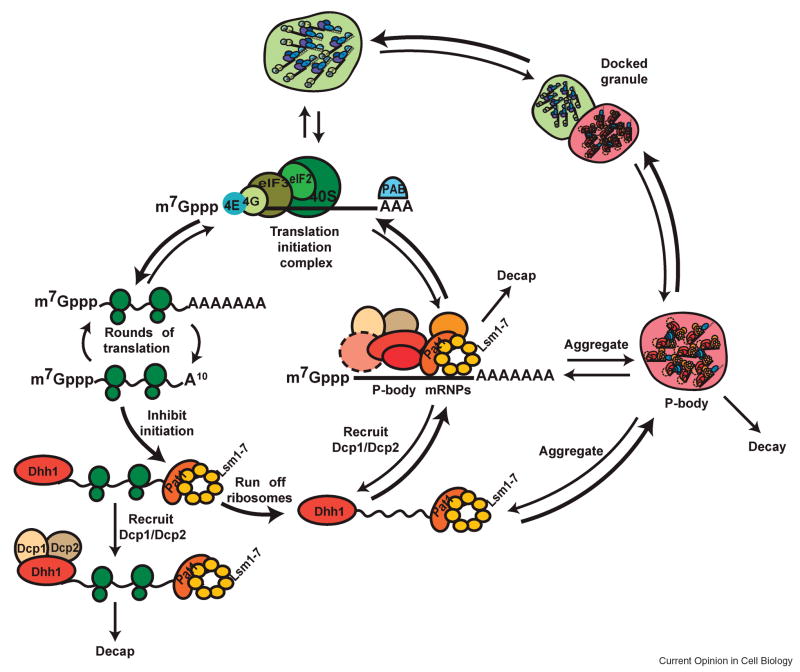
The analyses of P bodies and stress granules suggest a working model for the metabolism of cytoplasmic mRNA termed the mRNA cycle (Figure 1). In this model, mRNAs present in polysomes undergo repeated rounds of translation initiation, elongation and termination to produce polypeptides. In response to defects in translation initiation and/or termination, or through specific recruitment, mRNAs found in polysomes interact with proteins that repress translation initiation such as Dhh1/Rck and Pat1. At this stage, we envision a kinetic competition between run-off of the elongating ribosomes, recruitment of the remainder of the decapping machinery (including the decapping enzyme), decapping followed by transcript degradation, and aggregation of an individual mRNP into a P body.
Figure 1. The mRNA cycle.
mRNA can exist in at least 3 states in the cytoplasm- Polysomes, P bodies and Stress granules. Recruitment of translational repressors such as Dhh1 and Pat1 facilitate the movement of mRNA into a non-translating state. Kinetic competition ensues between ribosome run-off, recruitment of decapping machinery, decapping followed by transcript degradation and aggregation into a P body. mRNAs complexed with the decapping machinery can be degraded, aggregate into a P body, or enter back into translation. Limiting conditions of translation initiation may be overcome by aggregation into Stress granules where local concentration of translation apparatus is high.
The balance between these different rates may be influenced by elongating ribosomes, based on three observations suggesting that loss of elongating ribosomes from the mRNA enhances, but is not required for decapping. First, when elongating ribosomes are trapped on the mRNA by cycloheximide, decapping is slowed down significantly, but can still occur [48, 49]. Second, the rate of decapping in some cases is proportional to the length of the coding region, which might be explained by the continued presence of elongating ribosomes inhibiting decapping [50]. One possibility is that the decapping enzyme can associate with mRNAs once components of the decapping machinery have interacted with the transcript, but that the rate of decapping enzyme delivery is enhanced if the translationally repressed mRNA enters a P body, where local concentrations of the decapping enzyme is high. This possibility is suggested by the observation that Dcp2 addition to decapping mRNPs is a late step that occurs after Dhh1 and Pat1 interaction with the mRNA [12,24]. Further support comes from the fact that Dcp2 is much less dynamic in P bodies than other factors, suggesting it might exchange from one mRNA to another within these structures [51*].
mRNAs complexed with the decapping machinery can be degraded, aggregate into a P body, or undergo an mRNP rearrangement wherein the degradation machinery is exchanged for translation initiation factors. Such mRNAs could then go on to initiate translation and enter polysomes. However, if steps in initiation are limiting, these mRNPs could accumulate in a stress granule state before eventually entering polysomes. Further, the state at which translation initiation is limiting might define the composition of the stress granule and therefore the composition of the stress granule might vary in different organisms or in response to different stresses. One anticipates that specific mRNAs may preferentially accumulate in stress granules, P bodies, or polysomes depending on their relative rates of transitions between these different biochemical states.
Does the Aggregation of mRNPs into Granules have a Function?
An unresolved issue for both stress granules and P bodies is why the individual mRNPs aggregate into larger structures. To date, aggregation of mRNPs into P bodies has been shown to not be required for mRNA decapping in yeast [25**], translation repression during stress in both yeast and mammals [25,35,52,53**], and mRNA stability during stress, at least in yeast[35*]. In addition, in metazoans, depletion of microscopically visible P bodies do not seem to affect miRNA mediated repression, decay of messages containing ARE elements, or decay of transcripts subject to NMD [21*, 54, 55]. However, since aggregation into RNP granules is a conserved feature of eukaryotic cells it is anticipated to have some role. One possibility is that granules do have functional consequences for the control of translation and/or degradation but these functions are either limited to a subset of mRNAs or are carried out by granules below the detection limit of the light microscope.
More generally, the formation of RNP granules such as stress granules and P bodies are expected to have specific consequences both by increasing the local concentration of factors within granules, and by depleting them from the bulk cytosol. For comparison, Cajal bodies improve the assembly of spliceosomal small nuclear ribonucleoprotein particles (snRNPs) by increasing the local concentrations of U4/U6 [56]. By analogy, the concentration of Dcp2 in P bodies might facilitate its interaction with mRNAs when Dcp2 is limiting, or the concentration of translation initiation factors in stress granules might drive the formation of productive translation complexes. In addition, an important role of granules may be to remove factors from the cytosol. For example, a recent study argues that formation of stress granules sequester RACK1 away from MAP kinases, thereby limiting signal transduction and apoptosis [57**]. Moreover, the aggregation of mRNPs into stress granules and P bodies may provide a buffering system for maintaining a proper ratio of translation capacity to the pool of mRNAs that are translating (discussed in [5]). An excessive amount of mRNAs within the translating pool may compete for limiting translation factors and thereby prevent effective translation of many mRNAs.
Future Directions
Although considerable advances have been made in the understanding of the mechanisms of mRNA decapping and the subcellular distribution of different mRNPs, there are several outstanding questions that need to be addressed. One key issue will be to understand the molecular functions of decapping activators and how they affect translation mechanisms as well as the recruitment and stimulation of the decapping enzyme. A second important question will be to understand the significance of the aggregation of mRNPs into P bodies and stress granules, which is likely to contribute to our growing understanding of the importance of subcellular organization. Finally, it will be critical to understand the mechanisms and rates of the transitions of mRNPs between polysomes, P bodies, and stress granules. Here it will be critical to understand the frequency of these exchanges, the molecular mechanisms that move mRNAs from one state to another, and how they differ on individual mRNAs, thereby impacting the control of gene expression.
Acknowledgments
We thank all members of the Parker laboratory, especially A. Webb, for helpful discussions and technical support in the preparation of the manuscript. This work was supported by funds from the National Institutes of Health (grant R37 GM45443) and the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
*Special interest
**Outstanding interest
- 1.Parker R, Song H. The enzymes and control of eukaryotic mRNA turnover. NatStructMol Biol. 2004;11:121–127. doi: 10.1038/nsmb724. [DOI] [PubMed] [Google Scholar]
- 2.Garneau NL, Wilusz J, Wilusz CJ. The highways and byways of mRNA decay. NatRevMol Cell Biol. 2007;8:113–126. doi: 10.1038/nrm2104. [DOI] [PubMed] [Google Scholar]
- 3.Franks TM, Lykke-Andersen J. The control of mRNA decapping and P-body formation. Mol Cell. 2008;32:605–615. doi: 10.1016/j.molcel.2008.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Muhlrad D, Decker CJ, Parker R. Deadenylation of the unstable mRNA encoded by the yeast MFA2 gene leads to decapping followed by 5'-->3' digestion of the transcript. Genes Dev. 1994;8:855–866. doi: 10.1101/gad.8.7.855. [DOI] [PubMed] [Google Scholar]
- 5.Coller J, Parker R. Eukaryotic mRNA decapping. AnnuRev Biochem. 2004;73:861–890. doi: 10.1146/annurev.biochem.73.011303.074032. [DOI] [PubMed] [Google Scholar]
- 6.Schwartz DC, Parker R. Mutations in translation initiation factors lead to increased rates of deadenylation and decapping of mRNAs in Saccharomyces cerevisiae. MolCell Biol. 1999;19:5247–5256. doi: 10.1128/mcb.19.8.5247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schwartz DC, Parker R. mRNA decapping in yeast requires dissociation of the cap binding protein, eukaryotic translation initiation factor 4E. MolCell Biol. 2000;20:7933–7942. doi: 10.1128/mcb.20.21.7933-7942.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wickens M, Bernstein DS, Kimble J, Parker R. A PUF family portrait: 3'UTR regulation as a way of life. Trends Genet. 2002;18:150–157. doi: 10.1016/s0168-9525(01)02616-6. [DOI] [PubMed] [Google Scholar]
- 9.Shyu AB, Wilkinson MF, van Hoof A. Messenger RNA regulation: to translate or to degrade. EMBO J. 2008;27:471–481. doi: 10.1038/sj.emboj.7601977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Behm-Ansmant I, Rehwinkel J, Doerks T, Stark A, Bork P, Izaurralde E. mRNA degradation by miRNAs and GW182 requires both CCR4:NOT deadenylase and DCP1:DCP2 decapping complexes. Genes Dev. 2006;20:1885–1898. doi: 10.1101/gad.1424106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tharun S, Parker R. Targeting an mRNA for decapping: displacement of translation factors and association of the Lsm1p-7p complex on deadenylated yeast mRNAs. Mol Cell. 2001;8:1075–1083. doi: 10.1016/s1097-2765(01)00395-1. [DOI] [PubMed] [Google Scholar]
- 12.Coller J, Parker R. General translational repression by activators of mRNA decapping. Cell. 2005;122:875–886. doi: 10.1016/j.cell.2005.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pilkington GR, Parker R. Pat1 contains distinct functional domains that promote P-body assembly and activation of decapping. MolCell Biol. 2008;28:1298–1312. doi: 10.1128/MCB.00936-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Steiger M, Carr-Schmid A, Schwartz DC, Kiledjian M, Parker R. Analysis of recombinant yeast decapping enzyme. RNA. 2003;9:231–238. doi: 10.1261/rna.2151403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schwartz D, Decker CJ, Parker R. The enhancer of decapping proteins, Edc1p and Edc2p, bind RNA and stimulate the activity of the decapping enzyme. RNA. 2003;9:239–251. doi: 10.1261/rna.2171203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Parker R, Sheth U. P bodies and the control of mRNA translation and degradation. Mol Cell. 2007;25:635–646. doi: 10.1016/j.molcel.2007.02.011. [DOI] [PubMed] [Google Scholar]
- 17.Eulalio A, Behm-Ansmant I, Izaurralde E. P bodies: at the crossroads of post-transcriptional pathways. NatRevMol Cell Biol. 2007;8:9–22. doi: 10.1038/nrm2080. [DOI] [PubMed] [Google Scholar]
- 18.Teixeira D, Sheth U, Valencia-Sanchez MA, Brengues M, Parker R. Processing bodies require RNA for assembly and contain nontranslating mRNAs. RNA. 2005;11:371–382. doi: 10.1261/rna.7258505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu J, Valencia-Sanchez MA, Hannon GJ, Parker R. MicroRNA-dependent localization of targeted mRNAs to mammalian P-bodies. Nat Cell Biol. 2005;7:719–723. doi: 10.1038/ncb1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pillai RS, Bhattacharyya SN, Artus CG, Zoller T, Cougot N, Basyuk E, Bertrand E, Filipowicz W. Inhibition of translational initiation by Let-7 MicroRNA in human cells. Science. 2005;309:1573–1576. doi: 10.1126/science.1115079. [DOI] [PubMed] [Google Scholar]
- 21*.Eulalio A, Behm-Ansmant I, Schweizer D, Izaurralde E. P-body formation is a consequence, not the cause, of RNA-mediated gene silencing. MolCellBiol. 2007;27:3970–3981. doi: 10.1128/MCB.00128-07. The authors examine the role of multiple proteins in P body formation in S2 cells. They go on to show that depletion of visible P bodies does not affect processes such as mRNA decay, NMD or miRNA mediated gene silencing. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22*.Franks TM, Lykke-Andersen J. TTP and BRF proteins nucleate processing body formation to silence mRNAs with AU-rich elements. Genes Dev. 2007;21:719–735. doi: 10.1101/gad.1494707. This study shows that that dissociation of mRNA from polysomes is not sufficient to aggregate into P bodies. They also show evidence that TTP and BRF proteins can localize AU rich mRNA into P bodies. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Anderson P, Kedersha N. RNA granules. J Cell Biol. 2006;172:803–808. doi: 10.1083/jcb.200512082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Teixeira D, Parker R. Analysis of P-body assembly in Saccharomyces cerevisiae. Mol Biol Cell. 2007;18:2274–2287. doi: 10.1091/mbc.E07-03-0199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25**.Decker CJ, Teixeira D, Parker R. Edc3p and a glutamine/asparagine-rich domain of Lsm4p function in processing body assembly in Saccharomyces cerevisiae. JCell Biol. 2007;179:437–449. doi: 10.1083/jcb.200704147. Multiple ways of forming P bodies in yeast is discussed here. Co-deletion of Yjef domains of Edc3 and Q/N rich C terminus of Lsm4 is shown to completely eliminate visible P bodies suggesting self aggregation domains play an important role in the assembly of P bodies. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26**.Reijns MA, Alexander RD, Spiller MP, Beggs JD. A role for Q/N-rich aggregation-prone regions in P-body localization. JCellSci. 2008;121:2463–2472. doi: 10.1242/jcs.024976. This works identifies Q/N rich regions in several P body proteins and they appear to promote aggregation of P bodies. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27*.Mazzoni C, D'Addario I, Falcone C. The C-terminus of the yeast Lsm4p is required for the association to P-bodies. FEBS Lett. 2007;581:4836–4840. doi: 10.1016/j.febslet.2007.09.009. The authors express mutant Lsm4 (that lacks the C terminus) from Kluyveromyces lactis in S.cerevisiae to show that the C terminus of LSM4 (contains Q/N rich region) is important for association with P bodies. Similar to a decapping mutant, more P bodies are observed, suggesting a correlation between P body association and decapping. [DOI] [PubMed] [Google Scholar]
- 28.Yu JH, Yang WH, Gulick T, Bloch KD, Bloch DB. Ge-1 is a central component of the mammalian cytoplasmic mRNA processing body. RNA. 2005;11:1795–1802. doi: 10.1261/rna.2142405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Eulalio A, Rehwinkel J, Stricker M, Huntzinger E, Yang SF, Doerks T, Dorner S, Bork P, Boutros M, Izaurralde E. Target-specific requirements for enhancers of decapping in miRNA-mediated gene silencing. Genes Dev. 2007;21:2558–2570. doi: 10.1101/gad.443107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu J, Rivas FV, Wohlschlegel J, Yates JR, 3rd, Parker R, Hannon GJ. A role for the P-body component GW182 in microRNA function. Nat Cell Biol. 2005;7:1261–1266. doi: 10.1038/ncb1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Anderson P, Kedersha N. Stress granules: the Tao of RNA triage. Trends Biochem Sci. 2008;33:141–150. doi: 10.1016/j.tibs.2007.12.003. [DOI] [PubMed] [Google Scholar]
- 32.Gilks N, Kedersha N, Ayodele M, Shen L, Stoecklin G, Dember LM, Anderson P. Stress granule assembly is mediated by prion-like aggregation of TIA-1. Mol Biol Cell. 2004;15:5383–5398. doi: 10.1091/mbc.E04-08-0715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tourriere H, Chebli K, Zekri L, Courselaud B, Blanchard JM, Bertrand E, Tazi J. The RasGAP-associated endoribonuclease G3BP assembles stress granules. J Cell Biol. 2003;160:823–831. doi: 10.1083/jcb.200212128. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 34.Brengues M, Parker R. Accumulation of polyadenylated mRNA, Pab1p, eIF4E, and eIF4G with P-bodies in Saccharomyces cerevisiae. Mol Biol Cell. 2007;18:2592–2602. doi: 10.1091/mbc.E06-12-1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35**.Buchan JR, Muhlrad D, Parker R. P bodies promote stress granule assembly in Saccharomyces cerevisiae. JCell Biol. 2008;183:441–455. doi: 10.1083/jcb.200807043. Authors report that stress granules can form in yeast and their assembly is promoted by pre-existing P bodies. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36*.Hoyle NP, Castelli LM, Campbell SG, Holmes LE, Ashe MP. Stress-dependent relocalization of translationally primed mRNPs to cytoplasmic granules that are kinetically and spatially distinct from P-bodies. JCell Biol. 2007;179:65–74. doi: 10.1083/jcb.200707010. This work shows that granules similar to mammalian stress granules and distinct from P bodies can exist it yeast. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hoyle NP, Ashe MP. Subcellular localization of mRNA and factors involved in translation initiation. BiochemSoc Trans. 2008;36:648–652. doi: 10.1042/BST0360648. [DOI] [PubMed] [Google Scholar]
- 38.Kedersha N, Stoecklin G, Ayodele M, Yacono P, Lykke-Andersen J, Fritzler MJ, Scheuner D, Kaufman RJ, Golan DE, Anderson P. Stress granules and processing bodies are dynamically linked sites of mRNP remodeling. J Cell Biol. 2005;169:871–884. doi: 10.1083/jcb.200502088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kedersha N, Cho MR, Li W, Yacono PW, Chen S, Gilks N, Golan DE, Anderson P. Dynamic shuttling of TIA-1 accompanies the recruitment of mRNA to mammalian stress granules. J Cell Biol. 2000;151:1257–1268. doi: 10.1083/jcb.151.6.1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cougot N, Babajko S, Seraphin B. Cytoplasmic foci are sites of mRNA decay in human cells. J Cell Biol. 2004;165:31–40. doi: 10.1083/jcb.200309008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mollet S, Cougot N, Wilczynska A, Dautry F, Kress M, Bertrand E, Weil D. Translationally repressed mRNA transiently cycles through stress granules during stress. Mol Biol Cell. 2008;19:4469–4479. doi: 10.1091/mbc.E08-05-0499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Andrei MA, Ingelfinger D, Heintzmann R, Achsel T, Rivera-Pomar R, Luhrmann R. A role for eIF4E and eIF4E-transporter in targeting mRNPs to mammalian processing bodies. RNA. 2005;11:717–727. doi: 10.1261/rna.2340405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wilczynska A, Aigueperse C, Kress M, Dautry F, Weil D. The translational regulator CPEB1 provides a link between dcp1 bodies and stress granules. JCell Sci. 2005;118:981–992. doi: 10.1242/jcs.01692. [DOI] [PubMed] [Google Scholar]
- 44.Anderson P. A Place for RNAi. Dev Cell. 2005;9:311–312. doi: 10.1016/j.devcel.2005.08.003. [DOI] [PubMed] [Google Scholar]
- 45.Brengues M, Teixeira D, Parker R. Movement of eukaryotic mRNAs between polysomes and cytoplasmic processing bodies. Science. 2005;310:486–489. doi: 10.1126/science.1115791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bhattacharyya SN, Habermacher R, Martine U, Closs EI, Filipowicz W. Stress-induced reversal of microRNA repression and mRNA P-body localization in human cells. Cold Spring HarbSympQuant Biol. 2006;71:513–521. doi: 10.1101/sqb.2006.71.038. [DOI] [PubMed] [Google Scholar]
- 47.Hilliker A, Parker R. Stressed out? Make some modifications! Nat Cell Biol. 2008;10:1129–1130. doi: 10.1038/ncb1008-1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Beelman CA, Parker R. Differential effects of translational inhibition in cis and in trans on the decay of the unstable yeast MFA2 mRNA. JBiol Chem. 1994;269:9687–9692. [PubMed] [Google Scholar]
- 49.Cereghino GP, Atencio DP, Saghbini M, Beiner J, Scheffler IE. Glucose-dependent turnover of the mRNAs encoding succinate dehydrogenase peptides in Saccharomyces cerevisiae: sequence elements in the 5’ untranslated region of the Ip mRNA play a dominant role. Mol Biol Cell. 1995;6:1125–1143. doi: 10.1091/mbc.6.9.1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cao D, Parker R. Computational modeling and experimental analysis of nonsense-mediated decay in yeast. Cell. 2003;113:533–545. doi: 10.1016/s0092-8674(03)00353-2. [DOI] [PubMed] [Google Scholar]
- 51*.Aizer A, Brody Y, Ler LW, Sonenberg N, Singer RH, Shav-Tal Y. The dynamics of mammalian P body transport, assembly, and disassembly in vivo. MolBiolCell. 2008;19:4154–4166. doi: 10.1091/mbc.E08-05-0513. Authors conduct a quantitative analysis of P body mobility in mammalian cells in vivo. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52**.Kwon S, Zhang Y, Matthias P. The deacetylase HDAC6 is a novel critical component of stress granules involved in the stress response. Genes Dev. 2007;21:3381–3394. doi: 10.1101/gad.461107. This study localizes HDAC6 to stress granules and shows using deletion and mutants of the HDAC6 gene, that its deacetylase activity and the ubiquitin-binding function are important for the SG formation. This hints at the importance of post translational modification in formation of SGs. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53**.Ohn T, Kedersha N, Hickman T, Tisdale S, Anderson P. A functional RNAi screen links O-GlcNAc modification of ribosomal proteins to stress granule and processing body assembly. NatCell Biol. 2008;10:1224–1231. doi: 10.1038/ncb1783. The first large scale RNAi screen to identify proteins that affect assembly of P bodies and stress granules. Results showed that reversible O-GlcNAc modification is important for the foci formation especially SG. This result emphasizes the role of post translational modification in the regulation of granule formation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chu CY, Rana TM. Translation repression in human cells by microRNA-induced gene silencing requires RCK/p54. PLoS Biol. 2006;4:e210. doi: 10.1371/journal.pbio.0040210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stoecklin G, Mayo T, Anderson P. ARE-mRNA degradation requires the 5′-3′ decay pathway. EMBO Rep. 2006;7:72–77. doi: 10.1038/sj.embor.7400572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Klingauf M, Stanek D, Neugebauer KM. Enhancement of U4/U6 small nuclear ribonucleoprotein particle association in Cajal bodies predicted by mathematical modeling. Mol Biol Cell. 2006;17:4972–4981. doi: 10.1091/mbc.E06-06-0513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57**.Arimoto K, Fukuda H, Imajoh-Ohmi S, Saito H, Takekawa M. Formation of stress granules inhibits apoptosis by suppressing stress-responsive MAPK pathways. NatCell Biol. 2008;10:1324–1332. doi: 10.1038/ncb1791. Authors undertake a siRNA based genomic screen that identifies numerous proteins affecting the assembly of P bodies and stress granules. Results showed that reversible O-GlcNAc modification is important for the foci formation, especially SG. This result emphasizes the role of post translational modification in the regulation of granule formation. [DOI] [PubMed] [Google Scholar]